Abstract
Familial Mediterranean fever (FMF) is a recessive disorder characterized by episodes of fever and neutrophil-mediated serosal inflammation. We recently identified the gene causing FMF, designatedMEFV, and found it to be expressed in mature neutrophils, suggesting that it functions as an inflammatory regulator. To facilitate our understanding of the normal function of MEFV, we extended our previous studies. MEFV messenger RNA was detected by reverse transcriptase–polymerase chain reaction in bone marrow leukocytes, with differential expression observed among cells by in situ hybridization. CD34 hematopoietic stem-cell cultures induced toward the granulocytic lineage expressed MEFV at the myelocyte stage, concurrently with lineage commitment. The prepromyelocytic cell line HL60 expressed MEFV only at granulocytic and monocytic differentiation. MEFV was also expressed in the monocytic cell lines U937 and THP-1. Among peripheral blood leukocytes, MEFV expression was detected in neutrophils, eosinophils, and to varying degrees, monocytes. Consistent with the tissue specificity of expression, complete sequencing and analysis of upstream regulatory regions of MEFV revealed homology to myeloid-specific promoters and to more broadly expressed inflammatory promoter elements. In vitro stimulation of monocytes with the proinflammatory agents interferon (IFN) γ, tumor necrosis factor, and lipopolysaccharide induced MEFV expression, whereas the antiinflammatory cytokines interleukin (IL) 4, IL-10, and transforming growth factor β inhibited such expression. Induction by IFN-γ occurred rapidly and was resistant to cycloheximide. IFN- also induced MEFV expression. In granulocytes, MEFV was up-regulated by IFN-γ and the combination of IFN- and colchicine. These results refine understanding of MEFV by placing the gene in the myelomonocytic-specific proinflammatory pathway and identifying it as an IFN-γ immediate early gene.
We and others1,2 identified a novel human gene, designated MEFV, by positional cloning. Mutations in this gene cause the autosomal recessive disorder familial Mediterranean fever (FMF; MIM249100). FMF is characterized by periodic attacks of fever accompanied by serosal, synovial, or cutaneous inflammation. FMF attacks are self-limited, lasting 1 to 3 days, and include the presence of purulent neutrophil-rich aseptic exudates at sites of inflammation, which suggests that the disease-causing alleles of MEFV result in defects in control of granulocyte-mediated inflammation.3,4 Seventeen independent mutations inMEFV have been identified.5-8 These mutations are limited in scope, causing only single amino acid changes in the putative protein product. The facts that no null mutations have been identified and that relatively conservative amino acid substitutions can result in a disease phenotype suggest that MEFV has an important physiologic role.
The 3.7-kilobase (kb) MEFV complementary DNA (cDNA) is a member of a family of highly conserved genes that includes nuclear effector molecules and nucleic acid binding proteins that regulate inflammation, hematopoiesis, oncogenesis, and embryonic development.9-11One member of this family is the ribonuclear protein Ro52, which is a target of autoantibodies in systemic lupus erythematosus and Sjögren syndrome. Other family members are transcriptional regulators, including rpt-1, a murine gene that controls expression of interleukin (IL) 2; Staf50, an interferon (IF)-regulated gene that attenuates expression of the human immunodeficiency virus long-terminal-repeat promoter; and PML, a retinoic acid–dependent transactivator of the p21 WAF1/CIP1gene that is involved in host defense, myelomonocytic differentiation, and control of tumor growth. In a survey of normal human tissues,MEFV messenger RNA (mRNA) was detected only in peripheral blood leukocytes, and in a preliminary Northern analysis of fractionated leukocytes, expression was detected specifically in neutrophils, the principal cell type found in the inflammatory infiltrates characteristic of FMF.1 Taken together, these data suggest that MEFV encodes a leukocyte-specific inflammatory regulator, mutations that cause the autoinflammatory phenotype of FMF.
Although mutations in MEFV lead to illness, the function of the gene remains a mystery because no gross functional differences have been observed between neutrophils from families with FMF and those from healthy volunteers.12,13 Also, compared with normal cells, FMF neutrophils have neither morphologic changes nor differences in the release of reactive oxygen products.12Moreover, no changes in infection rates have been observed in patients with FMF.3 This is in marked contrast to most other congenital diseases involving mutations in phagocyte-specific genes, such as chronic granulomatous disease, leukocyte adhesion deficiency, and neutrophil-specific granule deficiency, which are associated with host defense defects and recurrent infections.14 In fact, in several distinct Mediterranean populations, FMF carrier frequencies are high, suggesting a selective advantage in carriers with respect to an unidentified pathogen.4 5 As a first step in understanding the role of the MEFV gene in leukocyte biology, we undertook a more detailed analysis of the expression and regulation of the gene during hematopoietic differentiation and in response to inflammatory stimuli.
Neutrophil differentiation has multiple discrete stages characterized by morphologic changes and the acquisition of stage-specific granules. Primary granules first appear early in differentiation, at the myeloblast stage, whereas specific granules do not appear until the myelocyte stage, concurrently with loss of proliferative potential and granulocytic lineage commitment.15 Elucidation of the specificity and kinetics of myelomonocytic-specific genes during differentiation provides insights into their functional roles. Late gene expression can be studied in the multipotent prepromyelocytic cell lines HL60 and NB4, which can be further differentiated along the granulocytic lineage in vitro.16-18 However, differentiation does not proceed normally in these cells, as illustrated partly by defects in secondary granule-gene expression and granule formation.19,20 Neutrophil differentiation can be more accurately recapitulated ex vivo by growth factor–directed granulocytic differentiation of CD34 hematopoietic precursor cells. Differentiating cells in these cultures maintain a striking morphologic and functional similarity to bone marrow precursors.21 22
Increasing evidence from studies of human mutations, functional knockout experiments, characterization of chromosome breakpoints in human leukemias, and direct biochemical analysis of myelomonocytic-specific promoters suggests that key steps in neutrophil differentiation and activation are transcriptionally regulated.19,23-26 The promoters of myelomonocytic-specific genes have common cis-acting elements, as well as stage-specific and inflammatory mediator–activated promoter elements,27-31 the identification of which can facilitate understanding of the biologic function of a given gene.32Although expression studies suggest that MEFV expression is myeloid specific, no description of the upstream regulatory region of the gene exists.
Leukocyte-specific mediators of granulocytic inflammation include reactive oxygen intermediates, eicosanoids, and cytokines, with the balance of proinflammatory and Th1 mediators (eg, IL-12, tumor necrosis factor [TNF], and IFN-γ) and antiinflammatory and Th2 cytokines (eg, IL-10, IL-4, and transforming growth factor [TGF] β), playing a key role in regulation of the response.33-37 To begin to ascertain the normal function of MEFV in the inflammatory cell, we reexamined the specificity of expression of the gene in fractionated leukocytes from bone marrow and peripheral blood. We also defined the kinetics of MEFV induction during differentiation and inflammatory-mediator activation of these cells.
Materials and methods
Purification of peripheral blood leukocytes
Granulocyte purifications from heparin-treated blood from healthy donors were done as described previously.38 Eosinophils were purified from the granulocyte preparations as described previously.39 Highly purified populations of lymphocytes and monocytes were isolated as follows. Healthy volunteers underwent leukapheresis, and purified lymphocytes and monocytes were obtained by elutriation.40 Elutriated cells were further purified by using antibody-conjugated supermagnetic beads (MACS).41 For lymphocytes, 2 rounds of purification were done with anti-CD3 and anti-CD19 MACS, according to the manufacturer's instructions (Mitenyi Biotech, Auburn, CA). For monocytes, cells were further purified with a combination of anti-CD3, anti-CD7, anti-CD19, anti-CD45RA, anti-CD56, and anti-IgE MACS, and the mixture was applied to a negative-selection column to remove residual T cells, natural killer cells, B cells, dendritic cells, and basophils. The purity of preparations was determined by fluorescence-activated cell sorting (FACS) and visual inspection of cytospin slides stained with Wright-Giemsa stain. In addition, the purity of monocyte preparations was determined by histologic staining for nonspecific esterase and myeloperoxidase activity on cytospin preparations.42
Purification and granulocytic differentiation of CD34+peripheral blood hematopoietic precursors (PBHP)
PBHP were isolated as described previously.21 Purified CD34+ cells were stored frozen in X-Vivo 10 medium (Bio Whittaker, Walkersville, MD) supplemented with 1% human serum albumin (Baxter Healthcare, Deerfield, IL) and 10% dimethyl sulfoxide (Sigma Chemical, St Louis, MO) until use. Cells were induced to differentiate along the myeloid lineage in suspension cultures at 37°C with 7% carbon dioxide (CO2) in Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum (FBS) and the following recombinant human growth factors: 50 ng/mL PIXY321 (IL-3 and granulocyte-macrophage colony-stimulating factor [GM-CSF] fusion protein), 100 ng/mL Flt 3 (gifts from Immunex Corp, Seattle, WA), 50 ng/mL stem-cell factor, and 100 ng/mL granulocyte colony-stimulating factor [G-CSF] (R&D Systems, Minneapolis, MN). Cells maintained viability for approximately 26 days and appeared to achieve maximal differentiation on or about day 21. Cells were harvested for analysis on days 3, 7, 10, 13, 17, and 21.
Isolation and fractionation of whole bone marrow
After informed consent was given, 20 to 40 mL of bone marrow was obtained from the posterior superior iliac crest of healthy volunteers. CD34+ cells from bone marrow were purified by using the Ceprate immunoaffinity CD34+ cell system (Cellpro, Bothell, WA). To enrich for leukocytes, erythrocytes were lysed in hypotonic solution.38 In addition, granulocytes and granulocytic precursors were isolated from purified bone marrow leukocytes by standard density-gradient centrifugation on Ficoll gradients, according to the manufacturer's instructions (Organon Teknika, Durham, NC).
Flow cytometry analysis
Cells were labeled with fluorescein isothiocyanate–conjugated, phycoerythrin-conjugated, or peridinin chlorophyll protein–conjugated monoclonal antibodies against CD3, CD14, CD16, CD19, CD20, CD56, or isotype-matched nonspecific control antibodies (Becton Dickinson, San Jose, CA). Before staining, Fcγ receptors on monocytes were blocked by supplementing phosphate-buffered saline–bovine serum albumin solution with 100 μg/mL human IgG.
Cell lines
HL60, THP-1, and U937 cell lines were obtained from the American Type Culture Collection (Rockville, MD). The basophilic cell line KU812 and the mast cell line HMC1 were provided by Dr J. Rivera. HMC1 cells were grown in Iscove media supplemented with α-thioglycerol, and 10% FBS. All other cell lines were grown in RPMI 1640 (Biofluids, Rockville, MD) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 200 mmol/L L-glutamine, and 10% FBS (“complete media”). HL60 cells were induced to differentiate toward the granulocytic lineage with 1 μmol/L or 10 μmol/L retinoic acid for 7 days in cultures containing 2 × 105cells/mL.43 Cells were induced to differentiate toward the monocytic lineage with 16 nmol/L or 160 nmol/L phorbol ester for 48 hours, followed by 2 washes in RPMI and a subsequent 48-hour incubation, as described previously.44 Adherent cells were removed by trypsinization and analyzed. Differentiation toward the eosinophilic lineage by using alkaline culture media was done as described previously.45 Morphologic assessment of cells was made on cytospin slides stained with Wright-Giemsa stain.
Leukocyte activation
Cells (107) were seeded with 3 mL of complete media and left for 2 hours at 37°C in 5% CO2. Monocytes and granulocytes were stimulated with IL-1α (100 pg/mL), IL-4 (20 ng/mL), IL-6 (100 units/mL), IL-10 (2.5 ng/mL), TNF-α (200 pg/mL), IFN-γ (1000 units/mL), IFN-α (1000 units/mL), TGF-β (10 units/mL), G-CSF (2 ng/mL), GM-CSF (5 ng/mL; R&D Systems), lipopolysaccharide (LPS; 200 ng/mL), and colchicine (30 ng/mL; Sigma Chemical) alone or in combination.
In situ hybridization
In situ hybridization was done as described previously.46 For in situ probes, a 435-base-pair (bp) section of MEFV spanning exons 3 to 5 (nucleotides 986-1420 inMEFV cDNA; accession no. AF018080) that had no important similarity to other human DNA sequences in the NR and EST databases (National Center for Biotechnology Information, National Institutes of Health [NIH]) was amplified by using the forward primer 5′-CCACGCCCAGGAAGGAGACCCAGTTG-3′ and the reverse primer 5′-TGCTTCAGCGCTTCAGTTTGTTTCAG-3′. The polymerase chain reaction (PCR) product was cloned directly into the pCRII TA-cloning vector (Invitrogen, Carlsbad, CA), and the resulting plasmid was designated pV75IE. Sense and antisense digoxigenin-labeled riboprobes were produced as run-off transcripts from EcoRV-linearized pV75IE and T7 RNA polymerase, and from BamHI-linearized pV75IE and SP6 RNA polymerase, respectively, in the presence of digoxigenin-labeled uridine triphosphate (UTP).
RNA extraction and reverse transcriptase (RT)-PCR
Total RNA was prepared from cells by using Trizol reagent, and cDNA was prepared from 2 μg of total RNA with oligo(dT) priming using the SuperScript Preamplification System (Life Technologies, Gaithersburg, MD). RT-PCR analyses were performed by using 1/40 of the reverse transcription reaction (the amount of cDNA derived from 50 ng of total RNA) as a template to maintain a constant amount of input cDNA for all samples analyzed. PCR amplification using AmpliTaq Gold (PE Biosystems, Foster City, CA) was carried out so that reactions were completed within the exponential cycling phase. The PCR conditions were 5 minutes at 94°C, followed by cycling for 30 seconds at 94°C, 15 seconds at 61.9°C, and 30 seconds at 68.0°C, then elongation for 10 minutes at 72°C. MEFV, myeloperoxidase (accession no. J02694), and lactoferrin (accession no.M83202) were amplified for 45 cycles, and β-actin (accession no.X00351) was amplified for 40 cycles. AmpliTaq Gold requires heat activation, and accordingly, more cycles for amplification than other thermal stable polymerases.
PCR products (8 μL) were fractionated on 4% to 20% polyacrylamide gels (Novex, San Diego, CA) and visualized after ethidium bromide staining. Because yields of RNA preparations can vary, equal amounts of RNA were used for cDNA preparations. For all samples, cDNA derived from 50 ng of total RNA was amplified and β-actin message levels were assessed. Oligonucleotide PCR primers (with final product sizes) were as follows: β-actin (650 bp), 5′-CTGGCCGGGACCTGACTGACTACCTC-3′ and 5′-AAACAAATAAAGCCATGCCAATCTCA-3′; lactoferrin (422 bp) and 5′-CGGGGCTGGAGACGTGGCTTTTATCA-3′, 5′-GCCGGGCAGCCACTTCCTCCTCACTT-3′; myeloperoxidase (728 bp), 5′-GAACCCAACCCCCGTGTCCCCCTCAG-3′ and 5′-GGCCAGCCCAGATATACCCCTCACT-3′; and MEFV (351 bp), 5′-GATTGGCGCTC-AGGCACATGCT- GTTA-3′ and 5′-GTCGGGGGAACGCTGGACGCCTGGTA-3′.
RNase protection assay
RNA from unstimulated and stimulated cells (107) was prepared with Trizol (Life Technologies). The MEFV probe used in this assay was generated from plasmid pV75IE. Labeled RNA probes were synthesized by using SP6 RNA polymerase and phosphorus 32–labeled UTP. DNA was digested with DNase I (Boehringer Mannheim, Indianapolis, IN), and RNA probes were extracted with phenol and chloroform and precipitated with ethanol. Labeled RNA probes were hybridized overnight with target RNA (5 μg) at 56°C) and digested with T1 RNase (Life Technologies). The protected mRNA fragment was extracted with phenol and chloroform, precipitated with ethanol, resolved on a 6% denaturing polyacrylamide gel, and subjected to autoradiography. Gene transcripts were identified by the length of the protected fragments. Equal loading of RNA was estimated from the amounts of protected fragments of 2 housekeeping genes, L32 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Signal intensities were measured from scanned images by using NIH Image software. Signals were normalized relative to GAPDHexpression and the relative values reported.
Results
Kinetics of MEFV expression during granulopoiesis
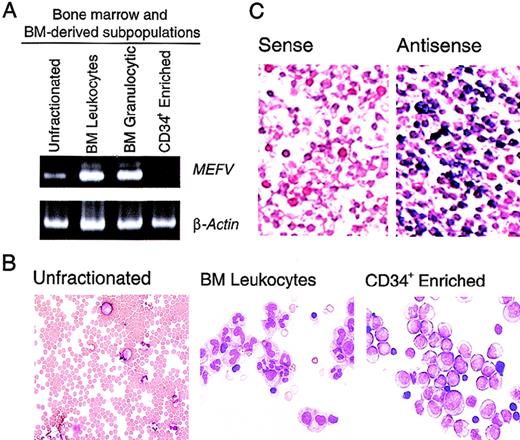
Previous studies of MEFV expression that used Northern analysis of panels of human tissues indicated that expression of this gene is limited to peripheral blood leukocytes.1 To gain more insight into the time course of MEFV expression in leukocytes, we performed the more sensitive RT-PCR assays. In whole bone marrow, which is composed predominantly of erythroid cells, a weakMEFV signal was detected by RT-PCR (Figure1A). Whole bone marrow was subjected to hypotonic lysis of red blood cells to produce enrichment for populations of bone marrow leukocytes and leukocyte precursors. Consistent with the leukocyte-specific expression previously observed, significantly more MEFV mRNA was detected in this population of cells (Figure 1A and B), suggesting that MEFV expression was not restricted to peripheral, and therefore mature, leukocytes. Enhanced MEFV expression was also detected in bone marrow granulocytes and their precursors purified by density-gradient fractionation (Figure 1A). MEFV expression was significantly diminished in populations of cells enriched in CD34+ hematopoietic precursor cells, indicating that expression is temporally restricted during hematopoiesis. To assess the possibility that the detection of MEFV message was due to contamination of bone marrow with peripheral blood, we further analyzedMEFV expression by using in situ hybridization. This analysis detected a large population of MEFV-expressing leukocytes in the bone marrow (Figure 1C).
MEFV expression in bone marrow leukocytes and precursor cells.
(A). Ethidium bromide (EtBr)–stained polyacrylamide gel electrophoresis (PAGE) of reverse transcriptase–polymerase chain reaction (RT-PCR) products of MEFV and β-actin messenger RNAs (mRNAs) from unfractionated bone marrow cells (lane 1), bone marrow leukocytes (lane 2), bone marrow granulocytic cells (lane 3), and an enriched population of CD34+ peripheral blood hematopoietic precursors (PBHP; lane 4). (B) Wright-Giemsa–stained cytospin preparations of the cells used for these analyses. Original magnification ×400. (C) Photomicrographs of Wright- Giemsa–stained in situ hybridizations of bone marrow leukocytes. Results with use of a gene-specific MEFV antisense riboprobe (right panel) and a nonspecific MEFV sense strand control riboprobe (left panel) are shown. Original magnification ×400.
MEFV expression in bone marrow leukocytes and precursor cells.
(A). Ethidium bromide (EtBr)–stained polyacrylamide gel electrophoresis (PAGE) of reverse transcriptase–polymerase chain reaction (RT-PCR) products of MEFV and β-actin messenger RNAs (mRNAs) from unfractionated bone marrow cells (lane 1), bone marrow leukocytes (lane 2), bone marrow granulocytic cells (lane 3), and an enriched population of CD34+ peripheral blood hematopoietic precursors (PBHP; lane 4). (B) Wright-Giemsa–stained cytospin preparations of the cells used for these analyses. Original magnification ×400. (C) Photomicrographs of Wright- Giemsa–stained in situ hybridizations of bone marrow leukocytes. Results with use of a gene-specific MEFV antisense riboprobe (right panel) and a nonspecific MEFV sense strand control riboprobe (left panel) are shown. Original magnification ×400.
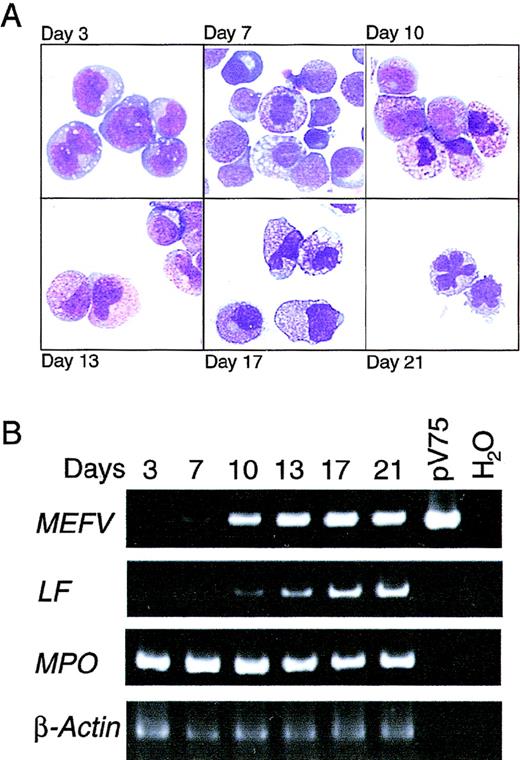
To define the temporal regulation of MEFV expression during granulopoiesis and to confirm the results obtained in bone marrow, mobilized CD34+ PBHP were isolated and induced to differentiate along the granulocytic lineage. Morphologic changes in the cells in culture were strikingly consistent with those observed in bone marrow (Figure 2A). Cells maintained a blast-like morphologic appearance for several days; promyelocytes (large cells containing small numbers of azurophilic granules) appeared on or about day 7, and increasing azurophilic granule expression was observed until about day 10. Day 10 also marked the appearance of myelocytes, ie, smaller cells containing specific granules. Changes in nuclear morphologic features and increased expression of specific granules, characteristic of metamyelocytes, occurred between days 13 and 17. Terminal differentiation of mature neutrophils, characterized by the appearance of cells with lobate nuclei, occurred on or about day 21 (Figure 2A).
Induction of MEFV expression during granulopoiesis ex vivo.
(A) Wright-Giemsa–stained cytospin preparations of cells from PBHP cultures induced to differentiate toward the granulocytic lineage. Representative images from cultures at 3, 7, 10, 13, 17, and 21 days are shown. Original magnification ×600. (B) EtBr-stained PAGE of RT-PCR products of the MEFV, lactoferrin (LF), myeloperoxidase (MPO), and β-actin mRNAs from PBHP cells collected on the days indicated. Positive and negative control PCR reactions using the MEFV-containing plasmid pV75-1 and water, respectively, are shown.
Induction of MEFV expression during granulopoiesis ex vivo.
(A) Wright-Giemsa–stained cytospin preparations of cells from PBHP cultures induced to differentiate toward the granulocytic lineage. Representative images from cultures at 3, 7, 10, 13, 17, and 21 days are shown. Original magnification ×600. (B) EtBr-stained PAGE of RT-PCR products of the MEFV, lactoferrin (LF), myeloperoxidase (MPO), and β-actin mRNAs from PBHP cells collected on the days indicated. Positive and negative control PCR reactions using the MEFV-containing plasmid pV75-1 and water, respectively, are shown.
MEFV expression as determined by RT-PCR was first detected weakly on day 7, with increased expression observed on day 10 and throughout the differentiation process (Figure 2B). Consistent with the kinetics of differentiation observed morphologically, lactoferrin gene expression was observed weakly on day 10, with increasing levels observed for the remainder of the experiment (Figure 2B). Induction ofMEFV expression therefore occurred near the myelocyte stage of differentiation, at or near lineage commitment.
MEFV expression in cell lines
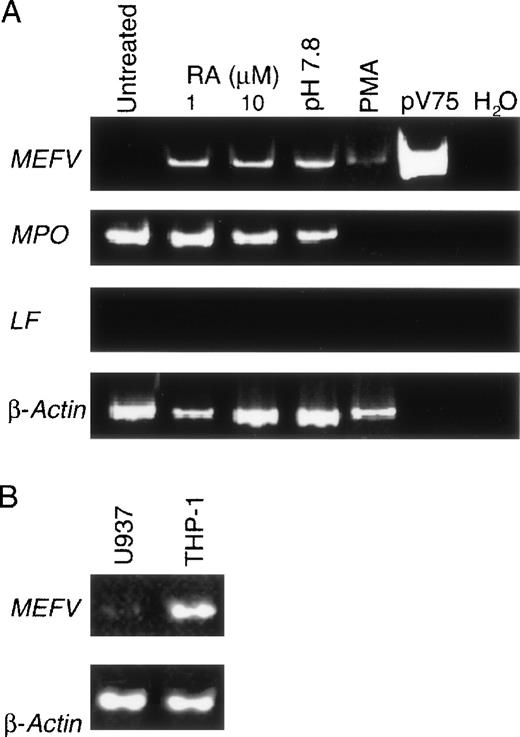
Specific granule genes are coordinately up-regulated during granulopoiesis. Because of the similarity of lactoferrin- andMEFV-gene induction in the CD34+ cultures, we further examined the stage specificity of MEFV expression in the prepromyelocytic cell line HL60. Although this multipotent cell line can be induced to differentiate along the granulocytic lineage so that several functional markers of terminal differentiation appear, specific granule genes are not expressed during maturation.19 As expected, lactoferrin was not detected in HL60 cells induced toward the neutrophilic lineage with retinoic acid or along the eosinophilic lineage with alkaline pH (Figure3A). Although MEFV expression was not detected before differentiation, it was observed after induction (Figure 3A), thereby confirming the findings on the kinetics ofMEFV expression determined in the CD34+ cultures and suggesting that MEFV expression is not under the common regulatory program described for secondary granule genes.
MEFV expression in unstimulated and in vitro differentiated myelomonocytic cell lines.
(A) EtBr-stained PAGE of RT-PCR products of the MEFV, LF, MPO, and β-actin RNAs from resting untreated HL60 cells (lane 1) and cultured HL60 cells induced to differentiate toward the neutrophilic lineage with 1 or 10 μmol/L retinoic acid (lanes 2 and 3), toward the eosinophilic lineage with alkaline pH (pH 7.8; lane 4), and toward the monocytic lineage with phorbol ester. Positive and negative control PCR reactions using the MEFV-containing plasmid pV75-1 and water, respectively, are shown. (B) EtBr-stained PAGE of RT-PCR products of the MEFV and β-actin mRNAs from the monocytic cell lines U937 and THP-1.
MEFV expression in unstimulated and in vitro differentiated myelomonocytic cell lines.
(A) EtBr-stained PAGE of RT-PCR products of the MEFV, LF, MPO, and β-actin RNAs from resting untreated HL60 cells (lane 1) and cultured HL60 cells induced to differentiate toward the neutrophilic lineage with 1 or 10 μmol/L retinoic acid (lanes 2 and 3), toward the eosinophilic lineage with alkaline pH (pH 7.8; lane 4), and toward the monocytic lineage with phorbol ester. Positive and negative control PCR reactions using the MEFV-containing plasmid pV75-1 and water, respectively, are shown. (B) EtBr-stained PAGE of RT-PCR products of the MEFV and β-actin mRNAs from the monocytic cell lines U937 and THP-1.
HL60 cells were also induced to differentiate along the monocytic lineage with phorbol esters. Because expression of MEFV was not previously detected in peripheral blood monocytes, we anticipated that no expression would be observed on commitment of HL60 cells to the monocytic lineage. Surprisingly, MEFV mRNA was observed in these cultures (Figure 3A) and in the monocytic cell lines U937 and THP-1 (Figure 3B). No expression was detected in the human mast cell line HMC-1 or in the human basophil line KU812 (data not shown).
MEFV expression in purified peripheral blood leukocytes
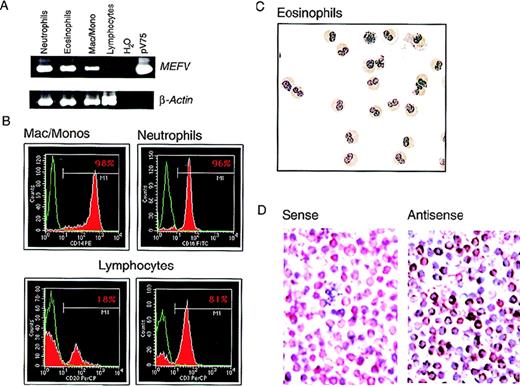
In purified populations of peripheral blood leukocytes from healthy donors, MEFV mRNA was observed in neutrophils and in eosinophils from all samples analyzed (Figure4A). The purity of the isolated cells was determined by FACS or histological staining (Figure 4B and C). Expression was also detected in populations of peripheral blood mononuclear cells (PBMC) enriched for monocytes by collecting cell culture plastic-adherent cells in samples from 3 of 4 healthy subjects (not shown). Expression of mRNA was assessed in highly purified monocyte preparations (> 98% CD14+ and nonspecific-esterase positive; Figure 4B) from 3 additional subjects.MEFV mRNA was detected in all 3 samples; a typical example is shown in Figure 4A. Message levels among individual subjects appeared highly variable compared with those observed for the housekeeping gene β-actin.
MEFV expression in peripheral blood leukocytes.
(A) EtBr-stained PAGE of RT-PCR products of the MEFV and β-actin mRNAs from highly purified populations of peripheral blood neutrophils (lane 1), eosinophils (lane 2), macrophages and monocytes (lane 3), and lymphocytes (lane 4). Negative and positive control PCR reactions using water and the MEFV-containing plasmid pV75-1, respectively, are shown (lanes 5 and 6). (B) Purity of populations analyzed in A, as determined by fluorescence-activated cell sorting. Cells were stained for a cell-type–specific marker (red) and nonspecific isotype control antibody (green). The antibodies used were monocytes (anti-CD14), neutrophils (anti-CD16), B cells (anti-CD20), and T cells (anti-CD3). (C) Wright-Giemsa–stained cytospin preparations of eosinophils used for these analysis. Original magnification ×400. (D) Photomicrographs of Wright-Giemsa–stained monocyte in situ hybridizations. Results with use of a gene-specificMEFV antisense riboprobe (right panel) and a nonspecificMEFV sense strand control riboprobe (left panel) are shown. Original magnification ×400.
MEFV expression in peripheral blood leukocytes.
(A) EtBr-stained PAGE of RT-PCR products of the MEFV and β-actin mRNAs from highly purified populations of peripheral blood neutrophils (lane 1), eosinophils (lane 2), macrophages and monocytes (lane 3), and lymphocytes (lane 4). Negative and positive control PCR reactions using water and the MEFV-containing plasmid pV75-1, respectively, are shown (lanes 5 and 6). (B) Purity of populations analyzed in A, as determined by fluorescence-activated cell sorting. Cells were stained for a cell-type–specific marker (red) and nonspecific isotype control antibody (green). The antibodies used were monocytes (anti-CD14), neutrophils (anti-CD16), B cells (anti-CD20), and T cells (anti-CD3). (C) Wright-Giemsa–stained cytospin preparations of eosinophils used for these analysis. Original magnification ×400. (D) Photomicrographs of Wright-Giemsa–stained monocyte in situ hybridizations. Results with use of a gene-specificMEFV antisense riboprobe (right panel) and a nonspecificMEFV sense strand control riboprobe (left panel) are shown. Original magnification ×400.
To confirm the results obtained with RT-PCR and rule out the possibility that the signals observed in monocyte preparations were due to granulocyte contamination, expression of MEFV was assessed at the single-cell level by in situ hybridization in the highly purified monocyte preparations shown in Figure 4B. MEFV mRNA was detected in a large population of monocytes, with individual cell-expression levels varying widely (Figure 4D), thus confirming the RT-PCR results. As previously reported,1 no MEFVmRNA was observed in lymphocytes purified to homogeneity (99% CD3+ or CD20+; Figure 4B) in samples from 3 healthy subjects (typical example shown in Figure 4A). The purity of the lymphocyte preparations was essential because signal was detected in populations of nonadherent PBMC.
The 5′ end of the MEFV transcript (accession no.AF018080) was defined by 5′ RACE,1 and the genomic sequences upstream of the transcription start site in the promoter region were delineated (accession no. AF111163). Sequence homologies to transcription factor binding sites in the TRANSFAC database were identified in the MEFV promotor region by using MatInspector software.47 The cis-acting elements necessary to confer myelomonocytic-specific expression were identified and, consistent with the observed expression profile of MEFV, these conserved DNA-sequence elements are just upstream of the transcription start site of MEFV (Table 1). These requirements include a TATA-less sequence containing a PU.1 transcription factor binding site adjacent to the transcription start site, as well as binding sites for the C/EBP and Runt/PEBP2/CBF families of transcription factors.32 A putative PU.1 site was identified 77 bp upstream of the transcription start site (−77). This sequence is identical to the canonical PU.1 binding sequence shown to bind PU.1 in vitro.48 In addition, a C/EBPα binding motif is present at position −57, and sites for several factors shown to mediate myelomonocytic-specific expression, including AML (acute myelogenous leukemia), c-Myb, MZF (myeloid zinc finger), and TAL1, were also present within 1 kb of the transcription start (Table 1). Interestingly, also present in the MEFVpromoter region were DNA-sequence identities to the proinflammatory mediator-specific cis-acting sites nuclear factor (NF) κb (−164), IF-stimulated response element (−105), IF regulatory factor (−496), and γ-IF activation sequence (GAS; −731), as well as the more ubiquitous AP-1 (−647). These observations are additional indications that MEFV expression is modulated by inflammatory mediators.
Sequence similarities to myelomonocytic and activation-specific cis-acting sites in 1 kilobase of genomic DNA upstream of MEFV
| Putative cis-acting Site . | Location . |
|---|---|
| Sites common in myeloid-specific promoters | |
| C/EBPα | −57 |
| PU.1 | −77, −320 |
| AML | −247 |
| TAL1 | −260 |
| c-myb | −403 |
| Sites required for cytokine activation | |
| Interferon-stimulated response element | −105 |
| Nuclear factor κB | −164 |
| Interferon regulatory factor 2 | −469 |
| Interferon γ-activation sequence | −731 |
| Putative cis-acting Site . | Location . |
|---|---|
| Sites common in myeloid-specific promoters | |
| C/EBPα | −57 |
| PU.1 | −77, −320 |
| AML | −247 |
| TAL1 | −260 |
| c-myb | −403 |
| Sites required for cytokine activation | |
| Interferon-stimulated response element | −105 |
| Nuclear factor κB | −164 |
| Interferon regulatory factor 2 | −469 |
| Interferon γ-activation sequence | −731 |
Regulation of MEFV mRNA levels by means of soluble inflammatory mediators
Because of the results described above, we stimulated peripheral blood monocytes from healthy control subjects with proinflammatory cytokines and mediators, including IL-1α, IFN-γ, TNF-α, and LPS; antiinflammatory cytokines IL-4, IL-10, and TGF-β; and G-CSF, GM-CSF, and IL-6. After stimulation, we quantified changes inMEFV expression by using a RNase protection assay.
In vitro stimulation of monocytes with the proinflammatory mediators IFN-γ, TNF-α, and LPS for 24 hours resulted in increased levels ofMEFV mRNA, whereas treatment with the antiinflammatory cytokines IL-4, IL-10, and TGF-β reduced MEFV message levels (Figure 5A). Moreover, all 3 antiinflammatory cytokines suppressed IFN-γ–induced and LPS-induced up-regulation of message levels after 24 hours; IL-4 treatment resulted in an almost complete attenuation of MEFV expression (Figure5B). No changes in gene expression were observed with the remaining inflammatory mediators (data not shown).
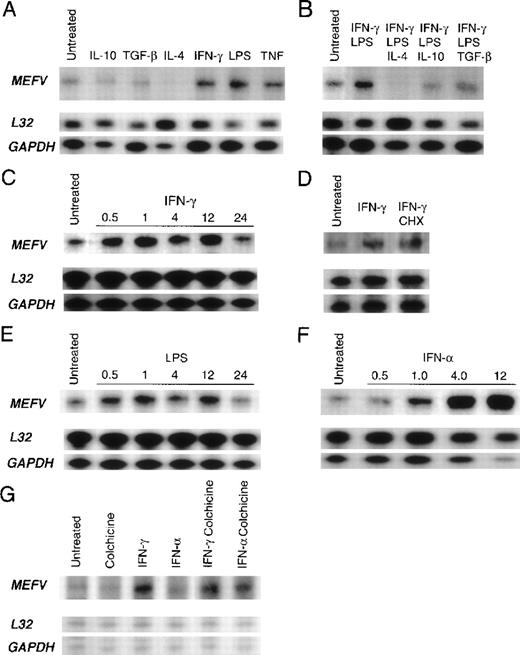
Quantitation of cytokine- and lipopolysaccharide (LPS)-mediated regulation of MEFV mRNA levels in peripheral blood myelomonocytic cells.
Autoradiograms of results obtained with RNase protection assay of total RNA derived from peripheral blood leukocytes by using an MEFVgene-specific riboprobe and 2 housekeeping gene–specific riboprobes (L32 and GAPDH) are shown. Baseline levels ofMEFV mRNA in resting cells before stimulation (untreated) are shown in each panel. (A) Products from monocytes treated for 24 hours in vitro with interleukin (IL) 10, transforming growth factor β (TGF-β), IL-4, interferon (IFN) γ, LPS, or tumor necrosis factor. (B) Products from monocytes treated with LPS and IFN-γ alone and with LPS and IFN-γ in combination with either IL-4, IL-10, or TGF-β. (C) Time course of MEFV induction in monocytes treated with IFN-γ for 0.5, 1, 4, 12, and 24 hours. (D) Products from untreated monocytes and monocytes treated with IFN-γ and both IFN-γ and cycloheximide. (E) Time course of MEFV induction in monocytes treated with LPS for 0.5, 1, 4, 12, and 24 hours. (F) Time course of MEFVinduction in monocytes treated with IFN-α for 0.5, 1, 4, and 12 hours. (G) Products from granulocytes treated with colchicine, IFN-γ, IFN-α, IFN-γ and colchicine, and IFN-α and colchicine.
Quantitation of cytokine- and lipopolysaccharide (LPS)-mediated regulation of MEFV mRNA levels in peripheral blood myelomonocytic cells.
Autoradiograms of results obtained with RNase protection assay of total RNA derived from peripheral blood leukocytes by using an MEFVgene-specific riboprobe and 2 housekeeping gene–specific riboprobes (L32 and GAPDH) are shown. Baseline levels ofMEFV mRNA in resting cells before stimulation (untreated) are shown in each panel. (A) Products from monocytes treated for 24 hours in vitro with interleukin (IL) 10, transforming growth factor β (TGF-β), IL-4, interferon (IFN) γ, LPS, or tumor necrosis factor. (B) Products from monocytes treated with LPS and IFN-γ alone and with LPS and IFN-γ in combination with either IL-4, IL-10, or TGF-β. (C) Time course of MEFV induction in monocytes treated with IFN-γ for 0.5, 1, 4, 12, and 24 hours. (D) Products from untreated monocytes and monocytes treated with IFN-γ and both IFN-γ and cycloheximide. (E) Time course of MEFV induction in monocytes treated with LPS for 0.5, 1, 4, 12, and 24 hours. (F) Time course of MEFVinduction in monocytes treated with IFN-α for 0.5, 1, 4, and 12 hours. (G) Products from granulocytes treated with colchicine, IFN-γ, IFN-α, IFN-γ and colchicine, and IFN-α and colchicine.
Our findings regarding the kinetics of induction revealed that both IFN-γ and LPS up-regulated MEFV message levels within 30 minutes, suggesting that these mediators act directly and independently (Figure 5C and 5E). Accordingly, IFN-γ induction was not inhibited by cycloheximide (Figure 5D), and MEFV can therefore be classified as an IFN-γ immediate early gene. IFN-γ treatment results in activation of the transcription factor STAT1, which up-regulates gene expression by binding to a GAS. The presence of the conserved GAS DNA sequence motif in the promoter region of MEFV further suggests that IFN-γ directly induces MEFV expression. Furthermore, the presence of DNA-sequence identities to NF-κB and AP-1 sites suggests that both LPS and TNF-α may also raise message levels by means of direct induction of transcription and, in this way, independently regulate MEFV expression (Table 1). Maximal induction ofMEFV mRNA levels by IFN-γ and LPS was similar both in magnitude and kinetics, with 5.7- and 6-fold induction, respectively, compared with results in untreated cells at 1 hour. IFN-α, which functions as an antiinflammatory agent in patients with FMF, was the most potent inducer in monocytes, increasing MEFV mRNA levels 58-fold in comparison with levels in untreated cells after 4 hours of stimulation (Figure 5F).
Soluble inflammatory mediators also modulated MEFV message levels in neutrophils. IFN-γ increased message levels (Figure 5G). Interestingly, the combination of colchicine and IFN-α up-regulatedMEFV mRNA levels in neutrophils (Figure 5G), thereby suggesting a role for MEFV in the antiinflammatory effects of both these agents. Colchicine alone did not change MEFV levels in neutrophils. Similarly, in monocytes, colchicine treatment alone did not up-regulate MEFV mRNA levels, nor did it act synergistically with IFN-α to up-regulate message levels above those observed with IFN-α alone (data not shown). Although IFN-γ treatment caused up-regulation of MEFV message levels in both monocytes and granulocytes, several inflammatory mediators that modulated MEFV mRNA levels in monocytes, including LPS, TNF, IFN-α, IL-10, and IL-4, did not change message levels in neutrophils. These findings indicate that MEFV is differentially regulated in the 2 cell types. In addition, no change in MEFV mRNA levels was observed in neutrophils cultured with C5a, GM-CSF, or G-CSF (data not shown). MEFV mRNA levels were highly variable in neutrophils and monocytes from healthy donors. Therefore, to detect cytokine-induced changes in MEFV expression, these analyses used monocytes and neutrophils from donors in whom MEFV mRNA levels were low relative to the detection limits of the assay.
Discussion
The data presented in this paper extend understanding of the expression of a novel gene, mutations in which cause a dramatic inflammatory phenotype. Through studies of in vitro differentiation of CD34+ hematopoietic precursor cells and leukemic cell lines, we demonstrated that MEFV expression begins approximately at lineage commitment, which is earlier in granulocytic differentiation than previously found.1 In addition toMEFV expression in neutrophils, we also observed MEFVexpression in monocytes and eosinophils and showed that monocyte expression is regulated by inflammatory cytokines and LPS. These findings place MEFV in the context of several important proinflammatory and antiinflammatory mediators and establish it as an IFN-γ immediate early gene. In addition, we showed that expression levels in mature granulocytes are under the control of cytokines and of pharmacologic agents.
Several lines of evidence strongly indicated that MEFV is expressed in bone marrow, including direct observation of message in fractionated bone marrow leukocytes by means of RT-PCR and in situ hybridization, induction of expression near the myelocyte stage in CD34+ hematopoietic stem-cell cultures induced to differentiate along the granulocytic lineage, induction of expression in the multipotent myelomonocytic cell line HL60 after induction toward the granulocytic or monocytic lineage, and the appearance of message in nonterminally differentiated monocytic cell lines. The overlap in analyses was necessary because collection of bone marrow can result in contamination by peripheral blood leukocytes, which are known to express MEFV message. The facts that no MEFV expression was observed early in the CD34+ cultures and that expression was induced before full maturation in these cultures and in the HL60 cell line indicate that MEFV is expressed in granulocyte precursors.
In the CD34+ cultures, MEFV expression occurred almost concurrently with the appearance of myelocytes, a stage of differentiation heralded by the production of specific granules; at this time, lineage commitment is clearly established.49Consistent with this observation, expression in the multipotent prepromyelocytic cell line HL60 was observed only after differentiation was induced. The myelocyte stage is characterized by the coordinate induction of specific granule-gene transcription.15 Because of the kinetics of MEFV expression, it was considered possible that MEFV is under the same developmental controls. However, the detection of MEFV mRNA in HL60 cells indicates that this hypothesis is unlikely to be correct because, although HL60 cells are capable of attaining a functionally mature stage of granulocyte differentiation, they do not express specific granule genes.19 Because MEFV does not share the common program of primary granule-gene expression, multiple independent regulatory controls must operate during this stage of differentiation.
In the periphery, MEFV expression was limited to eosinophils, monocytes, and neutrophils. Eosinophilia is present in a variety of pathologic states in which inflammatory lesions are present, including allergy, helminth infections, and inflammatory bowel disease.50 Although eosinophils have not been observed at sites of acute inflammation during attacks of FMF, a regulatory role for MEFV in these cells is suggested by a possible reduced prevalence of asthma in FMF carriers compared with ethnically matched control subjects and an increase in serum levels of eosinophil cationic protein in patients with FMF.51 52 These observations indicate that further analysis of the regulation of MEFVexpression in eosinophils is warranted.
The most prevalent inflammatory cell at sites of acute attacks in FMF is the neutrophil. However, mononuclear cells were observed in the synovial tissues of arthritic joints of patients with FMF,53 and monocytes from FMF patients had a decreased phagocytic and bactericidal capacity54 and increased spontaneous release of a thromboplastin-like procoagulant factor in unstimulated cultured cells.55 Thus, the activity of these cells may be affected by mutations in MEFV. However, as with neutrophils in FMF, functional analyses have so far provided no clear clues about the function of MEFV in monocytes.56
In response to inflammatory stimuli, neutrophils can regulate the inflammatory response by means of the production of soluble proinflammatory and antiinflammatory mediators, and it is clear that monocytes play a central role in coordinating inflammatory processes.57 58 Our data suggest that MEFV is a downstream element in cytokine-induced regulatory cascades. Proinflammatory activators, including the Th1 cytokine IFN-γ, TNF, and LPS, up-regulated monocyte MEFV message levels; and antiinflammatory cytokines, including the Th2 cytokines IL-4 and IL-10 and TGF-β, down-regulated monocytic MEFV expression. Up-regulation by IFN-γ occurred rapidly and the effect was not inhibited by cycloheximide, showing that MEFV is an IFN-γ immediate early gene and suggesting that MEFV plays a direct role in IFN-γ activation. It is intriguing that of the stimulators used singly in these studies, only IFN-γ up-regulated MEFVlevels in both monocytes and granulocytes. These results suggest thatMEFV may mediate common IFN-γ–specific mechanisms of action in these cells.
It should be noted that not all proinflammatory mediators up-regulatedMEFV levels in neutrophils. No changes were observed on stimulation of cells with C5a, IL-8, or TNF, indicating thatMEFV up-regulation is not a general property of activated cells. IFN-γ–mediated leukocyte activation functions primarily through the modulation of gene expression. More than 50 IFN-γ–induced genes have been described, including major histocompatibility complex (MHC) classes I and II transcriptional regulators (eg, p48, IRF1), apoptosis-related genes, and the 3MEFV homologs PML-1, acid finger protein, and 52-kd SSA/Ro autoantigen.59 The fact that IFN-γ directly and rapidly induced MEFV expression indicates thatMEFV plays a role in the early stages of IFN-γ–mediated cell activation, and perhaps, given the importance of this cytokine in host defense, regulates phagocyte response in general.
The effects of TNF and LPS on mature monocytes are also mediated by activation of transcription factors that control inflammatory regulator and effector genes. IFN-γ activates STAT-1, which in turn binds to a conserved cis-acting promoter element (the GAS), whereas LPS and TNF directly regulate transcription principally by activating NF-κB.60-64 Our examination of the DNA sequences upstream of the start of MEFV transcription revealed sequences that match the consensus GAS site at position −731 and the NF-κB binding site at position −164. Laboratory studies to test whether these factors regulate MEFVexpression directly are warranted.
The balance of production of Th1 and Th2 provides both a critical regulatory circuit for the adaptive immune response and communication to the innate immune response, as indicated by the profound effects of these classes of cytokines on monocytes and neutrophils. IFN-γ mediates up-regulation of a variety of effector functions, including phagocytosis, superoxide production, microbicidal activity, and antitumor activity. Th-2 cytokines (ie, IL-4 and IL-10) can have strong deactivating effects on these cells and directly antagonize the effects of proinflammatory agents.65,66 Repression of Th1 cytokine–induced gene expression by Th2 cytokines, as observed by us for MEFV, is common for several genes of consequence to the inflammatory response, including MHC class II, iNOS, and IRF-1.67-69 Although no direct data on MEFVmechanism of action were obtained from the study of purified leukocytes from patients with FMF, the facts that FMF is a recessive disease and that no cases in which a carrier has symptoms of the disease have been reported suggest that the role of MEFV is as an inhibitor of inflammation. If MEFV does play an antiinflammatory role, then the facts that MEFV message levels are increased by proinflammatory and Th1 mediators and diminished significantly by antiinflammatory and Th2 mediators suggest that the gene functions in a negative-feedback loop that is specific for Th1 and proinflammatory-mediator activation of myelomonocytic cells.
Given this hypothesis, it is intriguing that both IFN-α and colchicine, the only therapeutic agents known to ameliorate FMF attacks, also up-regulated MEFV message levels. IFN-α is known to have pleiotropic effects, suppressing symptoms in some autoinflammatory diseases70-72 but exacerbating inflammation in others.73,74 This agent may exert its therapeutic effect in FMF by causing overexpression of a functionally deficient MEFV-gene product by stimulating Th1 or proinflammatory pathways. Consistent with this idea, administration of IFN-α during an FMF attack increased levels of C-reactive protein and erythrocyte sedimentation rate while effecting a rapid, complete alleviation of abdominal pain.75 The effect of colchicine in suppressing inflammation in granulocyte-mediated diseases, including FMF, gout, and Behçet disease, has been thought to be due to either inhibition of leukocyte adhesion and migration or effects on inflammatory signaling.76-79 Our observation of up-regulation of MEFV mRNA levels by IFN-α and colchicine, which has not been reported before, suggests that MEFV may play a previously unrecognized role in the specific physiologic effects of these agents. In this context, it is of note that the effects of colchicine on MEFV expression in vitro were observed only in conjunction with IFN-α stimulation. However, when used alone therapeutically, colchicine blocks FMF attacks, suggesting that IFNs or similar Th1-inducing agents may be already be present at inflammatory sites in patients with FMF, although at levels insufficient to inhibit pathogenesis. If this model of therapeutic effect is correct, then up-regulation of MEFV mRNA by other agents, specifically IFN-γ, may be worth evaluating as an alternative treatment for FMF attacks.
Because mutations in MEFV result in a dysregulation of the inflammatory response, it was hypothesized before its identification that MEFV was likely to be an inflammatory regulator. The data presented here support this hypothesis. MEFV is expressed specifically in the primary cellular effectors and regulators of inflammation, and gene expression is controlled by characterized and fundamental cytokine-mediated pathways of the inflammatory cascade. Given the dramatic pathophysiologic consequences of MEFVmutations, our data suggest that MEFV is an inflammatory regulator of the phagocyte-mediated inflammatory response. In regard to etiologic aspects of FMF, defects in several inflammatory regulators have been hypothesized to underlie the periodic dysregulation of the inflammatory response, including changes in the activities or production of lipocortins,80 TNF,81 and a C5a inhibitor.82 Our data suggest an alternative hypothesis—that MEFV mediates a Th-1–responsive negative-feedback loop during proinflammatory activation of myeloid cells and that the pathophysiologic features of FMF result from defects in this inhibitory activity. Moreover, these data clearly establish an expanded context in which to study the function ofMEFV as it pertains to the normal function of the inflammatory response, perhaps with important implications for infectious, rheumatic, and autoinflammatory diseases.
Acknowledgments
We thank the Clinical Pathology Department and the Department of Transfusion Medicine of the Warren Magnuson Clinical Center, National Institutes of Health, for supplying cells from healthy donors and for advice regarding this work.
Supported by a grant (to M.A.) from the Max Kade Foundation.
Reprints:Michael Centola, Arthritis and Rheumatism Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Building 10, Room 9N210, National Institutes of Health, Bethesda, MD 20892-1820; e-mail: centolam@arb.niams.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal