Transgenic and gene targeted mice have contributed greatly to our understanding of the mechanisms underlying B-cell development. We describe here a model system that allows us to apply molecular genetic techniques to the analysis of human B-cell development. We constructed a retroviral vector with a multiple cloning site connected to a gene encoding green fluorescent protein by an internal ribosomal entry site. Human CD34+CD38− fetal liver cells, cultured overnight in a combination of stem cell factor and interleukin-7 (IL-7), could be transduced with 30% efficiency. We ligated the gene encoding the dominant negative helix loop helix (HLH) factor Id3 that inhibits many enhancing basic HLH transcription factors into this vector. CD34+CD38− FL cells were transduced with Id3-IRES-GFP and cultured with the murine stromal cell line S17. In addition, we cultured the transduced cells in a reaggregate culture system with an SV-transformed human fibroblast cell line (SV19). It was observed that overexpression of Id3 inhibited development of B cells in both culture systems. B-cell development was arrested at a stage before expression of the IL-7R. The development of CD34+CD38− cells into CD14+ myeloid cells in the S17 system was not inhibited by overexpression of Id3. Moreover, Id3+ cells, although inhibited in their B-cell development, were still able to develop into natural killer (NK) cells when cultured in a combination of Flt-3L, IL-7, and IL-15. These findings confirm the essential role of bHLH factors in B-cell development and demonstrate the feasibility of retrovirus-mediated gene transfer as a tool to genetically modify human B-cell development.
IT IS WELL-ESTABLISHED that the generation of lineage-committed cells from pluripotent stem cells is tightly controlled by transcription factors (TFs). Several TFs have been identified that contribute to the steering of B-cell differentiation in the mouse. Some of these factors control B-cell differentiation at a developmental stage before commitment to the B-cell lineage. For example, the Ikaros factor is essential for differentiation of lymphoid cells, including B cells.1 TFs that direct B-cell development include Pax5,2EBF,3 Sox4,4 and E12 and E47.5,6These latter factors, encoded by the E2A gene, belong to the family of basic helix loop helix (bHLH) proteins. This family is particularly interesting, because its members control lineage commitment of different cell types in organisms varying from yeast to mammals.7 Two highly conserved bipartite domains characterize these proteins. A motif of basic residues mediates binding to a consensus E-box (CANNTG) sequence present in promoters or enhancers of target genes.8,9 The helix-loop-helix domain, mainly consisting of hydrophobic residues, permits homodimerization or heterodimerization of these proteins. The involvement of the E2A gene products in B-cell differentiation in the mouse has been well-documented. Two groups generated E2A mutant mice and have shown that a severe block occurs at an early stage of B-cell differentiation before Ig gene rearrangements.5,6 More recently, Bain et al10 presented evidence that both E12 and E47 allow commitment to the B-cell lineage. Two other factors, HEB and E2-2, are also involved in B-cell development, because mice deficient for these factors demonstrate perturbations of B-cell development as well.11 It seems, therefore, that B-cell development is controlled by the combined dosage of many bHLH factors. Inhibitors of DNA binding (Id) proteins control transcriptional activity of bHLH factors. This family of HLH factors comprises 4 members (Id1-4) that are highly homologous in their HLH domains.12,13 Id proteins can heterodimerize with bHLH factors, but lack a basic DNA binding domain, therefore blocking transcriptional activity of bHLH factors. Sun14 has demonstrated that constitutive expression of the Id1 gene under control of the B-cell–specific mb-1 promotor impairs mouse B-cell development, and the cells exhibit a similar phenotype to the one described for the E2A-deficient mice.5,6 These investigators also documented that Id1 and Id2 mRNA levels are relatively high in tumor cell lines representative for early B-cell progenitors and low in cell lines representing late B-cell precursors.15 More recently, Li et al16reported that Id1 levels are high in an uncommitted B-cell progenitors in the bone marrow and sharply downregulate when the cells enter a B-cell committed stage, whereas at the same time, E12 and E47 are upregulated. These findings together have led to the hypothesis that B-cell differentiation is controlled by the relative proportions of E2A and Id proteins.
Transgenic technology has provided a powerful tool for the study of lymphoid development in the mouse. We have developed a novel model system that allows us to apply these molecular genetic techniques to the analysis of human T-cell development. This system makes use of retrovirus-mediated gene transfer to introduce genes into T-cell progenitors. The effects of the introduced genes on T-cell development are investigated in a fetal thymic organ culture. Using this system, we documented that overexpression of Id3 in human thymic progenitors by retroviral delivery resulted in a complete inhibition of T-cell differentiation and promotion of natural killer (NK) cell development.17 We demonstrate here that overexpression of Id3 into uncommitted CD34+CD38−CD10− fetal liver (FL) cells strongly inhibits their capacity to develop into B cells upon coculture with the murine stromal cell line S17 or with a human stromal cell line in a novel reaggregation assay. B-cell development was inhibited at a very early stage of development before acquisition of the interleukin-7 receptor α (IL-7Rα) chain and CD10.
MATERIALS AND METHODS
Construction of the vector.
We have constructed bicistronic vectors with a gene of interest linked to a downstream internal ribosomal entry site (IRES) and a marker gene that allow independent translation of the products of both genes in the transduced target cells.17 The IRES-GFP sequence was ligated into the LZRS vector, and a polylinker was placed downstream of the gag and upstream of the IRES sequence. The Id3 coding sequence was cloned from the pCDNAId3 plasmid (gift of Dr C. Murre, University of California at San Diego, San Diego, CA) by polymerase chain reaction (PCR) using oligonucleotide primers with appropriate linkers. The product was ligated between the Xho I and SnaBI site of the polylinker from our plasmid LZRS-linker-IRES-GFP to obtain the retroviral vector LZRS-Id3-IRES-GFP. As a control, we constructed into the LZRS-linker-IRES-GFP vector a form of murine Id3 with a mutation in the HLH domain (R72P; kindly provided by Dr G. Kato, Johns Hopkins University, Baltimore, MD) that has lost the capacity to dimerize with bHLH factors.18 The HLH domains of human and murine Id3 are identical. A modified version of GFP (enhanced GFP) was used in this study and was obtained from Clontech (Palo Alto, CA). A schematic representation of the vector is shown in Fig 1. Helper-free recombinant retrovirus was produced after transfection into a 293T-based amphotropic retroviral packaging cell line, Phoenix.19
Structure of the LZRS IRES-GFP vector. The important features of this vector are the polylinker upstream of the IRES sequence to insert the different genes of interest and the selection marker GFP downstream of the IRES sequence. The LZRS vector contains a puromycin resistance gene and the EBNA-1 nuclear retention sequence outside the LTRs.
Structure of the LZRS IRES-GFP vector. The important features of this vector are the polylinker upstream of the IRES sequence to insert the different genes of interest and the selection marker GFP downstream of the IRES sequence. The LZRS vector contains a puromycin resistance gene and the EBNA-1 nuclear retention sequence outside the LTRs.
Isolation of FL progenitor cells.
FL tissue was obtained from elective abortions. Gestational age was determined by measurement of the diameter of the skull and ranged from 14 to 17 weeks. The use of this tissue was approved by the Medical Ethical Committee of the Netherlands Cancer Institute and was contingent upon informed consent. Human FL cells were isolated by gentle disruption of the tissue, followed by density gradient centrifugation over Ficoll-Hypaque (Lymphoprep; Nycomed Pharma, Oslo, Norway). Subsequently, CD34+ cells were positively selected by immunomagnetic cell sorting (varioMACS; Miltenyi, GmbH, Germany), as described elsewhere.20 The resulting population was stained with anti-CD10 fluorescein isothiocyanate (FITC; Immunotech, Marseille, France), anti-CD38 phycoerythrin (PE; Becton Dickinson, San Jose, CA), and anti-CD34 Tricolor (Immunotech). CD34+CD38−CD10−, CD34+CD38+CD10−, and CD34+CD38+CD10+ cells were then sorted with a FACstar plus (Becton Dickinson). The purity of the populations used in this study was greater than 97%.
Reverse transcriptase-PCR (RT-PCR) assays.
RNA was isolated from fluorescence-activated cell sorting (FACS)-sorted human FL subpopulations using TRIzol reagent (GIBCO BRL, Grand Island, NY) according to the manufacturer’s instructions and reverse transcribed using a poly-dT15 oligonucleotide (Promega Corp, Leiden, The Netherlands) and 400 U of Moloney murine leukemia virus (M-MuLV) reverse transcriptase (GIBCO) at 37°C for 1 hour. Semiquantitative RT-PCR was performed essentially as described previously.21 PCR assays were performed in 30 mL reaction volumes using the appro- priate diluted amounts of cDNA template, 2 mmol/L MgCl2, 0.25 mmol/L each dNTP, 10 pmol/L of each primer, and 0.8 U Taq polymerase (GIBCO) in 1× buffer (10 mmol/L Tris-HCl, pH 8.5, 50 mmol/L KCl). Reaction conditions were a 5-minute denaturing step at 95°C followed by 31 cycles of 1 minute at 95°C, 1 minute at 55°C, and 1 minute at 72°C. PCR products were separated on 1% agarose gels, stained with ethidium bromide, and analyzed by video-densitometry using the Eagle Eye still video system and Eagle Sight software (Stratagene, La Jolla, CA). Three separate FL donor pools were analyzed and each PCR was repeated 4 to 6 times. The E2A-, E12-, and E47-specific Id1, Id3, RAG-1, and HPRT primers that were used are as follows: E2A-sense: 5′-AGCACGTTTGGTGGCCTGC-3′; E47-antisense: 5′-AGCACCTCGTCCGTACTGC-3′; E12-antisense: 5′-TCCTCGTCCTCGTCTGGGC-3′; Id1-sense: 5′-CGCTGTCTGTCTGAGCAGAGC-3′; Id1-antisense: 5′-ATCTCGCCGTTGAGGGTGCTG-3′; Id3-sense: 5′-TCGGAACGCAGTCTGGCCATC-3′; Id3-antisense: 5′-CTCGGCTGTCTGGATGGGAAG-3′; RAG-1 sense: 5′-GAACACACTTTGCCTTCTCTTTGG-3′; RAG-1 antisense: 5′-GCTTTGCCTCTTGCTTTCTCGTT-3′; HPRT-sense: 5′-TATGGACAGGACTGAACGTCTTGC-3′; and HPRT-antisense: 5′-GACACAAACATGATTCAAATCCCTGA-3′.
Retroviral transduction of human CD34+ FL cells.
The transduction procedure using recombinant human fibronectin fragments CH-296 was performed according to Hanenberg et al.22,23 The sorted CD34+CD38−CD10− FL cells were cultured overnight in the presence of 10 ng/mL human IL-7 and 20 ng/mL stem cell factor (SCF; both from R&D, Abingdon, UK). The cells were then transduced by 6 hours of incubation with virus supernatant in nontissue culture-treated Falcon petri dishes (3 cm; Becton Dickinson) precoated with 30 mg/mL recombinant human fibronectin fragment CH-29624 (RetroNectin; Takara, Otsu, Japan). After transduction, the cells were washed twice. The transduction efficiency was tested by determining the percentage of GFP+ cells 2 days after transduction using flow cytometry analysis.
Monolayer cell culture.
The monolayer cell culture assays were performed with the murine bone marrow stromal cell line S17 (a kind gift from Dr David Rawlings, UCLA, Los Angeles, CA). Two days before their use in coculture experiments, S17 cells were plated in 96-well flat-bottom plates (3 to 5 × 103 cells per well in 100 μL medium). Monolayer cultures were initiated by seeding 5 to 10 × 103 FL progenitor cells to the wells precoated with S17 cells. Half of the culture medium was replaced by fresh medium once a week. After incubation in Yssel’s medium25 supplemented with 5% fetal calf serum (FCS; BioWhittaker, Verviers, Belgium), to which we will refer as complete medium, cells were harvested, stained with PE- or Tricolor-labeled human specific antibodies, and analyzed by flow cytometry for cell surface phenotype and GFP expression. In some experiments, part of the recovered cells was assayed for their capacity to develop into NK cells.
Reaggregation assays.
The reaggregation assay is based on a similar culture system developed by Anderson et al26 to test the development of mouse T cells. The human SV19 stromal cell line27 was used in this study. Briefly, 3 to 5 × 104 FL progenitor cells were mixed with 10 to 15 × 104 stromal cells in a 1.5-mL Eppendorf tube and spun into a pellet. After removal of the supernatant, the cell pellet was vortexed in a remaining volume of 2 to 5 μL, gently expelled as a discrete standing drop on the surface of a nucleopore filter (0.8 μm; Millipore, Cork, Ireland) that was layered on a gelfoam raft (Upjohn Co, Kalamazoo, MI), and cultured in an air/liquid interphase with complete medium. Reaggregates were then harvested and gently dissociated into a homogeneous cell suspension for further cell surface analysis by flow cytometry.
Differentiation of NK cells.
After a period of 2 weeks of culture of transduced CD34+CD38−CD10− FL cells on a monolayer of S17 stroma, the cells were harvested, counted, and tested for their capacity to develop into NK cells, as previously described.20 Five thousand to 10,000 FL cells were then further cultured in U-bottom 96-well plates with complete medium in the presence of 25 U/mL Flt-3 ligand (a kind gift from Dr M.G. Roncarolo, DNAX, Palo Alto, CA), 10 ng/mL IL-7 (our own source), and 10 ng/mL IL-15 (Immunex, Seattle, WA). The cultures were maintained in a humidified atmosphere at 37°C in 5% CO2 for 10 days. Cells were then stained and their antigenic phenotype was analyzed by flow cytometry.
Flow cytometry analysis and monoclonal antibodies (MoAbs).
Flow cytometry analysis was performed using a FACScan (Becton Dickinson). FL cells were incubated for 10 minutes on ice with normal mouse serum Ig to prevent nonspecific binding of the MoAbs and then stained with fluorochrome-conjugated MoAbs. The following mouse MoAbs (FITC-, PE-, or Tricolor-coupled) were used: CD10-Tricolor (Caltag, San Francisco, CA); CD34-Tricolor, CD19-Tricolor, CD45-Tricolor, anti-c-kit-PE and CD10-FITC were purchased from Immunotech; CD19-PE, CD45RA-PE, CD38-PE, CD20-PE, CD22-PE, CD56-PE, and CD14-PE were obtained from Becton Dickinson. For detection of the IL-7Rα chain, an indirect staining was used with anti–IL-7Rα (Immunotech) and PE-labeled F(ab)2 fragments of a goat antimouse antibody (Zymed, San Francisco, CA). After staining, cells were washed twice in phosphate-buffered saline (PBS) supplemented with 2% bovine serum albumin (BSA) and 0.01% NaN3 (PBA) and analyzed by flow cytometry. All steps of the staining procedure were performed on ice, and appropriate fluorochrome-conjugated, isotype-matched control Igs were used in all experiments.
RESULTS
Expression of RAG-1, Id, and bHLH mRNAs in distinct CD34+FL populations.
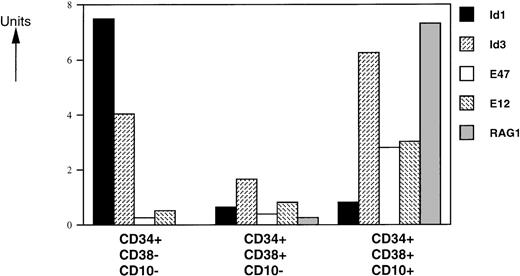
Human pluripotent stem cells are enriched in a subpopulation of CD34+ cells that express no or only low levels of CD38.28 Differentiation of these cells is accompanied by upregulation of the CD38 marker.28 Within the CD34+ compartment, CD10 is expressed on lymphoid progenitors,29,30 early B-cell precursors,31and T-cell precursors.32 To investigate the role of bHLH factors and Id proteins in human B-cell development, we analyzed the mRNA levels of 2 Id proteins (Id1 and Id3), the E2A gene products (E12 and E47), and RAG-1 in sorted CD34+CD38−CD10−, CD34+CD38+CD10−, and C34+CD38+CD10+ human FL cells by semiquantitative PCR (Fig 2). We found that RAG-1 mRNA is absent in the CD34+CD38−CD10−immature population, as expected. Low and high levels of RAG-1 mRNA were observed in CD34+CD38+CD10− and CD34+CD38+CD10+ cells, respectively. E12- and E47-specific messages were present at similar levels in CD34+CD38−CD10− and CD34+CD38+CD10− cells and were 2- to 3-fold higher in the CD34+CD38+CD10+ population. Expression of Id1 was high in the CD34+CD38−CD10− subset and was dramatically decreased with upregulation of CD38 and CD10. Id3 levels decreased 2-fold in the transition from the CD34+CD38−CD10− to CD34+CD38+CD10− stages and had the highest expression levels in the more mature CD34+CD38+CD10+ population.
Expression levels of HLH, bHLH, and RAG-1 mRNAs on CD34+ FL-sorted populations. RNA was isolated from FACS-sorted human FL subpopulations from 3 separate donor pools and analyzed by RT-PCR. mRNA levels were compared with HPRT expression and expressed in arbitrary units (HPRT = 1). One representative experiment from 1 donor pool is shown. All assays were performed 4 to 6 times. Similar data were obtained with analyses of 2 other donor pools.
Expression levels of HLH, bHLH, and RAG-1 mRNAs on CD34+ FL-sorted populations. RNA was isolated from FACS-sorted human FL subpopulations from 3 separate donor pools and analyzed by RT-PCR. mRNA levels were compared with HPRT expression and expressed in arbitrary units (HPRT = 1). One representative experiment from 1 donor pool is shown. All assays were performed 4 to 6 times. Similar data were obtained with analyses of 2 other donor pools.
Retroviral transduction of primitive CD34+CD38− FL hematopoietic progenitor cells.
It has been demonstrated that CD34+ cells of different origins can be efficiently transduced after culture in multiple cytokines. However, CD34+CD38− cells appear to be difficult to transduce.33 We isolated CD34+CD38− cells from FL using Tricolor-labeled anti-CD34 and PE-labeled anti-CD38 as described in Materials and Methods (Fig 3A). The use of an anti-CD38 antibody labeled with PE allows for a better depletion of CD38+ cells than when using an FITC-labeled antibody, due to the greater sensitivity of the PE fluorochrome (for a discussion, see Lanier and Recktenwald34). The CD34+ cells, rigorously depleted of CD38+ cells, were cultured overnight in a combination of SCF and IL-7 and incubated for 6 hours with cell-free virus-containing supernatants. Two facilitating reagents were used, the lipofectin reagent dotap17 and recombinant fibronectin-derived peptides (Retro- Nectin; Takara).22Transduction efficiencies in the presence of RetroNectin fragments were 32.7% ± 4.2% (n = 8; a representative experiment is shown in Fig3B), as determined by measuring GFP expression by flow cytometry. Transduction efficiencies using dotap were, in general, 2 to 3 times lower when performed in parallel with fibronectin fragments (results not shown).
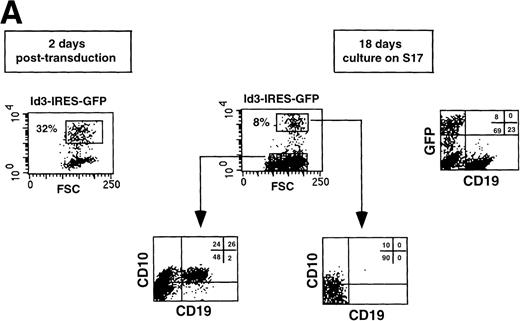
Retroviral transduction of CD34+CD38−CD10− FL cells does not affect their capacity to develop along the B-cell lineage. The CD34+CD38−CD10− FL population was isolated (A), transduced with virus harboring the IRES-GFP sequence, and cultured for 18 days on a stromal cell monolayer (B). Cell suspensions were stained with the indicated Tricolor- and PE-labeled antibodies and analyzed on a FACScan. Isotype-matched control antibodies were used in all experiments.
Retroviral transduction of CD34+CD38−CD10− FL cells does not affect their capacity to develop along the B-cell lineage. The CD34+CD38−CD10− FL population was isolated (A), transduced with virus harboring the IRES-GFP sequence, and cultured for 18 days on a stromal cell monolayer (B). Cell suspensions were stained with the indicated Tricolor- and PE-labeled antibodies and analyzed on a FACScan. Isotype-matched control antibodies were used in all experiments.
Retroviral transduction does not affect the B-cell differentiation pattern of CD34+CD38−CD10− FL progenitors.
We have observed that CD34+CD38−CD10− FL cells developed into B cells after 2 to 3 weeks of culture on a monolayer of S17 stromal cells, consistent with observations with CD34+ cord blood cells.35 To determine whether retroviral transduction per se would affect the capacity of CD34+CD38−CD10− FL precursors to differentiate along the B-lineage pathway, we sorted this population (Fig 3A), infected the cells with a retrovirus carrying the IRES-GFP sequences, and cocultured the transduced cells with S17. As shown in Fig 3B, 30% of the cells were GFP+ 2 days after transduction. After 18 days of coculture, 12% of the recovered cells were positive for GFP, and no significant differences could be detected in the phenotype of the B cells generated when compared with the GFP− fraction.
Overexpression of Id3 inhibits B-cell development from CD34+CD38−CD10−uncommitted FL precursors.
To investigate whether bHLH factors are involved in human B-cell development, we overexpressed Id3 in FL hematopoietic progenitors and monitored the fate of the transduced cells in a B-cell differentiation assay. Purified CD34+CD38−CD10− FL cells were cultured overnight with a combination of SCF and IL-7 and subsequently transduced with retrovirus harboring the Id3-IRES-GFP sequence. After 18 days of culture on a monolayer of the murine S17 stromal cell line, cell numbers were 10- to 12-fold increased and 8% of the cells expressed the transgene (Fig4A). The Id3− fraction developed into B cells and exhibited a phenotype similar to the control transduced cells (Figs 4A and 3B). Twenty-six percent of the cells were CD10+CD19+. In striking contrast, Id3+ cells lacked expression of CD19, demonstrating that enforced expression of Id3 in CD34+CD38−CD10− FL cells strongly blocked B-cell differentiation. In this experiment, some cells were CD10+CD19− (10%). However, we found that expression of CD10 was variable. In some experiments, we observed a small percentage of CD10+CD19−cells in the cultures with Id3-transduced CD34+CD38− cells, whereas in other experiments these cells were not detectable. We consider it therefore likely that the small proportion of CD10+ cells in Id3 cultures that we observed in some experiments is an artifact due to nonspecific staining of the Tricolor labeled anti-CD10. Cells recovered after culture of the FL progenitors in this culture system also contain a population of CD14+ cells. As shown in Fig 4B, the percentage of CD14+ cells present in both Id3− and Id3+ fractions was identical, indicating that the inhibitory effect of Id3 in this assay is restricted to the B-cell lineage.
Overexpression of Id3 inhibits differentiation of B cells from CD34+CD38−CD10− FL uncommitted precursors but does not affect development into monocytes. CD34+CD38−CD10− FL cells were sorted, incubated with retroviral supernatant harboring the Id3-IRES-GFP sequence, and subsequently cocultured with S17 stroma. After 18 days, the cells were harvested and expression of B-cell (A) and monocyte (B) antigens was analyzed by flow cytometry. Thirty-two percent of the cells were GFP+ 2 days after transduction. A representative experiment of 3 performed is shown.
Overexpression of Id3 inhibits differentiation of B cells from CD34+CD38−CD10− FL uncommitted precursors but does not affect development into monocytes. CD34+CD38−CD10− FL cells were sorted, incubated with retroviral supernatant harboring the Id3-IRES-GFP sequence, and subsequently cocultured with S17 stroma. After 18 days, the cells were harvested and expression of B-cell (A) and monocyte (B) antigens was analyzed by flow cytometry. Thirty-two percent of the cells were GFP+ 2 days after transduction. A representative experiment of 3 performed is shown.
It was recently reported that Id2 could induce apoptosis in a myeloid precursor cell line independent of its capacity to dimerize with bHLH factors.36 Therefore, we transduced CD34+CD38− FL cells with a mutant of Id3, ΔId3, that has no capacity to dimerize with bHLH factors as E4718 and tested the B-cell precursor activity of these cells. In these experiments, we performed a more extensive phenotypic analysis to characterize the Id3-arrested developmental stage in more detail. Figure 5 shows that ΔId3-transduced cells developed into CD19+ cells, whereas, in contrast, Id3-transduced cells fail to develop into CD19+ B cells. The ΔId3-transduced cells were somewhat less efficient in developing into B cells compared with the untransduced cells in the same sample, suggesting an Id3 effect independent of the dimerization domain. However, the inhibition by Id3 is much stronger than that of ΔId3, indicating that Id3 blocks B-cell development mainly by sequestering pertinent bHLH factors. Further analysis of the cells that were blocked by Id3 showed that Id3 also inhibited the appearance of IL-7Rα+ cells. It is also shown that the proportion of c-kit+CD45RA− cells in the Id3 cultures (30%) was higher than that in the control or ΔId3 cultures (9% to 12%). The percentages of c-kit−CD45RA+cells in the control and ΔId3 cultures were higher than in the Id3 culture, but there was no difference in the proportions of c-kitlowCD45RA+ in the Id3 and ΔId3 cultures. In the untransduced cultures, a higher proportion of ckitlowCD45RA+ was found. This could mean that a proportion of ckitlowCD45RA+ cells in the untransduced cultures is derived from precursors that are difficult to transduce. We also analyzed expression of CD38. It is shown in Fig 5that the control and untransduced cell populations contain a population of CD38high cells. Part of those CD38high cells react with an anti-CD19 MoAb. The anti-CD19 MoAb used in this staining was Tricolor labeled and stained less efficiently than the PE-labeled antibody used in the CD10/CD19 staining. Nonetheless, one can observe in the Id3 cultures a strong reduction of these CD38highcells and almost no CD38highCD19+ cells. Figure5 also shows that Id overexpression does not affect the percentage of CD34+ cells. Although the percentage of CD34+CD38high cells in the Id3-transduced cells is slightly lower than in the untransduced cells, a similar percentage of CD34+CD38high cells was present in the ΔId3-transduced cells. It seems, therefore, that Id3 inhibits B-cell development at a CD34+c-kit+/lowCD45RA±IL-7Rα−CD38highCD10−CD19−stage. The phenotype of these cells may correspond with that of an early lymphoid precursor, as proposed by Ryan et al.30
Id3 inhibits B-cell development at an early stage of development. The inhibition by Id3 is much stronger than by a mutant of Id3 that is unable to dimerize with bHLH factors. CD34+CD38−CD10− FL cells were sorted, transduced either with Id3-IRES-GFP or with Did3-IRES-GFP, and subsequently cocultured with S17 stroma. After 14 days, the cultures were analyzed with the indicated antibodies. This experiment is representative of 3 similar experiments.
Id3 inhibits B-cell development at an early stage of development. The inhibition by Id3 is much stronger than by a mutant of Id3 that is unable to dimerize with bHLH factors. CD34+CD38−CD10− FL cells were sorted, transduced either with Id3-IRES-GFP or with Did3-IRES-GFP, and subsequently cocultured with S17 stroma. After 14 days, the cultures were analyzed with the indicated antibodies. This experiment is representative of 3 similar experiments.
Id3+ FL cells recovered after 2 weeks of coculture with S17 stroma can develop into NK cells.
As shown in the previous experiments, the Id3+ population recovered after culture on a S17 stromal cell monolayer did not contain any CD19+ B cells, whereas generation of cells expressing CD14 was not affected. To determine whether the Id3+population contains precursor cells that had retained the capacity to develop into the NK cell lineage, they were cultured in a cytokine combination containing IL-15, as previously described.20Control cultures were established with cells recovered after culture of the IRES-GFP–transduced population with S17, as shown in Fig 3B. We observed that the cell numbers increased 3- to 4-fold and the levels of GFP expression were maintained during the 2-week culture period (Fig 6). As expected, no B cells developed from the Id3+ population, whereas 57% of the CD19+ cells were found in the control GFP progeny (Fig 6). This observation indicates that the culture conditions used in this experiment supported the proliferation and/or maturation of B-cell precursors present after culture of FL progenitors on a monolayer of S17 stroma. Most importantly, we observed that CD56+ NK cells could be detected in the progeny of both the Id3− and the Id3+ fractions (Fig 6). The absolute numbers of CD56+ Id3+ cells were 25% higher than the numbers of CD56+ cells found in the control cultures (Table 1).
Both Id3− and Id3+ FL cells recovered after culture on S17 stroma retain the capacity to develop into NK cells. Cells harvested after coculture of transduced FL cells on a monolayer of S17 stroma were further cultured in a cytokine combination consisting of Flt-3L, IL-7, and IL-15. Ten days later, cell suspensions were analyzed by flow cytometry for cell surface expression of CD19 and CD56 antigens. The markers were set to exclude greater than 98% of cells stained with irrelevant control antibodies.
Both Id3− and Id3+ FL cells recovered after culture on S17 stroma retain the capacity to develop into NK cells. Cells harvested after coculture of transduced FL cells on a monolayer of S17 stroma were further cultured in a cytokine combination consisting of Flt-3L, IL-7, and IL-15. Ten days later, cell suspensions were analyzed by flow cytometry for cell surface expression of CD19 and CD56 antigens. The markers were set to exclude greater than 98% of cells stained with irrelevant control antibodies.
Absolute Numbers of CD56+ NK Cells Upon Sequential Culture of Transduced CD34+CD38− FL Cells Upon S17 and Cytokines
| FL Cells Transduced With . | % GFP+ Cells . | No. of Recovered GFP+ Cells Per Input of 10,000 GFP+ Cells . | ||
|---|---|---|---|---|
| Before Culture . | After Culture . | Total . | CD56+ Cells . | |
| IRES-GFP | 12 | 13 | 43,000 | 10,400 |
| Id3-IRES-GFP | 8 | 6 | 30,000 | 12,900 |
| FL Cells Transduced With . | % GFP+ Cells . | No. of Recovered GFP+ Cells Per Input of 10,000 GFP+ Cells . | ||
|---|---|---|---|---|
| Before Culture . | After Culture . | Total . | CD56+ Cells . | |
| IRES-GFP | 12 | 13 | 43,000 | 10,400 |
| Id3-IRES-GFP | 8 | 6 | 30,000 | 12,900 |
Cells were harvested after coculture of transduced FL cells on a monolayer of S17 stroma and the percentage of GFP+ cells was determined to get the indicated percentage of GFP+cells before culture. The harvested cells were further cultured in a cytokine combination consisting of Flt-3L, IL-7, and IL-15. Two weeks later, cell suspensions were analyzed by flow cytometry for GFP expression (% GFP+ cells after culture) and cell surface expression of the CD56 antigen. The percentage of GFP+ cells and therefore the absolute numbers of the input were different. To allow for a meaningful comparison, we expressed the output numbers per input number of 10,000 cells.
Inhibition of human B-cell differentiation by overexpression of Id3 is not due to intrinsic characteristics of the in vitro model used.
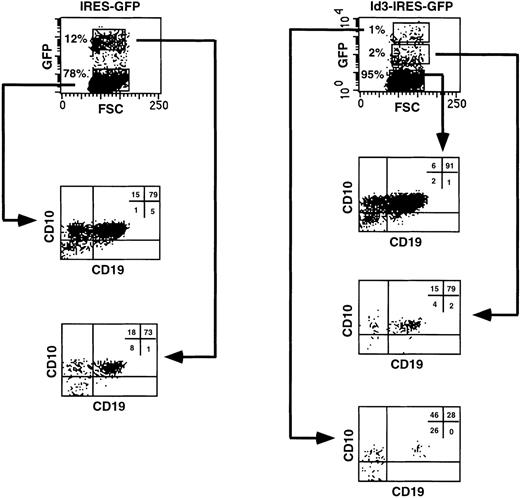
It is possible that the inhibitory effect on human B-cell differentiation observed when FL progenitors overexpressing Id3 were cocultured with the murine stromal cell line S17 was peculiar for this in vitro model. To investigate this point, we explored a different culture system that will be described in detail elsewhere (Jaleco et al, manuscript submitted) that efficiently supports B-cell development from uncommitted FL hematopoietic cells. CD34+CD38−CD10− FL cells, transduced with the IRES-GFP or the Id3-IRES-GFP sequences, expressed similar levels of GFP at the onset of the culture (28% and 30%, respectively). The transduced cells were reaggregated with the human SV19 stromal cell line and cultured for 2 to 3 weeks. In the control culture, 12% of the cells recovered expressed high levels of GFP, and the proportion of CD10+CD19+ B cells present was identical in both the GFP+ and the GFP− fractions (73% and 79%, respectively; Fig 7, left panel). In contrast, only 1% of the FL cells transduced with Id3-IRES-GFP had high expression of Id3, whereas 2% expressed low levels and the remaining cells were Id3− (Fig 7, right panel). These fractions were analyzed separately for expression of CD10 and CD19, and we found that the proportion of CD10+CD19+ cells decreased dramatically with increase in GFP expression (91%, 79%, and 28% in the Id3−, Id3low, and Id3highpopulations, respectively; Fig 7, right panel). This was not due to enhanced proliferation and differentiation of the transduced cells into other cell lineages. We determined the absolute numbers of Id3-GFPhighCD10+CD19+ cells recovered at the end of the culture period and compared these with the numbers of GFPhigh cells recovered from the cultures with control, GFP-transduced cells (Table 2). This analysis showed that the numbers of CD10+CD19+ cells found in the samples expressing high levels of Id3 were 18-fold lower than those recovered in the controls, confirming the strong inhibition of B-cell differentiation upon overexpression of Id3 on FL progenitors (Table 2). Although not shown, CD19+ cells were also generated in a reaggregate culture with CD34+CD38− FL cells transduced with ΔId3, whereas precursors overexpressing Id3, tested in parallel, generated much lower numbers of CD19+cells.
B-cell differentiation of Id3-transduced FL progenitors is strongly blocked but not completely inhibited in a reaggregation culture with human stromal cells. CD34+CD38−CD10− FL cells were sorted, transduced with the IRES-GFP or Id3-IRES-GFP vectors, and reaggregated with the SV19 human stromal cell line for 2 to 3 weeks. After the culture period, cells were harvested, stained with the indicated antibodies, and analyzed by flow cytometry. Quadrant limits defining positive and negative cells were set up using cells stained with isotype-matched control antibodies. Thirty percent and 28% of the cells were GFP+ 2 days after transduction in control- and Id3-transduced samples, respectively. The results shown are representative of 2 similar experiments.
B-cell differentiation of Id3-transduced FL progenitors is strongly blocked but not completely inhibited in a reaggregation culture with human stromal cells. CD34+CD38−CD10− FL cells were sorted, transduced with the IRES-GFP or Id3-IRES-GFP vectors, and reaggregated with the SV19 human stromal cell line for 2 to 3 weeks. After the culture period, cells were harvested, stained with the indicated antibodies, and analyzed by flow cytometry. Quadrant limits defining positive and negative cells were set up using cells stained with isotype-matched control antibodies. Thirty percent and 28% of the cells were GFP+ 2 days after transduction in control- and Id3-transduced samples, respectively. The results shown are representative of 2 similar experiments.
Absolute Numbers of B Cells Harvested From Reaggregate Cultures
| FL Cells Transduced With . | % GFP+ Cells . | No. of Recovered GFP+ Cells Per Input of 10,000 GFP+ Cells . | ||
|---|---|---|---|---|
| Before Culture . | After Culture . | Total . | CD10+CD19+ Cells . | |
| IRES-GFP | 28 | 12 | 43,000 | 31,000 |
| Id3-IRES-GFP | 30 | 1 | 6,000 | 1,680 |
| FL Cells Transduced With . | % GFP+ Cells . | No. of Recovered GFP+ Cells Per Input of 10,000 GFP+ Cells . | ||
|---|---|---|---|---|
| Before Culture . | After Culture . | Total . | CD10+CD19+ Cells . | |
| IRES-GFP | 28 | 12 | 43,000 | 31,000 |
| Id3-IRES-GFP | 30 | 1 | 6,000 | 1,680 |
CD34+CD38−CD10− FL cells were sorted and transduced with the IRES-GFP or Id3-IRES-GFP containing virus. Two days later, the percentage of GFP+cells was determined to get the percentage before culture. The cells were reaggregated with the SV19 human stromal cell line for 3 weeks. After the culture period, cells were harvested and analyzed for cell surface expression of GFP, CD10, and CD19 by flow cytometry.
DISCUSSION
In this report, we show that enforced expression of the dominant negative HLH Id3 factor in human FL progenitor cells severely impairs B-cell differentiation at an early stage. CD34+CD38−CD10− FL progenitors were transduced with Id3, followed by coculture with a monolayer of murine S17 stroma. The Id3+ cells recovered after 2 weeks of culture did not express the B-cell specific antigen CD19, in contrast to the Id3− cells and the cells generated in cultures with the control GFP- or with ΔId3-transduced CD34+CD38−CD10− cells. The effect of Id3 in the S17 system was B-cell–specific, because development of CD14+ myeloid cells was not affected. Moreover, Id3-GFP+ cells that accumulate in these cultures were able to develop into NK cells in the presence of Flt-3L, IL-7, and IL-15, confirming our previous findings that Id3-overexpression does not affect NK cell development.17 We also observed a strong inhibition of B-cell development in the reaggregate culture system, indicating that the inhibition of B-cell development by Id3 is not peculiar for the S17 culture system. It was recently reported that the related HLH factor Id2 could induce apoptosis in a myeloid precursor cell line independent of its dimerization motif.36 The observation that a mutant of Id3 that is unable to dimerize with bHLH factors18 is not affecting B-cell development in the S17 system indicates that inhibition of B-cell development is based mainly on its ability to sequester bHLH factors. This observation strongly suggests that E12 and E47, which are sequestered by Id3, are also essential for human B-cell development.
In the S17 assays (Fig 5), no IL-7Rα+ cells were detected, and the appearance of these cells was also inhibited in the reaggregation assay (results not shown). Examination by FACS of the phenotype of cells harvested from the S17 cultures that fell outside of the life gate and that are supposedly dying cells failed to provide evidence for the presence of dying IL-7Rα+ cells in the cultures with Id3-transduced cells, indicating a truly developmental block before acquisition of the IL-7Rα chain. A detailed analysis of the developmental stage that is inhibited by Id3 indicated that progression is arrested at the CD34+c-kitdimCD45RA±IL-7Rα−CD38highCD10−CD19−stage. In a model proposed by Ryan et al,30 cells with this phenotype represent early lymphoid progenitors. According to Ryan et al,30 these cells already express RAG-1. Unfortunately, S17 cells do not induce RAG-1 expression in human CD34+CD38− precursors,37 and we were therefore unable to verify whether overexpression of Id3 results in inhibition of RAG-1 expression. Our finding that Id3 inhibits development in the S17 system at an early progenitor stage could indicate that bHLH factors control induction of B-cell commitment. Indeed, data in the mouse support the notion that E12 and E47 are required for B-cell lineage commitment.10 In E2A-deficient mice, B-cell development was arrested at a stage in which cells express low levels of B220, and CD43 have germline μ0 transcripts and express IL-7Rα mRNA, but do not express RAG or have IgH gene rearrangements.5,6 10Interestingly, IL-7Rα+ cells could not be detected in the B-cell differentiation cultures with Id3-transduced cells, suggesting that the developmental block of human B-cell development in the S17 system imposed by Id3 overexpression could be earlier than that of E2A deficiency. The discrepancy in IL-7Rα expression in Id3 transduced S17 cultures and E2A-deficient mice could be due to the fact that in the mouse this was analyzed in vivo, and in human, it was analyzed in an in vitro assay. It is also possible that these discrepancies are due to species differences. Alternatively, our observations may mean that bHLH factors different from E12 and E47 are required for generation of IL7Rα+ precursor cells. It is interesting to note that CD34+ cells expressing Id3 develop normally into NK cells. This combined with the observation that Id3 inhibits appearance of IL-7Rα+ cells suggests that NK development can proceed without traversing an IL-7Rα+stage.
Previously, we have shown that overexpression of Id3 inhibits generation of T cells and promotes development of NK cells from CD34+ FL cells.17 Thus, Id3 inhibits development of both T and B cells but not of NK cells. One interpretation of these observations is that NK cells branch before T and B cells, implying that a common T/B-cell progenitor exists in the FL that is unable to develop into NK cells. Although this possibility cannot be formally excluded, this seems unlikely for several reasons. T and NK cells are much more similar to each other than to B cells with respect to expression of cell surface markers and function (discussed in Lanier et al38 and Spits et al39). In addition, it has been demonstrated that the murine thymus contains progenitor cells that can develop into T and NK cells but not into B cells.40 What could then be the reason that Id3 overexpression inhibits T- and B-cell but not NK cell development? It seems most logical to assume that this is due to an effect of Id3 overexpression on a process common to T and B cells. Gene rearrangements of their respective receptors are the most conspicuous common feature of T and B cells and are essential for the development and function of both cell types. It is possible that bHLH factors are contributing to control these gene rearrangements. Some observations in the mouse support this notion. When E47 is overexpressed in a pre-T–cell line, RAG expression and gene rearrangements at the IgH locus are induced.41 However, in the S17 system, inhibition of human B-cell development by Id3 occurs independently from RAG expression and IgH gene rearrangements, which should imply that bHLH factors are required for B-cell differentiation independently of their possible effects on IgH gene rearrangements.
It has been shown that the levels of the E12 and E47 mRNA increase and Id1 and Id2 decrease in cell lines representative for subsequent stages of B-cell differentiation, suggesting that the balance of activating bHLH factors and dominant negative Id proteins drive B-cell differentiation. Recently, Li et al16 identified subpopulations of early B-cell progenitors based on expression of AA4.1 and B220 and absence of CD24. The earliest progenitor (fraction Ao) is AA1+ and B220− and can differentiate into multiple hematopoietic lineages. Upon appearance of B220, μ0 mRNA is strongly upregulated and RAG1/2 mRNA become detectable. These cells (fraction A1) are developmentally committed to the B-cell lineage but have not yet initiated IgH gene rearrangements. In the mouse, Id1 expression is high in a subset of CD43+CD24−B220− cells (fraction Ao) and decreases when these cells acquire germline Ig transcripts and commit to the B-cell lineage (fractions A1and A2).16,42 E12 and E47 are relatively low in fraction Ao cells and increase upon progression to fraction A1 and A2 cells. Similarly, we observed the highest levels of E12 and E47 in the more mature population of CD34+CD38+CD10+ FL cells that includes pro-B and pre-B cells.30 In addition, Id1 expression was high in uncommitted CD34+CD38−CD10− cells and was decreased dramati- cally in the more mature CD34+CD38+CD10− and CD34+CD38+CD10+ populations. These data are consistent with findings in the mouse system.15 It is interesting to note that Id1 has been shown to inhibit RAG-1 activation, suggesting that the relative ratio of E2A to Id1 is possibly critical for expression of the RAG-1 gene in the B-cell lineage and consequently for the onset of the B-cell developmental pathway.14 However, Id3 mRNA expression, although decreasing when the cells transit from CD34+CD38− into CD34+CD38+ cells, increases upon further differentiation. The increasing levels of Id3 when cells differentiate from CD10− into CD10+ would present an apparent paradox, because enforced expression of Id3 in CD10− cells inhibited B-cell differentiation. However, it is possible that upregulation of Id3 drives downregulation of RAG expression when pre-B cells proliferate.43 On the other hand, one may assume that upregulation of E2A expression and probably also of other bHLH factors in the CD10+ subset is sufficient to neutralize the negative effects of Id3. It thus becomes important to further investigate the expression levels and functional relevance of other factors previously implicated in mouse B-cell differentiation, such as E2-2 and HEB.
This report demonstrates the feasibility of retrovirus-mediated gene transfer as a way to genetically modify human B-cell differentiation in vitro. This technology was applied here and in previous reports17 44 to investigate the role of bHLH factors in T-cell, B-cell, and NK cell development and can be used to elucidate the role of other transcription factors in human lymphoid cell development.
ACKNOWLEDGMENT
The authors thank the staff of the Bloemenhovekliniek in Heemstede (The Netherlands) for their cooperation in obtaining fetal tissue. We also thank E. Noteboom for expert technical assistance with flow cytometry. We are grateful to D. Rawlings for the S17 cell line, to G. Nolan for the LZRS vector, to M.G. Roncarolo for her gift of recombinant Flt-3L, to G. Kato for the gift of ΔId3, to Takara Shuzo Co, Ltd for the CH-296 fibronectin fragment, and to Immunex for recombinant IL-15. We thank Dr A. Kruisbeek for critical reading of the manuscript.
A.C.J. was supported by a PhD fellowship from Junta Nacional De Investigação Cientı́fica e Tecnológica, Lisbon, Portugal.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hergen Spits, PhD, Division of Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands; e-mail: hergen@nki.nl.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal