Abstract
Internal tandem duplication of the FLT3 gene and point mutations of the N-RAS gene are the most frequent somatic mutations causing aberrant signal-transduction in acute myeloid leukemia (AML). However, their prognostic importance is unclear. In this study, their prognostic significance was analyzed in 201 newly diagnosed patients with de novo AML except acute promyelocytic leukemia. Three patients had mutations in both genes, 43 had only theFLT3 gene mutation, 25 had only the N-RAS gene mutation, and 130 had neither. These mutations seemed to occur independently. Both mutations were related to high peripheral white blood cell counts, and the FLT3 gene mutation was infrequently observed in the French-American-British (FAB)-M2 type. AML cases with wild FLT3/mutant N-RAS had a lower complete remission (CR) rate than those with wild FLT3/wild N-RAS, whereas the presence of mutant FLT3 did not affect the CR rate. Univariate analysis showed that unfavorable prognostic factors for overall survival were age 60 years or older (P = .0002), cytogenetic data (P = .002), FAB types other than M2 (P = .002), leukocytosis over 100 ± 109/L (P = .003), and the FLT3 gene mutation (P = .004). However, the N-RAS gene mutation was only a marginal prognostic factor (P = .06). For the subjects under 60 years old, multivariate analysis showed that theFLT3 gene mutation was the strongest prognostic factor (P = .008) for overall survival. The FLT3 gene mutation, whose presence is detectable only by genomic polymerase chain reaction amplification and gel electrophoresis, might serve as an important molecular marker to predict the prognosis of patients with AML.
OUR UNDERSTANDING of the pathophysiology of acute myeloid leukemia (AML) has rapidly advanced over the past two decades. Cytogenetic studies have clarified the molecular mechanism of leukemogenesis. Chromosomal translocations, which are found in half of all AML cases and correlate with the French-American-British (FAB) types, target and deregulate the gene-coding transcriptional factors that are important to hematopoiesis.1-3Nonrandom chromosomal loss or deletion suggests that anti-oncogenes are also involved in AML.1,2 Mutations and/or deletion of theP53 gene were observed in AML,4 although the incidence of these events is far lower than that in solid tumors.5 Transfection studies using NIH/3T3 cells showed that activated RAS genes are associated with the pathogenesis of AML.6RAS gene mutations, the majority of which involve the N-RAS gene, are found in up to 30% of AML cases.7-9 These genetic alterations have been molecularly detectable and used for diagnosis, detection of minimal residual disease, and prediction of prognosis. For routine assessment, however, the following factors are required: clinical incidence and significance, time- and cost-saving measures, and specificity and sensitivity of the examination.
Recently an internal tandem duplication of the juxtamembrane (JM) domain-coding sequence of the FLT3 gene was found in 20% of AML.10,11 FLT3 is a class III receptor tyrosine kinase (RTK), along with KIT, FMS, and PDGFR.12,13 Since FLT3 preferentially expressed on hematopoietic stem cells and its ligand (FL) on bone marrow stroma cells,13-15 FLT3-FL interaction plays an important role in primitive hematopoiesis. Furthermore, most clinical samples from AML express functional FLT3, and the FLT3-FL interaction might also be associated with the proliferation of leukemia cells.16 The duplicated sequences of the mutantFLT3 consisted of exon 11, but sometimes involved intron 11 and exon 12.10,11 Although their location and length varied from sample to sample, the portion of tandem duplication was always readable in frame. Actually the transcripts with a long JM domain did not disrupt the downstream regions. The mutant FLT3 was ligand-independently phosphorylated when expressed in Cos 7 cells,17 indicating dominant positive mutation. Interleukin-3 (IL-3)–dependent myeloid progenitor cell lines, FDC/P1 and 32D, exhibited IL-3–independent growth when transfected with mutant FLT3 (H. Kiyoi, T. Naoe, unpublished data, 1998). The FLT3 gene mutation was found in all types of the FAB classification and in 3% of myelodysplastic syndrome, but never in chronic myeloid leukemia or in lymphoid malignancies.11 In acute promyelocytic leukemia (FAB-M3), the presence of the FLT3gene mutation was related to high peripheral white blood cell (WBC) counts, high peripheral leukemia cell counts, and high lactate dehydrogenase (LDH) level.18 These findings suggest that the FLT3 gene mutation plays an important role in leukemia progression rather than initiation.
Since the detection of the FLT3 gene mutation requires only polymerase chain reaction (PCR) amplification using genomic DNA followed by gel electrophoresis, we studied whether it could be used as a standard molecular marker for the prognosis of AML. Here we analyzed the prognostic significance of the FLT3 gene mutation together with the N-RAS gene mutations in a large number of patients with AML.
PATIENTS AND METHODS
Patients and treatments.
Two hundred one newly diagnosed patients with AML except for M3, who were treated with three protocols of the Japan Adult Leukemia Study Group, and whose leukemia cells were preserved with informed consent at initial diagnosis, were eligible for this study. Twenty-eight, 40, and 133 patients were treated by the AML-87,19AML-89,20 and AML-9221 protocols, respectively. AML was diagnosed according to the FAB classification, which was evaluated by the central review committee.
In the AML-87 study,19 the induction therapy consisted of daily behenoyl cytarabine (BHAC) 200 mg/m2, daily 6-mercaptopurine (6-MP) 70 mg/m2, daily prednisolone 40 mg/m2, and daunorubicin (DNR) 40 mg/m2 on days 1 to 3, and if necessary on days 7, 8, and 11. The therapy was continued for 10 to 12 days until the bone marrow became severely hypoplastic with less than 5% blasts. In the AML-89 study,20 patients were randomized to receive induction therapy that included BHAC (200 mg/m2 by 3-hour infusion) or cytarabine (AraC, 80 mg/m2 by continuous infusion). BHAC or AraC, and 6-MP 70 mg/m2 were administered for 10 to 12 days, and DNR 40 mg/m2 was administered on days 1 to 4, and if necessary, on days 10 to 12 in addition to the above schedule for AML-87. In the AML-92 study,21 patients were randomized to receive BHAC-DM similar to the AML-87 protocol with or without etoposide (ETP, 100 mg/m2 for 5 days). After achieving complete remission (CR), three courses of consolidation chemotherapy and six courses of intensification chemotherapy were administered. Patients 60 years or older received about two thirds of the dosage of each drug throughout the study period.
CR was determined when there were less than 5% blasts in normo-cellular bone marrow with normal levels of peripheral neutrophil and platelet counts. Overall survival (OS) was calculated from the first day of therapy to death. Disease-free survival (DFS) for patients who had achieved CR was measured from the date of CR to relapse or death. Patients who underwent bone marrow transplantation (BMT) were censored at the date of BMT.
Analysis of the internal tandem duplication of the FLT3gene.
High molecular weight DNA was extracted from AML cells as previously described.9 Because previous studies showed that the location of internal tandem duplication of the FLT3 gene was restricted to exons 11 and 12,11 18 genomic PCR amplification was performed using the primers 11F, 5′-GCAATTTAGGTATGAAAGCCAGC, and 12R, 5′-CTTTCAGCATTTTGACGGCAACC-3′. The PCR mixture contained 500 ng of genomic DNA, 50 pmol of 11F and 12R primers, 0.2 mmol/L of each deoxynucleotide triphosphate, 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 1.5 mmol/L MgCl2, 0.001% gelatin (wt/vol), 50 mmol/L tetramethylammonium chloride and 2.5 U ofTaq polymerase (Amplitaq; Perkin Elmer, Norwalk, CT). Denaturing, annealing, and extension steps were performed at 94°C for 30 seconds, 56°C for 1 minute and 72°C for 2 minutes, respectively, for 35 cycles on a GeneAmp PCR system 9600 (Perkin Elmer) including an initial 3 minutes denaturation step at 94°C and a final extension step at 72°C for 10 minutes. The amplified product was cut out from an agarose gel, purified with a Qiaex gel extraction kit (Qiagen Inc, Chatsworth, CA), and cloned into the pMOSBlue T-vector (Amersham, Buckinghamshire, UK) according to the manufacturer’s recommendation. Ten recombinant colonies were chosen and cultured in LB medium. Plasmid DNA was prepared using a QIAprep spin plasmid miniprep kit (Qiagen Inc), and both strands were sequenced using fluorescein-conjugated −21M13 and T7 primers on a DNA sequencer (377; Applied Biosystems, Foster City, CA).
N-RAS gene amplification and dot-blot hybridization.
To amplify the sequences spanning codons 12 and 13, and codon 61, the oligonucleotide primers were prepared as follows; 5′primer for codons 12, 13 (named NA12): 5′-GACTGAGTACAAACTGGTGG-3′, 3′primer for codons 12, 13 (NB12): 5′-CTCTATGGTGGGATCATATT-3′, 5′primer of codon 61 (NA61): 5′-GGTGAAACCTGTTTGTTGGA-3′, and 3′primer of codon 61 (NB61): 5′-ATACACAGAGGAAGCCTTCG-3′. The PCR was performed as described previously.9 Genomic DNA was subjected to 35 cycles of PCR amplification (denaturation for 60 seconds at 92°C, annealing for 60 seconds at 55°C, and elongation for 60 seconds at 72°C). The efficiency of amplification was evaluated by agarose gel electrophoresis, and only the reactions resulting in the appropriately sized band were further analyzed. Dot-blot and oligonucleotide probe hybridization were performed as previously described.9Briefly, either 100 ng DNA amplified by PCR was transferred to nylon filter membranes (Hybond-N+; Amersham) with a 96-well filtration manifold. Blotted DNA was crosslinked by UV illumination and the membranes were hybridized with [γ-32P] labeled oligonucleotide probes. The oligonucleotide panel included probes specific for the wild-type allele and all possible amino acid substitutions at codons 12, 13, and 61 of the N-RAS gene. The prehybridization, hybridization, and washing of membranes were performed under standard conditions.9 The membranes were exposed to Kodak XAR5 films (Eastman Kodak, Rochester, NY) at −70°C using intensifying screens.
Statistical methods.
The following clinical characteristics at diagnosis were analyzed: age, sex, FAB classification, peripheral WBC count, percentage of blasts in bone marrow, platelet count, serum LDH concentration, the presence of hepato-splenomegaly or extramedullary involvement, and cytogenetic findings. Analysis of frequencies was performed using the Fisher’s exact test for 2 × 2 tables or the Pearson’s χ2 test for larger tables. Differences in median variables in age, peripheral WBC counts, platelet counts, percentage of blasts in bone marrow, and LDH were also analyzed with the Wilcoxon rank-sum test. CR rates in the two groups were compared using the Fisher’s exact test. The logistic progression model was used for multivariate analysis.
Survival probabilities were estimated by the Kaplan-Meier method, and differences in the survival distributions between the mutation-positive and -negative groups were evaluated by the log-rank test. The prognostic significance of the clinical variables was assessed using the Cox proportional hazards model. These statistic analyses were performed with StatView software (Abacus Concepts Inc, Berkeley, CA) or SAS programs (SAS Institute Inc, Cary, NC). Because CR rates, OS, and DFS according to induction therapies (the AML-87, -89, and -92 studies) did not show any difference, the data of the three studies were combined and analyzed. For all analyses, the P values were two-tailed, and a P value of less than .05 was considered statistically significant.
RESULTS
A total of 201 newly diagnosed patients with AML were studied for theFLT3 and N-RAS gene mutations. FLT3 gene mutations were identified in 46 of the 201 patients. Translated into amino acids, the tandem duplications frequently involved a Y-rich stretch from codon 589 to 599 (data not shown), the same position as in previous studies.11 18 N-RAS gene mutations were detected in 28 of the 201 patients. The mutations at codons 12, 13, and 61 were observed in 12, 13, and 7 patients, respectively. Three of the 28 patients had multiple mutations at codon 12, one at codon 61, and three at codons 12 and 13. Of a total of 37 N-RAS gene point mutations, G to A transition was the most frequent (16/37).
In a total of 201 patients, 3 patients (1.5%) had mutations in both genes (mutant FLT3/mutant N-RAS), 43 (21.4%) had only mutant FLT3 gene (mutant FLT3/wild N-RAS), 25 (12.4%) had only mutant N-RAS gene (wildFLT3/mutant N-RAS), and 130 (64.7%) had neither (wild FLT3/wild N-RAS). Clinical characteristics were analyzed comparing these four groups (Table1). The presence of mutant FLT3 or mutant N-RAS was not related to age and sex (data not shown). WBC counts in the wild FLT3/wild N-RAS group were significantly lower than in other groups (P = .03 vmutant FLT3/mutant N-RAS, P < .0001v mutant FLT3/wild N-RAS, P = .002v wild FLT3/mutant N-RAS) (Table1). Serum LDH level in the wild FLT3/wild N-RAS group tended to be lower than that in the other groups. The occurrence of hepato-splenomegaly or extramedullary involvement was not significantly affected by these mutations. The incidence of theFLT3 gene mutation according to FAB classification was ranked as follows: M4/5 (22/62) > M1 (14/48) > M2 (8/83). Additionally, the FLT3 gene mutation was infrequently observed in the leukemia with t(8;21) (P = .02). The incidence of the N-RAS gene mutation was similarly ordered: M4/5 (11/62) > M1 (6/48) > M2 (10/83).
Clinical Characteristics of 201 Patients With De Novo AML Except M3
| . | Total (N = 201) . | Mutant FLT3/ Mutant N-RAS (N = 3) . | Mutant FLT3/ Wild N-RAS (N = 43) . | Wild FLT3/ Mutant N-RAS (N = 25) . | Wild FLT3/ Wild N-RAS (N = 130) . |
|---|---|---|---|---|---|
| Age | 49 | 60 | 56 | 45 | 48.5 |
| (41-64) | (15-77) | (16-82) | (15-85) | ||
| WBC (109/L) | 24.7 | 120* | 52.2‡ | 67.8† | 19.3 |
| (50.2-372) | (2.1-632) | (5.3-234) | (0.9-33.7) | ||
| FAB | |||||
| M0 | 3 | 0 | 0 | 1 | 2 |
| M1 | 48 | 2 | 12 | 4 | 30 |
| M2 | 83 | 1 | 7* | 9 | 66 |
| M4 | 47 | 0 | 17 | 11 | 19 |
| M5 | 15 | 0 | 5 | 0 | 10 |
| M6 | 4 | 0 | 1 | 0 | 3 |
| M7 | 1 | 0 | 1 | 0 | 0 |
| Cytogenetics | |||||
| t(8;21) | 28 | 0 | 2* | 1 | 25 |
| inv(16) | 6 | 0 | 2 | 0 | 4 |
| t(9;22) | 3 | 0 | 0 | 1 | 2 |
| del(5) or del(7) | 11 | 0 | 2 | 2 | 7 |
| Others | 41 | 0 | 5 | 7 | 29 |
| Normal | 82 | 3 | 20 | 12 | 47 |
| ND | 30 | 0 | 11 | 2 | 17 |
| Outcome | |||||
| CR | 147 | 2 | 30 | 13† | 102 |
| Failure | 51 | 0 | 13 | 12† | 26 |
| Unevaluable | 3 | 1 | 0 | 0 | 2 |
| . | Total (N = 201) . | Mutant FLT3/ Mutant N-RAS (N = 3) . | Mutant FLT3/ Wild N-RAS (N = 43) . | Wild FLT3/ Mutant N-RAS (N = 25) . | Wild FLT3/ Wild N-RAS (N = 130) . |
|---|---|---|---|---|---|
| Age | 49 | 60 | 56 | 45 | 48.5 |
| (41-64) | (15-77) | (16-82) | (15-85) | ||
| WBC (109/L) | 24.7 | 120* | 52.2‡ | 67.8† | 19.3 |
| (50.2-372) | (2.1-632) | (5.3-234) | (0.9-33.7) | ||
| FAB | |||||
| M0 | 3 | 0 | 0 | 1 | 2 |
| M1 | 48 | 2 | 12 | 4 | 30 |
| M2 | 83 | 1 | 7* | 9 | 66 |
| M4 | 47 | 0 | 17 | 11 | 19 |
| M5 | 15 | 0 | 5 | 0 | 10 |
| M6 | 4 | 0 | 1 | 0 | 3 |
| M7 | 1 | 0 | 1 | 0 | 0 |
| Cytogenetics | |||||
| t(8;21) | 28 | 0 | 2* | 1 | 25 |
| inv(16) | 6 | 0 | 2 | 0 | 4 |
| t(9;22) | 3 | 0 | 0 | 1 | 2 |
| del(5) or del(7) | 11 | 0 | 2 | 2 | 7 |
| Others | 41 | 0 | 5 | 7 | 29 |
| Normal | 82 | 3 | 20 | 12 | 47 |
| ND | 30 | 0 | 11 | 2 | 17 |
| Outcome | |||||
| CR | 147 | 2 | 30 | 13† | 102 |
| Failure | 51 | 0 | 13 | 12† | 26 |
| Unevaluable | 3 | 1 | 0 | 0 | 2 |
Mean (minimum to maximum) values are indicated in age and WBC. Number of cases are shown in FAB, cytogenetics, and outcome.
.1 > P > .01.
.01 > P > .001.
P < .001 compared with wild FLT3/wild N-RAS group.
Abbreviation: ND, not determined.
WBC counts were further analyzed in each FAB group. In M1 and M2, WBC counts in the wild FLT3/wild N-RAS group were lower than the mutant groups (P = .004 v mutantFLT3/wild N-RAS, P = .09 v wildFLT3/mutant N-RAS). In M2, those in the mutantFLT3/wild N-RAS group tended to be lower than other groups (P = .2 v mutant FLT3/wild N-RAS, P = .1 v wild FLT3/mutant N-RAS). In M4/5, however, there was no difference of WBC counts.
The CR rates by initial induction therapy were significantly different between the wild FLT3/mutant N-RAS group and the wildFLT3/wild N-RAS group (52.0% v 79.7%,P = .005). However, the presence of mutant FLT3 did not affect the CR rate. Chi-squared analysis showed that FAB types other than M2, the presence of N-RAS gene mutation, and leukocytosis (over 100 × 109/L) were unfavorable factors for achieving CR (P = .001, P = .04, andP = .07, respectively). Multivariate analysis using the logistic progression model showed that FAB types other than M2 (P = .001) and N-RAS gene mutation (P = .05) were independent unfavorable factors for achieving CR, whereas leukocytosis was not significant.
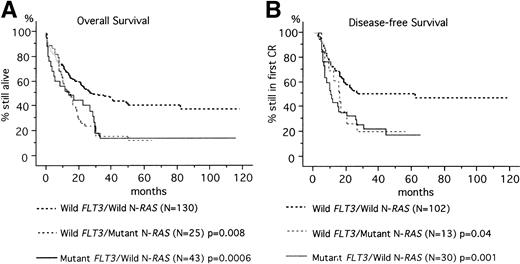
At a median follow-up time of 50 months (range, 3 to 118 months), 68 of 201 patients (33.8%) were alive. The predicted OS rates at 60 months were 14.0%, 16.0%, and 44.6% in the mutant FLT3/wild N-RAS, wild FLT3/mutant N-RAS, and wildFLT3/wild N-RAS groups, respectively (Fig1). Mutant FLT3/wild N-RASand wild FLT3/mutant N-RAS groups had worse prognosis than the wild FLT3/wild N-RAS group (P = .0006 and P = .008, respectively).
Kaplan-Meier curves according to the FLT3 and N-RAS gene mutations. (A) OS of 198 patients. (B) DFS of 145 patients who achieved CR. Three patients with both FLT3 and N-RAS gene mutations were excluded from the analysis because the number was small. Statistic difference was evaluated by the log-rank test.
Kaplan-Meier curves according to the FLT3 and N-RAS gene mutations. (A) OS of 198 patients. (B) DFS of 145 patients who achieved CR. Three patients with both FLT3 and N-RAS gene mutations were excluded from the analysis because the number was small. Statistic difference was evaluated by the log-rank test.
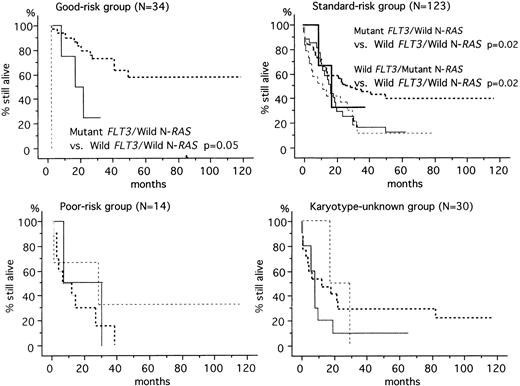
The prognosis of AML depends on factors such as age, initial leukocyte count, FAB classification, karyotype, immune phenotype, and response to remission-induction therapy.19-23 Among them, cytogenetic data is thought to be the most important prognostic factor for AML.1 Based on cytogenetic findings, the 201 patients were segregated into four groups: a good-risk group (n = 34) was defined by karyotype, t(8;21) or inv(16); a poor-risk group (n = 14) by t(9;22), 11q23 alterations, del(5) or del(7); a standard-risk group (n = 123) by normal or other karyotypes; and a karyotype-unknown group (n = 30). The predicted OS rates at 60 months were 57.1%, 33.6%, 13.3%, and 20.7% in the good-risk, standard-risk, poor-risk, and karyotype-unknown groups, respectively. In the good-risk and standard-risk groups, the FLT3 and N-RAS gene mutations were associated with unfavorable prognosis (Fig2). In the poor-risk and karyotype-unknown groups, however, there was no significant association between these mutations and prognosis.
OS according to the FLT3 and N-RAS gene mutations in each karyotype-risk group. In good-risk patients, the mutant FLT3/wild N-RAS group had worse prognosis than the wild FLT3/wild N-RAS group (P = .05). In standard-risk patients, the mutant FLT3/wild N-RASgroup and wild FLT3/mutant N-RAS group had worse prognosis than the wild FLT3/wild N-RAS group (P = .02).(), MutantFLT3/mutant N-RAS; (—), mutant FLT3/wild N-RAS; (···), wild FLT3/mutant N-RAS; (—·—·—·—·): wild FLT3/wild N-RAS.
OS according to the FLT3 and N-RAS gene mutations in each karyotype-risk group. In good-risk patients, the mutant FLT3/wild N-RAS group had worse prognosis than the wild FLT3/wild N-RAS group (P = .05). In standard-risk patients, the mutant FLT3/wild N-RASgroup and wild FLT3/mutant N-RAS group had worse prognosis than the wild FLT3/wild N-RAS group (P = .02).(), MutantFLT3/mutant N-RAS; (—), mutant FLT3/wild N-RAS; (···), wild FLT3/mutant N-RAS; (—·—·—·—·): wild FLT3/wild N-RAS.
Univariate analysis showed that unfavorable prognostic factors for OS were age 60 years or older (P = .0002), cytogenetic data (P = .002), leukocytosis over 100 × 109/L (P = .003), the FLT3 gene mutation (P = .004) (Table2). However, the N-RASgene mutation was only a marginal prognostic factor (P = .06). Multivariate analysis showed that age (60 years or older) was the strongest unfavorable factor (relative risk [RR], 1.9;P = .002), followed by cytogenetics (P = .004). FAB types, leukocytosis, and the FLT3 gene mutation were less important. It might be partly associated with the poor prognosis that the dosage of chemotherapy to the patients 60 years or older was reduced. If the subjects were limited to under 60 years old, theFLT3 gene mutation became the strongest prognostic factor (RR, 2.1; P = .008), with a second factor being cytogenetics (P = .07) and N-RAS gene mutation (RR, 1.7;P = .09) (Table 3).
Unfavorable Prognostic Factors for OS in 201 Patients With De Novo AML Except M3
| Prognostic Factors . | Univariate . | Multivariate . | |
|---|---|---|---|
| P Value . | P Value . | Relative Risk (95% CI) . | |
| Age, 60 yr or older | .0002 | .002 | 1.9 (1.2-2.6) |
| Cytogenetics* | .002 | .004 | 2.6† |
| FAB other than M2 | .002 | .2 | |
| WBC count >100 × 109/L | .003 | .2 | |
| FLT3 gene mutation | .004 | .1 | |
| N-RAS gene mutation | .06 | .6 | |
| Prognostic Factors . | Univariate . | Multivariate . | |
|---|---|---|---|
| P Value . | P Value . | Relative Risk (95% CI) . | |
| Age, 60 yr or older | .0002 | .002 | 1.9 (1.2-2.6) |
| Cytogenetics* | .002 | .004 | 2.6† |
| FAB other than M2 | .002 | .2 | |
| WBC count >100 × 109/L | .003 | .2 | |
| FLT3 gene mutation | .004 | .1 | |
| N-RAS gene mutation | .06 | .6 | |
Karyotypes were segregated into four groups: a good-risk group with t(8;21) or inv(16); a poor-risk group with t(9;22), 11q23 alterations, del(5) or del(7); a standard-risk group with normal or other karyotypes; and a karyotype-unknown group.
Comparison between the good-risk and poor-risk groups.
Unfavorable Prognostic Factors for OS in the 143 Patients Younger Than 60 Years Old
| Prognostic Factors . | Univariate . | Multivariate . | |
|---|---|---|---|
| P Value . | P Value . | Relative Risk (95% CI) . | |
| FLT3 gene mutation | .001 | .008 | 2.1 (1.2-3.4) |
| WBC count >100 × 109/L | .01 | .2 | |
| Cytogenetics | .03 | .07 | 1.53-150 |
| FAB other than M2 | .07 | .5 | |
| N-RASgene mutation | .09 | .09 | |
| Prognostic Factors . | Univariate . | Multivariate . | |
|---|---|---|---|
| P Value . | P Value . | Relative Risk (95% CI) . | |
| FLT3 gene mutation | .001 | .008 | 2.1 (1.2-3.4) |
| WBC count >100 × 109/L | .01 | .2 | |
| Cytogenetics | .03 | .07 | 1.53-150 |
| FAB other than M2 | .07 | .5 | |
| N-RASgene mutation | .09 | .09 | |
Comparison between the good-risk and poor-risk groups.
DFS was further analyzed in 147 patients who achieved CR. The predicted DFS rates at 60 months were 20.0%, 23.1%, and 53.9% in the mutantFLT3/wild N-RAS, wild FLT3/mutant N-RAS, and wild FLT3/wild N-RAS groups, respectively (Fig 1). According to univariate analysis, the following pretreatment variables showed statistical significance for DFS: age (P = .002), cytogenetic data (P = .004), leukocytosis (P = .004), the FLT3 gene mutation (P = .006) (Table 4). Multivariate analysis showed that age was the most unfavorable factor (RR, 2.0; P = .003), followed by cytogenetic data (P = .001), and leukocytosis (P = .04). For the subjects under 60 years old, leukocytosis was the sole important factor (RR, 2.6; P = .01) (Table 5).
Unfavorable Prognostic Factors for DFS in the 147 Patients Who Achieved CR
| Prognostic Factors . | Univariate . | Multivariate . | |
|---|---|---|---|
| P Value . | P Value . | Relative Risk (95% CI) . | |
| Age, 60 yr or older | .002 | .003 | 2.0 (1.3-3.3) |
| Cytogenetics | .004 | .001 | 4.14-150 |
| WBC count >100 × 109/L | .004 | .04 | 1.8 (1.0-3.3) |
| FLT3 gene mutation | .006 | .2 | |
| FAB other than M2 | .1 | .5 | |
| N-RAS gene mutation | .2 | .1 | |
| Prognostic Factors . | Univariate . | Multivariate . | |
|---|---|---|---|
| P Value . | P Value . | Relative Risk (95% CI) . | |
| Age, 60 yr or older | .002 | .003 | 2.0 (1.3-3.3) |
| Cytogenetics | .004 | .001 | 4.14-150 |
| WBC count >100 × 109/L | .004 | .04 | 1.8 (1.0-3.3) |
| FLT3 gene mutation | .006 | .2 | |
| FAB other than M2 | .1 | .5 | |
| N-RAS gene mutation | .2 | .1 | |
Comparison between the good-risk and poor-risk groups.
Unfavorable Prognostic Factors for DFS in the 106 Patients Who Achieved CR and Were Under 60 Years Old
| Prognostic Factors . | Univariate . | Multivariate . | |
|---|---|---|---|
| P Value . | P Value . | Relative Risk (95% CI) . | |
| WBC count >100 × 109/L | .003 | .01 | 2.6 (1.2-5.5) |
| FLT3 gene mutation | .02 | .1 | |
| Cytogenetics | .1 | .1 | |
| N-RAS gene mutation | .2 | .5 | |
| FAB other than M2 | .6 | .7 | |
| Prognostic Factors . | Univariate . | Multivariate . | |
|---|---|---|---|
| P Value . | P Value . | Relative Risk (95% CI) . | |
| WBC count >100 × 109/L | .003 | .01 | 2.6 (1.2-5.5) |
| FLT3 gene mutation | .02 | .1 | |
| Cytogenetics | .1 | .1 | |
| N-RAS gene mutation | .2 | .5 | |
| FAB other than M2 | .6 | .7 | |
DISCUSSION
In this study, we showed that the FLT3 gene mutation is significantly associated with clinical feature and prognosis of AML. In our previous analysis on M3,18 the FLT3 gene mutation closely correlated to leukocytosis but not significantly to prognosis. The reasons for the discrepancy between M3 and the others are the sample size and that the prognosis for M3 was favorable compared with other types of AML, especially after the clinical introduction of differentiation therapy with all-trans retinoic acid.24 The present study showed that the FLT3 gene mutation was significantly associated with leukocytosis in M1 and M2 but not in M4/5. However, the FLT3 gene mutation was associated with an unfavorable prognosis regardless of FAB type. MutantFLT3 might not be simply associated with cell proliferation but also with inhibition of apoptosis.25 The relevance of mutant FLT3 may be dependent on intracellular conditions, which are determined by cell lineage and gene alterations. Importantly, the difference of prognosis between mutant and wild FLT3 was more remarkable in the good- and standard-risk groups than in the poor-risk and karyotype-unknown groups. Furthermore, the FLT3 gene mutation was the strongest prognostic factor for subjects under 60 years old. Thus the FLT3 gene mutation is a useful molecular marker to identify high-risk patients who could not be characterized by conventional criteria.
The prognostic significance of the N-RAS gene mutation is a matter of controversy. Generally RAS gene mutation is associated with tumor progression and was reported to be associated with poor prognosis in solid tumors and acute lymphoblastic leukemia (ALL).26,27 In AML, however, there was no difference in survival between RAS mutation-positive and -negative patients.7,8 In this study, the presence of N-RASgene mutation was related to low CR rate and was marginally associated with unfavorable prognosis. One reason for the discrepancy between our data and previous reports is that we could exclude the influence ofFLT3 gene mutation in this study. When the cases with mutant N-RAS were compared with those with wild N-RAS, the prognostic difference was limited (OS, P = .06; DFS,P = .2). Because no ALL cases have mutant FLT3, their prognosis might be directly influenced by the presence of mutant N-RAS.27
It is particularly interesting to investigate the multiplicity of gene alterations associated with leukemia. Our results suggest that the mutations of FLT3 and N-RAS genes occurred independently (P = .07 by the Fisher’s exact test), although we could not entirely rule out the possibility that a weak adverse interaction exists between mutant FLT3 and mutant N-RAS. Because both gene alterations are associated with aberrant signal-transduction, these mutations may be additively or synergistically associated with leukemia progression. Our serial studies indicated that no leukemia cases carry both t(9;22)/BCR-ABL and mutant FLT3.11 Because p115CBL, which is activated by BCR-ABL,28 is one of the downstream proteins for FLT3,29FLT3 gene mutation may bring no growth advantage to leukemia clone withBCR-ABL. Aberrant signal-transduction through mutantFLT3 should be clarified to further characterize the functional significance of mutant FLT3.
Gain-of-function mutation of the FLT3 gene suggests that new strategies would be applicable for the treatment of AML. Inhibitors of FLT3-pathway may selectively inhibit leukemia cell proliferation. It has been reported that inhibition of Jak-2 activity by a specific tyrosine kinase blocker blocks leukemic cell growth in vitro and induces programmed cell death in vivo.30 In the future, the best choice of therapy may be established depending on an individual set of molecular alterations in each patient with leukemia.
ACKNOWLEDGMENT
We are grateful to Drs Akihisa Kanamaru, Junko Ohyashiki, Ritsuro Suzuki, and Masatomo Takahashi for sending patients’ samples.
Supported by a Grant-in-Aid from the Japanese Ministry of Health and Welfare.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Tomoki Naoe, MD, Department of Infectious Diseases, Nagoya University School of Medicine, Nagoya 466-8560, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal