Abstract
In addition to the Epstein-Barr virus (EBV) EBNA and LMP latency genes, there is a family of alternatively spliced BamHI-A rightward transcripts (BARTs). These latency transcripts are highly expressed in the EBV-associated malignancies nasopharyngeal carcinoma and Burkitt’s lymphoma, and are expressed at lower levels in latently EBV-infected B-cell lines. The contribution of the BARTs to EBV biology or pathogenesis is unknown. Resting B cells have recently been recognized as a reservoir for EBV persistence in the peripheral blood. In these cells, EBV gene expression is tightly restricted and the only viral gene known to be consistently expressed is LMP2A. We used cell sorting and reverse-transcriptase polymerase chain reaction (RT-PCR) to examine whether BARTs are expressed in the restricted form of in vivo latency. Our results demonstrated that RNAs with splicing diagnostic for transcripts containing the BART RPMS1 and BARFO open-reading frames (ORFs) were expressed in CD19+ but not in CD23+ B cells isolated from peripheral blood of healthy individuals. The product of the proximal RPMS1 ORF has not previously been characterized. The RPMS1 ORF was shown to encode a 15-kD protein that localized to the nucleus of transfected cells. Expression of the BARTs in peripheral blood B cells suggests that the proteins encoded by these transcripts are likely to be important for maintenance of in vivo latency.
EPSTEIN-BARR VIRUS (EBV) is a ubiquitous infectious agent that is associated with infectious mononucleosis and the human malignancies Burkitt’s lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease, and lymphoproliferative disease in immunocompromised subjects.1,2 However, in the majority of the population, EBV infection leads to a lifetime asymptomatic persistence, with the viral genome residing in circulating B lymphocytes in the peripheral blood.2-4 The pattern of EBV latency gene expression differs in different settings. The full spectrum of latency genes, namely, EBNA-1, 2, 3A, 3B, 3C, LP, LMP1, 2A, 2B and the polymerase III transcribed EBERs, are expressed in acute infectious mononucleosis (IM) and in lymphoblastoid cell lines (LCLs) established by in vitro outgrowth from peripheral blood.5This expression pattern has been designated latency III.6However, in EBV-associated tumors, a more restricted pattern of viral gene expression usually occurs. In Burkitt’s lymphoma, in endemic as well as sporadic and AIDS-related cases, only EBNA-1 is expressed.7 Nasopharyngeal carcinoma (NPC),8-10Hodgkin’s disease,11 and T-cell lymphoma12 13show an intermediate pattern characterized by expression of EBNA-1 and LMP2 and mixed expression of LMP1 (LMP1+/−).
EBV latency gene expression appears to be regulated in such a way as to allow escape from immune surveillance. The EBNA2 and EBNA3 family of proteins elicit a strong cytotoxic T-cell response and these proteins are expressed only on primary infection, in immunocompromised patients, or in cultured LCLs in vitro.14,15 EBNA-1, which is essential for replication of the latent EBV episome, contains sequences that prevent presentation to the immune system16,17 and EBNA-1 is expressed in all EBV-associated tumors. The differential expression of the EBNAs in the restricted forms of latency is mediated by a switch in viral promoter usage from Cp, which directs the synthesis of transcripts for all of the EBNAs, including those for the immunogenic EBNAs, to Qp, which exclusively directs synthesis of EBNA-1 mRNA.18-22
Two sites of EBV infection have been described: lymphoid cells and the mucosal epithelium. Although mucosal epithelial cells clearly can become infected as evidenced by the presence of EBV in the epithelial tumors nasopharyngeal carcinoma and gastric carcinoma, it is now believed that infection of the mucosal epithelium may be a relatively rare event, rather than an obligatory component of viral persistence.23,24 Evidence that the latently infected B lymphocyte forms the reservoir for EBV persistence in vivo came initially from the observation that EBV was eradicated in bone marrow transplant patients whose lymphoid system was destroyed by irradiation.25 In addition, abolition of lytic viral replication through acyclovir treatment does not eliminate EBV-infected B cells from the blood of IM patients, supporting the concept that these cells are latently infected.24 Recent studies indicate that the persistently infected peripheral B cell in which the EBV genome resides is a resting B cell in the G0 stage of the cell cycle with a phenotype of CD19+, CD23−, and CD80+.3,23,26,27The pattern of EBV gene expression in these cells is an important issue for the understanding of in vivo latency. Expression of LMP2A and the EBERs has been consistently detected in peripheral blood B cells28-30 and in that subset of cells selected for the resting B-cell phenotype.23 There is some question regarding the expression of EBNA-1 in the peripheral blood, but assays using B cells with the CD19+, CD23−, and CD80+ phenotype were negative for EBNA-1 expression.23
In addition to the spectrum of EBNA and LMP genes expressed during latency, there is a family of rightward transcripts derived from theBamHI-A region of the EBV genome that may also be latency related. The BamHI-A rightward transcripts (BARTs) were first described in NPC tumor tissues, where they are the most abundant viral transcripts.10,31,32 Subsequently, BARTs were shown to be expressed at a lower level in all three forms of latency in LCLs, after primary infection of B cells in culture8 and in other EBV malignancies.33-38 Several putative open-reading frames (ORFs) have been identified in the differentially spliced BART family of transcripts including a proximal ORF, RPMS1,39,40 a distal ORF BARF0, and an extension of BARF0, RK-BARF0.39Rabbit serum raised against an epitope within RK-BARF0 was recently used to show expression of this ORF in tumor samples.33 The function of the BARTs is unknown. Deletion of the BamHI-A region of the genome does not affect EBV immortalization of B cells in vitro,41 indicating that the BARTs are unlikely to contribute directly to immortalization. On the other hand, the consistent expression of these transcripts in EBV-associated epithelial and B-cell tumors suggests that they are making some contribution in vivo. In this report, we have selected CD19+ B cells from peripheral blood and show that the BARTs are expressed in this cell population. The BARTs may therefore play an important role in the maintenance of viral latency in vivo.
MATERIALS AND METHODS
Isolation and sorting of peripheral blood mononuclear cells.
Heparinized peripheral blood (100 to 200 mL) from an individual healthy adult volunteer or from a pool of healthy individuals was diluted with an equal volume of phosphate-buffered saline (PBS; 137 mmol/L NaCl, 2.7 mmol/L KCl, 4.3 mmol/L Na2HPO4 · 7H2O, 1.4 mmol/L KH2PO4 [pH 7.3]) and layered onto Ficoll-Hypaque (Pharmacia, Piscataway, NJ). After centrifugation at 3,000 rpm for 30 minutes, the peripheral blood mononuclear cell (PBMC)-containing layer was collected from the gradient and the cells were subjected to a second round of Ficoll-Hypaque separation. The PBMC collected from the second gradient were washed twice with RPMI 1640 culture medium lacking fetal bovine serum and then resuspended in RPMI 1640 (Mediatech, Herndon, VA).
CD19+ cells were positively selected using magnetic beads (Dynabeads M-450 CD19 [Pan B]; Dynal, New York, NY). A quantity of 1 × 107 beads (25 μL) was added to 1 × 108 PBMC in an Eppendorf tube and gently mixed on a bidirectional specimen mixer for 30 minutes at 4°C. The CD19+ B cells were isolated by placing the tube in a magnetic particle concentrator (Dynal MPC E-1) for 1 minute. The rosetted CD19+ cells attached to the wall of the tube, allowing the supernatant to be removed by pipetting. The remaining CD19-selected cells were washed three times with RPMI 1640.
CD23+ cells were positively selected from the CD19-depleted PBMC using magnetic beads coupled to anti-CD23 monoclonal antibody (MoAb) (Dako clone MHM6; Dako, Carpenteria, CA). A quantity of 1 × 107 (25 μL) sheep–anti-mouse IgG-coated beads (Dynabeads M-450) was added to 1 μg of mouse anti-CD23 MoAb and gently mixed for 30 minutes at 4°C. The beads coupled to anti-CD23 MoAb were collected using the magnetic particle concentrator and washed four times with phosphate-buffered saline (PBS) containing 0.3% bovine serum albumin. These beads were used to select CD23+ B cells following the procedure described above for CD19 selection.
RNA extraction, reverse-transcriptase polymerase chain reaction, and RNA analysis.
RNA from the CD19+- and CD23+-selected B cells was extracted and purified using a microprep mRNA kit (Pharmacia). The cells were lysed directly in extraction buffer and the samples processed according to the manufacturer’s instructions. For EBNA-1, BART (RPMS1 and BARF0), and CD23, cDNAs were synthesized from oligo-dT–enriched RNA by incubation for 1 hour at 42°C in a reaction mixture containing 1x reverse transcriptase (RT) buffer (Promega, Madison, WI), 0.4 mmol/L dNTPs, RNasin 40 U, and avian myeloblastosis virus (AMV) RT (5 U) and the specific primers listed in Table 1. The primers were selected to have a G + C content of 50% to 60% and to have a G or C at the 3′ end. EBER cDNA was prepared using random primers and total RNA treated with RNase free DNase. Polymerase chain reaction (PCR) was performed for 30 cycles with each cycle being 94°C for 1.5 minutes, 57°C for 1 minute, and 72°C for 2 minutes. For nested PCR, 2 μL of the product from the first round of PCR was subjected to 30 cycles of additional amplification. After PCR amplification, PCR products were electrophoretically separated on a 1.5% agarose gel. The gel was treated with 0.5 mol/L NaOH and 0.5 mol/L NaCl for 1 hour. The DNA was then transferred onto a nylon membrane and subjected to UV cross-linking (UV Stratalinker; Stratagene, La Jolla, CA). The membrane was preincubated for 2 to 3 hours at 50°C in hybridization buffer (1X Denhardt’s solution,42 10% dextran sulfate, 20 mmol/L NaH2PO4, 7% sodium dodecyl sulfate [SDS], 0.5X sodium chloride/sodium citrate buffer (SSC), and 100 μg denatured calf sperm DNA). Radiolabeled DNA oligonucleotide probes were then added to the hybridization buffer and the incubation was continued for a further 12 to 16 hours. After hybridization, the membranes were washed twice for 30 minutes at 50°C in 1X SSC plus 1% SDS and exposed to autoradiographic film. RNA extraction and RT-PCR of control EBV-positive (B95-8 and Raji) and EBV-negative (BJAB) cell lines were performed as described earlier.
Sequences and Coordinates of Primers
| Primers . | Sequences . | Coordinates . | PCR Products (bp) . |
|---|---|---|---|
| EBER1 | |||
| E1 | 5′-AGGACCTACGCTGCCCTAGAG-3′ | 6,649 | |
| E1 inner | 5′-AGAGGTTTTGCTAGGGAGG-3′ | 6,664 | |
| E2 | 5′-AAAACATGCGGACCACCAGC-3′ | 6,776 | |
| E2 nested | 5′-GACCACCAGCTGGTACTTG-3′ | 6,767 | 140 |
| Probe | 5′-AGACGTGTGTGGCTGTAG-3′ | 6,683 | |
| RPMS1 | |||
| P1 | 5′-CACGATGTCCTGGTCAGAGTG-3′ | 10,248 | |
| P1 nested | 5′-GGCTTGAGGAATACCTCGTTG-3′ | 10,282 | |
| P2 | 5′-CCTTCGATATGCAGTGTCTG-3′ | 155,847 | |
| P2 nested | 5′-ACCAACGAGGCTGACCTGATC-3′ | 155,801 | 296 |
| Probe | 5′-GCAGATATCCTGCGTCCTC-3′ | 155,781 | |
| BARF0 | |||
| P3 | 5′-GTGAGGGAAATAACCAGGATC-3′ | 157,086 | |
| P3 nested | 5′-CAGGACCAGAATGAGCATGC-3′ | 157,110 | |
| P4 | 5′-GCTTTCCTTTCCGAGTCTGC-3′ | 159,171 | |
| P4 nested | 5′-CTTCTCCTCGGACATCCAGTG-3′ | 159,121 | 237 |
| Probe | 5′-GGAGATGAAACCAGAGAC-3′ | 157,161 | |
| CD23 | |||
| C1 | 5′-GTTGTCAGGGAGTGAGTGC-3′ | ||
| C2 | 5′-GCTCGAAGTTCCTCCAGTTC-3′ | 271 | |
| Probe | 5′-CATCGGGAGAATCCAAGCAG-3′ | ||
| EBNA1 (U-K exon) | |||
| U1 | 5′-GAAGCGTTTCTTGAGCTTCC-3′ | 67,506 | |
| U1 nested | 5′-GTTTGGGAGAGCTGATTCTG-3′ | 67,537 | |
| K1 | 5′-CTCTTCTTTGAGGTCCACTG-3′ | 108,032 | |
| K1 nested | 5′-CTTCTGGTCCAGATGTGTCTC-3′ | 108,002 | 205 |
| Probe | 5′-ATGCCCTGAGACTACTCTCT-3′ | 67,548 |
| Primers . | Sequences . | Coordinates . | PCR Products (bp) . |
|---|---|---|---|
| EBER1 | |||
| E1 | 5′-AGGACCTACGCTGCCCTAGAG-3′ | 6,649 | |
| E1 inner | 5′-AGAGGTTTTGCTAGGGAGG-3′ | 6,664 | |
| E2 | 5′-AAAACATGCGGACCACCAGC-3′ | 6,776 | |
| E2 nested | 5′-GACCACCAGCTGGTACTTG-3′ | 6,767 | 140 |
| Probe | 5′-AGACGTGTGTGGCTGTAG-3′ | 6,683 | |
| RPMS1 | |||
| P1 | 5′-CACGATGTCCTGGTCAGAGTG-3′ | 10,248 | |
| P1 nested | 5′-GGCTTGAGGAATACCTCGTTG-3′ | 10,282 | |
| P2 | 5′-CCTTCGATATGCAGTGTCTG-3′ | 155,847 | |
| P2 nested | 5′-ACCAACGAGGCTGACCTGATC-3′ | 155,801 | 296 |
| Probe | 5′-GCAGATATCCTGCGTCCTC-3′ | 155,781 | |
| BARF0 | |||
| P3 | 5′-GTGAGGGAAATAACCAGGATC-3′ | 157,086 | |
| P3 nested | 5′-CAGGACCAGAATGAGCATGC-3′ | 157,110 | |
| P4 | 5′-GCTTTCCTTTCCGAGTCTGC-3′ | 159,171 | |
| P4 nested | 5′-CTTCTCCTCGGACATCCAGTG-3′ | 159,121 | 237 |
| Probe | 5′-GGAGATGAAACCAGAGAC-3′ | 157,161 | |
| CD23 | |||
| C1 | 5′-GTTGTCAGGGAGTGAGTGC-3′ | ||
| C2 | 5′-GCTCGAAGTTCCTCCAGTTC-3′ | 271 | |
| Probe | 5′-CATCGGGAGAATCCAAGCAG-3′ | ||
| EBNA1 (U-K exon) | |||
| U1 | 5′-GAAGCGTTTCTTGAGCTTCC-3′ | 67,506 | |
| U1 nested | 5′-GTTTGGGAGAGCTGATTCTG-3′ | 67,537 | |
| K1 | 5′-CTCTTCTTTGAGGTCCACTG-3′ | 108,032 | |
| K1 nested | 5′-CTTCTGGTCCAGATGTGTCTC-3′ | 108,002 | 205 |
| Probe | 5′-ATGCCCTGAGACTACTCTCT-3′ | 67,548 |
cDNA products generated by RT-PCR were ligated into the vector PCR2.1 (Invitrogen, Carlsbad, CA) and sequenced using the T7 primer.
Plasmids, Western blotting, and indirect immunofluorescence.
The RPMS1 ORF from a BART cDNA was amplified as a PCR fragment using the primers 5′ LGH1763 GCT AAG ATC TAT GGC CGG CGC TCG TGC and 3′ LGH2636 GAC AGA TCT TCA CCT TTG GCT GGT ACA GC and ligated into the BglII site of a modifed SG5 vector expressing the Flag epitope (pJH253) to generate pHC6.
For Western blot analysis, pHC6 (10 μg) was transfected by the standard calcium phosphate procedure into 293T cells that had been seeded at 1 × 106 cells into a 100-mm tissue culture plate 24 hours before transfection. After a 16-hour incubation at 35°C in a 3% CO2 atmosphere, the medium was replaced and the cells were incubated for a further 24 hours at 37°C with 5% CO2. Cells were lysed by heating at 100°C for 5 minutes in sample buffer (50 mmol/L Tris-HCl [pH 6.8], 100 mmol/L dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol) and the cell extract was fractionated through a 12% SDS-polyacrylamide gel. Proteins were detected by immunoblotting with anti-Flag MoAb (1:2,500; Kodak, Rochester, NY) and visualized using chemiluminescence (Amersham, Arlington Heights, IL).
Indirect immunofluorescence assays were performed using Vero cells seeded in two-well culture slides (Nunc, Denmark) at 5 × 104 cells per well. Cells were transfected as described earlier with pHC6 DNA (2 μg). Forty hours after transfection, the cells were fixed in methanol and stained with mouse anti-Flag MoAb (1:1,000) followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin antibody (1:100; Cappel, Durham, NC).
RESULTS
Specificity of the RT-PCR assay.
The BARTs are multispliced transcripts that contain a number of potential ORFs. The BART RK-BARF0 ORF has recently been shown to encode a membrane protein.33 This ORF is an extension of the BARF0 ORF and is generated by differential splicing. BARTs have also been described that contain a proximal ORF designated RPMS1.40Individual BART cDNAs have been characterized that contain both the RPMS1 ORF and the BARF0 ORF40 and both the RPMS1 ORF and RK-BARF0.39 The relative positions of the RPMS1, BARF0, and RK-BARF0 ORFs within BART transcripts and the location of the PCR primers used to detect RPMS1 and BARF0 are shown in Fig 1. We were particularly interested in BARTs that could potentially express the RPMS1 ORF. We therefore tested primers that would detect RPMS1 and also the more commonly described BARF0. The amplified products could be generated from individual transcripts containing both RPMS1 and BARF0 or separate transcripts containing the individual ORFs. The BARF0 primers used would not detect RK-BARF0. The EBERs are small noncoding polymerase III transcripts that are expressed at levels of approximately 1 × 107copies per cell in all latently EBV-infected cells. Their abundance makes them excellent markers in diagnostic assays for latent infection, and EBERs have consistently been detected in peripheral blood B cells of seropositive donors.29,30,43 Expression of EBER1 was therefore used as a positive control for EBV in our assays. CD23+ peripheral blood B cells have previously been described as being negative for EBV expression.27 PCR amplification of cellular CD23 mRNA was therefore chosen as the positive control for assays using these cells. The sequences of the primers used in this study are listed in Table 1.
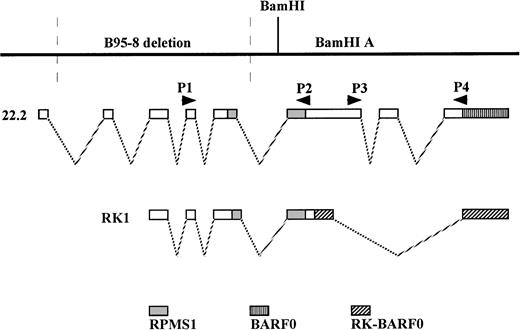
Illustration of the relative positions of the RPMS1, BARF0, and RK-BARF0 ORFs within BART transcripts and the location of the PCR primers used to detect RPMS1- and BARF0-containing cDNAs. The sequence and coordinates of the primers are listed in Table 1. The 22.2 and RK1 cDNA clones have been previously reported.39 40
Illustration of the relative positions of the RPMS1, BARF0, and RK-BARF0 ORFs within BART transcripts and the location of the PCR primers used to detect RPMS1- and BARF0-containing cDNAs. The sequence and coordinates of the primers are listed in Table 1. The 22.2 and RK1 cDNA clones have been previously reported.39 40
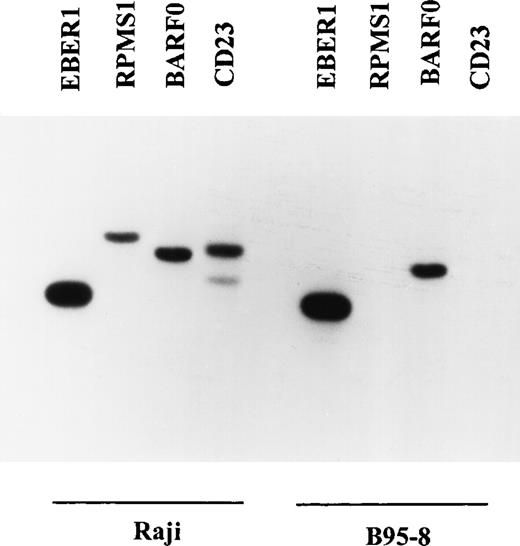
The EBV-positive B-cell lines B95-8 and Raji were used to verify the specificity of the RT-PCR primers and oligonucleotide probes (Fig2). The RT-PCR products were separated by gel electrophoresis and subjected to Southern blot analysis using end-labeled oligonucleotide probes for sequences within each RT-PCR product. The size of the product generated corresponded to the predicted size for each pair of primers. RT-PCR products for each of the four primer pairs were detected in Raji cells. Since B95-8 carries a large deletion that includes sequences from the BamHI-I region,44,45 no product was expected for the RPMS1 ORF in B95-8 and none was detected. The CD23 primers were based on the human CD23 sequence.46 B95-8 is a marmoset cell line and the CD23 primers were unable to amplify CD23 sequences from B95-8. The primers used for nested RT-PCR were also tested in Raji and B95-8 cells and showed the same specificity (data not shown).
Specificity of the RT-PCR primers and oligonucleotide probes for EBER1, RPMS1, BARF0, and CD23. RT-PCR and Southern blot analysis were performed as described in Materials and Methods. Raji is a human EBV+ lymphoblastoid cell line that expresses RPMS1, BARF0 and human CD23. B95-8 is a marmoset lymphoblastoid cell line that expresses simian CD23 and carries an EBV genome that is deleted in the BamHI-I region, which encodes RPMS1.
Specificity of the RT-PCR primers and oligonucleotide probes for EBER1, RPMS1, BARF0, and CD23. RT-PCR and Southern blot analysis were performed as described in Materials and Methods. Raji is a human EBV+ lymphoblastoid cell line that expresses RPMS1, BARF0 and human CD23. B95-8 is a marmoset lymphoblastoid cell line that expresses simian CD23 and carries an EBV genome that is deleted in the BamHI-I region, which encodes RPMS1.
Detection of BARTs in PBMC.
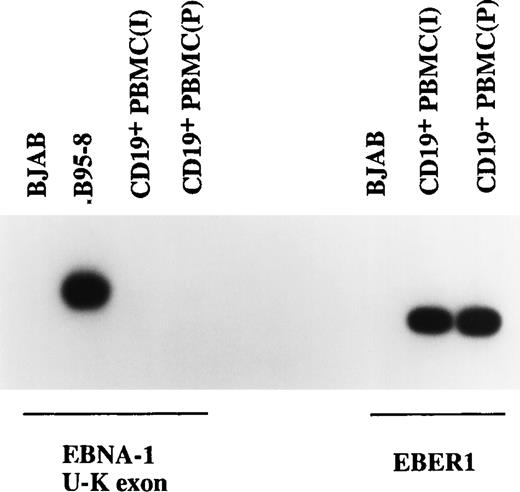
To determine whether BARTs were transcribed in latently EBV-infected cells in vivo, RT-PCR was used to test for expression of BARTs in PBMC prepared from a healthy seropositive individual and from pooled blood from healthy donors. PBMC prepared from both the individual sample (indicated by “I” in Fig 3) and the pooled blood sample (indicated by “P” in Fig 3) contained latently EBV infected cells as demonstrated by the presence of a positive signal for the abundant EBER1 RNA. Transcripts for EBNA-1 were not detected using primers across the BamHI-U andBamHI-K exon boundaries (Fig 3). The U-K splice is common to EBNA-1 transcripts originating from Wp, Cp, or Qp promoters. We also obtained negative results with primers spanning the Qp to BKRF1 (EBNA-1) exon boundaries (data not shown). Other studies have noted either a lack of detectable EBNA-1 expression or variable expression in peripheral blood from healthy donors.28 29 Expression of BARTs was detected in both individual (I) and pooled (P) PBMC samples with primers for the RPMS1 and BARF0 ORFs each giving a positive signal (Fig 4A).
Examination of EBNA-1 transcription in CD19+ PBMC by RT-PCR. No EBNA-1 transcripts were detected in two CD19+ PBMC samples using primers specific for the EBNA-1 U-K splice, although EBER1 RNA was easily detectable. BJAB and B95-8 cells served as the negative and positive controls.
Examination of EBNA-1 transcription in CD19+ PBMC by RT-PCR. No EBNA-1 transcripts were detected in two CD19+ PBMC samples using primers specific for the EBNA-1 U-K splice, although EBER1 RNA was easily detectable. BJAB and B95-8 cells served as the negative and positive controls.
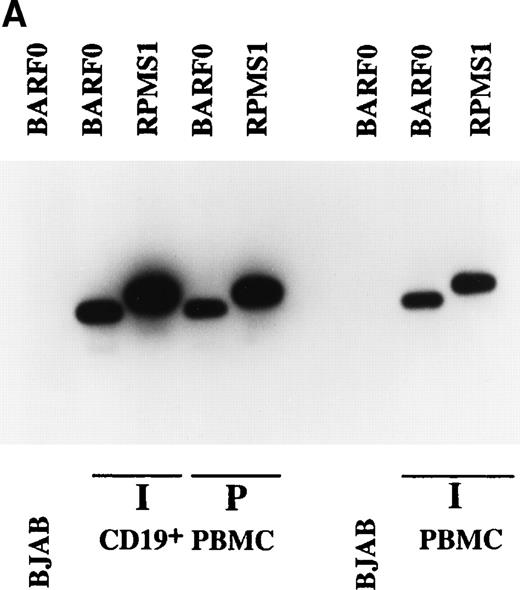
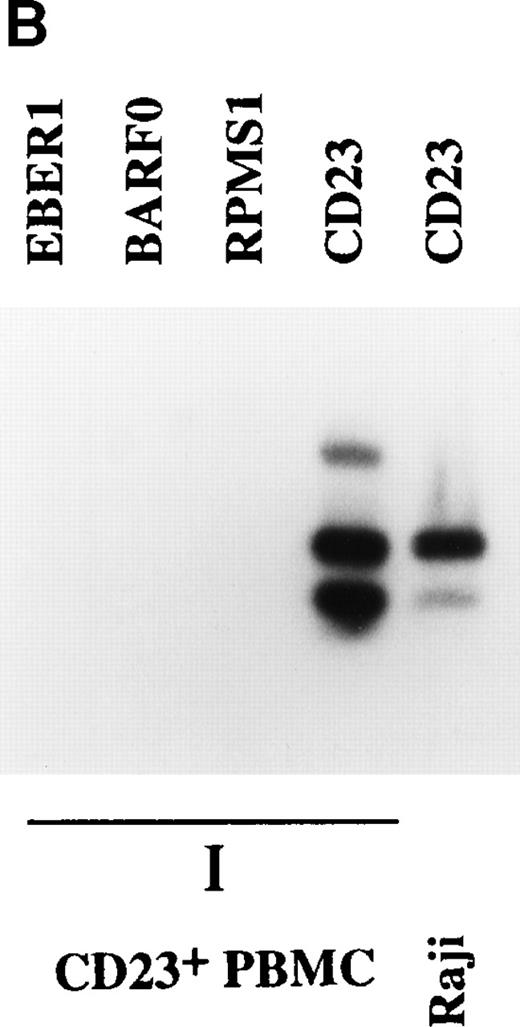
Expression of BARTs in PBMC. RT-PCR was used to examine BART expression in (A) unfractionated PBMC and CD19+-selected PBMC. RPMS1 and BARF0 were detected in CD19+-selected PBMC isolated from an individual donor (I) and from pooled blood (P), as well as in unfractionated PBMC from an individual (I). EBV− BJAB cells served as the negative control. (B) CD23+ PBMC. RPMS1, BARF0, and EBER1 transcripts were not detected in CD23+ PBMC isolated from an individual (I). Detection of CD23 transcripts in the individual PBMC sample was used to illustrate the quality of the mRNA in this sample. Raji was used as a positive control for CD23 RT-PCR.
Expression of BARTs in PBMC. RT-PCR was used to examine BART expression in (A) unfractionated PBMC and CD19+-selected PBMC. RPMS1 and BARF0 were detected in CD19+-selected PBMC isolated from an individual donor (I) and from pooled blood (P), as well as in unfractionated PBMC from an individual (I). EBV− BJAB cells served as the negative control. (B) CD23+ PBMC. RPMS1, BARF0, and EBER1 transcripts were not detected in CD23+ PBMC isolated from an individual (I). Detection of CD23 transcripts in the individual PBMC sample was used to illustrate the quality of the mRNA in this sample. Raji was used as a positive control for CD23 RT-PCR.
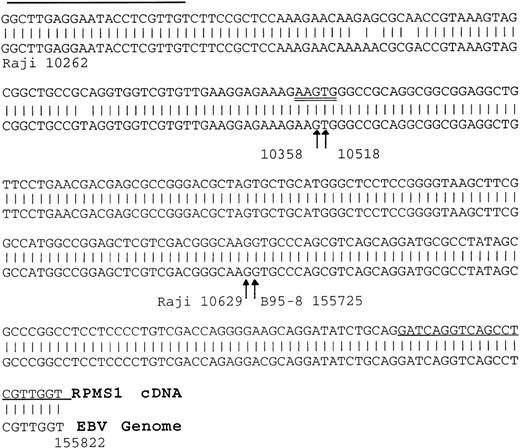
The site of EBV latency in the peripheral blood has recently been identified as a CD19+ resting B cell.27 The PBMC samples were fractionated into CD19+ and CD23+ cells as described in Materials and Methods. CD23 is a marker for activated B cells. As shown in Fig 4A, the CD19+ selected cells from the individual sample (I) and the pooled sample (P) were positive for BART expression. Both the RPMS1 and BARF0 primers amplified appropriately sized products. No expression of the BARTs or EBER RNA was detected in the CD23+-selected cells (Fig 4B). The RNA in these cells was of amplifiable quality as demonstrated by the detection of cellular CD23 transcripts. Thus, BART expression is specifically found in the CD19+ B-cell population and not in cells expressing the CD23 activation marker. To confirm the authenticity of the BART RT-PCR products, the RPMS1 cDNA was cloned into the PCR2.1 vector and sequenced. The cDNA sequence obtained was aligned with B95-8 genomic sequence or with Raji sequence for the region falling within the B95-8 deletion. The cDNA sequence matched that of the published RPMS1 cDNA40 with nucleotide variations at 4 positions in the noncoding 5′ leader region. The sequence contained a 5-bp insert relative to the cDNA described by Sadler and Raab-Traub39 (Fig5).
Verification of the identity of the RPMS1 RT-PCR product. Alignment of an RPMS1 RT-PCR sequence cloned from a CD19+PBMC sample to known EBV sequences. The 5′ part of the sequence matched to Raji sequences spanning the B95-8 major deletion and the 3′ part of sequence matched B95-8 sequences as shown. Arrows indicate the splice sites and the numbers represent the positions in the Raji or B95-8 genome. Single-underlined sequences represent the primers used to generate the RT-PCR clone and the double-underlined 5 bp indicates an insertion relative to a reported cDNA.39The splice acceptor site (115,725) is the same as that seen in RK139 and the same sequence as that seen in C22.2,40 although there is a discrepancy in the numbering, which is given as 115,730 in Smith et al.40
Verification of the identity of the RPMS1 RT-PCR product. Alignment of an RPMS1 RT-PCR sequence cloned from a CD19+PBMC sample to known EBV sequences. The 5′ part of the sequence matched to Raji sequences spanning the B95-8 major deletion and the 3′ part of sequence matched B95-8 sequences as shown. Arrows indicate the splice sites and the numbers represent the positions in the Raji or B95-8 genome. Single-underlined sequences represent the primers used to generate the RT-PCR clone and the double-underlined 5 bp indicates an insertion relative to a reported cDNA.39The splice acceptor site (115,725) is the same as that seen in RK139 and the same sequence as that seen in C22.2,40 although there is a discrepancy in the numbering, which is given as 115,730 in Smith et al.40
The RPMS1 ORF encodes a nuclear protein.
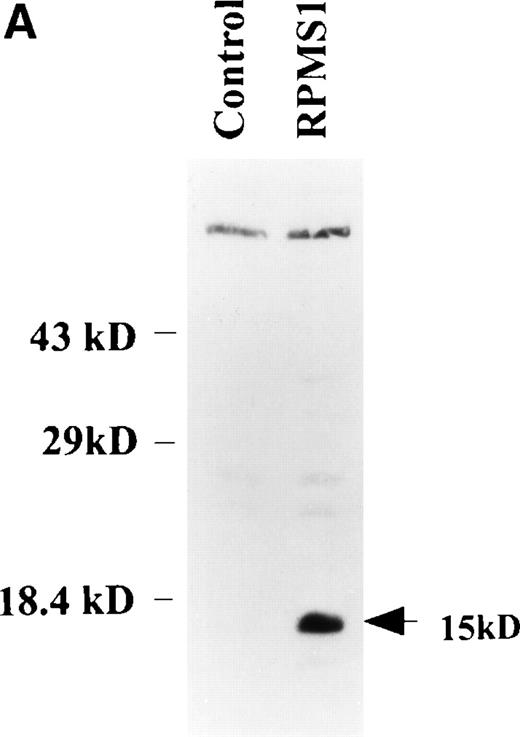
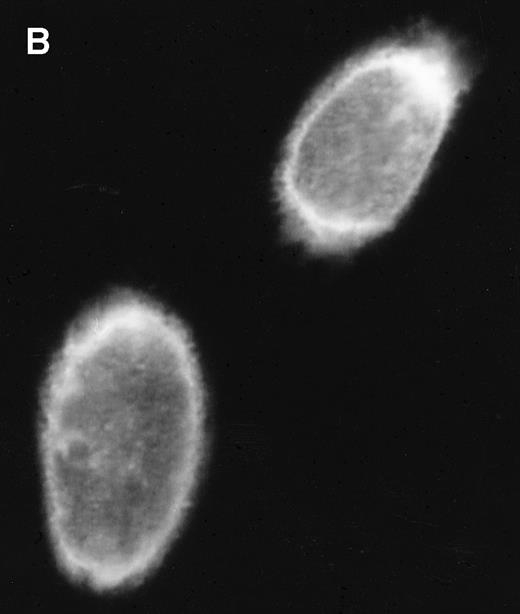
Several putative ORFs are identifiable in BART cDNAs. The distal RK-BARF0 ORF has recently been shown to express a 30-kD protein in transfected cells and the RK-BARF0 protein has been reported to be present in EBV-positive B-cell lines and in tumor specimens.33 A proximal ORF, RPMS1, has also been described40 and transcripts with splicing consistent with that described for the cDNA containing RPMS1 were detected in CD19+ peripheral blood B cells. The protein product of the RPMS1 ORF was examined using a vector that expressed RPMS1 tagged with the Flag epitope. In cells transfected with Flag-RPMS1, a protein of approximately 15 kD was detected by Western blotting with anti-Flag antibody (Fig 6A). Indirect immunofluorescence assays using anti-Flag antibody showed that the RPMS1 protein localized to the nucleus of transfected cells (Fig 6B). Thus, the BART family of RNAs has the potential to encode at least two independent proteins.
RPMS1 protein expression. (A) Western blot analysis of 293T cells transfected with an expression vector containing the Flag-tagged RPMS1 ORF. The Flag-RPMS1 protein band detected using anti-Flag mouse MoAb is indicated by an arrow. The relative positions of the molecular weight markers are shown on the left. Lysate of nontransfected 293T cells was used as the control. (B) Indirect immunofluorescence assay showing Flag-RPMS1 expression in transfected Vero cells. Flag-RPMS1 was detected using anti-Flag MoAb and FITC-conjugated secondary antibody.
RPMS1 protein expression. (A) Western blot analysis of 293T cells transfected with an expression vector containing the Flag-tagged RPMS1 ORF. The Flag-RPMS1 protein band detected using anti-Flag mouse MoAb is indicated by an arrow. The relative positions of the molecular weight markers are shown on the left. Lysate of nontransfected 293T cells was used as the control. (B) Indirect immunofluorescence assay showing Flag-RPMS1 expression in transfected Vero cells. Flag-RPMS1 was detected using anti-Flag MoAb and FITC-conjugated secondary antibody.
DISCUSSION
EBV infection leads to lifelong viral persistence with healthy seropositive individuals carrying between 1 and 50 per 106peripheral blood B cells latently infected with EBV.3,23,27Different programs of EBV latency gene expression are seen in different settings. The full spectrum of EBNA and LMP transcripts that is typical of latently infected LCLs in culture has been detected in vivo only in the setting of IM and in immunocompromised patients with lymphoproliferative disease. In the case of primary infection, virally induced B-cell proliferation may be necessary to establish a pool of latently infected cells. However, the EBNA2, EBNA3A, EBNA3C, and LMP-1 proteins that provide lymphoproliferative functions are also immunogenic and cells expressing these proteins elicit a strong cytotoxic T-cell response.15,47,48 How EBV maintains a latent infection in the face of active immune surveillance is an important issue for the understanding of EBV pathogenesis. Recent studies using sensitive RT-PCR technology have provided evidence that EBV persists in the peripheral blood in resting CD19+, CD23− B cells.27 The pattern of latency gene expression in these cells is highly restricted with only EBER and LMP2A (also called TP1) RNAs being consistently detected.28-30 The EBERs are small noncoding polymerase III transcripts. LMP2A contains an immunoreceptor tyrosine activation motif (ITAM). This motif also occurs in the B-cell and T-cell receptors and is critical for signal transduction initiated through these molecules.49 LMP2A blocks B-cell receptor-stimulated calcium mobilization, tyrosine phosphorylation, and induction of the EBV lytic cycle.50 51 Latently infected peripheral blood B cells may be exposed to ligands that could engage the B-cell receptor, and expression of LMP2A may be important in preventing lytic cycle induction of the endogenous EBV genomes. EBNA-1 has been detected in peripheral blood B cells in some studies, but not in others. EBNA-1 is essential for replication of the EBV episome. In resting B cells, EBNA-1 function would not be required, but if these resting cells are periodically activated into cell cycle, then EBNA-1 would become necessary to maintain the EBV genome copy number.
In this study, we addressed whether the multispliced BART transcripts are also contributors to in vivo latency. Deletion of theBamHI-A region of the genome does not impair the ability of EBV to immortalize B cells in vitro.41 However, the consistent detection of these transcripts in EBV-associated tumors implies an as yet undefined contribution in vivo. We detected BARTs with splicing consistent with RPMS1 encoding transcripts in CD19+, CD23− PBMCs. The presence of BARTs in this tightly latent cell population supports the argument that they play a role in viral persistence. Exactly what BARTs contribute to latency remains to be explored. A role in evasion of cytotoxic T-cell immune responses would be compatible with their expression both in peripheral blood B cells and in NPC and Burkitt tumors. Alternatively, or additionally, their role may complement that of LMP2A. Whereas LMP2A acts to prevent induction of EBV lytic gene expression, BART encoded proteins may function to prevent induction of the full latency program that includes the antigenic and growth stimulatory EBNAs. Neither the number of proteins encoded by the differentially spliced BARTs nor the function of these proteins is currently defined. We showed here that the RPMS1 ORF that is present in BARTs with the splicing pattern detected in CD19+, CD23− PBMCs gives rise to a nuclear protein in transfected cells. Thus, BARTs apparently encode at least two proteins, RPMS1 and RK-BARF0, and have the potential to facilitate a program of EBV latency that fosters viral persistence.
Supported by Grant No. R01 CA42245 from the National Institutes of Health. S.D.H. is the recipient of an American Cancer Society award (FRA 429). P.S. was supported by the Cancer Research Campaign (UK) and a Fulbright Cancer Fellowship.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to S. Diane Hayward, PhD, Department of Pharmacology and Molecular Science, and Department of Oncology, Johns Hopkins School of Medicine, 725 N Wolfe St, Baltimore, MD 21205.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal