Abstract
Targeted mutation of CCAAT/enhancer binding protein (C/EBP) ɛ in mice results in early death, primarily due to spontaneous infection with Pseudomonas aeruginosa. Functional analysis of C/EBPɛ-deficient neutrophils, in an in vivo model of peritoneal inflammation, shows multiple defects. Reduction of phagocytotic killing by C/EBPɛ-deficient neutrophils is a result of decreased uptake of opsonized bacteria as well as little to no expression of secondary granule proteins. Abnormalities in neutrophil migration detected in a chemical peritonitis model are likely secondary to abnormal CD11b integrin and L-selectin expression on C/EBPɛ-deficient neutrophils. Alterations in neutrophil cytokine expression in response to inflammation show decreased levels of interleukin-1 receptor antagonist (IL-1Ra) and increased levels of tumor necrosis factor- (TNF-) expression by C/EBPɛ-deficient neutrophils. Additionally, TNF- expression is increased in nonactivated, circulating C/EBPɛ-deficient neutrophils. Overall, C/EBPɛ-deficient neutrophils are severely functionally impaired, evoking an abnormal microenvironment, which may contribute to the loss of normal responses to inflammatory stimuli. Similarities between the C/EBPɛ-deficient mouse model and the human disease, specific granule deficiency, will be discussed.
CCAAT/ENHANCER BINDING proteins (C/EBPs) are basic-leucine zipper (bZIP) transcription factors, which dimerize, bind cognate DNA sequences, and effect transcriptional activation or repression via N-terminal domains.1-4C/EBPε, the newest member of the leucine zipper C/EBP family of transcription factors, plays a critical role in myelopoiesis.5-9 C/EBPε is unusual among C/EBPs due to its restricted expression in myeloid and T-cell lineages in contrast to other family members that are more ubiquitously expressed.8Also, induction of neutrophil differentiation in promyelocytic leukemia lines is associated with C/EBPε expression.8,10C/EBPε’s targeted expression suggests a role in myeloid cell function; however, it is not the only C/EBP with an apparent role in hematopoiesis. C/EBPα-deficient neutrophils develop no further than the myeloblast stage.11 Additionally, these cells lack primary and secondary granule proteins and have significant downregulation of granulocyte colony-stimulating factor (G-CSF) receptor expression.11 Macrophages deficient for C/EBPβ have altered cytokine expression and an impaired immune response towards challenged pathogens, Listeria monocytogenes andCandida albicans.12 13
Our initial work showed that mice deficient for C/EBPε are severely immunocompromised, surviving no longer than 5 months of age with 60% succumbing to systemic infection with Pseudomonas aeruginosa.5 Furthermore, C/EBPε-deficient neutrophils are functionally defective, lacking an oxidative burst in response to phorbol myristate acetate (PMA) stimulation.5 This defect, however, does not explain the pathogenicity of P aeruginosa in C/EBPε-deficient mice. Patients with chronic granulomatous disease (CGD) and mice engineered with similar genetic defects lack an effective oxidative burst; however, they do not develop spontaneous infections with P aeruginosa.14,15 Both X-linked gp91phox and autosomal recessive p47phox CGD mice develop spontaneous infections with Staphylococcus aureus and Aspergillus fumigatus, a phenotype similar to CGD patients.14 15 These correlations strongly suggest that the severe immunodeficiency seen in C/EBPε-deficient mice represents multiple defects, intrinsic to neutrophil function and possibly involving cytokine signaling and response to inflammatory stimuli.
Finally, C/EBPs have known regulatory effects on other C/EBP family members. C/EBPα, for example, has cognate recognition sequences for C/EBP dimers in its upstream elements and its hepatic expression is in part regulated by C/EBPβ and C/EBPδ.16 17 Analogous regulation of C/EBPs in the myeloid lineage is likely and may influence the phenotype of the C/EBPε-deficient neutrophil.
Here we show that C/EBPε-deficient neutrophils have both intrinsic and extrinsic defects. Phagocytotic and migratory function is abnormal as well as neutrophil cytokine response to an inflammatory challenge. Finally, neutrophil expression of other C/EBP family members is altered.
MATERIALS AND METHODS
Mice.
C/EBPε-deficient mice5 and wild-type littermates were bred in specific pathogen-free conditions. Genotype of all progeny was verified by Southern blot analysis, as described.18 All mouse experiments complied with the National Human Genome Research Institute (NHGRI) Animal Care and Use Committee and Association for Accreditation of Laboratory Animal Care (AALAC) regulations.
Thioglycollate challenge and peritoneal lavage.
Mice were injected intraperitoneally with 2 mL of 3% thioglycollate broth via a 27-gauge needle. Mice were euthanized by CO2inhalation and peritoneal exudate cells harvested using one intraperitoneal wash with 8 mL of phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA) and 20 μmol/L disodium EDTA. Peritoneal exudate cells were counted by Coulter Counter (Coulter, Hialeah, FL) and cytospin stained with Wright-Giemsa staining. Cells of myeloid lineage comprised the majority of exudate cells (>95%), including granulocytes, monocytes, and macrophage cells.
Phagocytosis assays.
Phagotest kit (Orpegen Pharma, Heidelberg, Germany) was used for quantitation of neutrophil phagocytosis of opsonized FITC-labeledEscherichia coli with the following modifications. Mouse whole blood collected into heparized tubes was incubated with 40 μL of precooled E coli bacteria (1 × 109/mL). Additionally, incubation periods were lengthened from 10 minutes to 1 hour at 37°C or 0°C. After incubation and quenching of external, nonphagocytosed bacteria, the percentage of cells ingesting bacteria and the fluorescent intensity was determined using FACScaliber flow cytometer and Cell Quest software (Becton Dickinson, San Jose, CA). Staphylocidal assay with lysostaphin (L-0761; Sigma, St Louis, MO) was performed as described,19 using 250 μL of whole blood. A total of 1.25 × 106 bacteria was added to each sample. Total cell counts were (mean ± standard error [SE]) 7.7 ± 1.0 (× 1,000/μL) for wild-type samples and 9.8 ± 2.4 (× 1,000/μL) for knock-out samples. Neutrophil counts were 932 ± 82 cells/μL for wild-type samples and 1,417 ± 649 cells/μL for knock-out samples. Aliquots at 0-, 30-minute, and 60-minute time points were osmotically lysed and dilutions plated onto trypticase soy agar (TSA) plates. Colony counts were recorded and used to derive statistical analysis (Mann-Whitney U test).
Northern blot and ribonuclease protection assays.
Total RNA was harvested from peritoneal lavage or bone marrow using Ultraspec RNA isolation system (Biotecx, Houston, TX). A total of 10 μg of bone marrow or peritoneal RNA was resolved on 1% agarose gel under denaturing conditions as described20 and transferred to nylon membrane. Hybridization was performed as described20 using probe concentrations of 2 × 106 cpm/mL buffer. Granule protein probes were the generous gift of Drs Atsushi Iwama and Daniel Tenen (Harvard University).
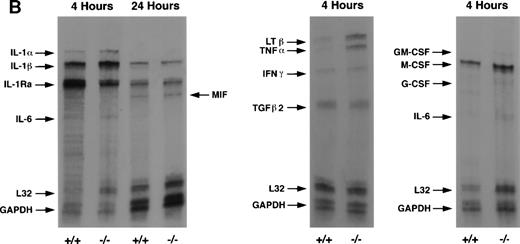
Peritoneal lavage total RNA (10 μg) from wild-type or knock-out mice was hybridized with ribonucleotide probes generated from multiprobe template sets and Riboquant, an in vitro transcription kit (Pharmingen, San Diego, CA) according to manufacturer’s instructions. Multiprobe template set mCK-2 contained the following templates, with probe and protected fragment sizes given (probe/protected fragment): interleukin (IL)-12p35 (388/359), IL-12p40 (349/321), IL-10 (314/287), IL-1α (284/257), IL-1β (255/229), IL-1Ra (231/202), macrophage inhibitory factor (MIF) (209/180), IL-6 (191/163), interferon (IFN)-γ (172/143), L32 (141/112), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (125/97). Multiprobe template set mCK-3 contained: tumor necrosis factor (TNF)-β (389/361), lymphotoxin β (Ltβ) (351/322), TNF-α (316/287), IL-6 (284/255), IFN-γ (257/228), IFN-β (232/203), transforming growth factor (TGF)β1 (208/179), TGFβ2 (191/162), L32 (141/112), and GAPDH (125/97). Multiprobe template set mCK-4 contained: IL-3 (389/360), IL-11 (351/322), IL-7 (315/286), granulocyte-macrophage colony-stimulating factor (GM-CSF) (283/257), M-CSF (257/231), G-CSF (230/202), leukemia inhibitory factor (LIF) (209/180), IL-6 (191/163), stem cell factor (SCF) (173/144), L32 (141/112), and GAPDH (125/97). Control lanes included yeast RNA with or without RNase digestion. HybSpeed Ribonuclease Protection Assay (RPA) (Ambion, Austin, TX) was used according to manufacturer’s instructions, hybridizing at 68°C for 10 minutes followed by RNase digestion at 37°C for 30 minutes. Resultant protected fragments were resolved on 6% polyacrylamide, 8 mol/L urea, denaturing gels (Novex, San Diego, CA).
Intracellular cytokine staining and in vitro stimulation of peritoneal neutrophils.
Protocol, antibodies, and recombinent mouse cytokines described herein were obtained from Pharmingen. Peritoneal exudate cells collected by lavage with Hanks’ balanced salt solution (HBSS) or peripheral blood cells (after lysis with Ack lysis buffer) were washed once with RPMI complete and incubated for 4 hours at 37°C with Golgistop (monesin) 4 μL/mL. Cells were washed twice with staining buffer, labeled with Gr-1, fixed with 4% paraformaldehyde for 20 minutes at 4°C, permeabilized with 0.1% saponin in staining buffer at 4°C, and labeled with R-phycoerythrin (PE)-anti–IL-6, PE-anti–TNF-α, or PE-anti–IL-10 (1 μg/106 cells.) Cells were immediately analyzed by flow cytometry. Neutrophils were gated by morphologic criteria or by double staining with Gr-1.
Surface antigen labeling.
Cells used for surface labeling experiments were washed twice in staining buffer (Dulbecco’s PBS without magnesium or calcium, 1% heat-inactivated fetal bovine serum (FBS), 0.1% sodium azide, filtered), resuspended in 100 μL staining buffer, and incubated with labeled antibody (Pharmingen) for 30 minutes at 4°C in the dark. Antibody concentrations used were 1 μg/106 cells. After incubation, cells were washed twice in staining buffer, resuspending in 0.5 mL staining buffer, and analyzed by flow cytometry.
RESULTS
Neutrophils deficient for C/EBPε phagocytize bacteria poorly.
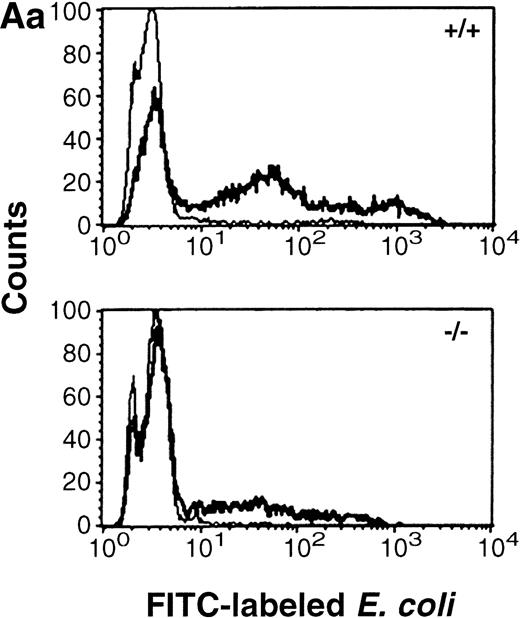
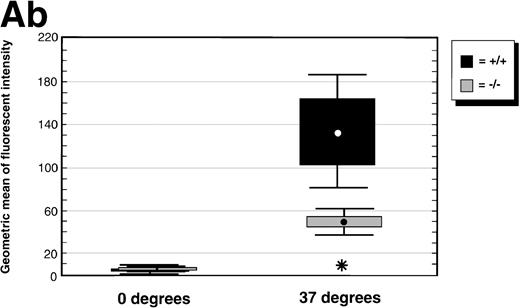
Mice deficient for C/EBPε were recently described.5Peripheral blood neutrophils from mice deficient for C/EBPε were assessed for uptake of fluorescein isothiocyanate (FITC)-labeled, opsonized E coli by flow cytometry. Both wild-type and C/EBPε-deficient neutrophils showed no evidence of phagocytosis after incubation at 0°C. Intracellular FITC-labeled bacteria were detected in neutrophils after 37°C incubation; however, both the number of cells harboring bacteria (mean no. phagocytosing neutrophils ± SE: wild-type 54.3% ± 6.2%, knock-out 25.7% ± 3.8%;P < .03, Mann Whitney U test) and the mean fluorescent intensity of the samples were significantly decreased in C/EBPε-deficient neutrophils compared with wild-type cells (P< .03; Mann Whitney U test). Figure 1A (panel a) shows representative histograms of wild-type and C/EBPε-deficient neutrophils, demonstrating decreased uptake of FITC-labeled bacteria by C/EBPε-deficient samples. Figure 1A (panel b) shows a box and whisker graph indicating mean, SE, and standard deviation (SD), summarizing the analysis of three separate experiments, each including nine samples (three wild-type and six knock-out).
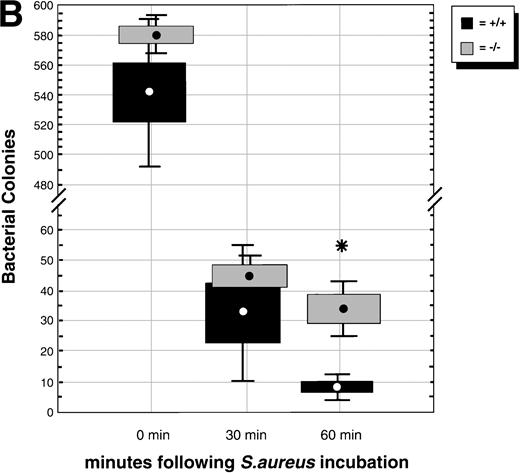
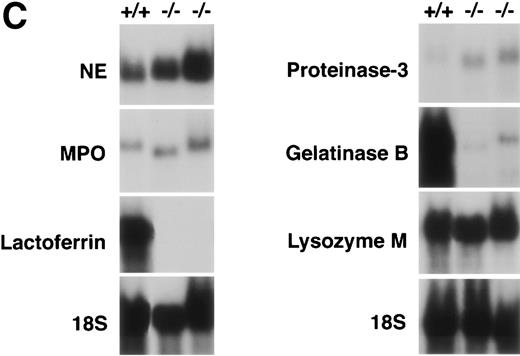
(A) Phagocytosis activity of peripheral blood neutrophils. Whole blood collected from wild-type and C/EBPɛ-deficient mice was incubated with FITC-labeled, opsonizedE coli at either 0°C or 37°C for 60 minutes, then accessed by flow cytometry. Panel a shows representative histograms of wild-type and C/EBPɛ-deficient neutrophil samples: thin lines, 0°C incubation; thick lines, 37°C incubations. Panel b summarizes results represented graphically by Box and Whisker graph showing standard deviation, standard error, and mean. (▪) Represent wild-type neutrophils, gray boxes represent C/EBPɛ-deficient neutrophils. An asterisk (*) indicates wild-type neutrophil phagocytosis, as determined by geometric mean of fluorescent intensity, is significantly greater than C/EBPɛ-deficient neutrophils (P< .03, Mann Whitney U). (B) Phagocidal activity of peripheral blood neutrophils. Whole blood collected from wild-type and C/EBPɛ-deficient mice was incubated with titered S aureus, followed by treatment with lysostaphin. At represented time points, aliquots were lysed osmotically, samples streaked on TSA plates, and incubated overnight. Results are represented by Box and Whisker graph showing standard deviation, standard error, and mean. (▪) Represent wild-type neutrophils, gray boxes represent C/EBPɛ-deficient neutrophils. An asterisk (*) indicates wild-type neutrophil bacterial killing is significantly greater than C/EBPɛ-deficient neutrophil killing at 60 minutes (P < .02, Mann Whitney U). (C) Granule protein expression in wild-type and C/EBPɛ-deficient bone marrow. Northern blot hybridization of total RNA harvested from bone marrow, resolved by electorphoresis and transferred to Nytran. Blots were hybridized with 32P-–dCTP-labeled granule protein probes. C/EBPɛ-deficient neutrophil RNA (−/−); wild-type (+/+). Equivalent sample loading is shown by 18S bands.
(A) Phagocytosis activity of peripheral blood neutrophils. Whole blood collected from wild-type and C/EBPɛ-deficient mice was incubated with FITC-labeled, opsonizedE coli at either 0°C or 37°C for 60 minutes, then accessed by flow cytometry. Panel a shows representative histograms of wild-type and C/EBPɛ-deficient neutrophil samples: thin lines, 0°C incubation; thick lines, 37°C incubations. Panel b summarizes results represented graphically by Box and Whisker graph showing standard deviation, standard error, and mean. (▪) Represent wild-type neutrophils, gray boxes represent C/EBPɛ-deficient neutrophils. An asterisk (*) indicates wild-type neutrophil phagocytosis, as determined by geometric mean of fluorescent intensity, is significantly greater than C/EBPɛ-deficient neutrophils (P< .03, Mann Whitney U). (B) Phagocidal activity of peripheral blood neutrophils. Whole blood collected from wild-type and C/EBPɛ-deficient mice was incubated with titered S aureus, followed by treatment with lysostaphin. At represented time points, aliquots were lysed osmotically, samples streaked on TSA plates, and incubated overnight. Results are represented by Box and Whisker graph showing standard deviation, standard error, and mean. (▪) Represent wild-type neutrophils, gray boxes represent C/EBPɛ-deficient neutrophils. An asterisk (*) indicates wild-type neutrophil bacterial killing is significantly greater than C/EBPɛ-deficient neutrophil killing at 60 minutes (P < .02, Mann Whitney U). (C) Granule protein expression in wild-type and C/EBPɛ-deficient bone marrow. Northern blot hybridization of total RNA harvested from bone marrow, resolved by electorphoresis and transferred to Nytran. Blots were hybridized with 32P-–dCTP-labeled granule protein probes. C/EBPɛ-deficient neutrophil RNA (−/−); wild-type (+/+). Equivalent sample loading is shown by 18S bands.
In addition, we tested for neutrophil bacteriocidal activity using the staphylocidal lysostaphin assay. Results are summarized in Fig 1B, from three separate experiments, each including six animals. Neutrophils from C/EBPε-deficient mice contained significantly more live bacteria 60 minutes after addition of lysostaphin, compared with wild-type neutrophils (P < .02; Mann Whitney U test). Taken together, these experiments show that the phagocytosis defect present in C/EBPε-deficient neutrophils includes both uptake of opsonized bacteria as well as intracellular killing.
Expression of granule protein mRNA was thus addressed to explore other tiers of bacteriocidal defects in C/EBPε-deficient neutrophils. Total RNA from bone marrow harvested from 3-week-old C/EBPε-deficient mice and wild-type littermates was fractionated by electrophoresis, transferred to nylon membranes, and hybridized with a variety of α32P deoxycytidine triphosphate (dCTP)-labeled granule protein cDNAs. Myeloid to erythroid (M:E) ratios were calculated from Wright-Giemsa–stained cytospins and were similar for all samples, ranging from 1.95 to 2.01. Bone marrow differential for myeloid cells in the wild-type sample was myeloblast, 7%; promyelocyte, 2%; myelocyte, 3%; metamyelocyte, 8%; band, 24%; and segmented, 57%. Similarly, the myeloid cell differentials for the two C/EBPε-deficient samples were myeloblast, 5% to 13%; promyelocyte, 2% to 10%; myelocyte, 9% to 17%; metamyelocyte, 22% to 27%; band, 10% to 23%; and segmented, 29% to 34%. Expression of primary granule proteins, myeloperoxidase, neutrophil elastase, and proteinase-3 was slightly upregulated in bone marrow mRNA of C/EBPε-deficient mice compared with wild-type. Expression of secondary granule protein lactoferrin and tertiary granule protein, gelatinase B, however, was severely downregulated in the C/EBPε-deficient neutrophils. Lysozyme M, present in both primary and secondary granules, was detected equally well in wild-type and C/EBPε-deficient bone marrows (Fig 1C).
C/EBPε-deficient neutrophils migrate poorly to site of inflammatory challenge.
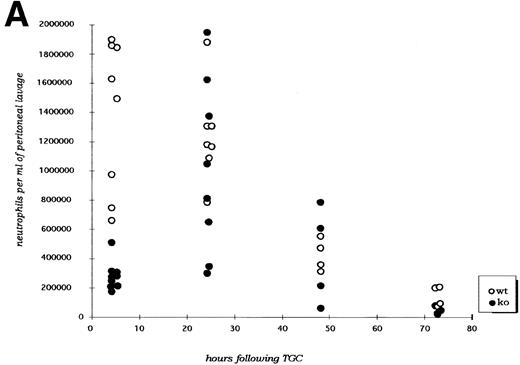
Mice deficient for C/EBPε and wild-type control mice received 3% thioglycollate broth intraperitoneally at time zero and were subsequently killed at specified time points and assessed for neutrophil migration into the peritoneal cavity, surface antigen expression, and cytokine expression. Figure2A shows that migration of C/EBPε-deficient neutrophils at 4 hours after thioglycollate challenge was significantly reduced (P < .001; Mann Whitney U test). By 24 hours, however, numbers of neutrophils within the peritoneal cavity are similiar for C/EBPε-deficient and wild-type mice. Additionally, peritoneal neutrophil numbers are similar at 48 and 72 hours after challenge.
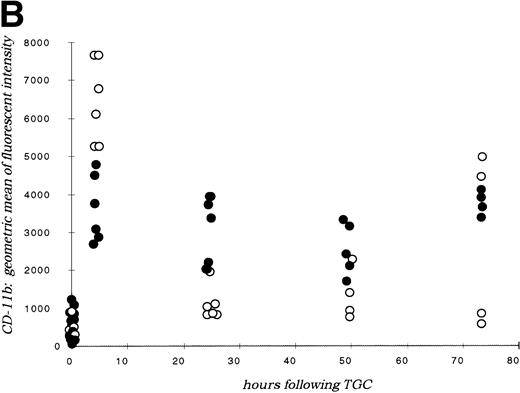
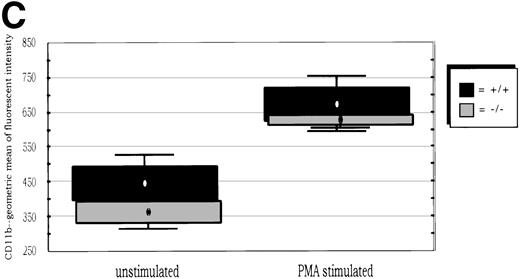
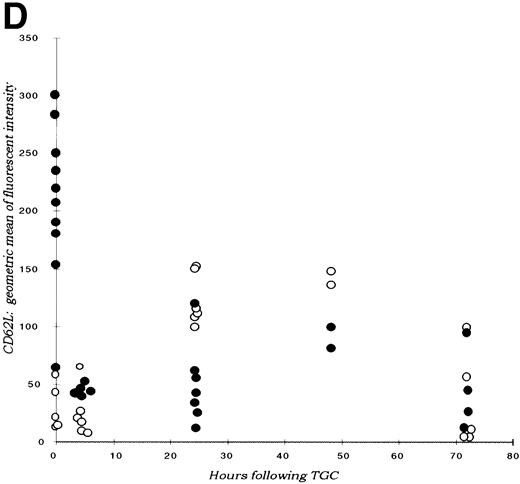
(A) Neutrophil migration into the peritoneal cavity after thioglycollate challenge. At indicated time points after intraperitoneal thioglycollate injection, peritoneal cells were harvested by lavage, counted, and cytospins stained for differential counting. Results are represented graphically, showing the migration of neutrophils into the peritoneal cavity over time after thioglycollate stimulus. (○) Represent wild-type neutrophils; (•) represent C/EBPɛ-deficient neutrophils. An asterisk (*) indicates significantly increased numbers of wild-type neutrophils at 4 hours, compared with C/EBPɛ-deficient neutrophils (P < .001, Mann Whitney U). (B) CD11β expression on peritoneal neutrophils after thioglycollate challenge. Mice received thioglycollate intraperitoneally, and neutrophils were harvested at indicated time points. Cells were doubly stained with Gr-1(FITC) and CD11b(PE) and assessed by flow cytometry. Results are represented graphically, by geometric mean of fluorescent intensity. (○) Represent wild-type neutrophils; (•) represent C/EBPɛ-deficient neutrophils. CD11β staining is significantly decreased on C/EBPɛ-deficient neutrophils compared with wild-type cells (P < .004, Mann Whitney U) at 4 hours. By 24 hours, however, CD11β staining is more intense on C/EBPɛ-deficient cells (P < .004), compared with wild-type. CD11b staining of peripheral blood neutrophils is shown at time = 0. (C) CD11b expression on PMA-stimulated peripheral blood neutrophils. Peripheral blood neutrophils were stimulated with PMA, stained with -Gr-1 (PE) and -CD11b (FITC), and examined by flow cytometry. Results are represented by box and whisker graph, showing mean, standard deviation, and standard error of fluorescent intensity. (▪) Represent wild-type neutrophils, gray boxes represent C/EBPɛ-deficient neutrophils. (D) L-selectin (CD62L) expression on peritoneal neutrophils after thioglycollate challenge. Peritoneal lavage cells were harvested at indicated time points and doubly stained with Gr-1(FITC) and anti-mouse CD62L(PE) and assessed by flow cytometry. Results are represented graphically, by geometric mean of fluorescent intensity. (○) Represents wild-type neutrophils; (•) represents C/EBPɛ-deficient neutrophils. Circles graphed at 0 hours represent data obtained from circulating neutrophils from nonchallenged mice (P < .001, Mann Whitney U). L-selectin staining is significantly increased on C/EBPɛ-deficient neutrophils compared with wild-type cells (P< .04, Mann Whitney U) at 4 hours. By 24 hours, however, L-selectin expression is higher on wild-type neutrophils (P < .02, Mann Whitney U) compared with C/EBPe-deficient neutrophils.
(A) Neutrophil migration into the peritoneal cavity after thioglycollate challenge. At indicated time points after intraperitoneal thioglycollate injection, peritoneal cells were harvested by lavage, counted, and cytospins stained for differential counting. Results are represented graphically, showing the migration of neutrophils into the peritoneal cavity over time after thioglycollate stimulus. (○) Represent wild-type neutrophils; (•) represent C/EBPɛ-deficient neutrophils. An asterisk (*) indicates significantly increased numbers of wild-type neutrophils at 4 hours, compared with C/EBPɛ-deficient neutrophils (P < .001, Mann Whitney U). (B) CD11β expression on peritoneal neutrophils after thioglycollate challenge. Mice received thioglycollate intraperitoneally, and neutrophils were harvested at indicated time points. Cells were doubly stained with Gr-1(FITC) and CD11b(PE) and assessed by flow cytometry. Results are represented graphically, by geometric mean of fluorescent intensity. (○) Represent wild-type neutrophils; (•) represent C/EBPɛ-deficient neutrophils. CD11β staining is significantly decreased on C/EBPɛ-deficient neutrophils compared with wild-type cells (P < .004, Mann Whitney U) at 4 hours. By 24 hours, however, CD11β staining is more intense on C/EBPɛ-deficient cells (P < .004), compared with wild-type. CD11b staining of peripheral blood neutrophils is shown at time = 0. (C) CD11b expression on PMA-stimulated peripheral blood neutrophils. Peripheral blood neutrophils were stimulated with PMA, stained with -Gr-1 (PE) and -CD11b (FITC), and examined by flow cytometry. Results are represented by box and whisker graph, showing mean, standard deviation, and standard error of fluorescent intensity. (▪) Represent wild-type neutrophils, gray boxes represent C/EBPɛ-deficient neutrophils. (D) L-selectin (CD62L) expression on peritoneal neutrophils after thioglycollate challenge. Peritoneal lavage cells were harvested at indicated time points and doubly stained with Gr-1(FITC) and anti-mouse CD62L(PE) and assessed by flow cytometry. Results are represented graphically, by geometric mean of fluorescent intensity. (○) Represents wild-type neutrophils; (•) represents C/EBPɛ-deficient neutrophils. Circles graphed at 0 hours represent data obtained from circulating neutrophils from nonchallenged mice (P < .001, Mann Whitney U). L-selectin staining is significantly increased on C/EBPɛ-deficient neutrophils compared with wild-type cells (P< .04, Mann Whitney U) at 4 hours. By 24 hours, however, L-selectin expression is higher on wild-type neutrophils (P < .02, Mann Whitney U) compared with C/EBPe-deficient neutrophils.
Next, the presence of surface antigens integral to neutrophil migration was assessed. Coincident with the delay in neutrophil migration, CD11b (integrin) expression is significantly decreased on C/EBPε-deficient neutrophils at 4 hours after thioglycollate challenge compared with wild-type neutrophils (P < .004; Mann Whitney U test) (Fig2B). By 24 hours after challenge, however, CD11b is more abundant on C/EBPε-deficient cells compared with wild-type neutrophils (P< .004; Mann Whitney U test). CD11b expression on circulating peripheral blood neutrophils, shown at time zero, is not statistically different between groups. Further, CD11b expression was examined on PMA-stimulated, circulating neutrophils to determine if the delay observed with thioglycollate challenge was due to deficient intracellular stores. Peripheral blood neutrophils from wild-type and knock-out animals were stimulated with PMA in vitro, then stained with α-Gr-1 (PE) and α-CD11b (FITC), and analyzed by flow cytometry. Results shown are representative of three experiments using eight samples each. Both wild-type and C/EBPε-deficient neutrophils showed the expected upregulation of CD11b expression after PMA stimulation, as shown on Fig 2C, with no statistical differences detectable (unstimulated, P = .135; stimulated, P = .263, independent t-test). These results suggest that intracellular stores of CD11b are present in C/EBPε-deficient neutrophils; however, surface expression is delayed with in vivo inflammatory challenge.
In contrast to CD11b integrin expression, L-selectin expression is higher on C/EBPε-deficient neutrophils at 4 hours after challenge (P < .04; Mann Whitney U test) and higher on wild-type cells 20 hours later (P < .02) (Fig 2D). L-selectin expression on peripheral, unstimulated neutrophils, shown on the y-axis, is significantly higher on C/EBPε-deficient neutrophils, compared with wild-type cells (P < .001; Mann Whitney U test).
Neutrophil-mediated cytokine expression is significantly altered in C/EBPε-deficient neutrophils.
Response of neutrophils to chemical peritonitis, in vivo, was measured by intracellular cytokine fluorescent antibody staining and flow cytometry. Summarized results comprise three separate experiments each using 6 to 10 animals. Additionally, total RNA harvested from peritoneal exudate cells was analyzed by RPA. Results shown are representative for four separate RPA experiments each using RNA from two wild-type and two knock-out animals at each time point (4 hours, 24 hours, 48 hours, and 72 hours).
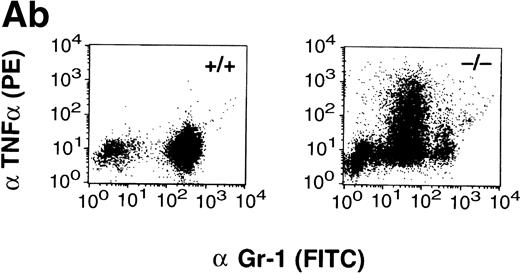
Intracellular cytokine staining of peritoneal exudate cells showed significantly increased levels of TNF-α in C/EBPε-deficient neutrophils compared with wild-type cells at 4 hours after thioglycollate challenge (Fig 3A, panel a). Neutrophil production of cytokines, IL-6 and IL-10, was also tested at 4 and 24 hours and found to be similarly elevated in activated peritoneal neutrophils from both C/EBPε-deficient and wild-type mice. (Geometric mean of fluorescent intensity at 4 hours: wild-type IL-6, 8.4 ± 1.1; knock-out IL-6, 6.6 ± 0.4; wild-type IL-10, 3.47 ± 0.32; knock-out IL-10, 3.43 ± 0.13. Geometric mean of fluorescent intensity at 24 hours: wild-type IL-6, 24.09 ± 2.13; knock-out IL-6, 20.02 ± 1.39; wild-type IL-10, 6.79 ± 0.24; knock-out IL-10, 9.27 ± 0.38.) Figure 3A (panel b) shows representative dot blots derived from fluorescence-activated cell sorting (FACS) analysis of wild-type and C/EBPε-deficient neutrophils.
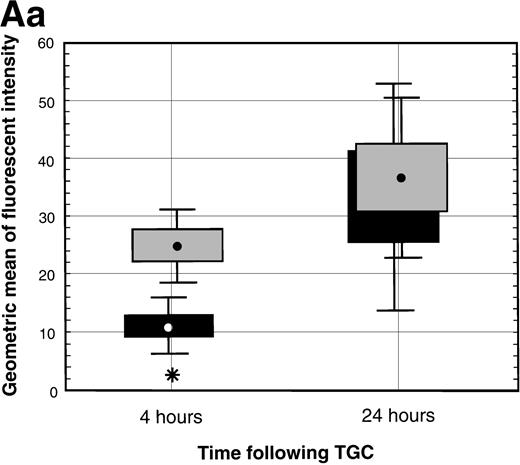
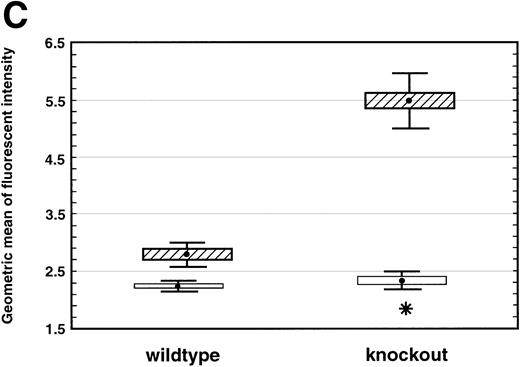
(A) TNF- expression by peritoneal neutrophils after thioglycollate challenge. Peritoneal lavage cells were incubated with monesin for 5 hours to block transport of proteins from the golgi apparatus, permeabilized, and stained with FITC-labeled Gr-1 and PE-labeled anti-mouse TNF-. Panel a shows box and whisker plot of geometric mean of fluoroscent intensity as determined by flow cytometry. (▪) Represent wild-type neutrophils, gray boxes represent C/EBPɛ-deficient neutrophils. An asterisk (*) indicates TNF- staining is significantly increased in C/EBPɛ-deficient neutrophils at 4 hours compared with wild-type neutrophils (P < .005, Mann Whitney U). Panel b shows representative dot blots of wild-type and C/EBPɛ-deficient neutrophil samples (+/+ wild-type; −/− C/EBPɛ-deficient). (B) Cytokine expression by ribonuclease protection assay in peritoneal neutrophils after thioglycollate challenge. Arrows show indicated cytokine transcripts as determined by size. L32 and GADPH bands represent controls for sample size. C/EBPɛ-deficient neutrophil RNA (−/−); wild-type (+/+). (C) TNF- expression in circulating, unstimulated neutrophils as determined by intracellular cytokine staining. Results are represented graphically by geometric mean of fluorescent intensity: (□), unlabeled neutrophils; (▨), TNF-–labeled neutrophils. Fluorescent intensity of labeled and unlabeled wild-type neutrophils was not statistically different (P = .999). Asterisk shows indicated statistical significance of TNF- labeling of peripheral blood C/EBPɛ-deficient neutrophils (P < .001, Student’s t-test).
(A) TNF- expression by peritoneal neutrophils after thioglycollate challenge. Peritoneal lavage cells were incubated with monesin for 5 hours to block transport of proteins from the golgi apparatus, permeabilized, and stained with FITC-labeled Gr-1 and PE-labeled anti-mouse TNF-. Panel a shows box and whisker plot of geometric mean of fluoroscent intensity as determined by flow cytometry. (▪) Represent wild-type neutrophils, gray boxes represent C/EBPɛ-deficient neutrophils. An asterisk (*) indicates TNF- staining is significantly increased in C/EBPɛ-deficient neutrophils at 4 hours compared with wild-type neutrophils (P < .005, Mann Whitney U). Panel b shows representative dot blots of wild-type and C/EBPɛ-deficient neutrophil samples (+/+ wild-type; −/− C/EBPɛ-deficient). (B) Cytokine expression by ribonuclease protection assay in peritoneal neutrophils after thioglycollate challenge. Arrows show indicated cytokine transcripts as determined by size. L32 and GADPH bands represent controls for sample size. C/EBPɛ-deficient neutrophil RNA (−/−); wild-type (+/+). (C) TNF- expression in circulating, unstimulated neutrophils as determined by intracellular cytokine staining. Results are represented graphically by geometric mean of fluorescent intensity: (□), unlabeled neutrophils; (▨), TNF-–labeled neutrophils. Fluorescent intensity of labeled and unlabeled wild-type neutrophils was not statistically different (P = .999). Asterisk shows indicated statistical significance of TNF- labeling of peripheral blood C/EBPɛ-deficient neutrophils (P < .001, Student’s t-test).
RPA analysis of peritoneal exudate neutrophils confirmed these results showing increased expression of TNF-α and LTβ in C/EBPε-deficient neutrophils compared with wild-type cells (Fig 3B). Also, comparison of cytokine mRNA showed higher levels of IL-1Ra expression in wild-type neutrophils compared with C/EBPε-deficient neutrophils at 4 hours after thioglycollate challenge (TGC).
Finally, expression of TNF-α was assessed by intracellular cytokine staining in circulating peripheral blood neutrophils from healthy nonchallenged mice. Results shown are representative of four experiments each using 15 to 20 samples. Wild-type neutrophils do not express detectable levels of TNF-α. Neutrophils deficient in C/EBPε, however, express detectable amounts of TNF-α compared with wild-type neutrophils (Fig 3C) (P < .001; Student’st-test). TNF-α expression is significantly lower in nonchallenged circulating neutrophils compared with peritoneal exudate cells; however, low-level expression was consistently found in C/EBPε-deficient, circulating neutrophils from both young as well as older mice.
Circulating peripheral blood neutrophils from C/EBPε-deficient mice express decreased levels of Gr-1.
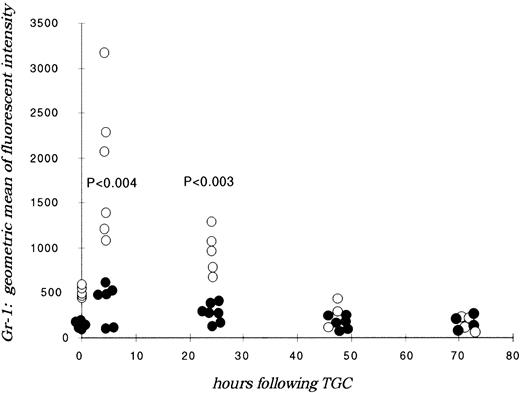
Wild-type peritoneal neutrophils collected at differing time points after thioglycollate-induced peritonitis showed abundant expression of Gr-1 at 4 hours, which decreased over time, consistent with local priming of cells within the peritoneal cavity and shedding of glycosylphosphatidylinositol (GP-1) anchored epitopes.21Alternatively, C/EBPε-deficient neutrophils maintained poor expression of Gr-1 throughout the time course of the thioglycollate challenge, significantly less than wild-type expression at all time points (P < .004; Mann Whitney U test) (Fig 4). Circulating peripheral blood neutrophils in C/EBPε-deficient mice also expressed significantly less Gr-1 compared with wild-type cells, as shown on Fig 4, at time zero (P < .001, Mann Whitney U test).
Gr-1 expression on peritoneal lavage neutrophils. Mice received thioglycollate intraperitoneally, and neutrophils were harvested at indicated time points. Cells were stained with Gr-1 (FITC) and assessed by flow cytometry. Results are represented graphically, by geometric mean fluorescent intensity. (○) Represent wild-type neutrophils; (•) represent C/EBPɛ-deficient neutrophils. Significant differences between samples are shown (Mann Whitney U test).
Gr-1 expression on peritoneal lavage neutrophils. Mice received thioglycollate intraperitoneally, and neutrophils were harvested at indicated time points. Cells were stained with Gr-1 (FITC) and assessed by flow cytometry. Results are represented graphically, by geometric mean fluorescent intensity. (○) Represent wild-type neutrophils; (•) represent C/EBPɛ-deficient neutrophils. Significant differences between samples are shown (Mann Whitney U test).
Expression of C/EBPδ is altered in C/EBPε-deficient neutrophils.
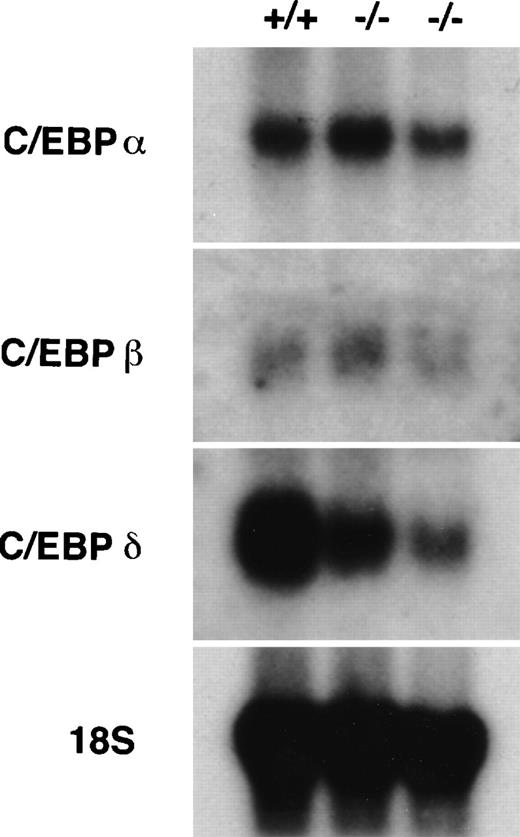
Northern hybridization of mRNA collected from peritoneal lavage cells showed differential expression of C/EBPδ (Fig 5). Altered morphology of C/EBPε-deficient neutrophils, as previously described,5did not permit density discrimination from macrophages by Ficol gradient, however, percent mature neutrophils in each sample were similar, ranging from 65.9% to 60.5%, the remainder of the cells being macrophages. C/EBPα and C/EBPβ are expressed equally well in C/EBPε-deficient neutrophils compared with wild-type cells. C/EBPδ expression, however, is downregulated in C/EBPε-deficient neutrophils relative to wild-type cells.
Expression of other C/EBP family members in C/EBPɛ-deficient neutrophils. RNA harvested from peritoneal lavage cells (4 hours after thioglycollate challenge) was resolved by electrophoresis, transferred to Nytran, and hybridized with C/EBP-specific 32P-–dCTP-labeled probes. C/EBPɛ-deficient neutrophil RNA (−/−); wild-type (+/+).
Expression of other C/EBP family members in C/EBPɛ-deficient neutrophils. RNA harvested from peritoneal lavage cells (4 hours after thioglycollate challenge) was resolved by electrophoresis, transferred to Nytran, and hybridized with C/EBP-specific 32P-–dCTP-labeled probes. C/EBPɛ-deficient neutrophil RNA (−/−); wild-type (+/+).
DISCUSSION
The results described in this study show that C/EBPε-deficient neutrophils have wide-ranging defects relating to phagocytosis and bacterial killing, migration, and cytokine production. Further, these neutrophils evoke an abnormal microenvironment, due to increased TNF-α production, analogous to disease states of malignancy or chronic inflammation. In its entirety, C/EBPε-deficient cells show the complex signaling network between cellular proliferation and terminal function and suggest the molecular basis for the severe immunodeficiency and myelodysplasia seen in C/EBPε-deficient mice.5
Northern blot analysis showed equivalent levels of C/EBPα and C/EBPβ expression in C/EBPε-deficient and wild-type neutrophils. Interestingly, C/EBPδ expression was clearly decreased in C/EBPε-deficient neutrophils compared with wild-type cells. C/EBP protein expression varies during myelopoiesis. Analysis of temporal expression of C/EBP factors during myeloid development shows C/EBPα and C/EBPδ are expressed early, with levels of C/EBPβ steadily increasing and C/EBPα quickly decreasing with cell maturation.22 Northern hybridization of peritoneal exudate neutrophils permits analysis of C/EBP expression in a mature, homogenous population, unlike bone marrow preparations. C/EBP recognition sequences are present in the neutrophil elastase promoter and in the G-CSF receptor promoter.23,24 Also, C/EBPβ has functional sites in the regulatory sequences of the genes for IL-1, IL-8, G-CSF, and GM-CSF.25 Given the varying transactivating and DNA binding potentials of the different C/EBP homodimers and heterodimers, the intricacies of transcriptional regulation within a single family of transcription factors is formidable.
Neutrophils are traditionally viewed as the mediators of nonspecific cellular immunity, migrating to sites of infection, phagocytizing bacteria, and releasing toxic oxidative products. Recent work, however, suggests that neutrophils are also metabolically active cells, expressing inflammatory cytokines and undergoing significant morphological change in response to infection.26-28 Severe neutropenia, as seen with cancer chemotherapy or ablative therapy preceding bone marrow transplantation, often precipitates gram-negative bacterial sepsis, the most frequent culprit being P aeruginosa. The appearance of systemic P aeruginosa infection in C/EBPε-deficient mice suggests, at first glance, that these mice have an early maturation block in myelopoiesis and lack segmented neutrophils. Analysis of peripheral blood differential smears, however, shows the presence of numerous, segmented neutrophils.5 The appearance of this specific organism in unchallenged mice affords the rare opportunity to assess neutrophil defects secondary to the loss of C/EBPε transcriptional activity.
Multiple components of the antibacterial armaenterium are defective in the absence of C/EBPε transactivation. Our previous work showed the complete absence of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase activity in C/EBPε-deficient neutrophils after PMA activation.5 CGD is the human genetic disease resulting from the lack of NADPH oxidase activity. Mice engineered to lack various components of the NADPH oxidase protein complex share phenotypic qualities with the human disease, specifically, susceptibility to catalase positive organisms, S aureus, C albicans, and others.14 15 Importantly, however, neither CGD patients nor these animal models develop spontaneous infections with P aeruginosa, suggesting that additional functional defects exist in C/EBPε-deficient neutrophils.
Phagocytosis is defective on several levels in C/EBPε-deficient neutrophils. Ingestion of opsonized bacteria is significantly decreased in C/EBPε-deficient neutrophils, possibly due to decreased receptor population. Additionally, intracellular killing of phagocytized bacteria is impaired. Inability to generate an oxidative burst certainly contributes to this defect; however, expression of secondary granule proteins in C/EBPε-deficient neutrophils is nearly undetectable. This is not without precedent, as C/EBPα-deficient neutrophils lack both primary and secondary granule proteins.11 Likely, the combined lack of toxic oxidative metabolites and secondary granule proteins renders the C/EBPε-deficient neutrophil ineffective against intracellular bacteria.
Neutrophil migration is also impaired in the absence of C/EBPε. C/EBPε-deficient neutrophils show delayed migration into the peritoneal cavity of thioglycollate-challenged mice. Coincident with this delay is the impaired early expression of neutrophil surface receptors CD62L (L-selectin) and CD11b on C/EBPε-deficient cells, which are integral to neutrophil adhesion and migration.29-31
C/EBPε-deficient neutrophils display an unusual pattern of selectin and integrin expression in response to inflammation. CD11b/CD18-deficient mice show decreased neutrophil adhesion, however, paradoxically increased numbers of neutrophils in peritoneal exudates after thioglycollate challenge.32L-selectin–deficient mice show delayed neutrophil migration after thioglycollate challenge compared with wild-type controls.31 The relatively high levels of L-selectin on circulating knock-out neutrophils may effect the normal response of selectin shedding with neutrophil activation. Interestingly, loss of CD11b/CD18 expression is also linked to the absence of neutrophil phagocytosis, reduction of oxidative burst, and diminished neutrophil apoptosis.32 Neutrophil phagocytosis and oxidative burst are blunted in C/EBPε-deficient neutrophils, as is neutrophil migration, despite normal or exaggerated neutrophil migration in the CD11b/CD18 knock-out model.32
CD11a expression is not effected by neutrophil activation,33 nor was differential expression of CD11a detected between C/EBPε-deficient and wild-type neutrophils (data not shown). These results suggest that the delay in CD11b upregulation in response to inflammation lies in an inappropriate response to priming signals, rather than intrinsic defects in surface receptor expression levels. Alternatively, an altered microenvironment, due to abnormal cytokine expression, such as TNF-α, may result in aberrant neutrophil responses.
Analysis of neutrophil cytokine expression over the course of an inflammatory response shows unexpected differences between C/EBPε-deficient and wild-type cells. Both intracellular cytokine antibody staining and steady-state mRNA expression show significantly increased expression of TNF-α and LTβ in C/EBPε-deficient neutrophils early in the response to an in vivo inflammatory stimulus. Additionally, IL-1Ra expression was downregulated in C/EBPε-deficient neutrophils.
TNF-α is a bifunctional regulator of the inflammatory response. Low and moderate doses of TNF-α prime neutrophils,34 enhance leukocyte recruitment to inflammatory sites,35,36 induce expression of GM-CSF,37 and prolong neutrophil survival.38 High-dose TNF-α, however, inhibits colony formation induced by G-CSF, downregulates CSF receptor expression,39 promotes cellular apoptosis,40and in the clinical setting, is associated with poor survival from sepsis or lymphoma.41 Lymphotoxin β enhances neutrophil respiratory burst and inhibits locomotion.42 Deciphering the effects of elevated TNF-α levels associated with thioglycollate-induced peritonitis in C/EBPε-deficient mice is confounded by the associated differences in IL-1Ra expression. One may speculate that high local levels of TNF-α at infectious foci in C/EBPε-deficient mice may set up a cycle of cell recruitment and cell degradation, with release of cytokines and further cell recruitment. Interestingly, chronic granuloma formation seen in challenged gp91phox-deficient (CGD) mice is associated with elevated levels of TNF-α and IL-1β.43 Dysregulation of the mechanisms governing acute inflammation and resolution may result in the localized accumulation of myeloid cells, seen in a high proportion of endstage C/EBPε-deficient mice.
IL-1Ra, an antagonist of IL-1β, is expressed in response to GM-CSF and TNF-α stimulation.44Yersinia enterocoliticainfection in mice promotes IL-1Ra expression in circulating neutrophils.45 IL-1Ra completely abrogates the priming effects on the respiratory burst induced by IL-1β and IL-1α.46 Decreased expression of IL-1Ra in C/EBPε-deficient neutrophils may be due to loss of C/EBPε transcriptional activity, particularly because a number of cytokines, including IL-1, IL-8, G-CSF, and GM-CSF have C/EBP cognate sequences within their regulatory regions.25 Interestingly, levels of IL-6, which induce IL-1Ra expression,45 appear normal in these mice by RPA and intracellular cytokine staining, suggesting complex signaling pathways of cytokine expression. Decreased IL-1Ra expression may fail to suppress ongoing acute inflammatory responses. In conjunction with elevated TNF-α levels, the lack of IL-1Ra may be, in part, responsible for the myeloid cell aggregations seen in end-stage C/EBPε-deficient mice, an aberrant response to low pathogenicity, chronic infections.
TNF-α expression in circulating neutrophils preceding thioglycollate challenge is also increased in C/EBPε cells compared with wild-type cells. Chronically elevated levels of TNF-α, even in mice as young as 3 weeks, provides explanation for the preactivated state of C/EBPε-deficient neutrophils, as suggested by decreased Gr-1 staining. Neutrophil priming is reported in human immunodeficiency virus (HIV)-infected patients, associated with increased CD11β/CD18 expression, increased actin polymerization, and decreased L-selectin expression.47 Elevated TNF-α levels are associated with adverse prognosis in patients with lymphoma.41 The possible role of C/EBPε in effecting disease states needs to be investigated.
Interestingly, C/EBPε-deficient mice share many phenotypic features with patients with specific granule deficiency (SGD). SGD patients are severely immunocompromised, developing frequent bacterial infections with S aureas, P aeruginosa, andKlebsiella.48,49 SGD neutrophils possess atypical, bilobed nuclei, similar in appearance to C/EBPε-deficient neutrophils.49 Additionally, SGD neutrophils, like C/EBPε-deficient neutrophils, lack lactoferrin and gelatinase B expression, display delayed chemotaxis, and decreased bacterial killing in vitro.48-54 These similarities suggest a regulatory role for C/EBPε transactivation in the disease pathology of SGD.
Analogous to the role of C/EBPα in linking hepatocyte and adipocyte function and differentiation, C/EBPε may also regulate both normal neutrophil function and the terminal steps of myeloid differentiation. C/EBPε-deficient mice have multiple neutrophil defects including depressed phagocytosis and phagocidal killing, impaired oxidative burst, deficient secondary and tertiary granule protein mRNAs, and delayed response to an inflammatory stimulus. The combination of increased TNF-α and decreased IL-1Ra expression further creates an abnormal microenvironment, likely affecting neutrophil recruitment, function, and resolution of inflammation. This abnormal microenvironment, secondary to loss of critical feedback loops necessary for resolution of inflammatory responses may, in part, result in uncontrolled myeloid proliferation. Alternatively, intrinsic neutrophil defects due to loss of C/EBPε expression may drive the myeloproliferation seen in knock-out animals. Work continues to decipher the processes driving the myeloproliferative phenotype in C/EBPε-deficient mice.
ACKNOWLEDGMENT
We are grateful to L. Garrett and T. Hernandez for excellent technical assistance and devoted animal care. We thank Drs D.G. Tenen and D. Bodine for expert advice and reagents, and Dr J.I. Gallin for critical input. We thank Dr R.M. Blaese for creating an inspiring environment and providing constant support.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Julie Lekstrom-Himes, MD, 10 Center Drive, 11N120, Bethesda, MD 20892; e-mail:jlekstrom@atlas.niaid.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal