Abstract
The retinoblastoma gene product (pRb) is involved in both cell cycle regulation and cell differentiation. pRb may have dual functions during cell differentiation: partly by promoting a cell cycle brake at G1 and also by interacting with tissue-specific transcription factors. We recently showed that pRb mediates differentiation of leukemic cell lines involving mechanisms other than the induction of G1 arrest. In the present study, we investigated the role of pRb in differentiation of human bone marrow progenitor cells. Human bone marrow cells were cultured in a colony-forming unit–granulocyte-macrophage (CFU-GM) assay. The addition of antisense RB oligonucleotides (-RB), but not the addition of sense orientated oligonucleotides (SO) or scrambled oligonucleotides (SCR), reduced the number of colonies staining for nonspecific esterase without affecting the clonogenic growth. Monocytic differentiation of CD34+ cells supported by FLT3-ligand and interleukin-3 (IL-3) was correlated to high levels of hypophosphorylated pRb, whereas neutrophilic differentiation, supported by granulocyte colony-stimulating factor (G-CSF) and stem cell factor (SCF), was correlated to low levels. The addition of -RB to liquid cultures of CD34+ cells, supported with FLT3-ligand and IL-3, inhibited monocytic differentiation. This was judged by morphology, the expression of CD14, and staining for esterase. Moreover, the inhibition of monocytic differentiation of CD34+ cells mediated by -RB, which is capable of reducing pRb expression, was counterbalanced by an enhanced neutrophilic differentiation response, as judged by morphology and the expression of lactoferrin. CD34+ cells incubated with oligo buffer, -RB, SO, or SCR showed similar growth rates. Taken together, these data suggest that pRb plays a critical role in the monocytic and neutrophilic lineage commitment of human bone marrow progenitors, probably by mechanisms that are not strictly related to control of cell cycle progression.
THE RETINOBLASTOMA protein (pRb) is believed to play a key role in the regulation of the cell cycle. The activity of pRb depends on the degree of phosphorylation, which is cell cycle phase dependent. When hypophosphorylated, pRb mediates a brake at the G1-phase of the cell cycle by inhibiting the E2F family of transcription factors, which activate genes for progression from G1- to S-phase (reviewed in Weinberg1). Recently, mechanisms by which hypophosphorylated pRb silences the transcription of mitotic genes were described. pRb recruits a histone deacetylase to E2F, thus creating a complex that probably by modulating local chromatin structure prevents expression from E2F-regulated genes.2,3 In addition to inhibiting cell proliferation, pRb also promotes specific differentiation programs.4-6 Mice with targeted retinoblastoma gene (RB) function show defective differentiation in tissues such as blood and brain. The hematopoietic abnormalities described in the Rb −/− mice include reduced formation of hepatic blood islands, coupled with an increased proportion of immature nucleated erythroid cells.7-9 A role for pRb in human hematopoiesis has been implicated; high levels of pRb are induced and sustained during erythroid differentiation, whereas pRb is downregulated during granulocytic maturation.10 The mechanisms governing hematopoietic lineage commitment and differentiation are not fully understood. However, increasing evidence indicates a role for transcription factors in orchestrating these processes. The functions of hematopoietic transcription factors seem to be modulated in a complex manner by a variety of signals and interactions (reviewed in Shivdasani and Orkin11 and Tenen et al12). In particular, pRb has been shown to interact with hematopoietic transcription factors, including NF-IL-6, PU1, and Elf1.13-15 Recently, we demonstrated that pRb is involved in differentiation responses in leukemic cell lines involving mechanisms distinct from those specifically linked to the control of cell cycle progression.16 Data indicating that pRb plays a critical role in the monocytic and neutrophilic lineage commitment of normal human bone marrow progenitors are presented.
MATERIALS AND METHODS
Oligonucleotides.
Two phosphorothioate-modified oligodeoxynucleotides were designed to target RB mRNA and designated antisense RB oligonucleotide 1 (AS1) and antisense RB oligonucleotide 2 (AS2), respectively. Three phosphorothioate-modified oligodeoxynucleotides were used as controls for unspecific or toxic effects of the antisense oligos: 2 sense-orientated oligonucleotides (SO1 and SO2) and a scrambled oligonucleotide (SCR). The sequence of AS1 read 5′ GGG GGT TTT GGG CGG CAT GAC 3′, complementary to 21 nucleotides starting from position −3 to +18 bp in the RB mRNA coding sequence.16 AS2 read 5′ GTG AAC GAC ATC TCA TCT AGG 3′, complementary to 21 nucleotides starting from position 465 to 485 bp in the RB mRNA coding sequence.10,17The sequence of SO1 read 5′ GTC ATG CCG CCC AAA ACC CCC 3′, corresponding to position −3 to +18 bp in the RB cDNA coding sequence. The sequence of SO2 read 5′ GGA TCT ACT CTA CAG CAA GTG 3′, corresponding to the reversed orientation of AS2. The SCR sequence read 5′ TAC TGG CTA AGC CTA GCA TGA 3′ and corresponded to a randomly scrambled AS2.10AS1, AS2, and SO1 were initially synthesized by Bio Molecular Resource Facility, Lund University (Lund, Sweden). AS2, SO2, and SCR were later synthesized by Cyber Gene AB (NOVUM, Huddinge, Sweden).
Hematopoietic growth factors.
The growth of unfractionated mononuclear bone marrow cells in clonogenic assay (colony-forming unit–granulocyte-macrophage [CFU-GM]) was supported by 10% conditioned medium from the bladder carcinoma cell line 5637. The liquid culture of CD34+ bone marrow cells was supported by purified recombinant human growth factors at predetermined optimal concentrations. Interleukin-3 (IL-3) and FLT3-ligand (FL; both available due to the generosity of Immunex, Seattle, WA) were used at 25 ng/mL and 50 ng/mL, respectively. Stem cell factor (SCF) and granuloctye colony-stimulating factor (G-CSF; Amgen Inc, Thousand Oaks, CA) were both used at 50 ng/mL.
Assay for granulocyte-macrophage progenitors.
Liquid culture of human CD34+ bone marrow cells.
CD34+ bone marrow cells were initially isolated as described elsewhere.20 Later, mononuclear bone marrow cells obtained by gradient centrifugation were positively selected for CD34 expression by MidiMACS (Miltenyi Biotech, Auburn, CA), followed by staining with antihuman CD34 fluorescein isothiocyanate (FITC; Becton Dickinson, San Jose, CA) and antihuman CD38 phycoerythrin (PE) or isotype control antibodies. CD34+CD38+ cells were then sorted on FACSVanatage (Becton Dickinson).
The purity of CD34+ cells was greater than 90%, as determined by flow cytometry. Isolated CD34+ cells were grown in a defined complete Iscove’s modified Dulbecco’s medium (IMDM) medium as previously described.20 Cells incubated with FL plus IL-3, promoting predominantly monocytic differentiation,20 were seeded at 5,000 cells/mL. Cells stimulated with G-CSF plus SCF, to promote mainly granulocytic differentiation,21 were seeded at 2,000 cells/mL. Cells were expanded in a volume of 1 mL for 14 days at 37°C, 5% CO2 in fully humidified air. Cells destined for determination of pRb levels were cultured in a volume of 20 to 100 mL.
Flow cytometric evaluation of cell surface phenotype.
Morphological evaluation of hematopoietic differentiation.
Cytospin preparations of cultured cells were stained with May-Grünwald-Giemsa and examined with light microscopy.
Immunocytochemistry.
Lactoferrin was detected as described,23 with the following modifications. Cytospin preparations of cultured cells were fixed in 4% formaldehyde in 0.1 mol/L phosphate buffer (pH 7.0) for 20 minutes. Cells were then permeabilized by incubation for 30 minutes in Tris-buffered saline (TBS; 50 mmol/L Tris-HCl, pH 7.6, 150 mmol/L NaCl) containing 1% Triton X-100. Unspecific binding was blocked by incubating cells with TBS containing 1% bovine serum albumin (BSA; Sigma, St Louis, MO). Polyclonal rabbit antibodies to human lactoferrin (A106; DAKO D 306, Glostrup, Denmark) were diluted to 0.86 μg/mL in TBS containing 0.25% BSA and allowed to bind during 1 hour of incubation. The slides were then washed 3 times in TBS, followed by incubation for 1 hour with alkaline phosphatase-conjugated swine antirabbit Igs (D306; DAKO D 306) diluted 60-fold in TBS containing 0.25% BSA. After 3 washes in TBS, alkaline phosphatase activity was detected by Fast Red/Naphtol AS-MS (Sigma Fast; Sigma) as a substrate. After washing in running tap water, slides were counterstained in Mayers hematoxylin. All steps were performed at room temperature.
Western blot analysis.
Cells were lysed at 4°C in a buffer consisting of 50 mmol/L Tris HCl (pH 8.0), 0.15 mol/L NaCl, 5 mmol/L EDTA (pH 8.0), and 0.5% Nonidet P40 (KEBO, Stockholm, Sweden), including 1 tablet of a protease inhibitor cocktail (Complete; Boehringer Mannheim, Mannheim, Germany) per 50 mL of lysis buffer. Incubation on ice for 1 hour followed, before approximately 10 seconds of vigorous vortexing. After lysis, the DNA was removed by centrifugation at 37,500g for 1 hour at 4°C. The lysates were then subjected to specific immunoprecipitation by the addition of 1 μg of mouse sc 102 anti-Rb MoAb (Santa Cruz Laboratories, Santa Cruz, CA). Immunocomplexes were allowed to form and adsorbed to a mixture of protein A- (Pharmacia, Uppsala, Sweden) and protein G-sepharose (Sigma) at 4°C overnight. After centrifugation, the precipitate was washed 3 times with lysis buffer. The immunoprecipitated proteins were separated on a 6% precast Tris-Glycine gel electrophoresis (Novex, San Diego, CA). Proteins were electroforetically transferred to Immobilon-P membranes (Millipore, Bedford, MA) in blotting buffert (39 mmol/L glycin [pH 9.2], 48 mmol/L Tris, 1.3 mmol/L sodium dodecyl sulfate [SDS], 20% methanol) at 20 V for 1 hour. After incubation in blocking buffert (5% dry milk powder in PBS) for 0.5 hour, the membrane was incubated for 2 hours with mouse sc 102 anti-Rb MoAb at 0.5 μg/mL in the following PBS buffer (pH 7.3 to 7.4): 0.137 mol/L NaCl, 8 mmol/L Na2HPO4 × 2 H2O, 2.7 mmol/L KCl, 1.5 mmol/L KH2PO4, and 0.05% Tween 20. The membrane was then probed with alkaline phosphatase-conjugated rabbit antimouse IgG (DAKO A/S, Copenhagen, Denmark) diluted 1:500 in PBS buffer for 1 hour. The specific proteins were then visualized with chromogenic substrates (5-bromo-4-chloro-3-indolyl phosphate-p-toluidine salt [ICN] at 0.05 mg/mL and nitro blue tetrazolium [Sigma] at 0.1 mg/mL) in a blocking buffert without BSA (10.6 mmol/L Na2CO3, 39.3 mmol/L NaHCO3) that contained 4 mmol/L MgCl2.
RESULTS
Effects of α-RB on growth and differentiation of unfractionated mononuclear human bone marrow cells.
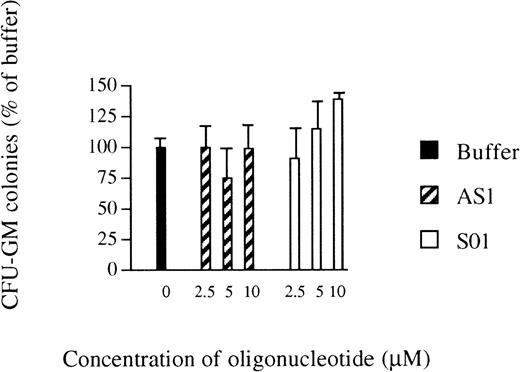
The inhibitory effects of AS1 on myelomonocytic differentiation of U-937 cells16 led us to investigate the effects of antisense RB oligonucleotides on the growth of normal human bone marrow progenitor cells. The total number of CFU-GM colonies formed was not affected by the addition of AS1 at 2.5 to 10.0 μmol/L, compared with the addition of oligo buffer only, except by a slight reduction at 5.0 μmol/L, indicating that AS1 did not exert toxic effects (Fig 1). However, reduced staining for unspecific esterase, a marker of monocytic differentiation,19 was observed after incubation with AS1, compared with the addition of oligo buffer. However, the addition of SO1 did not reduce staining for unspecific esterase compared with the addition of oligo buffer (Table 1). Similar data were obtained using another set of oligonucleotides; the addition of AS2 reduced staining for unspecific esterase compared with the addition of buffer, whereas, after the addition of SO2 or SCR, positive staining for unspecific estherase was comparable to addition of buffer alone (Table 1). These data suggested that suppression of pRb inhibits differentiation of myeloid progenitors along the monocytic lineage, without affecting their clonogenic growth.
Effects of oligonucleotides on the number of CFU-GM colonies. Bone marrow cells were obtained by gradient centrifugation and cultured in agar. Oligo buffer only (▪), antisense RB oligonucleotides (AS1; ▨), or sense orientated oligonucleotides (SO1; □) were added at indicated concentrations on days 0 and 7. Colonies (>40 cells) were scored on day 14. Values shown are the percentages of the number of colonies formed when oligo buffer only was added. Results shown represent the mean values from 3 separate experiments (bars ± SEM). The total number of colonies formed per dish with oligo buffer added was 63 ± 7 (SEM).
Effects of oligonucleotides on the number of CFU-GM colonies. Bone marrow cells were obtained by gradient centrifugation and cultured in agar. Oligo buffer only (▪), antisense RB oligonucleotides (AS1; ▨), or sense orientated oligonucleotides (SO1; □) were added at indicated concentrations on days 0 and 7. Colonies (>40 cells) were scored on day 14. Values shown are the percentages of the number of colonies formed when oligo buffer only was added. Results shown represent the mean values from 3 separate experiments (bars ± SEM). The total number of colonies formed per dish with oligo buffer added was 63 ± 7 (SEM).
Effects of Oligonucleotides on CFU-GM Staining for Unspecific Esterase
| Additive . | Positive . | Mixed Positive . | Mixed Negative . | Negative . |
|---|---|---|---|---|
| Oligo buffer | 3 ± 0.6 | 39 ± 4 | 23 ± 2 | 33 ± 6 |
| AS1 2.5 μmol/L | 0 ± 0.0 | 20 ± 4 | 40 ± 4 | 44 ± 6 |
| AS1 5.0 μmol/L | 0 ± 0.0 | 20 ± 1 | 31 ± 6 | 48 ± 5 |
| AS1 10.0 μmol/L | 0 ± 0.0 | 26 ± 1 | 26 ± 5 | 47 ± 6 |
| CO 2.5 μmol/L | 2 ± 0.5 | 44 ± 2 | 19 ± 2 | 44 ± 6 |
| CO 5.0 μmol/L | 0 ± 0.0 | 49 ± 3 | 22 ± 3 | 33 ± 3 |
| CO 10.0 μmol/L | 3 ± 0.7 | 48 ± 2 | 19 ± 1 | 30 ± 2 |
| Additive . | Positive . | Mixed Positive . | Mixed Negative . | Negative . |
|---|---|---|---|---|
| Oligo buffer | 3 ± 0.6 | 39 ± 4 | 23 ± 2 | 33 ± 6 |
| AS1 2.5 μmol/L | 0 ± 0.0 | 20 ± 4 | 40 ± 4 | 44 ± 6 |
| AS1 5.0 μmol/L | 0 ± 0.0 | 20 ± 1 | 31 ± 6 | 48 ± 5 |
| AS1 10.0 μmol/L | 0 ± 0.0 | 26 ± 1 | 26 ± 5 | 47 ± 6 |
| CO 2.5 μmol/L | 2 ± 0.5 | 44 ± 2 | 19 ± 2 | 44 ± 6 |
| CO 5.0 μmol/L | 0 ± 0.0 | 49 ± 3 | 22 ± 3 | 33 ± 3 |
| CO 10.0 μmol/L | 3 ± 0.7 | 48 ± 2 | 19 ± 1 | 30 ± 2 |
Mononuclear bone marrow cells were obtained by gradient centrifugation and cultured in agar for 14 days. At days 0 and 7, antisense RB oligonucleotides (AS1) or control oligonucleotides (CO), respectively, were added at the indicated concentrations. At day 14, agar dishes were fixated and stained for unspecific esterase as a marker of monocytic differentiation. A colony (>40 cells) was defined as positive if all of its cells stained and mixed positive if a majority but not all of its cells stained. A colony was defined as negative if none of its cells stained and mixed negative if a minority of its cells stained. Values shown are percentages of colonies. Results shown represent the mean values from 3 separate experiments ± SEM.
pRb levels during monocytic or neutrophilic differentiation of CD34+ progenitor cells, respectively.
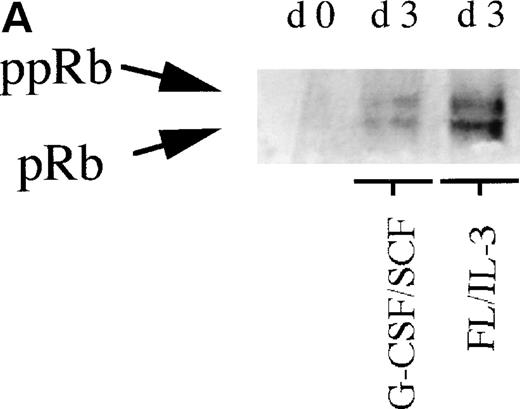
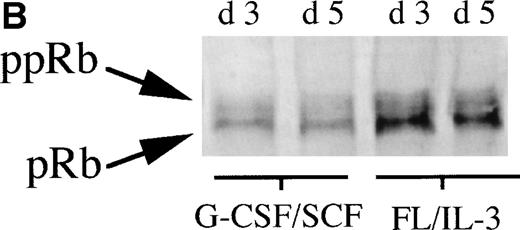
To further investigate the role of pRb in monocytic and neutrophilic differentiation, purified CD34+ bone marrow cells were cultured in the presence of FL plus IL-3, promoting predominantly monocytic differentiation20(Table 2) or, in the presence of G-CSF plus SCF, favoring mainly neutrophilic differentiation21 (Table2). On day 0, during uninduced conditions, the purified CD34+ bone marrow cells expressed very low levels of pRb, which is in line with previous findings10(Fig 2A). In cells stimulated towards neutrophilic differentiation with G-CSF plus SCF, a slight increase in the level of pRb was detected on days 3 and 5. In contrast, in CD34+ cells stimulated with FL plus IL-3, promoting monocytic differentiation, a considerable increase in pRb levels was detected at days 3 and 5 as compared with day 0 (Fig 2A and B). Moreover, at day 3, the ratio between hyperphosphorylated pRb and hypophosphorylated pRb seemed higher in cells stimulated with G-CSF plus SCF than in cells supported with FL plus IL-3 (Fig 2A and B). These data suggest that monocytic commitment, in contrast to neutrophilic commitment, is correlated to a considerable upregulation of levels of hypophosphorylated pRb. A similar, but less pronounced, difference in pRb level between cells incubated with G-CSF plus SCF and cells stimulated with FL plus IL-3 was observed also on days 7 and 14 (data not shown).
Effects of Oligonucleotides on Myeloid Differentiation of CD34+ Cells
| Cytokines/ Oligos . | Total Cell No. . | Monocytes . | Neutrophils . | Blasts . |
|---|---|---|---|---|
| FL, IL-3 | 269 ± 49 | 70 ± 8 | 18 ± 4 | 12 ± 6 |
| FL, IL-3/CO | 274 ± 44 | 73 ± 9 | 15 ± 2 | 13 ± 7 |
| FL, IL-3/AS2 | 257 ± 35 | 28 ± 5 | 62 ± 9 | 11 ± 3 |
| G-CSF, SCF | 303 ± 42 | 19 ± 4 | 68 ± 11 | 13 ± 4 |
| G-CSF, SCF/CO | 292 ± 50 | 22 ± 6 | 67 ± 8 | 11 ± 6 |
| G-CSF, SCF/AS2 | 286 ± 39 | 19 ± 6 | 74 ± 8 | 8 ± 4 |
| Cytokines/ Oligos . | Total Cell No. . | Monocytes . | Neutrophils . | Blasts . |
|---|---|---|---|---|
| FL, IL-3 | 269 ± 49 | 70 ± 8 | 18 ± 4 | 12 ± 6 |
| FL, IL-3/CO | 274 ± 44 | 73 ± 9 | 15 ± 2 | 13 ± 7 |
| FL, IL-3/AS2 | 257 ± 35 | 28 ± 5 | 62 ± 9 | 11 ± 3 |
| G-CSF, SCF | 303 ± 42 | 19 ± 4 | 68 ± 11 | 13 ± 4 |
| G-CSF, SCF/CO | 292 ± 50 | 22 ± 6 | 67 ± 8 | 11 ± 6 |
| G-CSF, SCF/AS2 | 286 ± 39 | 19 ± 6 | 74 ± 8 | 8 ± 4 |
CD34+ cells were incubated for 14 days in complete medium. Medium was supplemented with FL at 25 ng/mL plus IL-3 at 50 ng/mL to promote monocytic differentiation or with G-CSF plus SCF, both at 50 ng/mL, to promote neutrophilic differentiation. At days 0, 5, and 10, control oligonucleotides (CO) or antisense RB oligonucleotides (AS2), respectively, were added, each time at a concentration of 5.0 μmol/L. At day 14, cell morphology was determined after May-Grünwald-Giemsa staining of cytospin preparations. Results on morphology are the mean percentages of 200 counted cells from three separate experiments ± SEM. Total cell number was scored at day 14 and values are thousands of cells per milliliter ± SEM.
(A) Levels of pRb during neutrophilic and monocytic differentiation on days 0 and 3. Cells selected for CD34 expression were incubated on day 0 with cytokines favoring neutrophilic (G-CSF and SCF, both at 50 ng/mL) or monocytic (FL at 50 ng/mL and IL-3 at 25 ng/mL) differentiation, respectively. On day 0, 0.55 × 106 cells not stimulated with any cytokines were subjected to protein extraction, followed by specific immunoprecipitation with an anti-pRb antibody (sc-102; Santa Cruz), 6% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blot analysis. On day 3, 0.55 × 106 cells from cells incubated with G-CSF/SCF or FL/IL-3, respectively, were subjected to protein extraction, followed by specific immunoprecipitation with an anti-pRb antibody (sc-102; Santa Cruz), 6% SDS-PAGE, and Western blot analysis. pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated). (B) Levels of pRb during neutrophilic and monocytic differentiation on days 3 and 5. Cells selected for CD34 expression were incubated on day 0 with cytokines, favoring neutrophilic (G-CSF and SCF, both at 50 ng/mL) or monocytic (FL at 50 ng/mL and IL-3 at 25 ng/mL) differentiation, respectively. After 3 and 5 days of expansion, respectively, 1.5 × 106 cells were subjected to protein extraction, followed by specific immunoprecipitation with an anti-pRb antibody (sc-102; Santa Cruz), 6% SDS-PAGE, and Western blot analysis. pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated).
(A) Levels of pRb during neutrophilic and monocytic differentiation on days 0 and 3. Cells selected for CD34 expression were incubated on day 0 with cytokines favoring neutrophilic (G-CSF and SCF, both at 50 ng/mL) or monocytic (FL at 50 ng/mL and IL-3 at 25 ng/mL) differentiation, respectively. On day 0, 0.55 × 106 cells not stimulated with any cytokines were subjected to protein extraction, followed by specific immunoprecipitation with an anti-pRb antibody (sc-102; Santa Cruz), 6% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blot analysis. On day 3, 0.55 × 106 cells from cells incubated with G-CSF/SCF or FL/IL-3, respectively, were subjected to protein extraction, followed by specific immunoprecipitation with an anti-pRb antibody (sc-102; Santa Cruz), 6% SDS-PAGE, and Western blot analysis. pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated). (B) Levels of pRb during neutrophilic and monocytic differentiation on days 3 and 5. Cells selected for CD34 expression were incubated on day 0 with cytokines, favoring neutrophilic (G-CSF and SCF, both at 50 ng/mL) or monocytic (FL at 50 ng/mL and IL-3 at 25 ng/mL) differentiation, respectively. After 3 and 5 days of expansion, respectively, 1.5 × 106 cells were subjected to protein extraction, followed by specific immunoprecipitation with an anti-pRb antibody (sc-102; Santa Cruz), 6% SDS-PAGE, and Western blot analysis. pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated).
α-RB inhibit monocytic differentiation and enhance neutrophilic differentiation of human CD34+ bone marrow progenitor cells.
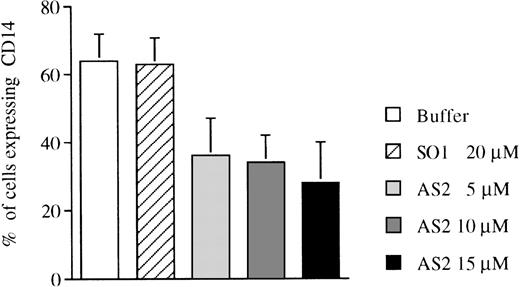
To analyze the indicated differences in pRb expression during monocytic and neutrophilic differentiation (Fig 2A and B) in a functional way, the effects of α-RB on the growth and differentiation of CD34+ cells were investigated. Constitutive suppression of pRb expression by transfection with an antisense-RB construct inhibits myelomonocytic differentiation of leukemic U-937 cells,16and AS2 has been shown to downregulate RB mRNA levels in liquid culture of human hematopoietic progenitors.10 Purified CD34+ bone marrow cells were cultured in the presence of FL plus IL-3 to promote monocytic differentiation. The addition of AS2, but not of SO1, reduced the expression of the monocyte-related surface antigen CD1422 compared with incubation with FL/IL-3 alone (Fig 3 and Table 2). The addition of AS2, SO2, or SCR (Fig 4) to CD34+ progenitors stimulated with FL plus IL-3 allowed a similar growth rate, as did the addition of oligo buffer (Table 2). Morphologically, FL/IL-3 stimulated predominantly the formation of large cells with vacuolated cytoplasm and with nonlobulized nuclei (hereafter referred to as monocytes; Fig 5A and Table 2), confirming previous findings.20 The addition of AS2, but not of SO2, SCR, or oligo buffer, to cells incubated with FL/IL-3 resulted not only in reduced formation of monocytes, but also in an increased fraction of cells with granulocytic morphology (Fig 5and Table 2). The addition of AS2 or SO1 to CD34+ cells supported by G-CSF/SCF resulted in cells with granulocytic morphology, which is not different from cells incubated with G-CSF/SCF alone (Table2). To further characterize the differentiation response mediated by AS2, but not by SO2 or SCR, as judged by morphology (Fig 5), cells were subjected to immunocytochemistry. The addition of AS2, but not of SO2 or SCR, to cells incubated with FL/IL-3 resulted in cells with positive staining for lactoferrin, which is a granulocyte-specific protein with iron-binding capacity stored in secondary granulae (reviewed in Levay and Viljoen24; Fig 6). The staining of normal neutrophils, used as positive control, was of comparable intensity (Fig 6). The addition of AS2, but not of SO2 or SCR, to CD34+ cells incubated with FL/IL-3 reduced positive staining for unspecific esterase of 200 counted cells from 64% of cells incubated with SO2 and 62% of cells incubated with SCR to 20% of cells incubated with AS2.
Effects of oligonucleotides on differentiation of CD34+ cells induced with FL plus IL-3 assayed by expression of CD14. CD34+ cells were seeded at a concentration of 5,000 cells/mL and incubated in medium supplemented with FL at 50 ng/mL plus IL-3 at 25 ng/mL to promote monocytic differentiation. On days 0, 5, and 10, antisense RB oligonucleotides (AS2) or sense-orientated oligonucleotides (SO1) were added, each time at a concentration of 5.0 μmol/L. After 14 days, cells were assayed for expression of CD14, a marker of monocytic differentiation. Values shown are the percentages of cells expressing CD14. Results shown represent the mean values from 3 separate experiments (bars ± SEM).
Effects of oligonucleotides on differentiation of CD34+ cells induced with FL plus IL-3 assayed by expression of CD14. CD34+ cells were seeded at a concentration of 5,000 cells/mL and incubated in medium supplemented with FL at 50 ng/mL plus IL-3 at 25 ng/mL to promote monocytic differentiation. On days 0, 5, and 10, antisense RB oligonucleotides (AS2) or sense-orientated oligonucleotides (SO1) were added, each time at a concentration of 5.0 μmol/L. After 14 days, cells were assayed for expression of CD14, a marker of monocytic differentiation. Values shown are the percentages of cells expressing CD14. Results shown represent the mean values from 3 separate experiments (bars ± SEM).
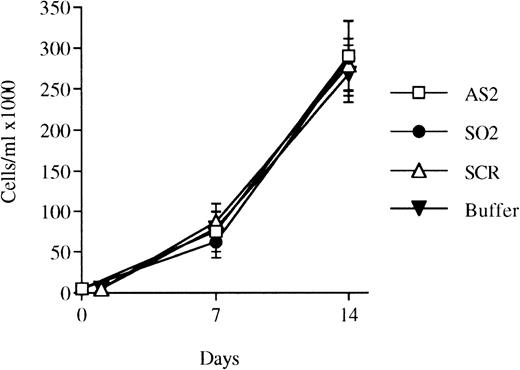
Effects of oligonucleotides on the proliferation of bone marrow cells selected for CD34 expression and incubated with FL plus IL-3. CD34+ cells were seeded at a concentration of 5,000 cells/mL and incubated in medium supplemented with FL at 50 ng/mL and IL-3 at 25 ng/mL to promote monocytic growth. On days 0, 5, and 10, antisense RB oligonucleotides (AS2), sense-orientated oligonucleotides (SO2), scrambled oligonucleotides (SCR), or oligo buffer, respectively, were added at a concentration of 5 μmol/L. On days 7 and 14, cells were counted. Results shown represent the mean values from 3 separate experiments (bars ± SEM). Results from SCR are from 2 independent experiments. Viability was always greater than 90% (data not shown).
Effects of oligonucleotides on the proliferation of bone marrow cells selected for CD34 expression and incubated with FL plus IL-3. CD34+ cells were seeded at a concentration of 5,000 cells/mL and incubated in medium supplemented with FL at 50 ng/mL and IL-3 at 25 ng/mL to promote monocytic growth. On days 0, 5, and 10, antisense RB oligonucleotides (AS2), sense-orientated oligonucleotides (SO2), scrambled oligonucleotides (SCR), or oligo buffer, respectively, were added at a concentration of 5 μmol/L. On days 7 and 14, cells were counted. Results shown represent the mean values from 3 separate experiments (bars ± SEM). Results from SCR are from 2 independent experiments. Viability was always greater than 90% (data not shown).
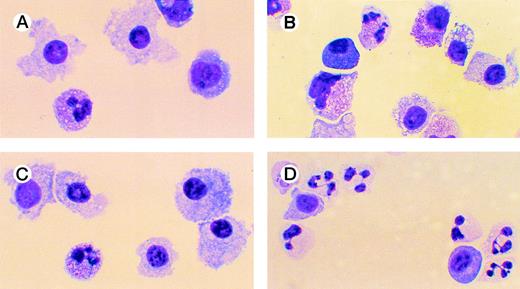
Effects of oligonucleotides on differentiation of CD34+ cells induced with FL plus IL-3 assayed by morphology. Cells at an initial concentration of 5,000/mL were incubated in medium supplemented with FL at 50 ng/mL and IL-3 at 25 ng/mL to promote monocytic differentiation. On days 0, 5, and 10, antisense RB oligonucleotides (AS2), sense-orientated oligonucleotides (SO2), or scrambled oligonucleotides (SCR) were added, each time at a concentration of 5.0 μmol/L. After 14 days, cytospin slides were prepared and stained with May-Grünwald-Giemsa. Cells incubated with FL, IL-3, and oligo buffer (A); with FL, IL-3, and SO2 (B); with FL, IL-3, and SCR (C); or with FL, IL-3, and AS2 (D).
Effects of oligonucleotides on differentiation of CD34+ cells induced with FL plus IL-3 assayed by morphology. Cells at an initial concentration of 5,000/mL were incubated in medium supplemented with FL at 50 ng/mL and IL-3 at 25 ng/mL to promote monocytic differentiation. On days 0, 5, and 10, antisense RB oligonucleotides (AS2), sense-orientated oligonucleotides (SO2), or scrambled oligonucleotides (SCR) were added, each time at a concentration of 5.0 μmol/L. After 14 days, cytospin slides were prepared and stained with May-Grünwald-Giemsa. Cells incubated with FL, IL-3, and oligo buffer (A); with FL, IL-3, and SO2 (B); with FL, IL-3, and SCR (C); or with FL, IL-3, and AS2 (D).
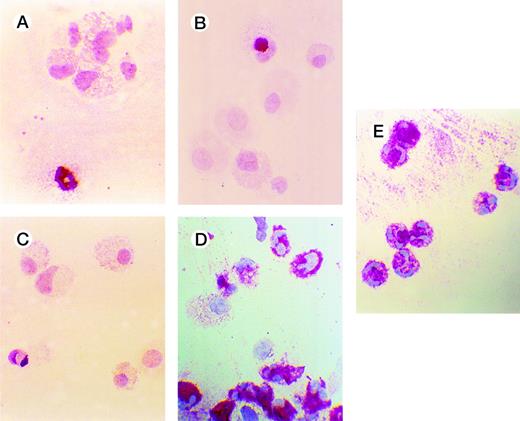
Effects of oligonucleotides on differentiation of CD34+ cells induced with FL plus IL-3 assayed by immunocytochemistry. Cells at an initial concentration of 5,000/mL were incubated in medium supplemented with FL at 50 ng/mL and IL-3 at 25 ng/mL to promote monocytic differentiation. On days 0, 5, and 10, antisense RB oligonucleotides (AS2), sense-orientated oligonucleotides (SO2), scrambled oligonucleotides (SCR), or oligo buffer were added, each time at a concentration of 5.0 μmol/L. After 14 days, cytospin slides were subjected to immunochemistry using antibodies to human lactoferrin, as described in Materials and Methods. Red-colored cytoplasm indicates the presence of lactoferrin, a granulocyte-specific protein. Cells incubated with FL, IL-3, and buffer (A); with FL, IL-3, and SO2 (B); with FL, IL-3, and SCR (C); with FL, IL-3, and AS2 (D); and human neutrophils (E).
Effects of oligonucleotides on differentiation of CD34+ cells induced with FL plus IL-3 assayed by immunocytochemistry. Cells at an initial concentration of 5,000/mL were incubated in medium supplemented with FL at 50 ng/mL and IL-3 at 25 ng/mL to promote monocytic differentiation. On days 0, 5, and 10, antisense RB oligonucleotides (AS2), sense-orientated oligonucleotides (SO2), scrambled oligonucleotides (SCR), or oligo buffer were added, each time at a concentration of 5.0 μmol/L. After 14 days, cytospin slides were subjected to immunochemistry using antibodies to human lactoferrin, as described in Materials and Methods. Red-colored cytoplasm indicates the presence of lactoferrin, a granulocyte-specific protein. Cells incubated with FL, IL-3, and buffer (A); with FL, IL-3, and SO2 (B); with FL, IL-3, and SCR (C); with FL, IL-3, and AS2 (D); and human neutrophils (E).
The analysis of pRb expression during granulocytic and monocytic differentiation (Fig 2A and B); the effects of AS2 or SO1 on the expression of CD14 (Fig 3); the effects of AS2, SO2, or SCR on morphology (Fig 5) and on staining for lactoferrin (Fig 6) and unspecific esterase; and also the results from the CFU-GM experiments supported the conclusion that pRb is involved in human myeloid differentiation. Taken together with the data on proliferation from this part of the study (Fig 4), a role for pRb in monocytic and neutrophilic lineage commitment of human bone marrow progenitor cells is indicated, probably by mechanisms that are at least partly distinct from those involved in cell cycle control.
Effects of AS2 on the production of pRb.
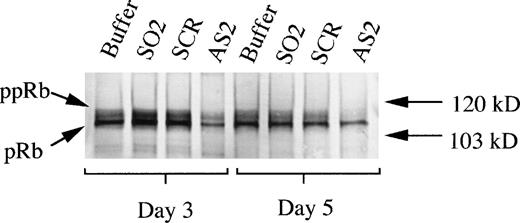
To determine whether the effects of AS2 on differentiation of CD34+ cells supported by FL/IL-3 were correlated to an inhibition of the upregulation of pRb levels, as demonstrated in Fig2A, pRb levels in these cells were assayed. The addition of AS2 was correlated to a lower level of pRb compared with addition of oligo buffer, SO2, or SCR, as judged by Western blot on days 3 and 5 (Fig 7). These results indicate that the effects of AS2 on the differentiation of CD34+ bone marrow progenitors were pRb specific. Effects of AS2 on pRb level were also determined on days 7 and 14. A similar, but generally less pronounced, difference in pRb level on days 7 and 14 between cells incubated with AS2 and cells incubated with SO1 was repeatedly documented (data not shown).
Effects of AS2 on the production of pRb. Cells at an initial concentration of 5,000/mL were incubated in medium supplemented with FL at 50 ng/mL and IL-3 at 25 ng/mL to promote monocytic differentiation. Oligo buffer, sense-orientated oligonucleotides (SO2), scrambled oligonucleotides (SCR), or antisense RB oligonucleotides (AS2) were added on days 0, 5, and 10, each time at a concentration of 5.0 μmol/L. On days 3 and 5, 1.5 × 106 cells from each incubation were subjected to protein extraction, followed by specific immunoprecipitation with an anti-pRb antibody (sc-102; Santa Cruz), 6% SDS-PAGE, and Western blot analysis. pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated).
Effects of AS2 on the production of pRb. Cells at an initial concentration of 5,000/mL were incubated in medium supplemented with FL at 50 ng/mL and IL-3 at 25 ng/mL to promote monocytic differentiation. Oligo buffer, sense-orientated oligonucleotides (SO2), scrambled oligonucleotides (SCR), or antisense RB oligonucleotides (AS2) were added on days 0, 5, and 10, each time at a concentration of 5.0 μmol/L. On days 3 and 5, 1.5 × 106 cells from each incubation were subjected to protein extraction, followed by specific immunoprecipitation with an anti-pRb antibody (sc-102; Santa Cruz), 6% SDS-PAGE, and Western blot analysis. pRb of different molecular weights are indicated with arrows (ppRb, hyperphosphorylated; pRb, hypophosphorylated).
DISCUSSION
The aim of the present study was to investigate the role of pRb in monocytic differentiation of human bone marrow cells. The data presented show that monocytic differentiation is correlated to high levels of hypophosphorylated pRb, whereas during neutrophilic differentiation, pRb levels are low. The low levels of pRb during neutrophilic differentiation are in line with previous findings.10 Antisense RB oligonucleotides (α-RB) inhibited the formation of monocytic colonies in a CFU-GM assay, with little effect on clonogenic growth, as well as inhibited monocytic differentiation of CD34+ bone marrow progenitors in liquid culture, again with no effect on cell growth. Furthermore, the α-RB–mediated inhibition of monocytic differentiation of CD34+ cells was counterbalanced by a neutrophilic differentiation response.
The exact mechanisms governing hematopoietic differentiation and lineage commitment are not clear. External signals, such as cytokines, are known to evoke responses affecting key cellular events such as proliferation, differentiation, and survival. These signals are mediated through complex regulation of specific genes, eg, genes controlling cell cycle progression and cell-type–specific genes. The molecular basis for regulation of hematopoiesis seems to depend on the activities of different hematopoietic transcription factors (reviewed in Shivdasani and Orkin11 and Tenen et al12). In this context, it is interesting to note that hematopoietic transcription factors, such as NF-IL6 (C/EBPβ), PU1, and Elf1, have been shown to interact with pRb.13-15
It has previously been demonstrated by both ourselves and other researchers that pRb is involved in monocytic differentiation of leukemic U937 cells.13 16 Our present results show that monocytic differentiation of normal hematopoietic progenitor cells is dependent on pRb. This notion is based on the findings that monocytic differentiation is associated with a strong upregulation of levels of hypophosphorylated pRb (Fig 2A) and that, when bone marrow cells were incubated with α-RB, the appearance of monocytic characteristics, such as morphology, the expression of CD14, and esterase positivity, was inhibited. Moreover, a role for pRb in the choice between neutrophilic and monocytic commitment was indicated. CD34+cells incubated with cytokines that otherwise promote predominantly monocytic differentiation showed neutrophilic maturation, such as neutrophilic morphology, and the appearance of lactoferrin as a response to the addition of α-RB.
How is it possible to understand that progenitors incubated with cytokines (FL plus IL-3) normally promoting predominantly monocytic differentiation not only respond with reduced monocytic differentiation, but also show a corresponding increase in neutrophilic differentiation when α-RB are added (Figs 3, 5, and 6 and Table 2)? Hypothetically, putative monocyte-specific pRb-dependent transcription factors are not able to activate their target genes optimally when upregulation of pRb is suppressed in the presence of α-RB. Given the importance of transcription factors of the C/EBP family in neutrophil differentiation,25-28 it is particularly interesting that pRb can interact with several members of this family.6,13For example it has been shown that pRb can interact physically with C/EBPs to increase binding to DNA and transactivation of target genes during terminal adipocyte differentiation.6 Recently, it was reported that the transcription factor C/EBPα is upregulated during granulocytic differentiation and rapidly downregulated during the alternative monocytic pathway.29 This finding indicates that C/EBPα serves as a myeloid differentiation switch, acting on bipotential precursors and directing them to mature granulocytes. In the present study, the levels of pRb during granulocytic commitment and differentiation were low (Fig 2A and B), which has also been reported by others.10 This may contradict an activating role for pRb in C/EBP-mediated transcription during granulocytic differentiation. Data presented in this study rather suggest that pRb suppresses activation of neutrophilic transcription factors, in analogy to the ability of pRb to suppress activity of other transcription factors.1-3
We did not find any effect on proliferation of progenitor cells mediated by the addition of antisense Rb oligonucleotides (Fig4). In line with this, it was recently reported that the main regulation of the E2F family of transcription factors in hematopoietic cells may be mediated by the pRb-related pocket protein p130.30 This report lends support to our finding that pRb may be primarily involved in other processes than cell cycle control in hematopoietic cells, eg, lineage commitment and cell differentiation.
Antisense techniques have been successfully used for functional analysis of various genes, including RB.17 However, antisense strategies suffer from an inborn vagueness. Any observed biological effect correlated to the use of antisense oligonucleotides might be nonspecific, ie, not due to sequence-specific targeting of the mRNA in question.31 However, in the present study, AS2 seemed to act in a sequence specific way, insofar as it was able to reduce pRb levels in the cells investigated (Fig 7). However, it cannot be formally excluded that AS2 mediates neutrophilic commitment through pRb-independent mechanisms and that the observed reduction of pRb in cells incubated with α-RB compared with cells incubated with oligo buffer, SO2, or SCR (Fig 7) merely reflects pRb levels when neutrophils are formed (Fig 2A and B and Table 2). Nonantisense mechanisms, including cytotoxic breakdown effects of antisense oligonucleotides, have recently been demonstrated to be the cause of the antiproliferative effects of antisense oligonucleotides, which has been shown in a number of studies.32 However, the absence of antiproliferative effects or toxic effects of α-RB on progenitor cells in this study argue in favor of a sequence-specific action of the α-RB. In this work, α-RB affected the differentiation, but not the proliferation, of CD34+ cells supported by FL/IL-3 (Figs 3,4, 5, and 6 and Table 2). This suggests that the proliferation and the differentiation induced by FL/IL-3 on hematopoietic cells may be mediated through separate signaling pathways. AS2 was not able to inhibit the minor monocytic growth of CD34+ cells supported by G-CSF/SCF (Table 1). This may reflect that the minor population of monocytic progenitors recruited by G-CSF/SCF is not dependent on pRb. In partial contrast to our results, growth-stimulatory effects of α-RB on early, but not on late, hematopoietic progenitor cells have been reported.17 The reason for these differences in effects of α-RB on growth is unclear, but could be that the growth stimulation used in the present work is not favoring very early progenitors.
Williams et al33 have reported extensive contribution of pRb-deficient cells to the tissues of adult RB −/− RB +/+ chimeric mice, including mature erythrocytes and neurons. This observation questions an intrinsic cell autonomous requirement for pRb in the differentiation of the tissues targeted in the RB −/− mice, eg, erythrocytes and the central nervous system. Instead, it suggests that the phenotype of the RB −/− mice is due to extrinsic effects of pRb-deficient cells other than precursors of erythroid or neuronal cells, eg, surrounding stromal cells. However, the results in the present study argue for cell autonomous intrinsic effects of pRb in myeloid differentiation, because nonmyeloid cells, eg, stromal cells, are probably absent in our experimental settings.
The present study suggests that pRb is crucial for monocytic commitment and differentiation in humans. The RB −/− mice show hematopoietic defects: a high proportion of nucleated immature erythroid cells compared with the wild-type mouse. However, no specific defect in monocytic maturation in RB −/− mice has been reported. Lee et al7 report that myeloid cells are present in mutant embryos. Monocytes constitute a minor proportion of the total blood cell population in the wild-type animal and the nucleated blood cell population of the mutated animal is totally dominated by immature erythrocytes.7 These circumstances may bring about difficulties in demonstrating a putative specific defect in monocytic differentiation in the pRb-deficient mice. Also, pRb deficiency targets different cells in different species; humans are highly susceptible to development of retinoblastoma, whereas mice develop pituitary tumors.9
To conclude, our data suggest that pRb is involved in the monocytic and neutrophilic lineage commitment of human bone marrow progenitor cells, probably by mechanisms not directly linked to pRb-mediated cell cycle regulation.
ACKNOWLEDGMENT
The authors thank Ellinor Johnsson for skillfully and enthusiastically making the most beautiful immunocytochemistry stainings and Gunilla Naumann for help and expert guidance with esterase stainings.
Supported by grants from the Swedish Cancer Society, the Swedish Society of Paediatric Cancer, The Swedish Medical Research Council (Project No. 11546), the Georg Danielsson Foundation, the Gunnar, Arvid and Elisabeth Nilsson Foundation, the John Persson Foundation, the John and Augusta Persson Foundation, Funds of Lunds Sjukvårdsdistrikt, the Swedish Society for Medical Research, the Tobias Foundation, the Thelma Zoegas Foundation, the Crafoord Foundation, the Foundation of Greta and Johan Kock, the Alfred Österlund Foundation, and the Hipple Cancer Research Center (Dayton, OH).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Gösta Bergh, MD, Research Dept. 2., E-block, University Hospital, S-221 85 Lund, Sweden.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal