Abstract
The hallmark of T- and B-lymphocyte development is the rearrangement of variable (V), diversity (D), and joining (J) segments of T-cell receptor (TCR) and immunoglobulin (Ig) genes to generate a diverse repertoire of antigen receptor specificities in the immune system. The process of V(D)J recombination is shared in the rearrangement of all seven antigen receptor genes and is controlled by changes in chromatin structure, which regulate accessibility to the recombinase apparatus in a lineage- and stage-specific manner. These chromatin changes are linked to transcription of the locus in its unrearranged (germline) configuration. To understand how germline transcription of the TCRβ-chain gene is regulated, we determined the structure of germline transcripts initiating near the Dβ1 segment and identified a promoter within this region. The Dβ1 promoter is active in the presence of the TCRβ enhancer (Eβ), and in this context, exhibits preferential activity in pro-T versus mature T-cell lines, as well as T- versus B-lineage specificity. These studies provide insight into the developmental regulation of TCRβ germline transcription, one of the earliest steps in T-cell differentiation.
ADAPTIVE IMMUNITY in vertebrates depends on the generation of a vast repertoire of antigen receptor specificities among lymphocytes. During development of T and B cells, variable region segments of the antigen receptor genes undergo somatic rearrangement to create a unique primary structure in the antigen recognition domains of these proteins in each developing lymphocyte.1-3 The mechanism of variable (V) diversity (D) joining (J) recombination appears to be identical for genes encoding T-cell receptor (TCR)α, β, γ, and δ, as well as for IgH, Igκ, and Igλ.1,2 Recognition signals (RS), comprised of a conserved heptamer, a 12 or 23 bp spacer, and an AT-rich nonamer, flank the coding sequences of recombining gene segments and target DNA scission by the Rag-1 and Rag-2 proteins to generate a blunt RS end and a sealed hairpin structure at the coding end.4,5This step is lymphoid-specific, while subsequent processing and rejoining of coding and RS ends is mediated by DNA repair mechanisms present in all cells.6 7
V(D)J recombination occurs in an ordered fashion and involves only the antigen receptor genes, which are appropriate for a given lineage and developmental stage. Thus, in the B-cell lineage, IgH genes are rearranged before Igκ and Igλ, while in the major T-cell lineage, the TCRβ locus is rearranged before TCRα. The rearrangement process itself controls lymphocyte development, in that the protein products of rearranged antigen receptor genes generate signals, which mediate the major transitions in T- and B-cell differentiation.3 Cells expressing Rag-1 and Rag-2 are competent to rearrange extrachromosomal recombination substrates, but chromosomal DNA is not a substrate for the cleavage reaction unless it is in a developmentally appropriate configuration, indicating that V(D)J recombination is controlled at the level of accessibility of the recombinase to the appropriate locus.1,3,6,8,9 A major clue to the mechanism of accessibility regulation is provided by the observation that antigen receptor loci are transcribed in their unrearranged (germline) state before rearrangement,8,10-16 a process known to be associated with changes in chromatin patterns.17 Thus, developmental regulation of chromatin structure at antigen receptor loci and germline transcription are intimately linked, forming an attractive hypothesis to explain the stage- and lineage-specific control of V(D)J recombination. A number of experiments using minigene recombination substrates, as well as targeted deletion studies in mice show that transcriptional enhancer elements are required both for efficient and developmentally appropriate rearrangement of antigen receptor genes and for germline transcription of these loci.8,18-25 Relatively little is known regarding the regulation of germline transcription, which may initiate from several points across antigen receptor loci. While promoters directing this process have been identified for several antigen receptor genes,14,26-29 the transcription factors that bind these elements have not been characterized. In the αβ T-cell lineage, germline TCRβ transcripts can be recognized at the earliest identifiable stage of T-cell development in the thymus,12,30 31 indicating that activation of this locus for rearrangement is among the first steps in differentiation of this lineage. To understand the regulation of TCRβ germline transcription, we have determined the structure of transcripts initiating within the first D-J-C complex near Dβ1 and have identified a promoter that directs Dβ1 transcription in pro-T cells. The Dβ1 promoter interacts with the Eβ enhancer, and in this context, appears to contribute to stage- and lineage-specific regulation of germline transcription. These studies form the basis for the identification of trans-acting factors which interact with these regulatory elements to control germline TCRβ transcription in pro-T cells.
MATERIALS AND METHODS
Cell lines, cell culture, and transfections.
The p5424 and p4980 thymoma cell lines were derived from p53−/− mice deficient in Rag-1 and Rag-2, respectively (provided by Jianzhu Chen, MIT, Cambridge, MA). Cell lines BW5147, WEHI 231, EL-4, and A20 were obtained from American Type Culture Collection (ATCC; Manassas, VA). Cells were cultured in either Dulbecco’s modified Eagle medium (DMEM) (p4980, p5424, and BW5147) or RPMI-1640 (EL-4, WEHI 231, and A20), supplemented with 10% fetal calf serum (FCS), pen/strep, and L-gln. Eight million cells were transfected in 0.2 mL phosphate-buffered saline (PBS) with 20 μg of the reporter construct by electroporation at 220 V, 900 μF, and 13 ohms using a BTX apparatus (San Diego, CA). After electroporation, cells were cultured for 24 hours in 6 cm dishes, then collected, and assayed for luciferase activity using reagents from Promega (Madison, WI). Transfection efficiency was measured by inclusion of 2 μg of an expression vector for either human growth hormone32 or Renilla luciferase (Promega). Culture supernatants were assayed for growth hormone using a radioimmunoassay (Nichols Institute, San Juan Capistrano, CA), or for Renilla luciferase using the Dual-Luciferase assay system (Promega).
Plasmid construction and cloning.
A TCRβ genomic clone was isolated from a 129-strain mouse genomic library (Stratagene, La Jolla, CA). A fragment spanning from −2184 to +151 relative to the first base coding for Dβ1 was subcloned and deletions made either by Exonuclease II and Mung Bean nuclease or by polymerase chain reaction (PCR) amplification. The 550-bp HpaI to NcoI Eβ core fragment33was cloned from mouse genomic DNA by PCR amplification. Luciferase reporter constructs were based on the pGL-3 basic, pGL-3 enhancer, or pGL-3 control vectors (Promega). The fidelity of all constructs were confirmed by sequence analysis. The Rag-2−/−thymocyte cDNA library was made using the Zap II vector (Stratagene). Total RNA was collected from Rag-2−/− thymocyte suspensions, and first-strand cDNA synthesized using superscript RT (GIBCO, Gaithersburg, MD) and oligo-dT. Subsequent steps for cDNA synthesis and library construction were performed with reagents from Stratagene. Site-directed mutagenesis was performed to alter the Ikaros/Lyf-1 site at −35 (m35 construct, ATGGGAGGG to ATGTCAGGG) and the GATA binding sequence at −74 (m74 construct, CCAGATAAGC to CCATCCGAGC) according to the U.S.E. mutagenesis protocol from Pharmacia Biotech (Piscataway, NJ). Mutant constructs were sequenced in their entirety to confirm sequence fidelity.
RNA analysis.
For Northern blots, 10 μg of total RNA was run on a denaturing agarose gel then transferred to a nylon membrane. Cβ1-containing transcripts were detected with a random primed 32P-labeled genomic fragment spanning the last two exons of the constant region. Primer extension analysis was performed using a downstream primer, 5′-GGTGGTCTGTTTTATGGACGTTGGCAGAAGAGGAT-3′ (+345 to +311 relative to Dβ1), and an upstream primer, 5′-TCCCATAGAATTGAATCACCGTGGCCCCCTGTCCC-3′ (+35 to +1). These were end-labeled with 32P and gel purified before hybridization with 100 μg total RNA and reverse transcribed using Superscript II polymerase (GIBCO). Samples were digested with RNAse and purified before running on a denaturing sequencing gel. Gels were fixed and dried before exposing to film. Ribonuclease protection assays were performed using an RNA probe corresponding to bases −202 to +285 surrounding Dβ1, cloned into the pGEM5ZF+ vector (Promega), and synthesized by in vitro transcription. Reagents for synthesis and hybridization were obtained from PharMingen (San Diego, CA). The labeled probe was hybridized with 8 μg of total RNA from the cells indicated before being digested and analyzed by electrophoresis on a 6% polyacrylamide gel under denaturing conditions.
RESULTS
Germline TCRβ transcription in pro-T cells and Rag-deficient T-cell lines.
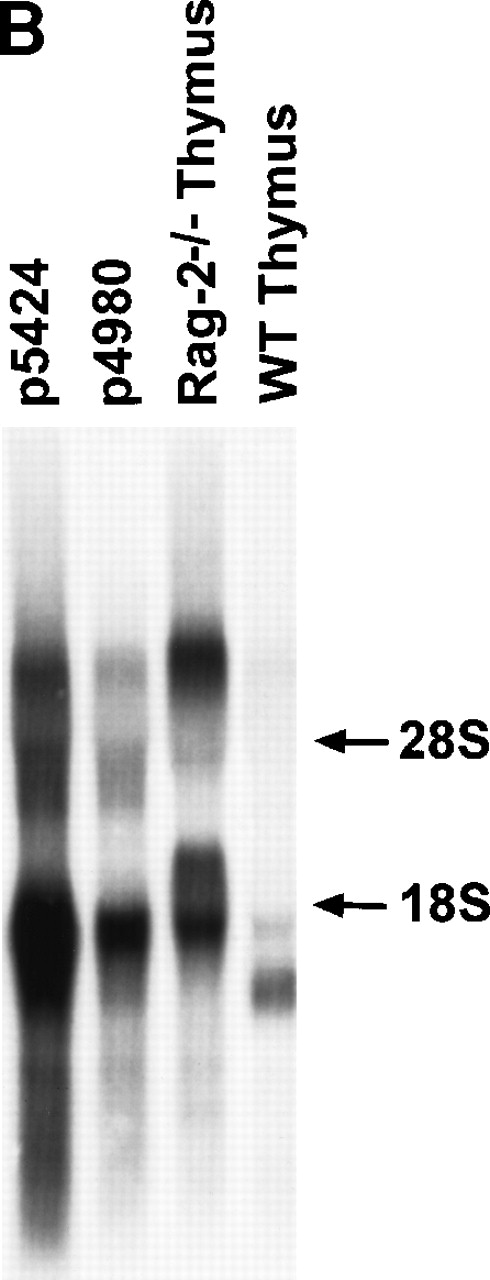
The constant (C) regions of the TCRβ locus lie in the germline as a tandem duplication, with each constant region located 3′ of one D segment and a cluster of J segments (Fig1A).34 Transcription of the unrearranged D-J-C cluster occurs in pro-T cells, a subset of CD4−CD8− double-negative thymocytes comprising 2% to 5% of normal thymocytes.3,12,30,31T-cell development is blocked in Rag-1 or Rag-2–deficient mice, and thymi from these strains contain a small population of pro-T cells, which necessarily have TCRβ genes in the germline configuration.35,36 As previously shown, Northern blot analysis of thymocyte RNA from Rag-2–deficient mice indicates abundant levels of transcripts hybridizing with a probe spanning Dβ1, which does not hybridize to V(D)J rearranged alleles (Fig 1B).23Two major bands were observed, including a high Mr species, which likely represents a precursor transcript and a family of lower Mr species presumably representing processed mRNAs species. The broad banding pattern, consistently seen in multiple Northern blots, was not due to RNA degradation, as detected by ethidium bromide stain, or in blots probed for other messages. In comparison, the same probe detected a lower Mr family of transcripts in normal thymus, most likely representing transcripts from DJ rearranged alleles, although a very faint band corresponding to germline transcripts was also recognized. Thus, the relative enrichment for rare early thymocyte populations in Rag-2–deficient versus normal thymocytes corresponds with high-level expression of germline TCRβ transcripts in this cell population. The Rag-deficient thymoma cell lines, p4980 and p5424 express CD4, CD8, CD28, CD43, CD95, and low levels of CD25, CD44, and CD45, but do not express CD3, CD19, CD69, or interleukin (IL)-2Rβ (data not shown), thus exhibiting several surface characteristics of pro-T cells. Germline TCRβ transcripts were also abundant in these cell lines (Fig 1B), indicating that they behave as Rag-deficient pro-T cells with respect to regulation of the TCRβ locus. Compared with Rag-2−/−thymocytes, there was less of the large Mr transcript and relatively less heterogeneity of the processed transcripts, with a bias toward smaller-sized transcripts. Together, these data indicate that germline Dβ1 transcripts are heterogeneous in vivo, and that Rag-2–deficient thymocytes, as well as the p4980 and p5424 lines, represent useful reagents for studying germline TCRβ transcription.
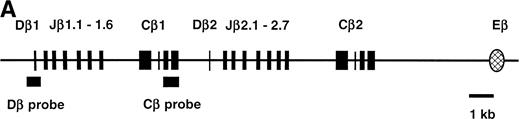
Northern blot analysis of TCRβ germline transcription. (A) Map of the D-J-C region of the TCRβ locus showing the location of the Dβ1 probe used in Northern analysis, as well as the Cβ1 probe used to isolate cDNA clones depicted in Fig 2. (B) Northern blot using Dβ1 probe to detect germline TCRβ transcripts in total RNA from the p5424 and p4980 cell lines and from thymocytes from either Rag-2–deficient or normal mice as indicated. Lane loading was equivalent as determined by ethidium bromide staining. The position of the 28S and 18S ribosomal RNA bands are indicated by arrows.
Northern blot analysis of TCRβ germline transcription. (A) Map of the D-J-C region of the TCRβ locus showing the location of the Dβ1 probe used in Northern analysis, as well as the Cβ1 probe used to isolate cDNA clones depicted in Fig 2. (B) Northern blot using Dβ1 probe to detect germline TCRβ transcripts in total RNA from the p5424 and p4980 cell lines and from thymocytes from either Rag-2–deficient or normal mice as indicated. Lane loading was equivalent as determined by ethidium bromide staining. The position of the 28S and 18S ribosomal RNA bands are indicated by arrows.
Structure of TCR Dβ1 germline transcripts.
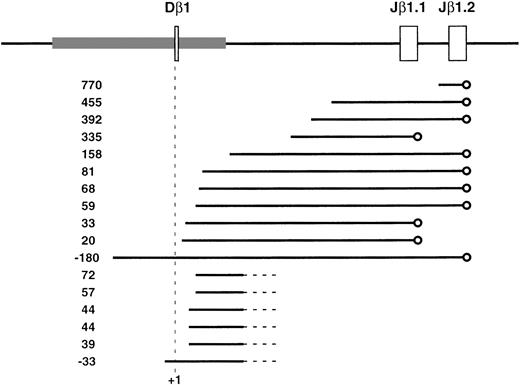
Although it is known that early T cells synthesize TCRβ transcripts hybridizing with gene segments deleted during V(D)J recombination,3,12,30,31 the structure of these transcripts has not been reported. To determine the structure of transcripts involving Dβ1 in vivo, we synthesized a cDNA library from Rag-2–deficient thymocytes and isolated cDNA clones using a Cβ1 probe (Fig 1A). The 5′ ends of the clones were quite heterogeneous and ranged from 180 bp upstream through 770 bp downstream of Dβ1, suggesting that the Dβ1 transcripts may initiate from multiple sites (Fig 2). The cDNA clones represented processed transcripts and contained normal splice junctions between either Jβ1.1 (in about a third of transcripts) or Jβ1.2 and constant region exons. The alternative Cβ0 exon was used in four transcripts, consistent with the frequency of this exon found in expressed TCRβ cDNAs.37 The Cβ1 probe used to screen the cDNA library cross-reacts with Cβ2, and cDNAs containing the second constant region were also isolated (data not shown). These had a structure similar to Dβ1 germline transcripts, but were shorter, with 5′ ends clustering near the Jβ2.1 segment. Spliced transcripts containing both D-Jβ1 and Cβ2 were not seen among the cDNA clones we sequenced. Thus, germline transcription of the duplicated D-J-C complex of the TCRβ locus appears to initiate independently within each iteration of the complex.
Genomic sequence analysis and structure of cDNA clones representing Dβ1 germline transcripts. The genomic structure of the Dβ1 region is indicated at the top of the figure, with the area of sequence conservation (72% identity) between mouse and human indicated by the shaded box. cDNA clones representing germline TCRβ transcripts were obtained from a Rag-2−/− thymocyte library screened with a Cβ1 probe and analyzed by DNA sequencing. Individual clones are depicted by a solid horizontal line, with the position of the 5′ end relative to the first base of Dβ1 (+1) indicated on the left. Splice junctions are indicated by circles and unsequenced regions by dashed lines.
Genomic sequence analysis and structure of cDNA clones representing Dβ1 germline transcripts. The genomic structure of the Dβ1 region is indicated at the top of the figure, with the area of sequence conservation (72% identity) between mouse and human indicated by the shaded box. cDNA clones representing germline TCRβ transcripts were obtained from a Rag-2−/− thymocyte library screened with a Cβ1 probe and analyzed by DNA sequencing. Individual clones are depicted by a solid horizontal line, with the position of the 5′ end relative to the first base of Dβ1 (+1) indicated on the left. Splice junctions are indicated by circles and unsequenced regions by dashed lines.
The 5′ ends of the multiple cDNA clones suggested that germline transcription initiated near Dβ1. Therefore, we isolated a genomic clone containing the TCR Dβ1 region and sequenced from approximately 2 kb upstream of Dβ1 through to 2 kb downstream of Dβ1. The sequence of our clone is identical to the sequence in the Genbank database (AE000665). DNA sequence alignment between the human and murine β chain loci showed a region of 72% identity spanning from 350 bp upstream through 150 bp downstream of Dβ1 (Fig 2). A similar conserved region was also identified proximal to the Dβ2 region. Other regions of the TCRβ locus share 40% to 60% identity between mouse and human, suggesting this highly conserved region proximal to Dβ1 contains functionally important sequences, potentially including regulatory elements for germline Dβ1 transcription.
Initiation of Dβ1 transcription from multiple sites.
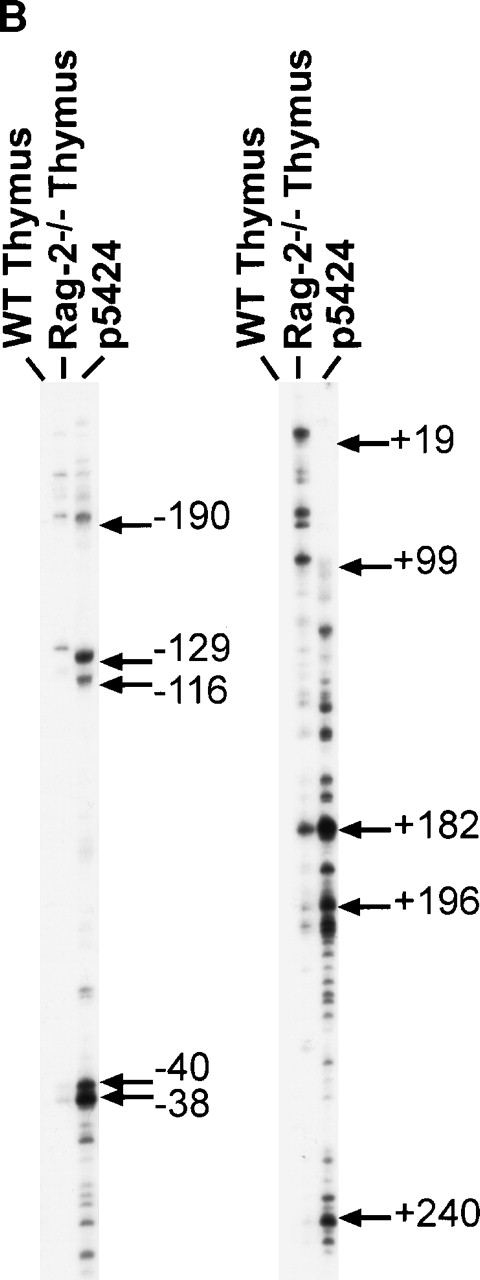
The heterogeneous pattern of Dβ1 germline transcripts on Northern analysis (Fig 1B), coupled with our observation that the 5′ end of the germline transcript cDNA clones was quite variable (Fig2), suggested that transcriptional initiation occurs at multiple sites. We performed primer extension analysis using primers designed to detect transcripts initiating either upstream or downstream of Dβ1 (Fig 3A and B). Multiple start sites were detected in both Rag-2–deficient thymocytes and in the p5424 cell line. As expected, primer extension products were not seen in normal thymus RNA, as the majority of cells have already undergone DJ rearrangement, which deletes the segments corresponding to the primers. There was substantial overlap in the bands observed in the Rag-2–deficient thymocytes and p5424 cells, although some differences were apparent. In particular, there was a bias toward downstream initiation sites in the p5424 cells, consistent with the finding of somewhat shorter Dβ1 transcripts on Northern blots in these cells compared with Rag-2−/− thymocytes (Fig 2). To exclude artifacts due to incomplete processivity of reverse transcriptase, we also performed RNAse protection assays using a probe, which spanned −202 to +285 bp (Fig 3). The protection assay indicated the presence of major transcription start sites at approximately +32, which was not prominent on the primer extension assay, probably due to the distance from the DS primer. Protection of the full-length probe was also noted in pro-T cells, indicating transcriptional start sites upstream of −202. A protected band seen in normal thymus likely represents transcripts from DJ rearranged alleles, also initiating upstream of −202. Taken together, these results indicate that germline Dβ1 transcription initiates over a broad region upstream and downstream of the Dβ1 element, and suggests that cis-regulatory elements may be relatively diffuse.
Analysis of Dβ1 germline transcriptional start sites. (A) Location of upstream (US) or downstream (DS) primers used in the primer extension assay, as well as probe used for ribonuclease protection assay. (B) Primer extension analysis of RNA from either normal or Rag-2–deficient thymocytes or the p5424 pro-T cell line using US (left-hand gel) or DS (right-hand gel) primers. The positions of several of the major bands are indicated by arrows, with numbering relative to the first base in the Dβ1 element. (C) Ribonuclease protection assay for Dβ1 germline transcripts. A major transcriptional start site is indicated, which maps to approximately +32. The full-length protected fragment (FL) is 478 bases long and indicates the presence of transcripts initiating upstream of −202. The undigested probe is 578 bases long. Marker sizes are as indicated.
Analysis of Dβ1 germline transcriptional start sites. (A) Location of upstream (US) or downstream (DS) primers used in the primer extension assay, as well as probe used for ribonuclease protection assay. (B) Primer extension analysis of RNA from either normal or Rag-2–deficient thymocytes or the p5424 pro-T cell line using US (left-hand gel) or DS (right-hand gel) primers. The positions of several of the major bands are indicated by arrows, with numbering relative to the first base in the Dβ1 element. (C) Ribonuclease protection assay for Dβ1 germline transcripts. A major transcriptional start site is indicated, which maps to approximately +32. The full-length protected fragment (FL) is 478 bases long and indicates the presence of transcripts initiating upstream of −202. The undigested probe is 578 bases long. Marker sizes are as indicated.
A functional promoter for Dβ1 germline transcription.
The presence of germline transcripts initiating in the proximity of Dβ1 indicates the presence of nearby promoter elements. To characterize these elements, we used a reporter gene assay in the p5424 cell line, which expresses high levels of endogenous Dβ1 germline transcripts (Fig 1B), and therefore contains the necessary factors to transcribe from the Dβ1 promoter. We identified promoter activity within a genomic fragment extending from 2184 bp upstream through 151 bp downstream of Dβ1 (Fig 4A and B), which encompasses many of the transcriptional initiation sites for Dβ1 germline transcripts. This construct had low-level reporter expression in the absence of enhancer sequences, which was not significantly greater than the vector- only control. However, this activity was consistently twofold to threefold higher than constructs containing the same fragment in the reverse orientation. The Eβ enhancer, which activates transcription of rearranged TCRβ genes from promoters located upstream of the V regions,33,38 is also required for TCRβ gene rearrangement and germline transcription in vivo.23 24 When Eβ was included in the reporter construct containing Dβ1 genomic fragment, reporter gene expression was increased 10-fold over the promoter alone. Orientation-dependent transcription from the Dβ1 fragment was maintained under these circumstances. Interestingly, the enhancement of Dβ1 promoter activity by Eβ was specific, as the SV40 enhancer had no effect, even though the SV40 enhancer was functional in p5424 cells in the context of the SV40 promoter (Fig 4B).
Analysis of Dβ1 promoter activity in reporter gene constructs in p5424 pro-T cells. (A) Depiction of the D-J-Cβ1 region and the ▵2184 genomic fragment used in the reporter assay, which corresponds to −2184 to +151 relative to the first base of Dβ1. (B) Orientation-specific promoter activity of the ▵2184 fragment in the presence or absence of the Eβ or SV40 (Esv) enhancers. (C) Promoter activity of nested deletions from the 5′ end of the ▵2184 fragment in the presence of the Eβ enhancer. (D) Promoter activity in constructs containing 3′ deletions of the ▵524 genomic fragment, subfragments corresponding to −147 to +6 and −303 to −147, and site-directed mutations of putative Ikaros/Lyf-1 (m35) and GATA (m74) transcription factor sites. Constructs containing genomic fragments in the reverse orientation are indicated by a left-hand arrow. Luciferase activity measured 24 hours after transfection was corrected for transfection efficiency and expressed as a percentage of the pGL-3 control SV40 promoter/enhancer construct. The mean and standard error for four independent transfections are given.
Analysis of Dβ1 promoter activity in reporter gene constructs in p5424 pro-T cells. (A) Depiction of the D-J-Cβ1 region and the ▵2184 genomic fragment used in the reporter assay, which corresponds to −2184 to +151 relative to the first base of Dβ1. (B) Orientation-specific promoter activity of the ▵2184 fragment in the presence or absence of the Eβ or SV40 (Esv) enhancers. (C) Promoter activity of nested deletions from the 5′ end of the ▵2184 fragment in the presence of the Eβ enhancer. (D) Promoter activity in constructs containing 3′ deletions of the ▵524 genomic fragment, subfragments corresponding to −147 to +6 and −303 to −147, and site-directed mutations of putative Ikaros/Lyf-1 (m35) and GATA (m74) transcription factor sites. Constructs containing genomic fragments in the reverse orientation are indicated by a left-hand arrow. Luciferase activity measured 24 hours after transfection was corrected for transfection efficiency and expressed as a percentage of the pGL-3 control SV40 promoter/enhancer construct. The mean and standard error for four independent transfections are given.
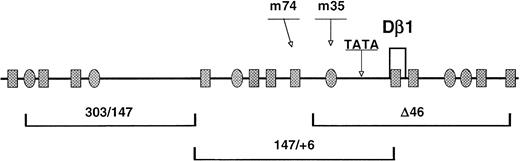
These findings indicate that the −2184 to +151 Dβ1 genomic fragment contains a promoter, which is functional in the context of Eβ. To localize this activity, we made a series of 5′ nested deletions and tested promoter activity in the presence of Eβ (Fig4C). Promoter activity was retained in a construct (Δ46) containing a 200-bp segment spanning from −46 to +151 relative to Dβ1. Similar findings were obtained using constructs lacking Eβ, with the overall level of transcription being substantially less (not shown). A series of 3′ deletions was also made, based on the Δ524 construct containing Eβ. Truncation of the construct within the Dβ1 region (6Δ) resulted in a significant increase in transcription, suggesting there may be inhibitory sequences within the +6 to 151 interval. Deletion of an additional 150 bp (147Δ) returned the activity to approximately the same level as the Δ524 construct, indicating that functional promoter elements lie both upstream and downstream of −147. Further, 3′ deletion to −303 abolished promoter activity. Thus, positive regulatory elements for Dβ1 germline transcription are contained within the region −303 to +6. Because some of the transcriptional start sites were 3′ of the region analyzed in these studies, it remains possible that additional regulatory elements lie beyond +151. Constructs representing separate segments of the Δ524 construct were then compared for reporter activity. Each of the fragments representing −303 to −147, −147 to +6, and −46 to +151 were sufficient to drive luciferase expression independently in the context of Eβ. In contrast, a fragment representing −524 to −303 had no activity. These data indicate that the Dβ1 promoter functions in the context of Eβ and contains several spatially distinct elements, which are sufficient for transcriptional initiation. Taken with the finding that transcriptional initiation is heterogeneous in vivo, these findings are consistent with diffuse or TATA-independent transcriptional regulation.
The Dβ1 promoter region defined by the reporter analysis correlates with the area of sequence conservation between mouse and human. The sequence from this region (−350 to +150) was analyzed for sites characteristic of known transcription factors using the TFSEARCH utility and TRANSFAC database (Fig5).39 A consensus TATA sequence is within the 5′ RS flanking Dβ1. However, this element is not conserved in the homologous human sequence, and the 303/147 construct in which this element was absent had high-level transcriptional activity (Fig 4D), indicating that the TATA element is not a required component for Dβ1 promoter activity. Among potential transcription factor sites, the most notable with respect to regulation of hematopoietic cells are multiple sites for transcription factors of the GATA and Ikaros families, which play critical roles in lymphoid development.40-44 To determine the functional significance of putative Ikaros/Lyf-1 and GATA binding sites for Dβ1 promoter activity, consensus sites for each of these factors within the 147/+6 construct (Fig 5) were modified by site-directed mutagenesis. The binding sequence for Ikaros/Lyf-1 at −35 (m35 construct) was mutated from ATGGG AGGG to ATGTCAGGG, while the GATA binding sequence at −74 (m74 construct) was mutated from CCAGATAAGC to CCATCCGAGC. Each of these mutations dramatically reduced reporter gene expression (Fig 4), suggesting that transcription factors binding to these putative Ikaros/Lyf-1 and GATA sites contribute to regulation of germline transcription from the Dβ1 promoter.
Analysis of potential transcription factor binding sites within the functional Dβ1 promoter region. The sequence corresponding to −350 to +150 relative to Dβ1 was analyzed using the TFSEARCH program.39 The results were notable for a number of potential binding sites for Ikaros/Lyf-1 (shaded ovals) and GATA (shaded rectangles) transcription factors. Site-directed mutations of two of these sites (corresponding to m35 and m74 constructs in Fig 4) are indicated. In addition, a potential TATA motif is shown. The genomic fragments exhibiting promoter activity in the context of Eβ are indicated beneath.
Analysis of potential transcription factor binding sites within the functional Dβ1 promoter region. The sequence corresponding to −350 to +150 relative to Dβ1 was analyzed using the TFSEARCH program.39 The results were notable for a number of potential binding sites for Ikaros/Lyf-1 (shaded ovals) and GATA (shaded rectangles) transcription factors. Site-directed mutations of two of these sites (corresponding to m35 and m74 constructs in Fig 4) are indicated. In addition, a potential TATA motif is shown. The genomic fragments exhibiting promoter activity in the context of Eβ are indicated beneath.
Stage- and lineage-specificity of Dβ1 promoter activity.
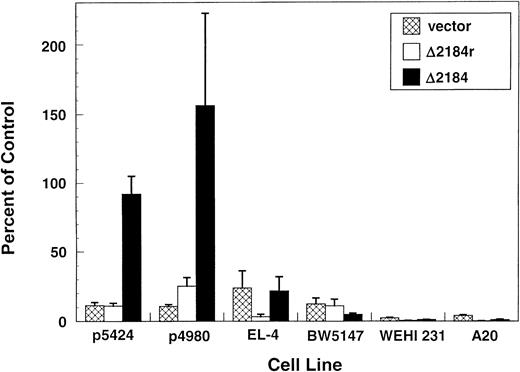
Germline transcription of TCRβ occurs at high levels specifically in early thymocytes. We therefore examined the stage- and lineage-specificity of the Dβ1 promoter element in the context of Eβ by comparing activity in T- and B-cell lines representing different stages of development (Fig 6). For each cell line, activity of the following constructs was compared: luciferase vector only, Dβ1 promoter (−2184 to +151), Dβ1 promoter reverse orientation. In addition, a control construct containing the SV40 promoter/enhancer was used for comparison of activity between cell lines. Relative to the SV40 promoter/enhancer, the activity of the Dβ1 promoter was greatest in the two Rag-deficient progenitor T-cell lines, p5424 and p4980, (92% and 156%, respectively) and significantly less active in the EL-4 and BW5147 lines, which represent more mature stages of the T-cell lineage (22% and 4.8%, respectively). Essentially no transcriptional activity was observed in the B-cell lines WEHI 231 or A20. Transfection of T and B lines with the Δ175 and the Δ46 constructs showed very similar results as those obtained with the longer genomic fragment (data not shown). These results indicate that in the context of the Eβ, the Dβ1 promoter exhibits strict T- versus B-lineage specificity, and relative specificity for early as opposed to mature T cells, similar to the pattern observed in vivo.
Lineage- and stage-specific activity of the Dβ1 promoter. Luciferase activity in lymphoid cell lines is shown for the luciferase vector alone and for the ▵2184 (−2184 to +151) Dβ1 genomic fragment in the reverse or forward orientation in constructs containing Eβ. p5424 and p4980 are Rag-deficient pro-T cell lines; EL-4 and BW5147 are characteristic of more mature stages of the T-cell lineage; WEHI 231 and A20 are B-cell lines. Results are expressed as percentage of the activity of the pGL-3 control vector containing SV40 promoter and enhancer as in Fig 4. Mean and standard error for four independent transfections are given. Differences in promoter activity between the p5424 or p4980 pro-T lines and each of the other T- and B-cell lines were statistically significant (P < .05).
Lineage- and stage-specific activity of the Dβ1 promoter. Luciferase activity in lymphoid cell lines is shown for the luciferase vector alone and for the ▵2184 (−2184 to +151) Dβ1 genomic fragment in the reverse or forward orientation in constructs containing Eβ. p5424 and p4980 are Rag-deficient pro-T cell lines; EL-4 and BW5147 are characteristic of more mature stages of the T-cell lineage; WEHI 231 and A20 are B-cell lines. Results are expressed as percentage of the activity of the pGL-3 control vector containing SV40 promoter and enhancer as in Fig 4. Mean and standard error for four independent transfections are given. Differences in promoter activity between the p5424 or p4980 pro-T lines and each of the other T- and B-cell lines were statistically significant (P < .05).
DISCUSSION
Transcription of unrearranged antigen receptor loci invariably occurs before V(D)J rearrangement, exhibiting a pattern of lineage and stage-specific regulation, which parallels this key developmental process.3,8,10,11,13,14,16 Given that both transcription17 and V(D)J recombination1,8 9 are regulated by changes in chromatin structure, understanding the regulation of germline antigen receptor transcription is likely to provide important insights into key steps in lymphoid development. For the TCRβ locus, germline transcription is known to occur at the earliest recognized stage in thymic development; however, little is known regarding the regulation of this process. As an initial approach, we have determined the structure and transcriptional start sites of germline transcripts originating near Dβ1 in pro-T cells. In addition, we have characterized a functional promoter, which interacts with the Eβ enhancer to direct germline TCRβ transcription in a manner recapitulating the lineage- and stage-specificity of this process observed in vivo. Further studies of this cis-acting element should lead to an understanding of DNA binding factors, which control transcription, and may also provide insight into how accessibility of this locus to V(D)J recombination is induced.
Dβ1 germline transcript cDNAs exhibited marked heterogeneity in size (Fig 2). Alternative splicing at either Jβ1.1 or Jβ1.2 and inclusion or exclusion of the Cβ0 exon may generate transcripts varying by up to 212 bp in size. However, the size range of Dβ1 transcripts seen in Rag-2 thymocyte RNA was much larger than this (Fig1B), suggesting that a third variable, heterogeneity in the position of transcriptional initiation is in large measure responsible for the size differences observed. The major transcription start sites mapped near +32 are consistent with the participation of a TATA element located in the 5′ RS flanking Dβ1. However, the presence of other transcripts, including those initiating upstream of Dβ1 indicate that elements in addition to the TATA motif determine sites of transcriptional initiation. This interpretation is supported by our reporter gene analysis, which indicates that promoter activity is distributed throughout the conserved region encompassing Dβ1, and that several subfragments, including one lacking the TATA motif, were sufficient to drive reporter gene transcription in the context of Eβ. Sikes et al45 have recently described promoter activity from a genomic segment containing Dβ1, corresponding to −377 to +70 in our scheme. In their study, the TATA motif was required for maximal promoter activity in pre-T cell lines using constructs that lacked Eβ, suggesting that the enhancer may influence the relative strengths of individual promoter elements in the Dβ1 region.
It is striking that the functional Dβ1 promoter region contains at least 11 potential binding sites for GATA family transcription factors and six potential sites for Ikaros factors (Fig 5). Among GATA factors, GATA-2 is required for development of all hematopoietic lineages,46 while GATA-3 regulates several T-cell–specific genes,41 and is required for T-cell development beyond the pro-thymocyte stage.42 The Ikaros gene, which encodes a family of transcription factors generated by alternative mRNA splicing, contributes to the regulation of a number of genes involved in lymphoid development, and targeted mutation experiments confirm that Ikaros factors play a crucial role in the developmental program of lymphocytes.43,44 Our data indicate that consensus binding sites for Ikaros and GATA factors located at −35 and −74 are important for Dβ1 promoter activity. Sikes et al45have also demonstrated by mutation analysis that GATA sites at −74 and −104 contribute to Dβ1 promoter activity in pre-T cell lines. Taken together, these findings support the notion that GATA and Ikaros factors contribute to the regulation of Dβ1 germline transcription.
The Dβ1 promoter can be compared with the promoter/enhancer, which directs germline transcription from the DQ52 segment of the IgH locus in early B cells.26,27,47 The location of these promoters is homologous, in that each resides within a region of mouse to human sequence conservation located upstream of the D element closest to the J cluster. As the D-J-C complex is reiterated in the TCRβ locus, this would imply that an additional promoter is located in an area of sequence conservation near Dβ2, a notion supported by our isolation of cDNA clones representing Dβ2 germline transcripts. Both the Dβ1 and DQ52 promoters interact with the respective enhancers, Eβ and Eμ.27 However, our data indicate that the Dβ1 promoter was highly dependent on Eβ and could not be replaced by a heterologous SV40 enhancer, even though this enhancer was active in the context of the SV40 promoter in p5424 cells (Fig 4B). In contrast, the DQ52 element contains associated enhancer activity. This difference in enhancer-dependence could explain why targeted deletion of Eβ produces a complete block in TCRβ rearrangement,23,24while deletion of Eμ has only a partial effect on inhibiting IgH rearrangement.21,22 The DQ52 promoter/enhancer exhibits lineage- and stage-specificity, being preferentially active in B versus T cells, and in early versus mature B cells, suggesting that this element may contribute to the developmental regulation of germline transcription. In our study, the Dβ1 promoter was only active in the context of Eβ; thus, whether this element contributes in an enhancer-independent way to stage and lineage-specificity could not be directly determined. However, given that Eβ is active in mature T cells,33,38 our results suggest that the Dβ1 promoter exhibits a relative preference for early T cells, similar to the pattern of Dβ1 transcription observed in vivo. Sikes et al45 found that when placed in the context of the Eμ enhancer, the Dβ1 promoter was also active in B-lineage cells. This result is consistent with what has been observed with in vivo replacement of Eβ with Eμ in TCRβ minigenes or by gene targeting, where transcription of the TCRβ locus also occurred in B cells.23,48 49 While these studies suggest lineage plasticity in the control of TCRβ germline transcription by the Dβ1 promoter, our data nevertheless indicate this combination of regulatory elements in large measure recapitulates the regulatory pattern for germline TCRβ germline transcription observed in vivo.
The Eβ enhancer and other enhancers within antigen receptor loci are required for efficient V(D)J recombination.8,18-25Relatively few studies have directly addressed how promoter elements for germline transcription participate in regulating V(D)J recombination. In mice transgenic for a chicken Igλ minigene, rearrangement of the substrate in B cells was impaired by mutation of either enhancer or promoter elements.50 In the TCRα locus, targeted deletion of the TEA element, which directs germline transcription at the 5′ end of the Jα cluster, prevented rearrangement to the most 5′ Jα segments. While these studies suggest that germline transcription may be necessary for V(D)J rearrangement, transcription itself is probably not sufficient to control rearrangement. Experiments replacing the Eβ with Eμ in TCRβ minigenes or by gene targeting resulted in redirection of TCRβ transcription in B cells without inducing rearrangement in that lineage.23,49 Moreover, in the Igκ locus, a cis element critical for rearrangement was identified by gene targeting studies, which had no effect on germline transcription.51 Taken together, these studies suggest that locus accessibility to V(D)J recombination is regulated both by germline transcription and by additional elements.
Germline transcription of the TCRβ and IgH loci are among the first lineage-specific differentiation steps in T and B cells, respectively.12,30 52 Our identification of promoter elements directing TCRβ germline transcription in pro-T cells points to important similarities with the cis-acting elements regulating activation of the IgH locus in pro-B cells. Identification of transcriptional regulatory factors interacting with these control elements in T- versus B-cell progenitors may offer important insights into how these lineages are specified.
ACKNOWLEDGMENT
The authors thank Jianzu Chen and Steve Collins for gifts of materials and Barry Sleckman and Steve Collins for helpful discussions. The authors are grateful to Gene Oltz for sharing unpublished data and to Lee Rowen and Leroy Hood for sharing TCRβ sequences before publication. Chris Wilson and Steve Collins provided critical comments on the manuscript.
Supported in part by National Institutes of Health Hematology Training Grant No. HL07093, the American Heart Association, Washington Affiliate, and by the Research Resources Program for Medical Schools from the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Dennis M. Willerford, MD, Puget Sound Blood Center, 921 Terry Ave, Seattle, WA 98104.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal