Abstract
Recent studies have identified several populations of progenitor cells in the human thymus. The hematopoietic precursor activity of these populations has been determined. The most primitive human thymocytes express high levels of CD34 and lack CD1a. These cells acquire CD1a and differentiate into CD4+CD8+ through CD3−CD4+CD8− and CD3−CD4+CD8+β− intermediate populations. The status of gene rearrangements in the various TCR loci, in particular of TCRδ and TCRγ, has not been analyzed in detail. In the present study we have determined the status of TCR gene rearrangements of early human postnatal thymocyte subpopulations by Southern blot analysis. Our results indicate that TCRδ rearrangements initiate in CD34+CD1a− cells preceding those in the TCRγ and TCRβ loci that commence in CD34+CD1a+ cells. Furthermore, we have examined at which cellular stage TCRβ selection occurs in humans. We analyzed expression of cytoplasmic TCRβ and cell-surface CD3 on thymocytes that lack a mature TCRβ. In addition, we overexpressed a constitutive-active mutant of p56lckF505 by retrovirus-mediated gene transfer in sequential stages of T-cell development and analyzed the effect in a fetal thymic organ culture system. Evidence is presented that TCRβ selection in humans is initiated at the transition of the CD3−CD4+CD8− into the CD4+CD8+β− stage.
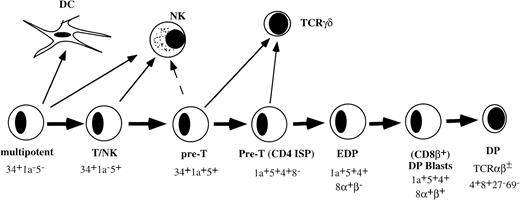
THE EARLY STAGES of T-cell development in the human thymus with respect to cell-surface phenotye and developmental activities have been well documented. The most primitive hematopoietic progenitor in the thymus expresses high levels of CD34 and CD45RA, low levels of CD38, and lacks CD2 or CD5 (Fig 1).1,2 These cells have the ability to develop into multiple hematopoietic lineages including T cells, dendritic cells (DC), natural killer (NK) cells, and monocytes.1-3 Upon further differentiation, these cells acquire CD2 and CD5. The CD34+CD2+CD5+ thymic population contains bipotential T/NK precursors.4 The next stage is characterized by acquisition of CD1a and is paralleled by greatly diminished NK precursor activity.4 The CD34+CD1a+ cells upregulate first CD4 followed by CD8α before acquisition of CD8β.5-8 Small CD4+CD8+ double-positive (DP) thymocytes that express low levels of CD3/TCR complex are subjected to positive selection. Positively selected cells acquire first the activation antigen CD69 followed by CD27.9
Model of early stages of human T-cell development in the thymus before positive selection.
Model of early stages of human T-cell development in the thymus before positive selection.
Despite the detailed information of the functional and phenotypic characteristics, information about the status of TCR gene rearrangements in these T-cell progenitor subsets is relatively scarce. Studies in T-cell acute lymphoblastic leukemias (T-ALL) suggest that TCRδ, TCRγ, and TCRβ rearrangements precede rearrangements of the TCRα gene.10-12 However, the exact sequence of TCRδ and TCRγ rearrangements in normal human thymocytes has not yet been determined. With respect to TCRβ rearrangements in purified thymic subsets, contradictory results have been published. Recently it was reported that the TCRβ locus is in germline configuration in early CD34+CD1a− thymocytes.13,14However, another group detected TCRβ D-J and V-DJ in part of the CD34+CD1a− subset using polymerase chain reaction (PCR) analysis.15 TCRβ gene rearrangements were detectable in the CD4 immature single-positive (ISP) population, while rearrangements at the TCRα locus were present only in CD4+CD8+ DP cells and subsequent stages of development.13
Productive rearrangements at the TCRβ locus are assumed to be effectively checked by signaling through a receptor formed by the protein product of a rearranged TCRβ gene, the pre-Tα (pTα), and the CD3 proteins. This receptor complex transmits a signal that induces survival and expansion of cells expressing the complete pre-TCR complex, ensuring selection of those cells that have productively rearranged their TCRβ genes.16,17 Cells with nonproductive TCRβ rearrangements do not express a pre-TCR complex and die. This process is referred to as TCRβ selection. At which stage in human T-cell development this complex is functional is not clear. Recent studies have shown that in humans the highest expression of the pTα mRNA is detectable at the CD4 ISP stage,18while at this stage the TCRβ protein is only expressed in 5% to 10% of the cells.13 Much higher levels of cytoplasmic TCRβ chains were found in large DP thymocytes. A complex of TCRβ and possibly pTα could be immunoprecipitated from these DP cells.13 However, the study of Ramiro et al13left unresolved the developmental status of a previously described intermediate DP population, which expresses CD8α but lacks CD8β.5 These early DP (EDP) cells could in fact constitute an important intermediate, because CD4−CD8− double-negative (DN) thymocytes cultured in vitro with interleukin-7 (IL-7) develop into CD4+CD8α+β− cells and then die.5 These data raise the possibility that development from CD4+CD8α+β− to CD4+CD8α+β+ thymocytes serves as the pre-TCR mediated control point in human T-cell development.
In the present report we have analyzed the status of TCR gene rearrangements in purified human thymic subsets using Southern blot analysis to obtain detailed information about the stage of initiation of TCR rearrangements. Southern blot analysis, compared with PCR analysis, may provide a more unambiguous assessment of the status of TCR gene rearrangements. Moreover, to gain more insight in the expression and functioning of the pre-TCR complex, we analyzed expression of CD3 and TCRβ proteins in different thymic subpopulations. Furthermore, we have used retrovirus-mediated gene transfer to express a constitutive active mutant of the tyrosine kinase p56lck in human thymic precursors and analyzed its effect on development of TCRαβ+ and TCRγδ T cells in a fetal thymic organ culture (FTOC).
MATERIALS AND METHODS
Isolation and cell sorting of cell samples from thymus.
Normal human thymocytes were obtained from thymus fragments removed during cardiac surgery of patients aged 1 month to 2 years. Thymic lobes were gently minced as described previously14 in RPMI-1640 (GIBCO-BRL Life Technologies Ltd, Paisley, UK) containing 2% (vol/vol) fetal calf serum (FCS; BioWhittaker, Verviers, Belgium) and antibiotics (penicillin 500 IU/mL, streptomycin 100 mg/mL; Boehringer Mannheim Biochemicals, Mannheim, Germany). Cells were isolated by Ficoll density gradient centrifugation (Lymphoprep, 1.077 g/mL; Nycomed Pharma, Oslo, Norway). Thymocytes recovered from the Ficoll interface were enriched for CD34+ cells using a CD34 separation kit (varioMACS; Miltenyi Biotec Inc, Sunnyvale, CA), according to the manufacturer’s instructions. Cells were sorted after staining with monoclonal antibodies (MoAbs) against molecular epitopes different to those recognized by the MoAbs used during preceding purification procedures. For cell sorting of CD34+CD1a− and CD34+CD1a+ subpopulations, cells were incubated with 1 μL/106 cells of CD34-fluorescein isothiocyanate (FITC) (HPCA-2; Becton Dickinson, San Jose, CA) and 0.5 μL/106 cells of CD1a-PE (T6-RD1; Coulter Clone, Miami, FL) at 4°C for 30 minutes. Cells were isolated by flow cytometric activated cell sorting on a FACStar Plus (Becton Dickinson, San José, CA), equipped with an argon laser emitting at 488 nm. During sorting procedure, cells were kept at 4°C until use. All sorted fractions were reanalyzed after sorting, and only those that contained more than 99% of the cells in the selected sort gates were used for further experiments.
To enrich for CD4 ISP cells, total thymocytes were depleted twice after incubating the cells with CD8 MoAb (RPA-TA, kindly provided by Dr G. Aversa, DNAX, Palo Alto, CA), CD19 and CD27 MoAbs (CLB-CD19 and 9F4, respectively; gift from Dr R. van Lier, Central Laboratory of The Blood Transfusion Service [CLB], Amsterdam, The Netherlands), CD69 MoAb (L78; provided by Dr J.H. Phillips, DNAX, Palo Alto, CA), CD3 MoAb (SVB-T3b) and glycophorin A MoAb (10F7 MN, obtained from the American Type Culture Collection [ATCC], Rockville, MD) followed by removal of the labeled cells with magnetic beads (Dynabeads, Dynal, Norway). For cell sorting, the depleted thymocytes were stained with CD4-PE (Leu3a; Becton Dickinson), CD3-FITC (Leu4a), and CD8α-tricolor (TRC; 3B5; Caltag Laboratories, South San Francisco, CA) and the sorted CD3−CD4+CD8− (CD4 ISP) cells were used for further experiments.
To enrich for CD8α+β− cells, total thymocytes were depleted three times after incubating the cells with CD8β MoAb (2ST8-5H7; kindly provided by Dr E.L. Reinherz, Dana-Farber Cancer Institute, Boston, MA), CD27 MoAb (9F4), CD19 MoAb (CLB-CD19), CD69 MoAb (L78), CD3 MoAb (SPV-T3b), and glycophorin A MoAb (10F7 MN), followed by removal of the labeled cells with magnetic beads (Dynabeads, Dynal, Norway). The depleted cells were then reincubated with anti-CD8β followed by incubation with phycoerythrin (PE)-labeled goat anti-mouse antibody. The remaining PE+ cells were then removed by cell sorting using a FACStar plus. This depleted subpopulation contained 30% CD8α+β−cells. The remaining 70% were CD4 ISP and CD3−CD4−CD8−cells. To obtain CD3−CD4+CD8α+β−thymocytes for reverse transcriptase (RT)-PCR experiments, the population depleted with magnetic beads was further purified by cell sorting after staining with CD4-PE (Leu3a), CD3-FITC (Leu4a), and CD8α-tricolor (TRC; 3B5).
It is well established that positive selection of thymocytes is accompanied by upregulation of CD69 and CD27.9,19 20CD27−CD69− cells can therefore be considered to be preselection thymocytes. This population was obtained by depletion of CD27+ and CD69+ cells using the CLB-3A12 and Leu-23 MoAbs and magnetic beads (Dynal). Both depleted and bead-coated thymic cells, which contain the more mature positive selected CD27+CD69+ thymocytes, were obtained by magnetic separation. The CD27−CD69− thymocyte subpopulation was greater than 98% pure (ie, hardly contained CD27+CD69+ thymocytes) and the majority of these cells were small CD4+CD8+ thymocytes. The cells that remained attached to the beads (the CD27+CD69+ subpopulation) were enriched for CD27+CD69+ cells but were contaminated with almost all immature and mature thymocyte subpopulations.
Cytoplasmic staining.
Cytoplasmic TCRβ-PE (anti-Cβ1; Ancell Corp, Bayport, MN) staining of sorted cells was done after fixation (>2 hours) with 2% paraformaldehyde at 4°C and after permeabilization in 0.1% Triton X-100 (Sigma, St Louis, MO) for 40 minutes on ice. For three-parameter analysis, cells were stained before fixation with CD8α-TRC (3B5; Caltag Laboratories), anti-TCRαβ-FITC (Immunotech, Marseille, France), anti-TCRγδ-FITC (Immunotech), and FITC-conjugated F(ab′)2 goat anti-mouse IgG (H+L) (Zymed, San Francisco, CA).
Southern blot analysis and quantification.
DNA was isolated from total thymocytes and the human adenocarcinoma cell line HeLa, which was used as a germline control. Subsequently, DNA was digested with the restriction enzyme EcoRI, size fractionated in agarose gels (0.7%), and transferred to Nytran-13N nylon membranes (Schleicher and Schuell, Dassel, Germany) as previously described.21 TCRδ, TCRγ, and TCRβ rearrangements were studied by hybridization of the filters using the previously described TCRDJ1 (Jδ1) and Jγ1.3 DNA probes12,21 and the TCRBJ1 (Jβ1) and TCRBJ2 (Jβ2) DNA probes (Langerak T, Verschuren MCM, Van Dongen JJM, unpublished observations, 1997), respectively, which were 32P random oligonucleotide labeled. To control for the amount of DNA loaded per lane, hybridization with the IGKDE (kappa deleting element, Kde)22 probe was performed. Hybridizations were analyzed by exposure to Fuji NIF-RX films (Fuji Photo Film Co, Tokyo, Japan) and semi-quantification of rearrangements was done with a phosphor-imager (Phospho-Imager; Molecular Dynamics, Sunnyvale, CA) using ImageQuaNT analysis software. The retained hybridization signal for the germline band (in percentage) was estimated as follows23:
Estimated Percentage of Signal Retained = Signal for Probe of Interest in Thymic Subset /Signal for Germline Control in Thymic Subset × Signal for Germline Control in HeLa/Signal for Probe of Interest in HeLa × 100.
The “estimated percentage of signal retained” was further corrected for the total amount of hybridization signal in one lane (because of different exposure times). For each lane this percentage was set at 100%.
DNA-PCR analysis.
TCR gene rearrangements were also analyzed in a genomic DNA-PCR assay with DNA of 1 to 2 × 104 cells. DNA was isolated as described.24 TCRβ PCR was performed in a total volume of 100 μL consisting of 1 μmol/L of each primer set, 100 μmol/L each dNTP, 1.5 mmol/L MgCl2, 1X PCR buffer, 1 U Taq DNA polymerase, 1 mg/mL bovine serum albumin (BSA), and 10 μL of the DNA and covered with 50 μL paraffin oil. To determine TCRβ Dβ1.1-Jβ1.3-1.4 rearrangement, samples were heated to 94°C for 5 minutes followed by amplification for 30 cycles of 1 minute at 94°C, 1 minute at 56°C, and 1 minute at 72°C. To determine TCRβ Vβ8-DJβpan or Vβ9-DJβ1.2 rearrangements, samples were heated to 94°C for 5 minutes followed by amplification for 5 cycles of 1 minute at 94°C, 1 minute at 30°C, and 2 minutes at 72°C, and 35 cycles of 1 minute at 94°C, 1 minute 45°C, and 2 minutes at 72°C. After the last cycle a final extension step at 72°C for 10 minutes was done. Gel electrophoresis (2.5% agarose) of 15 μL of the PCR products was followed by blotting to nylon filters, which were hybridized with a Dβ- or a Vβ-specific internal oligonucleotide probe: Dβ1.1: 5′-TGGTGGTCTCTCCCAGGCTCT-3′; Jβ1.3-1.4: 5′-CCAGCTGTCCAGCCTTGACTT-3′; Vβ8: 5′-ATTTACTTTAACAACAACGTTCCG-3′; Vβ9: 5′-ATTATAAATGAAACAGTTCCAAATCGC-3′; Jβpan: 5′-AGCAC(T/G/C)GTGAGCC(T/G)GGTGCC-3′; Jβ1.2: 5′-TACAACGGTTAACCTGGT-3′; Vβ8 probe: 5′-TGAGGAAAGCAGTCACCCTGAAC-3′; Vβ9 probe: 5′-CCTAAATCTCCAGACAAAGCTCAC-3′.
To control for the amount of DNA in the PCR, genomic amplification of the RAG-2 gene was performed with the primer pair: RAG2 sense: 5′-TGTGAATTGCACAGTCTTGCCAGG-3′; RAG2 antisense: 5′-GGGTTTGTTGAGCTCAGTTGAATAG-3′.
Of each control PCR reaction, 10 μL was separated in a 2.5% agarose gel and transferred onto a nylon filter, which was hybridized with the32P-endlabeled RAG-2 probe: 5′-CAAGATATGGTTTGGAAGCAACATGGGAAA-3′.
Retroviral constructs and transduction.
An LZRS retroviral vector was constructed comprising the cDNAs encoding the constitutive activated form of mouse p56lck(p56lckF505) (obtained from Dr A. Venkitaraman, MRC, Cambridge, UK) and the enhanced green fluorescent protein (GFP) (obtained from Clontech, Palo Alto, CA) as a marker gene.25The cDNAs were separated by the sequence of the internal ribosomal entry site (IRES) to allow individual translation of the bicistronic mRNA containing both p56lckF505 and GFP (p56lckF505-IRES-GFP) without generating a fusion protein.26,27 In addition, as a control we made a retroviral vector containing the IRES-GFP sequence only (IRES-GFP). Helper-virus–free recombinant retroviruses (titer 106/mL, as determined by transduction of 3T3 cells) were produced after transfection of the retroviral constructs into the 293T-based ΦNX-A amphotropic packaging cell line (kindly provided by Dr G. Nolan, Stanford University, Palo Alto, CA)28 and selection on the selectable marker puromycin. Progenitor cells were purified from thymus and cultured overnight in the presence of 10 ng/mL IL-7 (R & D Systems, Abingdon, UK) and 10 ng/mL SCF (R & D Systems) followed by incubation for 7 to 8 hours or overnight with virus supernatant in plates coated with 30 μg/mL recombinant human fibronectin fragment CH-296 (RetroNectin; Takara, Otsu, Japan).29-31
Hybrid human/mouse fetal thymic organ cultures.
The in vitro development of human T cells from CD34+ or CD4 ISP progenitor cells was studied using the hybrid human/mouse FTOC described before.2 Fetal thymuses were obtained from embryos of RAG-1–deficient mice on day 15-16 of gestation. The lobes were treated with deoxyguanosine for 5 days before incubation with human progenitor cells in a hanging drop culture for 2 days followed by incubation on an air/liquid interface. Culture medium consisted of Yssel’s medium32 supplemented with 2% normal human serum and 5% FCS. To analyze differentiation of human cells, the mouse thymi were dispersed into single-cell suspensions and stained with MoAbs specific for human cell-surface antigens.
RESULTS
Rearrangements of TCRδ and TCRγ genes in human thymocyte subpopulations.
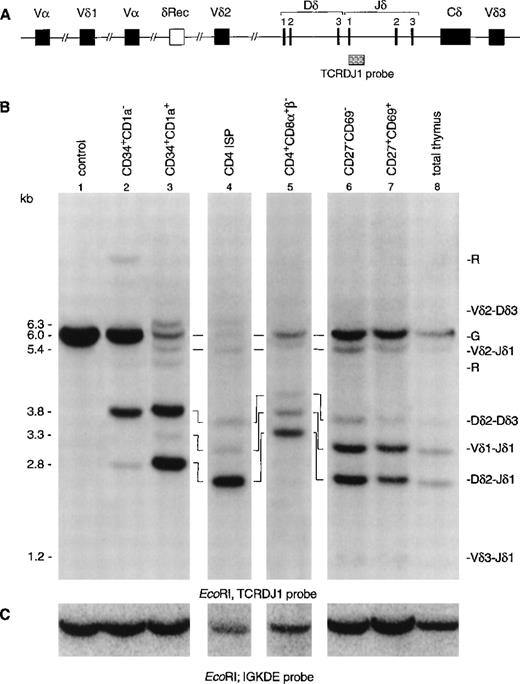
To gain more insight in the order of TCR rearrangements, we analyzed six well-defined thymic subpopulations for both TCRδ and TCRγ rearrangements using Southern blot analysis with TCRδ- and TCRγ-specific probes (Fig 2). CD34+ cells were enriched by positive selection with magnetic beads followed by flow cytometric cell sorting of CD34+CD1a− and CD34+CD1a+ cells (>99% pure). CD4 ISP cells were sorted from CD8α-depleted thymocytes (>99% pure). Because the subsequent CD4+CD8α+β−EDP population is small and not enough cells could be obtained for Southern blot analysis, these cells were enriched by depletion with CD3 and CD8β MoAbs using magnetic beads and sorting. This enriched population contained 30% EDP cells, the remainder being a mixture of CD4 ISP and CD3−CD4−CD8−cells. The human cell line HeLa served as negative control, while total thymus was used as positive control. Hybridization of DNA isolated from the CD34+CD1a− cells with the TCRDJ1 probe showed a strong TCRδ germline band after hybridization (Fig 2B, lane 2). We estimate that more than 60% of the germline band was retained (see Materials and Methods), which suggested that the majority of the cells still have their TCRδ locus in germline configuration. More interestingly, we were able to detect smaller-sized rearranged bands in this immature thymocyte population. These rearranged bands were previously assigned as incomplete Dδ2-Dδ3 and Dδ2-Jδ1 rearrangements (Fig 2B), based on the size of the bands.12In the more mature CD34+CD1a+ cells, most of the TCRδ germline band had disappeared (Fig 2B, lane 3): the estimated percentage of the retained germline band was 15%. While the most prominent rearranged bands consisted of incomplete Dδ2-Dδ3 and Dδ2-Jδ1 rearrangements, complete Vδ1-Jδ1 and Vδ2-Jδ1 rearrangements were also detectable in this CD34+CD1a+ cell population. Analysis of the CD4 ISP subset showed that more than 90% of these cells had rearranged TCRδ genes (Fig 2B, lane 4). The percentage of germline signal retained in the CD3 and CD8β-depleted cells was 25% (Fig 2B, lane 5). The increase of the germline percentage compared with the CD4 ISP cell stage is due to the fact that the CD3- and CD8β-depleted cells consist of several immature cell populations (see Materials and Methods). Low levels of complete Vδ3-Jδ1 rearrangements were only observed from the more mature CD27−CD69− subset onward, and the CD27+CD69+ subset gave comparable results to the total thymocyte cell sample (Fig 2B, lanes 6, 7, and 8).
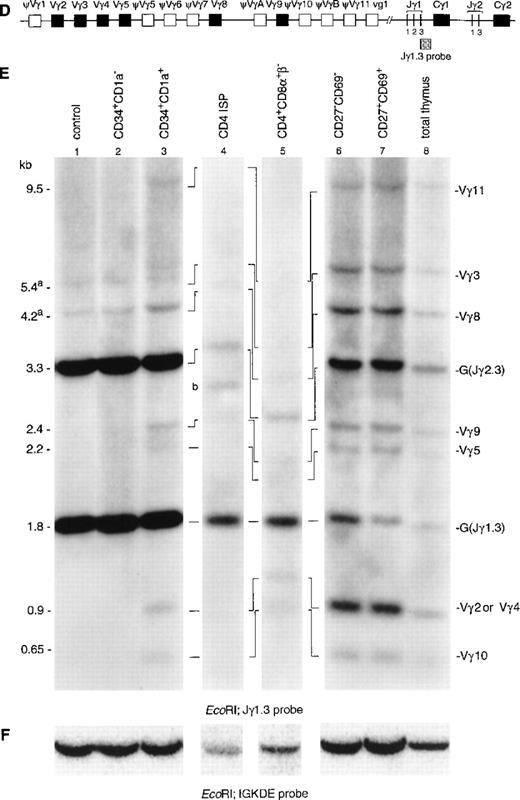
Southern blot analysis of TCRδ and TCRγ gene rearrangements. The schematic diagrams of the germline configuration of the human TCRδ (A) and TCRγ (D) genes show the position of the TCRDJ1 and Jγ1.3 probes, respectively. Exons and pseudo genes are depicted as solid and open boxes, respectively. (B and E) Southern blot analysis of filters containing EcoRI-digested DNA of HeLa cells (lane 1), six human thymocyte subpopulations (lanes 2 to 7), and total human thymocytes (lane 8) hybridized with the TCRDJ1 and Jγ1.3 probes, respectively. Rearranged bands were assigned based on the size of the bands.21 33 Rehybridization of the filters with the IGKDE probe was performed as a control for loading of the lanes (C and F). G and R indicate germline and as yet unidentified rearrangements, respectively. (a) Underdigestion of the DNA leads to two additional bands in the control DNA. (b) Band caused by plasmid contamination.
Southern blot analysis of TCRδ and TCRγ gene rearrangements. The schematic diagrams of the germline configuration of the human TCRδ (A) and TCRγ (D) genes show the position of the TCRDJ1 and Jγ1.3 probes, respectively. Exons and pseudo genes are depicted as solid and open boxes, respectively. (B and E) Southern blot analysis of filters containing EcoRI-digested DNA of HeLa cells (lane 1), six human thymocyte subpopulations (lanes 2 to 7), and total human thymocytes (lane 8) hybridized with the TCRDJ1 and Jγ1.3 probes, respectively. Rearranged bands were assigned based on the size of the bands.21 33 Rehybridization of the filters with the IGKDE probe was performed as a control for loading of the lanes (C and F). G and R indicate germline and as yet unidentified rearrangements, respectively. (a) Underdigestion of the DNA leads to two additional bands in the control DNA. (b) Band caused by plasmid contamination.
Hybridization of DNA from the CD34+CD1a−subset with a TCRγ-specific probe (Jγ1.3) showed the two expected germline bands for Jγ1.3 (1.8 kb) and Jγ2.3 (2.3 kb) (Fig 2E, lane 2).21 In addition, two faint additional bands were detected in this population (Fig 2E, lane 1). Although the size of these two bands seem to be identical to those of Vγ3 and Vγ8 rearrangements, we believe that these bands are caused by underdigestion of the DNA: firstly, because bands of the same intensity are also present in the Hela control, and secondly, because there are no rearrangements involving the other Vγ gene segments in the CD34+CD1a− population. There is no information indicating that Vγ3 and Vγ8 would rearrange earlier than the other Vγ gene segments. However, to further confirm that the CD34+CD1a− population lacks TCRγ gene rearrangements, we repeated the Southern blot experiment usingHindIII-digested DNA of the immature thymocyte populations. This analysis did not reveal TCRγ gene rearrangements in the CD34+CD1a− population (results not shown). Therefore, we believe that our data are consistent with the notion that the great majority of the TCRγ alleles in the CD34+CD1a− subpopulation are in the germline configuration. TCRγ rearrangements were detectable from the CD34+CD1a+ subset onwards (Fig 2E, lane 3). Based on the size of the rearranged bands, they could be assigned as rearrangements of Vγ11, Vγ3, Vγ8, Vγ9, Vγ2, or Vγ4, and Vγ10 to Jγ1.3 or Jγ2.3.21,33 It was estimated that more than 90% of the germline band was retained in the CD34+CD1a+ subset. Analysis of DNA isolated from the CD4 ISP and CD4+CD8α+β− subsets showed an almost complete absence of the Jγ2.3 germline band (Fig 2E, lanes 4 and 5). These two thymocyte subsets were isolated from a patient with a polymorphic EcoRI restriction site at the 5′ side of the Jγ2.3 gene segment, which results in comigration of the Jγ1.3 and Jγ2.3 germline bands.21 Quantification of this combined germline band revealed that 40% of the Jγ1.3/ Jγ2.3 germline signal is retained in the CD4 ISP subpopulation. The differences in the densities of the Jγ1.3 and Jγ2.3 germline bands in the more mature thymocyte subsets clearly show that rearrangements to the Jγ1.3 gene segment occur more frequently than to the Jγ2.3 gene segment (Fig 2E, lane 6, 7, and 8).
Our data indicate that CD34+CD1a− clearly contain TCRδ rearrangements in 40% of the alleles but that TCRγ rearrangements are present in less than 5% of the alleles (the detection limit of the Southern blot analysis). We conclude that TCRδ gene rearrangements precede those at the TCRγ locus.
The TCRβ genes are in the germline configuration in CD34+CD1a− cells and complete Vβ-DJβ rearrangements do not become prominent before the CD4+CD8α+β− population.
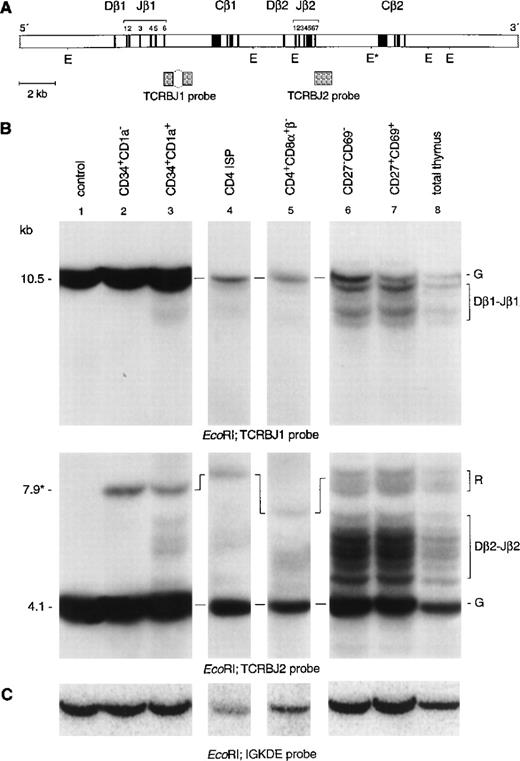
To clarify ambiguities concerning initiation of TCRβ gene rearrangements in immature thymocytes, we analyzed the TCRβ genes in the same subpopulations as described above by use of Southern blot analysis with specific TCRβ probes (Fig3). Hybridization of DNA from CD34+CD1a−cells with the TCRBJ1 probe showed a strong germline band, also after longer exposure times (Fig 3B, lane 2). Rehybridization with the TCRBJ2 probe showed a strong germline band and a weaker 7.9-kb band, owing to underdigestion of the DNA at a partial resistant EcoRI restriction site.21 Therefore, we can conclude that CD34+CD1a− cells have not started to rearrange their TCRβ locus. Hybridization of DNA isolated from the more mature CD34+CD1a+ thymocytes with the TCRDBJ1 and TCRBJ2 probes showed faint rearranged bands (Fig 3B lane 3, upper and middle panel). Rehybridization of DNA from CD34+CD1a+ thymocytes with Dβ1- and Dβ2-specific probes identified these bands as incomplete Dβ1-Jβ2 and Dβ2-Jβ2 rearrangements (data not shown). PCR analysis confirmed the presence of TCRβ D-J rearrangement in the CD34+CD1a+ subset (Fig 4, top panel). The estimated percentage of germline band retained in CD34+CD1a+ thymocytes was more than 90% in this subpopulation (see Materials and Methods for calculations). We also calculated that the CD4 ISP subpopulation still retained 85% of the germline signal, whereas only 65% of germline signal was retained in cells completely depleted of CD8β+ cells. Only 30% of the CD8β-depleted cells are CD3−CD4+CD8α+β−cells, the remainder being a mixture of CD4 ISP and CD3−CD4−CD8−cells. Because the latter two populations have their TCRβ genes mostly in the germline configuration, these two subsets should account for most, if not all, of the 65% germline signal detected in the CD8β-depleted cells, which implies that EDP cells have extensive TCRβ gene rearrangements. Thus, our data indicate that although incomplete rearrangements at the TCRβ locus are already initiated in the CD34+CD1a+ stage, the majority of TCRβ rearrangements occur at the transition of CD4 ISP to EDP cells. Hybridization of DNA isolated from the more mature cell populations with either a Jβ1 (TCRJB1) or a Jβ2 (TCRJB2) specific probe showed many different nongermline bands, representing extensive TCRβ gene rearrangements (Fig 3B, lanes 6, 7, and 8, top and middle panels, respectively). Furthermore, these hybridizations show that the density of the germline and rearranged bands of the Jβ1 locus is essentially lower as compared with the Jβ2 locus, indicating deletion of the Jβ1 locus and rearrangements to the Jβ2 locus in most cells of these subsets.
Southern blot analysis of TCRβ gene rearrangements. (A) Schematic diagram of the germline configuration of the human TCRβ locus showing the position of the TCRBJ1 and TCRBJ2 probes. (B) Southern blot analysis of filters containing EcoRI–digested DNA of HeLa cells (lane 1), six human thymocyte subpopulations (lanes 2 to 7), and total human thymocytes (lane 8) hybridized with the TCRBJ1 (top panel) and TCRBJ2 (bottom panel) probes. Rearranged bands were assigned based on the size of the bands.21 (C) Rehybridization of the filters with the IGKDE probe was performed as a control for loading of the lanes. G and R indicate germline and as yet unidentified rearrangements, respectively. *Partial resistantEcoRI restriction site in the TCRCB2 region. Underdigestion of this restriction site results in a 7.9-kb germline band.
Southern blot analysis of TCRβ gene rearrangements. (A) Schematic diagram of the germline configuration of the human TCRβ locus showing the position of the TCRBJ1 and TCRBJ2 probes. (B) Southern blot analysis of filters containing EcoRI–digested DNA of HeLa cells (lane 1), six human thymocyte subpopulations (lanes 2 to 7), and total human thymocytes (lane 8) hybridized with the TCRBJ1 (top panel) and TCRBJ2 (bottom panel) probes. Rearranged bands were assigned based on the size of the bands.21 (C) Rehybridization of the filters with the IGKDE probe was performed as a control for loading of the lanes. G and R indicate germline and as yet unidentified rearrangements, respectively. *Partial resistantEcoRI restriction site in the TCRCB2 region. Underdigestion of this restriction site results in a 7.9-kb germline band.
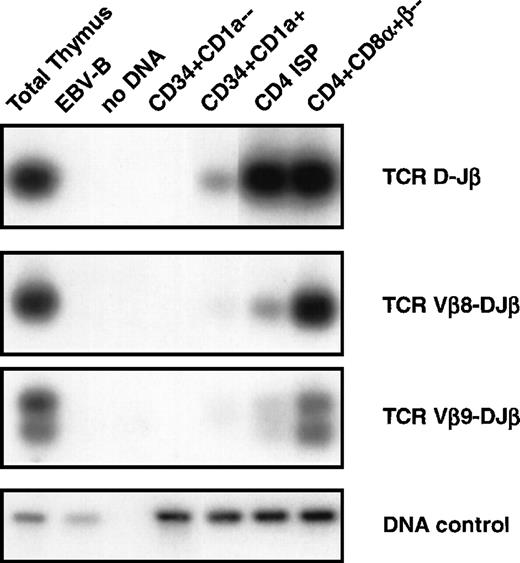
DNA-PCR analysis of TCRβ gene rearrangements. DNA was isolated from sorted CD34+CD1a−, CD34+CD1a+, CD4 ISP, and CD4+CD8+β− human thymic subsets were sorted as indicated in Materials and Methods and subjected to PCR. Primer combinations were used specifically recognizing TCRβ D-J (first panel), Vβ8-DJ (second panel), and Vβ9-DJ (third panel) as described in Materials and Methods. Amplification of the RAG2 gene was performed to control for the amount of DNA used in the PCR (DNA control, fourth panel). DNA isolated from total thymocytes was used as a positive control and from an EBV-B cell line as a negative control. PCR products were blotted onto nylon filters, and hybridized with endlabeled oligoprobes recognizing sequences different from the primers used for PCR. Filters were washed with 2X SSC, 0.1% sodium dodecyl sulfate and exposed to an autoradiographic film.
DNA-PCR analysis of TCRβ gene rearrangements. DNA was isolated from sorted CD34+CD1a−, CD34+CD1a+, CD4 ISP, and CD4+CD8+β− human thymic subsets were sorted as indicated in Materials and Methods and subjected to PCR. Primer combinations were used specifically recognizing TCRβ D-J (first panel), Vβ8-DJ (second panel), and Vβ9-DJ (third panel) as described in Materials and Methods. Amplification of the RAG2 gene was performed to control for the amount of DNA used in the PCR (DNA control, fourth panel). DNA isolated from total thymocytes was used as a positive control and from an EBV-B cell line as a negative control. PCR products were blotted onto nylon filters, and hybridized with endlabeled oligoprobes recognizing sequences different from the primers used for PCR. Filters were washed with 2X SSC, 0.1% sodium dodecyl sulfate and exposed to an autoradiographic film.
It is impossible to identify rearrangements of Vβ to DJ by using Southern blot analysis of thymocyte subpopulations because of the large amount of Vβ gene segment present. Therefore, we used DNA-PCR analysis to asses whether Vβ-DJβ rearrangements were present in the different subpopulations (Fig 4). We focused our PCR analyses on rearrangements of Vβ8 and Vβ9 to DJβ segments and also analyzed D to Jβ rearrangements. None of these rearrangements could be detected in the CD34+CD1a− (Fig 4, second panel). These results confirm those obtained with Southern blot analysis, and together these data indicate that TCRβ gene rearrangements are absent in CD34+CD1a− thymocytes. A D-Jβ band and faint bands Vβ8-DJβ and Vβ9-DJβ were observed in the CD34+CD1a+ subpopulation. The bands corresponding to TCR β rearrangements are a little more pronounced in the CD4 ISP cells but become clearly visible in the purified CD4+CD8α+β− cells with intensities comparable to those in total thymocytes. These data support the notion that although TCRβ gene rearrangements are already occurring in the CD34+CD1a+ population, the bulk of complete TCRβ gene rearrangements occur at the transition of CD4 ISP to EDP cells.
The pre-TCR complex is expressed in the CD4+CD8α+β− population.
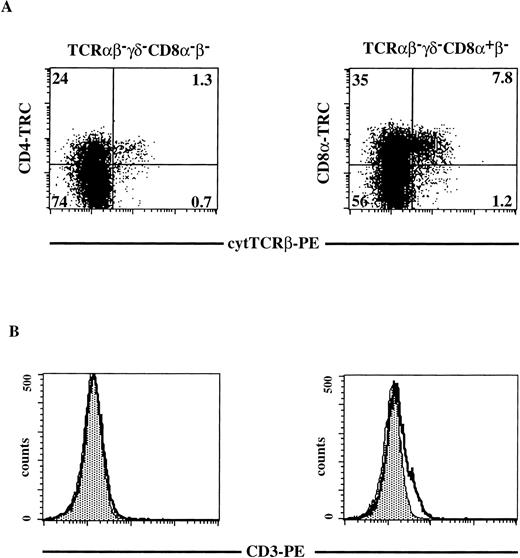
To determine where the pre-TCR complex is expressed during human T-cell development, we analyzed expression of components of this complex. It has been reported that CD4 ISP stage express high levels of pTα mRNA and downregulate in DP cells.18 Using a semiquantitative RT-PCR, we have confirmed that when the cells move from the CD4 ISP to the EDP stage they partly downregulate pTα mRNA (results not shown). Because mature TCRβ rearrangements are detectable in the CD4 ISP, we analyzed the CD4 ISP and the subsequent CD4+CD8α+β− stages for expression of TCRβ and CD3 (Fig 5). Total thymocytes were depleted of all mature and CD8β+thymocytes by magnetic bead depletion and cell sorting as described in Materials and Methods. Three-color flow cytometric analysis was performed on the sorted thymocytes (Fig 5). Cells that were negative for any residual CD8β and TCRαβ- and TCRγδ-bearing cells were stained with CD8α and for cytoplasmic TCRβ (Fig 5A, right panel). In addition, we analyzed cells that were completely negative for CD8α, CD8β, TCRαβ, and TCRγδ, for expression of CD4 and cytoplasmic TCRβ (Fig 5A, left panel). We observed in this representative experiment (out of three) that 25% of the CD4+CD8α+β− cells expressed cytoplasmic TCRβ, whereas only 5% of the CD4 ISP cells stained positive with the anti-TCRβ antibody (Fig 5A). This finding is consistent with the rearrangement data as determined by Southern blot analysis and PCR analysis, demonstrating that the majority of the CD4 ISP cells have the TCRβ in the germline configuration. Weak expression of cell-surface CD3 could be detected on a small fraction of the CD4+CD8α+β− cells, while CD3 was undetectable on CD4 ISP cells (Fig 5B, right and left panels, respectively).
Cytoplasmic TCRβ and membrane CD3 expression on CD8β-depleted, TCR-negative thymocytes. Total thymocytes were depleted and sorted for CD8β−CD27−CD69− cells as described in Materials and Methods. Three-color flow cytometric analysis was done after staining the sorted cells with FITC-conjugated MoAbs against TCRβ, TCRγδ, CD8 and with CD4-TRC (left panels) or after staining the sorted cells with FITC-conjugated MoAbs against TCRβ, TCRγδ and with CD8-TRC (right panels). Dot plots shown are electronically gated on FITC− cells. Histograms shown are electronically gated on FITC−, TRC+ cells (left panels TCRβ−TCRγδ−CD8−β−; right panels TCRβ−TCRγδ−CD8+β−). (A) cytoplasmic anti-TCRβ-PE staining on TCRβ−TCRγδ−CD8−β− thymocytes (left panel) and TCRβ−TCRγδ−CD8+β− thymocytes (right panel). The quadrants were chosen in such a way that greater than 99% of the dots of cells stained with isotype-matched control antibodies fell in the lower left quadrant (negative for both PE and FITC). Numbers in the quadrants represent percentages of cells. (B) Membrane CD3-PE staining (bold line) on TCRβ−TCRγδ−CD8−β−thymocytes (left panel) and TCRβ−TCRγδ−CD8+β−thymocytes (right panel). The shaded area in the histogram represents staining with the negative control (irrelevant IgGs). A representative analysis out of three independent experiments is shown.
Cytoplasmic TCRβ and membrane CD3 expression on CD8β-depleted, TCR-negative thymocytes. Total thymocytes were depleted and sorted for CD8β−CD27−CD69− cells as described in Materials and Methods. Three-color flow cytometric analysis was done after staining the sorted cells with FITC-conjugated MoAbs against TCRβ, TCRγδ, CD8 and with CD4-TRC (left panels) or after staining the sorted cells with FITC-conjugated MoAbs against TCRβ, TCRγδ and with CD8-TRC (right panels). Dot plots shown are electronically gated on FITC− cells. Histograms shown are electronically gated on FITC−, TRC+ cells (left panels TCRβ−TCRγδ−CD8−β−; right panels TCRβ−TCRγδ−CD8+β−). (A) cytoplasmic anti-TCRβ-PE staining on TCRβ−TCRγδ−CD8−β− thymocytes (left panel) and TCRβ−TCRγδ−CD8+β− thymocytes (right panel). The quadrants were chosen in such a way that greater than 99% of the dots of cells stained with isotype-matched control antibodies fell in the lower left quadrant (negative for both PE and FITC). Numbers in the quadrants represent percentages of cells. (B) Membrane CD3-PE staining (bold line) on TCRβ−TCRγδ−CD8−β−thymocytes (left panel) and TCRβ−TCRγδ−CD8+β−thymocytes (right panel). The shaded area in the histogram represents staining with the negative control (irrelevant IgGs). A representative analysis out of three independent experiments is shown.
Constitutive active p56lck expression in CD34+ thymocytes strongly blocks development of TCRαβ and TCRγδ T cells in an FTOC.
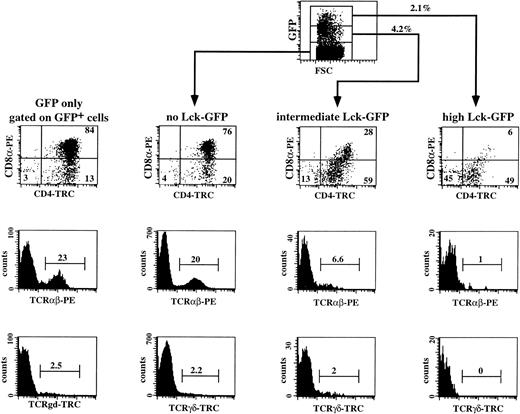
It is now commonly assumed that pre-TCR signaling involves the tyrosine kinase p56lck. It has been shown previously in mice that p56lck is required for thymic αβ T-cell development.34 Disruption of the function of p56lck either by deletion or by introduction of a dominant negative p56lck transgene in the mouse genome leads to accumulation of the cells in the CD3−CD4−CD8−stage in the thymus.34,35 Overexpression of a constitutive active form of p56lck in transgenic mice resulted in a complete inhibition of differentiation of DN into DP thymocytes.36 We analyzed the effects of a constitutive active Lck mutant (p56lckF505) on human T-cell development. This was done to identify more precisely the stage where the pre-TCR complex is functional in humans. We expressed the mutated p56lck in immature CD34+ thymocytes by using retroviral-mediated gene transfer. To allow tracing of the transduced cells, we linked the p56lckF505 cDNA to the GFP marker gene by means of the IRES sequence. We transduced sorted CD34+ thymocytes with the control IRES-GFP and the p56lckF505-IRES-GFP retroviruses and the combination of transduced and untransduced cells were cultured in the FTOC for 3 weeks. A representative experiment (out of four) is shown in Fig 6. The transduction efficiency of the CD34+ cells before the FTOC was 9.3%; after 3 weeks of incubation in the FTOC 9.6% of the cells expressed GFP, showing the stability of expression of GFP during the FTOC. We observed that development of untransduced CD34+ thymocytes and IRES-GFP–transduced thymocytes (GFP only) was identical (Fig 6; compare the GFP+ cells of the control with the GFP− cells of the active-Lck transduced sample). The majority (76% and 80%, respectively) of untransduced populations (that expressed no active Lck) and control IRES-GFP–transduced cells (GFP only) recovered from FTOC were DP, whereas 20% and 23% expressed TCRαβ, and 2.2% and 2.5% expressed TCRγδ (Fig 6). In contrast, expression of constitutive active p56lck in CD34+ thymocytes inhibited their T-cell development in a dose-dependent manner. The transduction efficiency before the FTOC was 6.7% and the percentage of p56lckF505-IRES-GFP+ cells after the FTOC was 6.3%. These data indicate that expression of p56lckF505did not lead to death of the cells. Twenty-eight percent of the intermediate p56lckF505-IRES-GFP+–expressing cells were DP, and 6.6 % were TCRαβ+ (Fig 6). Intermediate levels of expression of the mutant p56lck had no or little effect on the development of γδ+ T cells. Only 6% of the p56lckF505-IRES-GFPhigh cells were DP (Fig 6), of which 1% is TCRαβ+ (Fig 6). Although not shown, the few DP cells that were present in high p56lckF505-GFP+ cells contained both CD8α+β− and CD8α+β+ DP cells. Development of γδ+ T cells was completely blocked by high levels of the constitutive active p56lck (Fig 6).
Three-color flow cytometric analysis of IRES-GFP and p56lckF505-IRES-GFP–transduced CD34+thymocytes cultured in FTOC. CD34+ thymocytes were sorted, transduced with the retrovirus expressing either the control IRES-GFP or the p56lckF505-IRES-GFP construct, and cultured in FTOC for 3 weeks. Cell suspensions were stained with PE- and tricolor (TRC)-conjugated antibodies against different surface antigens. Flow cytometric analysis of the control was performed after gating on the GFP+ cells (GFP only). Analysis of the FTOC incubated with the p56lckF505 transduced cells was done following gating on untransduced cells (no Lck-GFP), p56lckF505-IRES-GFP+ cells (intermediate Lck-GFP), and p56lckF505-IRES-GFP++ cells (high Lck-GFP). Numbers in the dot plots and histograms represent percentages of cells. The input was 3 × 104 cells, and after 3 weeks FTOC we harvested 106 cells from the control IRES-GFP transduced and 5 × 105 cells from the p56lckF505-IRES-GFP–transduced FTOC. This is an expansion of 33-fold and 17-fold, respectively. The recoveries of transduced cells in absolute numbers were 95,000 IRES-GFP+ (9.5% GFP+) and 32,500 p56lckF505-IRES-GFP+ (6.3% GFP+), respectively.
Three-color flow cytometric analysis of IRES-GFP and p56lckF505-IRES-GFP–transduced CD34+thymocytes cultured in FTOC. CD34+ thymocytes were sorted, transduced with the retrovirus expressing either the control IRES-GFP or the p56lckF505-IRES-GFP construct, and cultured in FTOC for 3 weeks. Cell suspensions were stained with PE- and tricolor (TRC)-conjugated antibodies against different surface antigens. Flow cytometric analysis of the control was performed after gating on the GFP+ cells (GFP only). Analysis of the FTOC incubated with the p56lckF505 transduced cells was done following gating on untransduced cells (no Lck-GFP), p56lckF505-IRES-GFP+ cells (intermediate Lck-GFP), and p56lckF505-IRES-GFP++ cells (high Lck-GFP). Numbers in the dot plots and histograms represent percentages of cells. The input was 3 × 104 cells, and after 3 weeks FTOC we harvested 106 cells from the control IRES-GFP transduced and 5 × 105 cells from the p56lckF505-IRES-GFP–transduced FTOC. This is an expansion of 33-fold and 17-fold, respectively. The recoveries of transduced cells in absolute numbers were 95,000 IRES-GFP+ (9.5% GFP+) and 32,500 p56lckF505-IRES-GFP+ (6.3% GFP+), respectively.
The CD4 ISP population contain few if any cells that have completed β selection.
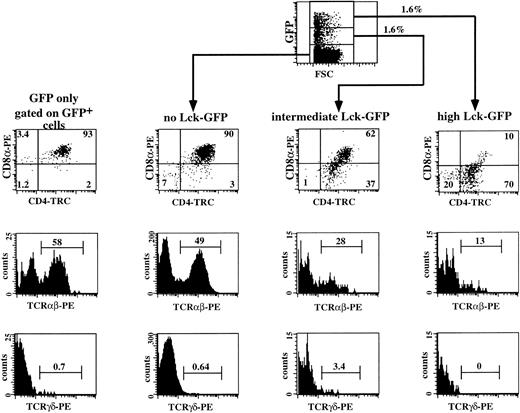
The results of the previous experiments show that the proportion of CD4+CD8α− cells gradually increased with increasing p56lckF505 expression, which suggests that the CD4 ISP cells accumulate as a result of expression of p56lckF505. To confirm that overexpression of p56lckF505 indeed prevents further development of CD4 ISP, we sorted CD4 ISP thymocytes and transduced these cells with either p56lckF505-IRES-GFP or the control IRES-GFP. Both transduced and untransduced cells were cultured in the FTOC for 14 days. Two independent experiments were performed with purified CD4 ISP thymocytes. In the representative experiment shown in Fig 7, the percentage of GFP+cells in the CD4 ISP transduced with lckF505-IRES-GFP cells before the FTOC was 3.6% and after the FTOC 3.2%, indicating that expression of p56lckF505 in CD4 ISP did not induce cell death. Figure 7 shows a flow cytometric analysis of a representative experiment. As expected, the control, IRES-GFP–transduced CD4 ISP thymocytes (Fig 7, GFP only) and the untransduced cells (not shown) developed similarly in the FTOC. The majority of the cells, both in control IRES-GFP–transduced and untransduced samples, developed to DP thymocytes, of which 50% to 60% expressed TCRαβ (Fig 7). Less than 1% of these cells were γδ+ T cells. We observed a dose-dependent inhibition by p56lckF505 of development of CD4 ISP thymocytes. Only 62% of DP cells developed from the intermediate p56lckF505-IRES-GFP+ CD4 ISP thymocytes; 37% of the cells remained at the CD4 ISP stage (Fig 7). Expression of the TCRαβ is reduced approximately twofold (28%; Fig7) compared with the control and untransduced cells. The amount of γδ+ T cells appears to be increased by intermediate levels of p56lckF505 expressed in developing CD4 ISP thymocytes (Fig 7). High levels of p56lckF505 expression led to a block in the development of αβ+ T cells (Fig7) and abrogation of γδ+ T-cell development (Fig 7).
Three-color flow cytometric analysis of IRES-GFP and p56lckF505-IRES-GFP–transduced CD4 ISP thymocytes cultured in FTOC. CD4 ISP thymocytes were sorted and transduced with the retrovirus expressing either the control IRES-GFP or the p56lckF505-IRES-GFP construct (both 2.5 × 104 cells input) and cultured in FTOC for 12 days. Flow cytometric analysis of the control was performed after gating on the GFP+ cells (GFP only). Analysis of the FTOC incubated with the p56lckF505 transduced cells was done following gating on untransduced cells (no Lck-GFP), p56lckF505-IRES-GFP+ cells (intermediate Lck-GFP), and p56lckF505-IRES-GFP++ cells (high Lck-GFP). Numbers in the dot plots and histograms represent percentages of cells. Comparable percentages of GFP+cells were recovered from the FTOC (4.7% IRES-GFP+ and 3.2% p56lckF505-IRES-GFP+). The recoveries in absolute numbers were 4.1 × 105 cells from the IRES-GFP and 6.9 × 105 cells from the p56lckF505-IRES-GFP seeded cultures, respectively. We harvested comparable numbers of GFP+ cells (16,400 IRES-GFP+ cells and 20,700 p56lckF505-IRES-GFP+ cells, respectively).
Three-color flow cytometric analysis of IRES-GFP and p56lckF505-IRES-GFP–transduced CD4 ISP thymocytes cultured in FTOC. CD4 ISP thymocytes were sorted and transduced with the retrovirus expressing either the control IRES-GFP or the p56lckF505-IRES-GFP construct (both 2.5 × 104 cells input) and cultured in FTOC for 12 days. Flow cytometric analysis of the control was performed after gating on the GFP+ cells (GFP only). Analysis of the FTOC incubated with the p56lckF505 transduced cells was done following gating on untransduced cells (no Lck-GFP), p56lckF505-IRES-GFP+ cells (intermediate Lck-GFP), and p56lckF505-IRES-GFP++ cells (high Lck-GFP). Numbers in the dot plots and histograms represent percentages of cells. Comparable percentages of GFP+cells were recovered from the FTOC (4.7% IRES-GFP+ and 3.2% p56lckF505-IRES-GFP+). The recoveries in absolute numbers were 4.1 × 105 cells from the IRES-GFP and 6.9 × 105 cells from the p56lckF505-IRES-GFP seeded cultures, respectively. We harvested comparable numbers of GFP+ cells (16,400 IRES-GFP+ cells and 20,700 p56lckF505-IRES-GFP+ cells, respectively).
DISCUSSION
In the present report we show that in the human thymus the TCRδ, γ, and β loci rearrange in an ordered way; TCRδ rearrange first followed by TCRγ and TCRβ. We observed, after Southern blot analysis (detection limit: ∼5%), that while part of the immature CD34+CD1a− thymic subset (∼40%) have initiated incomplete Dδ2-Dδ3 and Dδ2-Jδ1 rearrangements, these cells have not rearranged the TCRγ and TCRβ loci. The results of the Southern blot analysis of TCRβ rearrangements in CD34+CD1a− cells were confirmed by PCR analysis and are consistent with those presented in another report.13 Calculations on the percentage of germline signal retained of the TCR genes showed a strong decrease of the TCRδ gene germline signal when the cells mature from the CD34+CD1a− (60% germline signal retained) to the CD34+CD1a+ (14% germline signal retained) cell stage, which suggests that most TCRδ gene rearrangements occur during this transition. TCRγ and TCRβ rearrangements were observed in CD34+CD1a+thymocytes. However, the Southern blot analysis strongly suggest that most CD34+CD1a+ cells have their TCRγ and β genes in the germline configuration. A strong decrease in germline signal of the TCRγ genes was observed when thymocytes mature from the CD34+CD1a+ (±90% germline signal retained) to the CD4 ISP cell stage (40% germline signal retained). Finally, most cells start to rearrange their TCRβ genes when they move from the CD4 ISP to the EDP stage. These data indicate a hierarchy in TCR rearrangements in which the TCRδ locus rearranges first followed by TCRγ and then TCRβ. This notion is consistent with results of studies with T-ALL that identified a few CD3− T-ALL that had rearranged TCRδ genes and germline TCRβ and TCRγ genes, and a few T-ALL with rearranged TCRδ and γ genes but germline TCRβ genes.37,38 A similar sequence of TCR rearrangements has recently been observed in the adult mouse thymus.39
The finding that the majority of CD34+CD1a− cells have their TCR genes in germline configuration is consistent with the fact that most cells within this population are able to differentiate into NK cells4 and, thus, are not yet committed to the T-cell lineage. We have observed that TCRγδ cells appear in IL-7–supported cultures of CD34+CD1a+ but not of CD34+CD1a− progenitor cells (results not shown). As this culture condition promotes maturation but not differentiation of early T-cell progenitors, these results indicate the presence of complete TCRδ and TCRγ rearrangements in the CD34+CD1a+ but not in the CD34+CD1a− cells, consistent with the Southern blot analyses in this study.
Although TCRβ gene rearrangements are initiated in the CD34+CD1a+ population, TCRβ protein is not detectable before the CD4 ISP stage, and even in that stage expression of this protein is limited to a very small proportion of cells. A more explicit expression of TCRβ protein was observed in the EDP cells, although also in this stage not all cells express the TCRβ protein. No cell-surface expression of CD3 was detectable in the CD4 ISP cells, while weak expression was found in the EDP cells. These observations suggest that CD4 ISP cells do not express a functional pre-TCR, despite the presence of high levels of pTα mRNA in this population,18 and raise the possibility that β selection occurs at the transition of CD4 ISP to EDP. This notion is consistent with the finding that RAG and pTα mRNAs are downregulated in the EDP population (results not shown) and is supported by our observations with constitutive active p56lck. It has been shown in numerous reports that mutations in the tyrosine kinase p56lck block T-cell development at the pre-TCR checkpoint.34-36,39 In this study we exploited these observations to investigate at which stage of human T-cell development the pre-TCR is active. Development of T cells into DP cells in the FTOC from both p56lckF505-IRES-GFP transduced CD34+and CD4 ISP cells was inhibited in a dose-dependent manner, as was also shown for the p56lckF505 transgenic mice.36 We recovered dramatically less DP cells expressing a TCRαβ, starting from p56lckF505-IRES-GFP–transduced CD34+thymocytes, and observed that CD4 ISP cells accumulate in these cultures. More importantly, overexpression of constitutive active p56lck in CD4 ISP cells also inhibited generation of DP and TCRαβ+ cells. Inhibition of development of CD4 ISP into DP cells is not due to cell death caused by overexpression of p56lckF505 because the percentages of GFP+cells before and after the FTOC were similar. Recently it has been determined that in the murine thymus β selection occurs in the CD4−CD8− population that expresses CD25 and low levels of CD44. This population could be subdivided into two subsets on the basis of their size. Approximately 75% of the CD25+CD44− population (denoted the E subset) is small in size and 25% (denoted L) is large. The majority of cells in the L subset have productive TCRβ rearrangements while the TCRβ gene rearrangements in the E subset are random.40 As a consequence of mutations that disrupt β selection in mice, E cells accumulate and L cells are absent. These observations indicate that the E to L transition is the starting point of β selection in the mouse. That disruption of β selection by overexpression of p56lckF505 leads to accumulation of CD4 ISP and disappearance of DP cells strongly suggests that the transition of the CD4 ISP into the EDP stage in the human thymus is comparable to the E into L cell transition in the mouse. Therefore, we propose that the pre-TCR commences to function at the time that CD8α is upregulated. Trigueros et al41 very recently reported that cell-surface expression of pTα as detected with a rabbit anti-human pTα serum was not detectable before the DP stage. Based on the expression of cytoplasmic TCRβ, which is absent on a proportion of CD3−DP cells, these investigators position the β-selection point after upregulation of CD8α rather than in parallel with CD8α upregulation, as proposed here. They suggest that cytoplasmic TCRβ− DP cells that lack pre-TCR on their cell surface are precursors of cytoplasmic TCRβ+pre-TCR+DP cells. This possibility cannot be dismissed by our data. However, because we observed accumulation of CD4 ISP in the FTOCs with p56lckF505, we hypothesize that the cytoplasmic TCRβ−EDP cells are dead-end cells. Importantly, both our study and that of Trigueros et al41 provide evidence that β-selection does not occur in the CD4 ISP.
In this study we have not analyzed TCRα gene rearrangements. This is not possible by the Southern blot technique, because the TCRα locus is composed of too many Vα and Jα gene segments. TCRα gene rearrangements and transcripts are especially found in CD3+T-ALL, whereas most CD3− T-ALL have germline TCRα genes. This suggests that TCRα gene rearrangements occur late during T-cell development and are most probably immediately followed by TCR-αβ expression on the cell surface. Because the TCRδ locus is nested within the TCRα locus, it has been suggested that deletion of the TCRδ locus initiates TCRα rearrangements. Two so-called TCRδ-deleting elements, δRec and ΨJα, flank the major part of the human TCRδ gene.37 The nonproductive rearrangement of these elements deletes the intermediate germline and/or rearranged TCRδ gene segments.10,37,42,43 Recently we found that the nonproductive δREC-ΨJα rearrangement starts at the CD4+CD8α+β−stage.44 Furthermore, using RT-PCR, we found that Vα-Cα transcripts were present in the CD27−CD69− subset, but absent in the CD4+CD8α+β−subset.44
ACKNOWLEDGEMENT
We thank Prof Dr Ad J.J.C. Bogers (Department of Thoracic Surgery, Erasmus University Rotterdam and University Hospital Rotterdam) for collecting postnatal thymus samples. We are grateful to Drs Garry Nolan and Ashok Venkitaraman for providing us with the ΦNX-A packaging cell line and the p56lckF505 cDNA, respectively. We thank Dr Pieter C.M. Res and Franka Couwenberg for help with the FTOC system. We express our gratitude to Eric Noteboom for cell sorting and to the people of the mouse facilities for maintaining the pregnant RAG-1–deficient mice. We thank Drs Ada M. Kruisbeek and Sander A.P. Stegmann for critical review of the manuscript.
Supported by a grant from The Dutch Cancer Society (M.C.M.V.: EUR 95-1015).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hergen Spits, PhD, Division of Immunology, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands; e-mail: hergen@nki.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal