Abstract
The methylation status of p15INK4b(MTS2), p16INK4a (MTS1) andp14ARF (p16β) was analyzed in 56 lymphomas by restriction-enzyme related polymerase chain reaction (PCR) (REP), methylation-specific PCR (MSP), and bisulfite genomic sequencing (BGS). Methylation of the p15 andp16 genes was detected, respectively, in 64% and 32% of the B-cell lymphomas, in 44% and 22% of the T-cell lymphomas, and in none of the 5 reactive lymph nodes analyzed. Both p15 andp16 genes were methylated more often in the high-grade (78% and 50%, respectively) than in the low-grade B-cell lymphomas (55% and 21%, respectively). For 5 cases, mapping of the methylated CpGs of the p16 promoter region confirmed the results of REP and MSP. In addition, a large variation in the methylation patterns ofp16 exon 1 was observed, not only from one lymphoma to another, but also within a given tumor. Methylation of p15 andp16 was associated with an absence of gene expression, as assessed by reverse transcription-PCR. The p14 gene was unmethylated and normally expressed in all 56 tumors. We found no mutations of p15, p16, or p14 in any of the 56 lymphomas. Our results suggest a role for p15 and p16gene methylation during lymphomagenesis and a possible association between p15 and p16 inactivation and aggressive transformation in B-cell and T-cell lymphomas.
UNCONTROLLED PROLIFERATION of tumor cells in lymphoma, as in other malignancies, is thought to result from a deregulation of 2 main pathways of cell-cycle control: the p53/p21 and p16/Rb pathways. Wild-type p53 induces transcription of the cyclin-dependent kinase (CDK) inhibitorp21WAF1.1 The lack of functional p53 protein, due to mutation or deletion of the gene, results in reduced p21 protein levels, preventing the association of p21 with cyclin-CDK complexes, which leads to cell-cycle activation. The Mdm2 protein can bind to and inactivate the transcriptional activity of p53, abrogating its antiproliferative effect.2In addition, Mdm2 targets the p53 protein for rapid degradation.3 Therefore, Mdm2 overexpression may have the same biological consequences that p53 mutations have on cell proliferation.4,5Mdm2, p53, and p21 gene alterations are relatively rare in hematological malignancies,6-13 suggesting that deregulation of the p53/p21 pathway does not play a central role in the genesis of these tumors.
Phosphorylation of pRb by cyclin D-CDK 4/6 complexes or Rb gene mutations lead to dissociation of the Rb-E2F complex and subsequent entry of the cell into the S phase. Thep16INK4a (MTS1) tumor suppressor gene product14,15 acts as a negative regulator of cellular proliferation by interacting with CDK4 and inhibiting its kinase activity.16 In the absence of functional p16 protein, CDK4 binds to cyclin D and phosphorylates pRb, which stimulates entry into the S phase. The p16 gene is inactivated by mutations, homozygous deletions, or gene methylation in many tumors of diverse origin.17-19 The p16 locus encodes a second protein, referred to as p14ARF, p19ARF (mice), ORF2, or p16β, resulting from a distinct exon 1 (exon 1β) spliced to the second shared exon of p16.20-22
The overexpression of p14 protein induces cell-cycle arrest in mammalian fibroblasts.20 The recently characterizedp14 promoter23 is located within a CpG island and has been found methylated in several colon cancer cell lines.p16 and p14 (p19ARF) null mice develop diverse types of cancer including, notably, lymphomas.15,24 Recently, it has been shown that p14 promotes the rapid degradation of Mdm2, leading to stabilization and accumulation of p53.5 25 Thus, p14 gene inactivation might be one way to deregulate cell-cycle control through disruption of the p53/p21 pathway.
Another mechanism deregulating the cell cycle in neoplasia involves thep15INK4b (MTS2) gene. p15displays high homology to p16, maps to the same region of chromosome 9, acts as inhibitor of CDK4 and 6,26 and has been proposed to have a tumor suppressor role.27 It is transcriptionally activated by transforming growth factor-β (TGF-β). Homozygous deletions of the p15/p16 locus are found in a high proportion of acute lymphoblastic leukemias,6,7,28-35 adult T-cell leukemias,36,37 and chronic myeloid leukemias.38 Only a few studies have analyzed p15and p16 gene alterations in lymphomas. Homozygous deletions involving p15 and/or p16 are found in a low proportion (0% to 19%) of B-cell lymphomas,39-45 where they seem to be associated with progression toward high-grade lesions.46,47 Hemizygous allele loss (loss of heterozygosity) of these 2 genes is somewhat more frequent than complete loss,46,48 but mutations are rarely observed.29,39,41-43,47,48 Methylation of p15and/or p16 has been reported in several types of lymphomas,45 particularly in mucosa-associated lymphoid tissue (MALT) lymphomas, multiple myelomas, mantle cell lymphomas, Burkitt’s lymphomas, and anaplastic large cell lymphomas.49-54
In the present work, we studied p15, p16, and p14methylation status in human lymphomas to verify the role of methylation as a gene-silencing mechanism involved in lymphomagenesis. We specifically investigated the association of gene methylation with tumor grade and analyzed intra- and inter-tumor variations in the patterns of methylation.
MATERIALS AND METHODS
Histopathological analysis.
Biopsy material from 56 non-Hodgkin’s lymphomas (NHLs), as well as 5 nonspecific reactive lymphadenopathies from the files of the Department of Pathology of the University Hospital of Lausanne (Switzerland), was studied. Snap-frozen tissue was available for DNA extraction of all cases.
Morphological and immunophenotypical analysis was performed on formalin-fixed paraffin-embedded material. The cases were classified according to the Revised European American Lymphoma (REAL) classification55 and to the updated Kiel classification for NHLs.56 The series comprised of 47 B-cell lymphomas (BCL) and 9 T-cell lymphomas (TCL). The BCL included 8 chronic lymphocytic leukemia/small lymphocytic lymphomas, 6 mantle cell lymphomas, 1 splenic marginal zone lymphoma with villous lymphocytes, 14 follicle center cell lymphomas, 13 diffuse large B-cell lymphomas (9 centroblastic, 2 immunoblastic, 2 primary mediastinal), 1 T-cell rich BCL, 2 high-grade B-cell Burkitt-like lymphomas, and 2 Burkitt’s lymphomas. According to the updated Kiel classification, 29 BCL were low grade and 18 were high grade. The TCL included 7 peripheral TCL (PTCL) and 2 anaplastic large cell lymphomas (ALCL).
For the assessment of methylation status and mRNA expression ofp15, p16, and p14, snap-frozen tissue was used. Hematoxylin and eosin (H&E) sections of the frozen specimens used for DNA and RNA extraction were examined to ascertain representativity.
Methylation analysis of p15, p16, and p14.
The methylation status of p15, p16, and p14 exon 1 was determined by restriction enzyme–related polymerase chain reaction (PCR) (REP), as previously described.57 Genomic DNA samples were individually digested with 4 methyl-sensitive (Kspl,Hpall, Nael, and Eagl) and with 1 non–methyl-sensitive (Mspl) restriction enzyme, extracted with chloroform/phenol, and precipitated with ethanol before PCR. Undigested DNA was included as a positive control. A 150-bp fragment ofp16 exon 1 containing 2 Hpall and 1 Kspl site, a 316-bp fragment of p14 exon 1 containing 1 Hpall and 1 Kspl site, and a 384-bp fragment of p15 exon 1 containing 1 Kspl, 1 Nael, 1 Eagl, and 5Hpall sites were amplified by PCR. The primer sets and annealing temperatures used for PCR are given in Table1 (H through J). Cases were considered positive for p15, p16, or p14 methylation by REP when PCR amplification was obtained after digestion with 1 of the methyl-sensitive restriction enzymes used. By REP, 3 CpG dinucleotides from p16 exon 1, 10 from p15 exon 1, and 2 fromp14 exon 1 were evaluated.
Primers Used for PCR-SSCP, REP, MSP, BGS, and RT-PCR Analyses
| Primer Set . | Application and Specificity . | Template DNA . | Forward Primer (5′ → 3′) . | Reverse Primer (5′ → 3′) . | Product Size (bp) . | Annealing Temperature (°C) . |
|---|---|---|---|---|---|---|
| PCR-SSCP | ||||||
| A | p16 exon 1 | G | GGGAGCAGCATGGAGCCG | AGTCGCCCGCCATCCCCT | 203 | 58 |
| B | p16 exon 2A | G | CTGGCTCTGACCATTCTGT | AGCACCACCAGCGTGTCC | 171 | 58 |
| C | p16 exon 2B | G | GACCCCGCCACTCTCACC | AGGTACCGTGCGACATCGC | 170 | 58 |
| D | p16 exon 2C | G | GATGCCTGGGGCCGTCT | CAGGGTACAAATTCTCAGAT | 169 | 55 |
| E | p16 exon 3 | G | GTAGGGACGGCAAGAGA | ACCTTCGGTGACTGATG | 159 | 55 |
| F | p14 exon 1 | G | GCCTGCGGGGCGGAGAT | GCGGCTGCTGCCCTAGA | 316 | 58 |
| G | p15 exon 1 | G | TTCCCAGAAGCAATCCAG | GTTGACTCCGTTGGGATC | 384 | 55 |
| REP | ||||||
| H | p16 exon 1 | G, RE+ | GGGAGCAGCATGGAGCCG | CTGGATCGGCCTCCGACCGTA | 159* | 57 |
| I | p14 exon 1 | G, RE+ | GCCTGCGGGGCGGAGAT | GCGGCTGCTGCCCTAGA | 316† | 58 |
| J | p15 exon 1 | G, RE+ | TTCCCAGAAGCAATCCAG | GTTGACTCCGTTGGGATC | 384‡ | 55 |
| MSP and BGS | ||||||
| K | p16 exon 1, M | G, Bis+ | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA | 150 | 65 |
| L | p16 exon 1, M | G, Bis+ | TTATTAGAGGGTGGGGCGGATCGC | CCACCTAAATCGACCTCCGACCG | 234 | 65 |
| M | p16 exon 1, U | G, Bis+ | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 151 | 60 |
| N | p16 exon 1, U | G, Bis+ | TTATTAGAGGGTGGGGTGGATTGT | CCACCTAAATCAACCTCCAACCA | 234 | 60 |
| BGS | ||||||
| O | p16 promoter, M/U | G, Bis+ | GTGATTTTAGGGGTGTTA | TTCCAATTCCCCTACAAA | 483 | 48 |
| P | p14 exon 1, M/U | G, Bis+ | GGGTTTTAGTTTGTAGTT | CCTCAATAACATCAACAC | 255 | 48 |
| Q | p15 exon 1, M/U | G, Bis+ | TTTTTTAGGAAGGAGAGA | TAAAACCCCAACTACCTA | 285 | 48 |
| RT-PCR | ||||||
| R | p16 | cDNA | GGTGCGGGCGCTGCTGGA | AGCACCACCAGCGTGTCC | 210 | 58 |
| S | p14 | cDNA | GTGGCCCTCGTGCTGATG | AGCACCACCAGCGTGTCC | 207 | 58 |
| T | p15 | cDNA | GATCCCAACGGAGTCAAC | AGCACCACCAGCGTGTCC | 182 | 53 |
| U | GAPDH | cDNA | TTGTCAAGCTCATTTCCTG | AGGCCCCTCCCCTCTTC | 238 | 55 |
| Primer Set . | Application and Specificity . | Template DNA . | Forward Primer (5′ → 3′) . | Reverse Primer (5′ → 3′) . | Product Size (bp) . | Annealing Temperature (°C) . |
|---|---|---|---|---|---|---|
| PCR-SSCP | ||||||
| A | p16 exon 1 | G | GGGAGCAGCATGGAGCCG | AGTCGCCCGCCATCCCCT | 203 | 58 |
| B | p16 exon 2A | G | CTGGCTCTGACCATTCTGT | AGCACCACCAGCGTGTCC | 171 | 58 |
| C | p16 exon 2B | G | GACCCCGCCACTCTCACC | AGGTACCGTGCGACATCGC | 170 | 58 |
| D | p16 exon 2C | G | GATGCCTGGGGCCGTCT | CAGGGTACAAATTCTCAGAT | 169 | 55 |
| E | p16 exon 3 | G | GTAGGGACGGCAAGAGA | ACCTTCGGTGACTGATG | 159 | 55 |
| F | p14 exon 1 | G | GCCTGCGGGGCGGAGAT | GCGGCTGCTGCCCTAGA | 316 | 58 |
| G | p15 exon 1 | G | TTCCCAGAAGCAATCCAG | GTTGACTCCGTTGGGATC | 384 | 55 |
| REP | ||||||
| H | p16 exon 1 | G, RE+ | GGGAGCAGCATGGAGCCG | CTGGATCGGCCTCCGACCGTA | 159* | 57 |
| I | p14 exon 1 | G, RE+ | GCCTGCGGGGCGGAGAT | GCGGCTGCTGCCCTAGA | 316† | 58 |
| J | p15 exon 1 | G, RE+ | TTCCCAGAAGCAATCCAG | GTTGACTCCGTTGGGATC | 384‡ | 55 |
| MSP and BGS | ||||||
| K | p16 exon 1, M | G, Bis+ | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA | 150 | 65 |
| L | p16 exon 1, M | G, Bis+ | TTATTAGAGGGTGGGGCGGATCGC | CCACCTAAATCGACCTCCGACCG | 234 | 65 |
| M | p16 exon 1, U | G, Bis+ | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 151 | 60 |
| N | p16 exon 1, U | G, Bis+ | TTATTAGAGGGTGGGGTGGATTGT | CCACCTAAATCAACCTCCAACCA | 234 | 60 |
| BGS | ||||||
| O | p16 promoter, M/U | G, Bis+ | GTGATTTTAGGGGTGTTA | TTCCAATTCCCCTACAAA | 483 | 48 |
| P | p14 exon 1, M/U | G, Bis+ | GGGTTTTAGTTTGTAGTT | CCTCAATAACATCAACAC | 255 | 48 |
| Q | p15 exon 1, M/U | G, Bis+ | TTTTTTAGGAAGGAGAGA | TAAAACCCCAACTACCTA | 285 | 48 |
| RT-PCR | ||||||
| R | p16 | cDNA | GGTGCGGGCGCTGCTGGA | AGCACCACCAGCGTGTCC | 210 | 58 |
| S | p14 | cDNA | GTGGCCCTCGTGCTGATG | AGCACCACCAGCGTGTCC | 207 | 58 |
| T | p15 | cDNA | GATCCCAACGGAGTCAAC | AGCACCACCAGCGTGTCC | 182 | 53 |
| U | GAPDH | cDNA | TTGTCAAGCTCATTTCCTG | AGGCCCCTCCCCTCTTC | 238 | 55 |
Abbreviations: M, specific for methylated bisulfite-modified DNA; U, specific for unmethylated bisulfite-modified DNA; G, genomic DNA; RE+, restriction enzyme-digested DNA; Bis+, bisulfite-treated DNA.
Fragment containing 2 HpaII/MspI and 1 KspI site.
Fragment containing 1 HpaII/MspI and 1 KspI site.
Fragment containing 5 HpaII/MspI, 1 KspI, 1NaeI, and 1 EagI site.
In addition to REP, 2 other approaches were used to analyze the methylation status of p16 exon 1: methylation-specific PCR (MSP)58 and bisulfite genomic sequencing (BGS).59 In both techniques, DNA is chemically modified by sodium bisulfite, which changes the unmethylated but not the methylated cytosines into uracil. In MSP, the bisulfite-treated DNA is subjected to PCR amplification using primers designed to anneal specifically to the methylated bisulfite-modified DNA within a given gene. Thus, a PCR product is obtained when the sequence covered by the primers is methylated. MSP was performed as described by Herman et al,58 with some modifications. Briefly, genomic DNA (0.5 μg) was denatured in 0.3 mol/L NaOH (volume, 20 μL) for 20 minutes at 42°C. After the addition of 80 μL of a freshly prepared solution containing 1 mmol/L hydroquinone and 3.8 mol/L sodium bisulfite (pH 5.0), the samples were incubated overnight (16 hours) at 55°C. The bisulfite-treated DNA was purified on Wizard DNA Clean-Up columns (Promega, Madison, WI) according to the manufacturer’s specifications and eluted with 50 μL of water. The DNA was treated with 0.3 mol/L NaOH for 20 minutes at 37°C, precipitated by ethanol, and resuspended in 20 μL of water. Bisulfite-modified DNA was amplified by PCR using 2 primer sets specific for methylated and 2 primer sets specific for unmethylated p16 sequence (Table 1, K through N), as described.58 Because tumor cells are always admixed with reactive cells in uncultured lymphomas, the PCR amplifications using primers specific for unmethylated DNA were considered as positive controls. Eleven CpG dinucleotides from p16 exon 1 were covered by MSP analysis.
In BGS, bisulfite-treated DNA is amplified by PCR using primers designed to recognize either methylated, or both methylated and unmethylated, bisulfite-modified sequences, then cloned and sequenced. This enables a precise mapping of the methylated CpG dinucleotides present in a given DNA fragment. First, we cloned the 234-bp PCR products obtained by MSP, either with primers specific for the methylated, or with those specific for the unmethylated, p16exon 1 sequence (Table 1, L and N). This fragment contains 28 CpG dinucleotides. In some cases, we also cloned a 483-bp PCR fragment of the bisulfite-modified p16 promoter region obtained with primers recognizing both the methylated and the unmethylatedp16 exon 1 sequence (Table 1, O). This fragment contains 42 CpG dinucleotides. Cloning of PCR products was performed using the pGEM-T vector system (Promega) according to the manufacturer’s recommendations. Five to 10 clones from each PCR product were sequenced on an ALF Automatic DNA Analysis System (Amersham Pharmacia Biotech, Uppsala, Sweden).
Mutation analysis by PCR-single-stranded conformation polymorphism (SSCP).
Reverse transcription (RT)-PCR analysis of p15, p16, andp14 mRNA expression.
Representative tissue from 16 lymphomas (15 BCL and 1 ALCL) and 2 reactive lymph nodes was suitable for mRNA analysis. In all 16 tumors, the proportion of neoplastic cells in the analyzed sample was more than 80%. Total RNA was extracted from tissue sections using the method of Chomczynski and Sacchi.61 Five hundred nanograms of total RNA was reverse transcribed using Moloney-murine leukemia virus (M-MLV) RT (Promega) and oligo dT primer (2.5 μmol/L) in 75 mmol/L KCl, 3 mmol/L MgCl2, 10 mmol/L dithiothreitol, 50 mmol/L Tris/HCl, pH 8.3, containing 1 mmol/L each dNTP and 0.5 U/μL RNasin (Promega). A 210-bp fragment from exon 1 to exon 2 of p16, a 207-bp fragment from exon 1 to exon 2 ofp14, and a 182-bp fragment from exon 1 to exon 2 of p15cDNA were amplified by PCR using the primer sets and annealing conditions indicated in Table 1 (R through T). In parallel, a 238-bp fragment at the 3′ end of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH ) gene cDNA was amplified as a positive control (Table 1, U). Twenty-five cycles were performed for cDNA amplification, and the PCR product was analyzed on a 2% agarose gel. Under these conditions, and provided that their proportion was under 20%, p16 and p14 mRNA expression from reactive cells contaminating the tumor was not detectable (data not shown). Thus, absence of visible RT-PCR product for p15, p16, orp14 (with strongly positive GAPDH control) was considered to indicate absence of expression of these genes.
RESULTS
Methylation of the p15, p16, and p14 genes was analyzed in 56 lymphomas and 5 reactive lymph nodes by 3 different technologies: REP, MSP, and BGS (see Materials and Methods). In addition, thep15, p16, and p14 genes were analyzed by PCR-SSCP for mutation, and their mRNA expression was assessed by RT-PCR. The results are displayed in Table 2.
p15, p14, and p16 Gene Methylation, Mutation, and Expression in 56 BCL and TCL
| Histology . | Case . | p15 . | p14 . | p16 . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REP . | MUT . | EXP . | REP . | MUT . | EXP . | REP . | MSP1 . | MSP2 . | BGS . | MUT . | EXP . | ||
| BCL | |||||||||||||
| Lymphocytic | 1 | + | − | − | − | − | − | − | − | ||||
| 2 | − | − | − | − | − | − | − | − | |||||
| 3 | − | − | − | − | − | − | − | − | |||||
| 4 | − | − | − | − | − | − | − | − | |||||
| 5 | + | − | − | − | − | − | − | − | |||||
| 6 | + | − | − | − | − | − | − | − | |||||
| 7 | − | − | − | − | − | − | − | − | |||||
| 8 | − | − | − | − | − | − | − | − | |||||
| Mantle cell | 9 | + | − | − | − | − | − | − | − | ||||
| 10 | − | − | − | − | − | − | − | − | |||||
| 11 | − | − | − | − | − | − | − | − | |||||
| 12 | + | − | − | − | + | + | + | − | |||||
| 13 | − | − | − | − | − | − | − | − | |||||
| 14 | + | − | − | − | − | − | − | − | |||||
| Marginal zone | 15 | + | − | − | − | − | − | − | − | ||||
| Follicle center | 16 | − | − | + | − | − | + | − | − | − | − | + | |
| 17 | + | − | − | − | − | + | + | + | + | − | − | ||
| 18 | + | − | − | − | − | + | + | − | − | − | − | ||
| 19 | + | − | − | − | − | − | − | − | |||||
| 20 | − | − | − | − | − | − | − | − | |||||
| 21 | − | − | + | − | − | + | − | − | − | − | + | ||
| 22 | − | − | − | − | − | − | − | − | |||||
| 23 | + | − | − | − | − | − | − | − | |||||
| 24 | + | − | − | − | − | + | + | + | − | − | − | ||
| 25 | + | − | − | − | − | + | + | − | − | − | − | ||
| 26 | + | − | − | − | + | − | − | − | |||||
| 27 | − | − | − | − | − | − | − | − | |||||
| 28 | + | − | − | − | − | − | − | − | |||||
| 29 | + | − | − | − | − | − | − | − | |||||
| Large cell, centroblastic | 30 | − | − | + | − | − | + | − | − | − | − | + | |
| 31 | − | − | − | − | − | − | − | − | |||||
| 32 | + | − | − | − | − | − | − | − | |||||
| 33 | + | − | − | − | − | + | + | + | + | HD | − | − | |
| 34 | + | − | − | − | − | − | − | − | |||||
| 35 | + | − | − | − | − | + | + | + | + | − | − | ||
| 36 | + | − | − | − | − | + | + | − | − | LD | − | + | |
| 37 | + | − | − | − | − | − | − | − | |||||
| 42 | + | − | − | − | − | + | + | + | + | HD | − | − | |
| Large cell, immunoblastic | 40 | + | − | − | − | + | + | + | − | ||||
| 41 | + | − | − | − | − | + | + | + | + | − | − | ||
| Large cell, mediastinal | 38 | − | − | − | − | − | − | − | − | ||||
| 39 | + | − | − | − | − | − | − | − | |||||
| Large cell, T-cell rich | 43 | + | − | − | − | + | + | + | − | ||||
| Burkitt-like | 44 | + | − | − | − | − | + | + | + | + | HD | − | − |
| 45 | + | − | − | − | − | − | − | − | |||||
| Burkitt’s | 46 | − | − | + | − | − | + | − | − | − | − | + | |
| 47 | + | − | − | − | − | + | + | − | − | LD | − | − | |
| TCL | |||||||||||||
| ALCL | 48 | + | − | − | − | − | + | + | + | + | − | − | |
| 49 | + | − | − | − | − | − | − | − | |||||
| PTCL | 50 | − | − | − | − | − | − | − | − | ||||
| 51 | − | − | − | − | − | − | − | − | |||||
| 52 | − | − | − | − | − | − | − | − | |||||
| 53 | − | − | − | − | − | − | − | − | |||||
| 54 | − | − | − | − | − | − | − | − | |||||
| 55 | + | − | − | − | − | − | − | − | |||||
| 56 | + | − | − | − | + | + | + | − | |||||
| Histology . | Case . | p15 . | p14 . | p16 . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REP . | MUT . | EXP . | REP . | MUT . | EXP . | REP . | MSP1 . | MSP2 . | BGS . | MUT . | EXP . | ||
| BCL | |||||||||||||
| Lymphocytic | 1 | + | − | − | − | − | − | − | − | ||||
| 2 | − | − | − | − | − | − | − | − | |||||
| 3 | − | − | − | − | − | − | − | − | |||||
| 4 | − | − | − | − | − | − | − | − | |||||
| 5 | + | − | − | − | − | − | − | − | |||||
| 6 | + | − | − | − | − | − | − | − | |||||
| 7 | − | − | − | − | − | − | − | − | |||||
| 8 | − | − | − | − | − | − | − | − | |||||
| Mantle cell | 9 | + | − | − | − | − | − | − | − | ||||
| 10 | − | − | − | − | − | − | − | − | |||||
| 11 | − | − | − | − | − | − | − | − | |||||
| 12 | + | − | − | − | + | + | + | − | |||||
| 13 | − | − | − | − | − | − | − | − | |||||
| 14 | + | − | − | − | − | − | − | − | |||||
| Marginal zone | 15 | + | − | − | − | − | − | − | − | ||||
| Follicle center | 16 | − | − | + | − | − | + | − | − | − | − | + | |
| 17 | + | − | − | − | − | + | + | + | + | − | − | ||
| 18 | + | − | − | − | − | + | + | − | − | − | − | ||
| 19 | + | − | − | − | − | − | − | − | |||||
| 20 | − | − | − | − | − | − | − | − | |||||
| 21 | − | − | + | − | − | + | − | − | − | − | + | ||
| 22 | − | − | − | − | − | − | − | − | |||||
| 23 | + | − | − | − | − | − | − | − | |||||
| 24 | + | − | − | − | − | + | + | + | − | − | − | ||
| 25 | + | − | − | − | − | + | + | − | − | − | − | ||
| 26 | + | − | − | − | + | − | − | − | |||||
| 27 | − | − | − | − | − | − | − | − | |||||
| 28 | + | − | − | − | − | − | − | − | |||||
| 29 | + | − | − | − | − | − | − | − | |||||
| Large cell, centroblastic | 30 | − | − | + | − | − | + | − | − | − | − | + | |
| 31 | − | − | − | − | − | − | − | − | |||||
| 32 | + | − | − | − | − | − | − | − | |||||
| 33 | + | − | − | − | − | + | + | + | + | HD | − | − | |
| 34 | + | − | − | − | − | − | − | − | |||||
| 35 | + | − | − | − | − | + | + | + | + | − | − | ||
| 36 | + | − | − | − | − | + | + | − | − | LD | − | + | |
| 37 | + | − | − | − | − | − | − | − | |||||
| 42 | + | − | − | − | − | + | + | + | + | HD | − | − | |
| Large cell, immunoblastic | 40 | + | − | − | − | + | + | + | − | ||||
| 41 | + | − | − | − | − | + | + | + | + | − | − | ||
| Large cell, mediastinal | 38 | − | − | − | − | − | − | − | − | ||||
| 39 | + | − | − | − | − | − | − | − | |||||
| Large cell, T-cell rich | 43 | + | − | − | − | + | + | + | − | ||||
| Burkitt-like | 44 | + | − | − | − | − | + | + | + | + | HD | − | − |
| 45 | + | − | − | − | − | − | − | − | |||||
| Burkitt’s | 46 | − | − | + | − | − | + | − | − | − | − | + | |
| 47 | + | − | − | − | − | + | + | − | − | LD | − | − | |
| TCL | |||||||||||||
| ALCL | 48 | + | − | − | − | − | + | + | + | + | − | − | |
| 49 | + | − | − | − | − | − | − | − | |||||
| PTCL | 50 | − | − | − | − | − | − | − | − | ||||
| 51 | − | − | − | − | − | − | − | − | |||||
| 52 | − | − | − | − | − | − | − | − | |||||
| 53 | − | − | − | − | − | − | − | − | |||||
| 54 | − | − | − | − | − | − | − | − | |||||
| 55 | + | − | − | − | − | − | − | − | |||||
| 56 | + | − | − | − | + | + | + | − | |||||
Abbreviations: REP, methylation status analyzed by restriction enzyme–related PCR (+, methylated; −, unmethylated); MSP1 and MSP2, methylation status analyzed by methylation-specific PCR using respectively the primer sets indicated under K and L in Table 1 (+, methylated; −, unmethylated); BGS, density of methylated CpG dinucleotides analyzed by bisulfite genomic sequencing (HD, high density; LD, low density); MUT, mutation analysis by PCR-SSCP (−, wild type); EXP, gene mRNA expression analyzed by RT-PCR (+, expression; −, no expression).
Methylation status of the p15, p16, and p14 genes and correlation with lymphoma type.
Methylation of the p16 gene was found in 17 of 56 (30%) lymphomas: 15 of 47 (32%) BCL and 2 of 9 (22%) TCL. Of the BCL, 6 of 29 (21%) low-grade and 9 of 18 (50%) high-grade (Kiel classification) showed p16 methylation. The incidence of p16 gene methylation was higher in B-follicle center cell lymphomas (5 of 14) than in other low-grade lymphomas (1 of 15). Of the high-grade BCL, 6 of 13 diffuse large BCL (2 of 2 immunoblastic, 4 of 9 centroblastic, 0 of 2 mediastinal), 1 of 1 T-cell–rich BCL, 1 of 2 Burkitt-like, and 1 of 2 Burkitt’s lymphomas were p16 methylated. The only PTCL showing p16 gene methylation corresponded to a pleomorphic T-cell lymphoma, predominantly large cell, high-grade (Kiel classification). One ALCL was shown to have a methylated p16gene (Table 3).
Incidence of p15 and p16 Gene Methylation in BCL and TCL According to the Histopathological Subtypes
| Histology . | n . | p15 Methylated . | p16Methylated . |
|---|---|---|---|
| BCL | 47 | 30/47 (64%) | 15/47 (32%) |
| REAL | |||
| Lymphocytic | 8 | 3/8 (37%) | 0/8 |
| Mantle cell | 6 | 3/6 (50%) | 1/6 (17%) |
| Marginal zone | 1 | 1/1 | 0/1 |
| Follicle center | 14 | 9/14 (64%) | 5/14 (36%) |
| Diffuse large B-cell | 13 | 10/13 (77%) | 6/13 (46%) |
| Immunoblastic | 2 | 2/2 | 2/2 |
| Centroblastic | 9 | 7/9 | 4/9 |
| Mediastinal | 2 | 1/2 | 0/2 |
| T-cell–rich B-cell | 1 | 1/1 | 1/1 |
| High-grade Burkitt-like | 2 | 2/2 | 1/2 |
| Burkitt’s | 2 | 1/2 | 1/2 |
| Kiel | |||
| B-cell, low grade | 29 | 16/29 (55%) | 6/29 (21%) |
| B-cell, high grade | 18 | 14/18 (78%) | 9/18 (50%) |
| TCL | 9 | 4/9 (44%) | 2/9 (22%) |
| PTCL, unspecified | 7 | 2/7 (28%) | 1/7 (14%) |
| ALCL | 2 | 2/2 | 1/2 |
| Total | 56 | 34/56 (61%) | 17/56 (30%) |
| Histology . | n . | p15 Methylated . | p16Methylated . |
|---|---|---|---|
| BCL | 47 | 30/47 (64%) | 15/47 (32%) |
| REAL | |||
| Lymphocytic | 8 | 3/8 (37%) | 0/8 |
| Mantle cell | 6 | 3/6 (50%) | 1/6 (17%) |
| Marginal zone | 1 | 1/1 | 0/1 |
| Follicle center | 14 | 9/14 (64%) | 5/14 (36%) |
| Diffuse large B-cell | 13 | 10/13 (77%) | 6/13 (46%) |
| Immunoblastic | 2 | 2/2 | 2/2 |
| Centroblastic | 9 | 7/9 | 4/9 |
| Mediastinal | 2 | 1/2 | 0/2 |
| T-cell–rich B-cell | 1 | 1/1 | 1/1 |
| High-grade Burkitt-like | 2 | 2/2 | 1/2 |
| Burkitt’s | 2 | 1/2 | 1/2 |
| Kiel | |||
| B-cell, low grade | 29 | 16/29 (55%) | 6/29 (21%) |
| B-cell, high grade | 18 | 14/18 (78%) | 9/18 (50%) |
| TCL | 9 | 4/9 (44%) | 2/9 (22%) |
| PTCL, unspecified | 7 | 2/7 (28%) | 1/7 (14%) |
| ALCL | 2 | 2/2 | 1/2 |
| Total | 56 | 34/56 (61%) | 17/56 (30%) |
All the 17 lymphomas with a methylated p16 gene also had a methylated p15 gene. Overall, methylation of the p15gene was found in 34 of 56 (61%) lymphomas: 30 of 47 (64%) BCL and 4 of 9 (44%) TCL. Of the BCL, 16 of 29 (55%) low grade and 14 of 18 (78%) high grade (Kiel classification) showed p15 gene methylation: 3 of 8 lymphocytic, 3 of 6 mantle cell, 1 of 1 marginal zone, 9 of 14 follicle center cell, 10 of 13 diffuse large B-cell (2 of 2 immunoblastic, 7 of 9 centroblastic, 1 of 2 mediastinal), 1 of 1 T-cell rich, 2 of 2 Burkitt-like, and 1 of 2 Burkitt’s lymphomas. Two of the 7 PTCL were positive for p15 gene methylation; both were large cell lymphomas, high grade (Kiel classification). Both ALCL exhibited methylation of the p15 gene (Table 3).
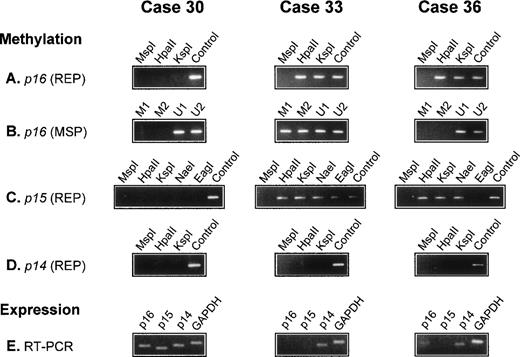
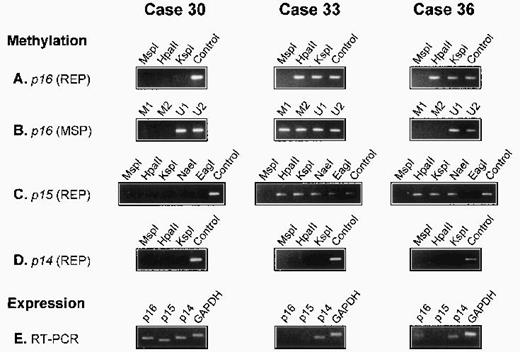
There was no detectable methylation of the p14 gene in any of the 56 lymphomas. In all 5 reactive lymph nodes, the p15, p16,and p14 genes were unmethylated. The results of three cases are illustrated in Fig 1.
Analysis of p16, p15, and p14 gene methylation and mRNA expression in 3 diffuse large BCL. The methyl-sensitive restriction enzymes used for REP are indicated (HpaII, KspI, NaeI, and EagI); digestion with the non–methyl-sensitive enzyme MspI serves as a negative control; undigested DNA (Control) serves as a positive control. For MSP, 2 primer sets specific for the methylated (M1 and M2) and 2 for the unmethylated (U1 and U2) bisulfite-modified p16gene are used. Expression of p16, p15, and p14 mRNA is analyzed by RT-PCR. The p16 gene is methylated in cases 33 and 36 but unmethylated in case 30. In case 36, p16 methylation is detected by REP but not by MSP (A and B). The p15 gene is methylated in cases 33 and 36 but not in case 30. In case 33, all assessed restriction sites are methylated. In case 36, all sites exceptEagI are methylated (C). The p14 gene is unmethylated in all 3 cases (D). GAPDH and p14 mRNA is present in all cases; p15 mRNA is detectable in case 30 but not in cases 33 and 36, whereas p16 mRNA is present in cases 30 and 36 but not in case 33 (E).
Analysis of p16, p15, and p14 gene methylation and mRNA expression in 3 diffuse large BCL. The methyl-sensitive restriction enzymes used for REP are indicated (HpaII, KspI, NaeI, and EagI); digestion with the non–methyl-sensitive enzyme MspI serves as a negative control; undigested DNA (Control) serves as a positive control. For MSP, 2 primer sets specific for the methylated (M1 and M2) and 2 for the unmethylated (U1 and U2) bisulfite-modified p16gene are used. Expression of p16, p15, and p14 mRNA is analyzed by RT-PCR. The p16 gene is methylated in cases 33 and 36 but unmethylated in case 30. In case 36, p16 methylation is detected by REP but not by MSP (A and B). The p15 gene is methylated in cases 33 and 36 but not in case 30. In case 33, all assessed restriction sites are methylated. In case 36, all sites exceptEagI are methylated (C). The p14 gene is unmethylated in all 3 cases (D). GAPDH and p14 mRNA is present in all cases; p15 mRNA is detectable in case 30 but not in cases 33 and 36, whereas p16 mRNA is present in cases 30 and 36 but not in case 33 (E).
Methylation patterns of p16 exon 1.
Of the 56 lymphomas analyzed for p16 gene methylation by REP and MSP, 17 were methylated. In 12 cases, methylation was shown by both methods (REP+/MSP+). In the 5 other cases, it was detected only by REP, but not by MSP (REP+/MSP−), indicating partial methylation in p16 exon 1 (Fig 1). To document this, 5 lymphomas (3 REP+/MSP+ and 2 REP+/MSP−) were analyzed by BGS, which enables a precise mapping of the methylated CpGs in the analyzed DNA fragment.
We first cloned and sequenced PCR products obtained by MSP with primers specific for either the methylated (REP+/MSP+cases), or the unmethylated (REP+/MSP− cases) bisulfite-modified p16 exon 1 sequence. In both REP+/MSP− cases, we also cloned and sequenced a PCR product obtained with primers recognizing both methylated and unmethylated sequences. In the 3 REP+/MSP+lymphomas, the different p16 exon 1 methylation patterns exhibited methylation at all or almost all of the CpG dinucleotides (Table 4, clones 33.a through 33.e, 42.a through 42.e, and 44.a through 44.e). In both REP+/MSP− lymphomas, all the sequenced clones were completely or almost completely unmethylated (Table 4, clones 36.a through 36.m and 47.a through 47.o); of the 11 CpG sites covered by MSP analysis, 8 were unmethylated in all clones, explaining the negative results obtained by MSP with the methylated primer sets.
Methylation Patterns of p16 Exon 1 Region Analyzed by BGS in Five Lymphomas
Individual clones obtained from 3 REP+/MSP+ (cases 33, 42, and 44) and 2 REP+/MSP− lymphomas (cases 36 and 47) were sequenced. The clones from REP+/MSP+ cases display complete or almost complete methylation. The clones from REP+/MSP− cases are either sparsely methylated or completely unmethylated.
Abbreviations: +, methylated CpG dinucleotides; −, unmethylated CpG dinucleotides.
Fourty-two CpG dinucleotides (numbered 1 to 42) contained in a 483-bp fragment of p16 exon 1. The numbers in gray boxes correspond to the 3 CpG sites assessed by digestion with KspI (K) andHpaII (H). The 11 CpG sites covered by the primers used for MSP-analysis are underlined by asterisks (forward primer) or small circles (reverse primers).
Expression of p15, p16, and p14 mRNA and correlation with methylation status.
Suitable tissue for mRNA extraction and RT-PCR analysis was available in 18 cases (16 lymphomas and 2 reactive lymph nodes). All tissues were strongly positive for GAPDH and p14 mRNA expression. The p16 mRNA was present in both reactive lymph nodes and in the 4 p16-unmethylated and 1 p16-methylated lymphomas. There was no detectable p16 mRNA expression in 11 of 12 lymphomas with a methylated p16 gene (Table 2).
Mutation analysis of the p15, p16, and p14 genes by PCR-SSCP.
All 56 lymphomas and 5 reactive lymph node tissues were screened by PCR-SSCP for mutations of p16 exon 1 to 3, p15 exon 1, and p14 exon 1. No aberrant migration patterns, indicative of mutation, were observed (Table 2).
DISCUSSION
Inactivation of p15INK4b andp16INK4a by homozygous deletion or gene methylation is probably one of the most common molecular events in hematological malignancies.52 Homozygous deletion of the 9p21 region containing the p15, p16, and p14 genes is frequently found in acute lymphoblastic leukemias of both B-cell and T-cell lineage, but is uncommon in lymphomas. In agreement with other studies,29,34,41,54 we did not find any mutations in the coding regions of the p15 and p16 genes among the 56 lymphomas analyzed. Thus, neither homozygous deletion nor mutation appears to be a relevant mode of inactivation of these genes during lymphomagenesis.29,39,41,45,54 Our results indicate that the principal mechanism of p15 and p16 silencing in lymphomas is gene methylation.45 A large proportion of lymphomas showed p15 and/or p16 gene methylation (61% and 30%, respectively), confirming previously reported results.49,50 52-54 Both genes were inactivated more frequently in BCL than in TCL, suggesting that they are more important in B than in T lymphomagenesis. High-grade (according to the Kiel classification) BCL were more often p15/p16 methylated (78% and 50%, respectively) than low-grade B-cell lymphomas (55% and 21%, respectively). This suggests that p15 and p16 gene silencing might be associated with aggressive transformation in lymphoma, and that p15 and p16 gene methylation might be a useful marker to predict aggressive behavior.
Both p16 and p14 (p19ARF) null mice develop lymphomas.15,24 However, given the fact that the p14 gene was also disrupted in the p16 null mice, it has been proposed that the major oncogenic event might be loss of p14 rather than of p16.24 We observed absence of p14 gene mutations or methylation and a high level of p14 mRNA expression in all lymphomas analyzed, which does not suggest a role for p14 inactivation in human lymphomagenesis. Our results indicate that one of the principal oncogenic events in human lymphomagenesis is inactivation of thep15 gene. Therefore, it could be of interest to determine whether p15 knockout mice develop lymphomas. In our series, all lymphomas with a methylated p16 gene were also p15methylated. It is striking that p14, a gene that is localized between p15 and p16, was always unmethylated even when both p15 and p16 genes were methylated. A possible mechanism protecting p14 from de novo methylation might be binding of the Sp1 transcription factor at Sp1 sites present in thep14 promoter, as proposed recently.23
The minimal sequence required by the methylation apparatus is a CpG dinucleotide.62 The present results are the first demonstrating variation in methylated CpGs within p16 exon 1 from one lymphoma to another. Three cases positive by MSP and REP displayed a high proportion of methylated and a few scattered unmethylated CpG dinucleotides by BGS. In contrast, 2 cases positive by REP but negative by MSP had only a few scattered methylated CpGs withinp16 exon 1. The significance of such heterogeneity of methylation patterns remains unclear, but suggests a complex mechanism for de novo methylation. Even those tumors with a few methylated CpGs within p16 exon 1 had no detectable p16 mRNA expression, indicating that low-level methylation is sufficient to repress p16 transcription. Recently, it has been shown that methyl-CpG-binding protein 2 (MeCP2), which is involved in transcriptional repression of methylated genes by forming complexes with the transcriptional repressor Sin3A and histone deacetylase (HD), may bind to DNA sequences containing only a single methylated CpG dinucleotide.63 64 However, it remains to be shown that transcriptional repression of methylated p15 and p16involves the MeCP2/Sin3A/HD-complex/pathway. By BGS, we found severalp16 methylation patterns coexisting in DNA extracted from a primary (uncultured) lymphoma, an observation for which several explanations might be considered. First, consecutive waves of clonal progression might yield heterogeneous p16 methylation patterns coinciding with different tumor regions. This could be confirmed by methylation analysis of microdissected tumor samples. Secondly, the 2 (or more) p16 alleles in a single tumor cell might have different methylation patterns. The latter possibility is supported by observations that several methylation patterns of p16 exon 1 coexist in established colon cancer cell lines (P.C., unpublished data).
ACKNOWLEDGMENT
The authors acknowledge M.M. Bertholet, M. Correvon, S. Burki, and J. Maillardet for technical assistance.
Supported by the Swiss Cancer League (P.C.)
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Pascal Chaubert, MD, Institut Universitaire de Pathologie, Bugnon 25, CH-1011 Lausanne, Switzerland.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal