BEGINNING AT THE BEDSIDE: A CASE OF T-CELL LEUKEMIA
THIS STORY BEGINS with a seminal case report describing a 16-year-old boy who presented with acute lymphoblastic leukemia (ALL).1 He had an elevated white blood cell (WBC) count, thymic enlargement, and leukemic cells with an early T-cell phenotype (CD2 and CD7 positive; CD3, CD4 and CD8 negative). Standard chemotherapeutic agents failed to induce remission, and treatment commenced with the adenosine deaminase inhibitor 2′-deoxycoformycin. Over the next 7 days, the leukemic cells underwent an abrupt and dramatic transformation. The lymphoid leukemic cells acquired morphological features of myeloid cells, gained myeloid antigens, and lost CD7 expression. Unfortunately, the patient died of extensive tissue infiltration with cells of promyelocyte morphology. This “stem cell leukemia” phenotype was also evident in vitro because exposure of the leukemic cells to 2′-deoxycoformycin stimulated multi-lineage differentiation. Before and after the phenotypic conversion, the leukemic cells carried a translocation between chromosome 1p32-33 and the T-cell receptor (TCR) α/δ locus on chromosome 14q11.
The investigators established a cell line that carried the same chromosomal translocation and recapitulated the stem cell phenotype during in vitro culture.2 They further showed that other cases of ALL, with a similar cell surface phenotype, did not show this specific cytogenetic abnormality.3 Although rearrangements of chromosome 1p32-33 are now recognized to occur frequently in T-cell ALL, there have been no reports of these leukemias being treated with similar agents, nor reports of such a striking morphological conversion. Based on what is now known about the molecular genetics of this event, it seems reasonable to speculate that other cases may respond in a similar manner and that ultimately this might be exploited for therapeutic advantage. However, there is still no clear explanation for the mechanism by which 2′-deoxycoformycin induced this dramatic change.
At that time, the recent successes in identifying genes involved in B-cell tumors as a result of their translocation into the Ig gene loci inspired a search for the gene responsible for the stem cell phenotype. By analogy with B-cell tumors, it seemed likely that the responsible gene was located on chromosome 1 and, as a result of the translocation into the TCR locus, it was aberrantly activated when transcription of the TCR locus was attempted. However, analysis of the translocation first required characterization of the TCR α and δ loci and recognition that the α locus contained the δ locus embedded within it.4-8 Once probes to the human TCR δ locus became available, the translocation breakpoint was isolated and a new gene on chromosome 1 was identified.9,10 The name “stem cell leukemia” gene or SCL was assigned, a name that has turned out to be remarkably appropriate given the essential function of this gene in normal hematopoietic stem cells (see below). The same gene was independently identified by several other groups.11-13
RECONSTRUCTING THE CRIME: A CASE OF MISTAKEN IDENTITY
The t(1;14) translocation was the first example of what has since transpired to be a common theme in a significant proportion of T-ALL cases; the aberrant activity of recombinase enzymes with abnormal expression of SCL as a result.
The leukemic cells were captured at a very early stage of T-cell development and frozen while in the process of rearranging/deleting both their TCR δ loci. On one chromosome 14 the cells were caught in the process of attempting to delete the entire TCR δ locus.14 Deletion of the TCR δ locus is a necessary prerequisite for TCR α rearrangement15 and although this transient intermediate state had been predicted to occur (ie, δ deleted but incomplete α rearrangement),16 it had not actually been previously observed.
The second chromosome 14 was the site of the translocation event.9 Here the recombinase enzymes had successfully juxtaposed two TCR δ diversity (or D region) genes and completed the required excision of the intervening DNA. However, during or immediately after this event, chromosome 1 had been inadvertently involved in this otherwise normal recombination process. The presence of additional, nontemplated nucleotides provided circumstantial evidence implicating enzymes such as terminal deoxytidyl transferase (TDT) as also being active at that moment.
There are mitigating circumstances that in part explain the “error” on the part of the recombinase enzymes. The recombinases normally recognize specific DNA sequences that flank the segments of DNA to be rearranged. These serve as specific signals that allow the enzymes to correctly identify and juxtapose segments of DNA in the process of assembling a functional TCR (or Ig) gene. Very similar sequences are present in several regions of the human SCL gene, but intriguingly are not seen in equivalent regions of the mouse gene.17-19 Their normal function in the human SCL gene, if any, is unknown. However, the presence of “signal-sequences” in the human SCL gene does provide an explanation for the case of mistaken identity in this chromosomal translocation. There is a second reason that perhaps also partly explains this event. It is now known that SCL is crucially important at the earliest stages of hematopoietic stem cell development and differentiation. Therefore, the SCL gene was likely to be hypomethylated in an “open,” chromatin configuration, thus making it vulnerable to the recombinase enzymes.20
This patient’s translocation disrupted the 3′ untranslated region of the SCL gene, preserving the SCL protein intact and thus allowing production of the normal protein to be inappropriately regulated by elements within the TCR δ locus. As a consequence, SCL expression was readily detected within the leukemic T cells while SCL is not detected in the vast majority of normal thymocytes.10 21-24
Other investigators had also noted rare cases of T-ALL with translocations between chromosomes 1 and 14 occurring in about 3% of cases,25-27 and Bernard et al13 went on to show that SCL was also dysregulated in some of these cases. At least 10 chromosomal translocations involving SCL have since been defined, with the majority of these occurring into the TCR α/δ locus.12,28-31 However, translocations involving SCL and chromosomes 3, 5, and 7 (the TCR β locus) have also been documented.32-34 These translocations all have in common the errant expression of SCL in a T-cell environment. In addition, the fact that breakpoints within the SCL gene lie at the site of signal sequences strongly suggests that the recombinase enzyme complex is again likely to be the primary culprit. Because of the frequent involvement of SCL in leukemias with a T-cell phenotype, the names TCL-5 and TAL-1 were also proposed.11,12 30
VARIATIONS ON THIS THEME: SCL DYSREGULATED BY UPSTREAM DELETIONS RESULTING FROM ABERRANT RECOMBINASE ACTIVITY
In addition to chromosomal translocations, there is a second mechanism by which the SCL gene can be disrupted and aberrantly activated in leukemic T lymphocytes. This involves deletions of approximately 90 kb of DNA from chromosome 1 that brings SCL under the control of the upstream, ubiquitously expressed SIL (for SCL interrupting locus) gene.35-38 Again, recombinase enzymes are the primary suspect in the genesis of the SIL/SCL deletions (also known as Tald): the presence of specific recombination signal sequences in both the SIL and SCL genes ensure that a limited number of SIL/SCL deletional events are seen recurrently in independent leukemic samples. Although this “limited number” could theoretically be as high as 18, with 3 recombination sites observed within the SIL gene and at least 6 different recombination sites observed within SCL, only two types of deletion are commonly observed. The SIL/SCL deletional event in T-cell leukemia occurs with a frequency estimated to be between 6% and 26% of cases.35,39-43 The generation of unique sequences at the site of the SIL/SCL junction can serve as a useful leukemia-specific and patient-specific marker for polymerase chain reaction (PCR)-based techniques to detect residual leukemic cells.44 More recently it was proposed that SCL may be aberrantly expressed in up to 60% of childhood T-ALL samples in many cases without detectable rearrangements of the gene.45 This potentially provided an explanation for earlier studies that failed to identify a significant clinical, phenotypic, or prognostic subgroup of T-ALL associated with SCL rearrangement. However, it has been suggested that SCL expression observed in the absence of SCL rearrangements was probably caused by contamination of the leukemic blasts with normal hematopoietic cells.46
Despite the lack of a clearly distinct clinical subgroup of T-ALL associated with SCL rearrangement, there is one characteristic that is highly correlated with the presence of the SIL/SCL deletion; deletion of one or both TCR δ gene(s). Therefore, SCL rearrangements are observed predominantly, but not exclusively, in CD3 positive, TCR α/β positive T-ALL.41,42,47 The striking concordance with deletion of the TCR δ locus, a prerequisite for TCR α rearrangement, raises the possibility that SCL may even play a role in regulating deletion of the TCR δ loci and in commitment to the TCR α/β T-cell lineage.20 47
Although the structural alterations of the SCL gene are seen in T-ALL, they are not seen in B-lineage ALL, acute myeloid leukemia (AML), T-cell non-Hodgkin’s lymphomas, or in a range of other solid tumors.43,44,48 Although expression of SCL has been observed in samples of AML and may even correlate with poor outcome,49 this probably reflects malignant transformation of myeloid progenitor/stem cells that normally express SCL.23,24 50
ILLICIT LIAISONS: HOW DOES SCL CONTRIBUTE TO T-ALL?
Although the genetic lesions outlined above result in ectopic expression of SCL in a T-cell environment, the precise mechanism(s) by which this transcription factor contributes to leukemia development remains uncertain.
There is clear evidence that SCL can provide a proliferative advantage to cells in a number of different contexts. Expression of antisense SCL in a multipotent human cell line caused a significant decrease in cellular proliferation, decreased progression of cells through the cell cycle, and resulted in a 50-fold decrease in self-renewal potential, thus suggesting that SCL normally influences these functions.51 Similarly, aberrant SCL expression in a T-cell environment increased clonogenic ability.52 In murine myeloid cells, retrovirally enforced SCL expression allowed cells to “escape” differentiation induction and overcome growth-factor–induced suppression of clonogenicity.53-55Likewise, SCL expression as a consequence of an insertion of a retroviral-like element, an intra-cisternal A particle, conferred a growth advantage on myeloid cells and resistance to differentiation-inducing agents.56,57 Thus, in these various contexts enforced SCL expression stimulates proliferation and opposes differentiation. Within a different cellular context, erythroid cells, enforced SCL expression also enhances cell proliferation, although in this setting differentiation is stimulated.58-60 In addition to its action to stimulate proliferation in a variety of contexts, SCL expression delayed apoptosis55,61,62; again, this action may depend on the cell type in which SCL is acting.55 Therefore, as with other oncogenes, it is likely that SCL contributes to leukemogenesis by multiple mechanisms that include an increase in proliferation, cell cycling, and self-renewal potential with a decrease in cell death.
These actions of SCL translate into increased leukemia development and subsequent death in a variety of animal models. In some transgenic models expression of SCL alone did not cause leukemia to occur, presumably as a consequence of the regulatory elements that were used to direct SCL expression.63-65 However, several groups have demonstrated cooperation in vivo between SCL and a variety of additional genes including ABL,52 casein kinase II,66 p53,62,67 and RAS.67 Whether any of these cooperating genes contribute to human T-ALL is unclear. However, SCL also collaborates with the LIM domain proteins LMO-1 and LMO-2 to generate tumors.64,65 This interaction is particularly intriguing because SCL, LMO-1, and LMO-2 are implicated in human T-ALL. However, all the mouse models suggest that additional events are still needed, and in the context of SCL and the LIM domain proteins, defects in the DNA mismatch repair gene MSH2 are probably important.68
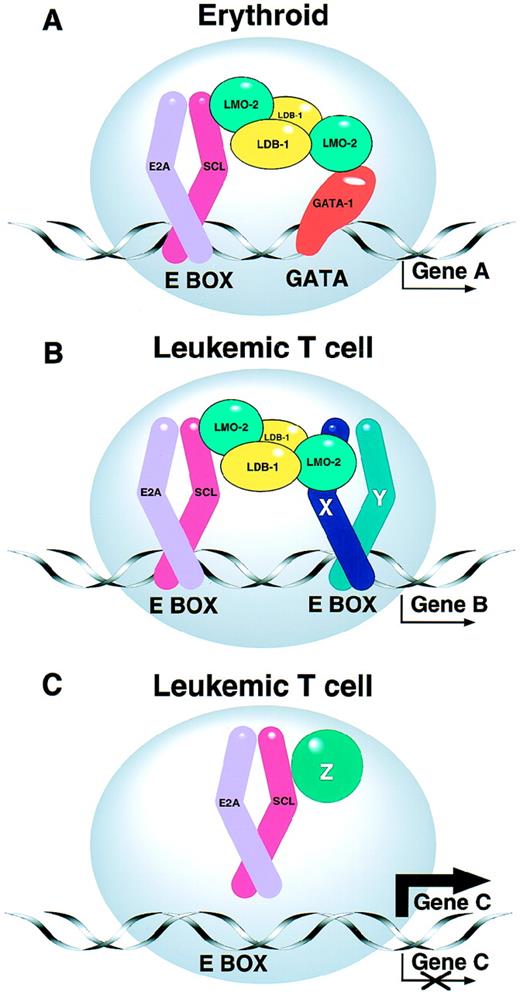
The biochemical basis for the tumorigenic cooperation described above remains unclear. However, it is known that SCL interacts with the products of the E2A gene69-71 and related proteins72 to form heterodimeric DNA-binding complexes. The heterodimer can form part of a larger protein complex in which LMO-2 acts as a molecular bridge between the zinc finger transcription factor GATA-1 and the SCL/E2A heterodimer (Fig1).73,74 Using a chromatin immuno-precipitation approach, Cohen-Kaminsky et al75 have recently identified a novel gene of unknown function that is expressed in erythroid cells and that is directly regulated by a complex containing SCL and GATA-1. Although these interactions have been characterized primarily in erythroid cells, where SCL is normally expressed, a large protein complex occurs in T cells, but with altered DNA binding specificity.76 The existence of such multi-component protein complexes is an emerging theme in the area of transcriptional regulation. The constituents of the complex are likely to vary in different cell types, differentiation states, or cell cycle stages, perhaps explaining in part the multiplicity of functions ascribed to SCL and other transcription factors.
(A) SCL forms part of a multiprotein complex. SCL normally interacts with other transcription factors in erythroid cells to form a multiprotein complex that contains transcription factors such as GATA-1, LMO-2, LDB-1, and E2A proteins.74 As a result, “Gene A” is appropriately regulated. The precise stoichiometry of the complex is not known. (B and C) Models for the leukemogenic effect of SCL expression in T cells. (B) The leukemogenic effect of ectopic SCL expression in T cells may involve aberrant activation of target genes (Gene B), perhaps by a complex involving additional bHLH proteins (eg, X and Y).76 (C) Alternatively ectopic SCL expression may sequester E2A and/or other T-cell proteins (eg, Z), thereby upregulating or downregulating expression of their normal target genes (Gene C).
(A) SCL forms part of a multiprotein complex. SCL normally interacts with other transcription factors in erythroid cells to form a multiprotein complex that contains transcription factors such as GATA-1, LMO-2, LDB-1, and E2A proteins.74 As a result, “Gene A” is appropriately regulated. The precise stoichiometry of the complex is not known. (B and C) Models for the leukemogenic effect of SCL expression in T cells. (B) The leukemogenic effect of ectopic SCL expression in T cells may involve aberrant activation of target genes (Gene B), perhaps by a complex involving additional bHLH proteins (eg, X and Y).76 (C) Alternatively ectopic SCL expression may sequester E2A and/or other T-cell proteins (eg, Z), thereby upregulating or downregulating expression of their normal target genes (Gene C).
The leukemogenic effect of SCL expression in T cells may reflect either activation of target genes that should normally be silent, or alternatively the sequestration of other components of the multi-protein complex (Fig 1). Several lines of circumstantial evidence support the latter view: (1) SCL can sequester E2A proteins, thereby inhibiting transactivation of genes that normally require E2A proteins.77-79 (2) Mice lacking a functional E2A gene develop T-cell tumors.80 (3) As mentioned above, casein kinase II cooperates with SCL to hasten T-cell tumors,66and casein kinase II is also known to inhibit activity of proteins derived from the E2A gene.81 (4) Mice carrying an SCL transgene lacking its transactivation domain still develop lymphoid tumors65 (although it remains possible that SCL/E2A heterodimers aberrantly transactivate critical target genes via the E2A transactivation domain).
If SCL does act as a molecular sink in T-ALL, one would predict that this property would not be unique to SCL and that other transcription factors capable of interacting with E2A proteins would also be leukemogenic if ectopically expressed in T cells. The proteins LYL-1 and TAL-2 proteins share this characteristic and are indeed involved in translocations in T-ALL, but only rarely.82 Therefore, the particular prevalence of SCL rearrangements in T-ALL probably reflects the presence of the specific signal sequences in the human SCL gene that are recognized by the recombinase complex and dictate the site of translocation and rearrangement events.
JEKYLL AND HYDE: SCL NORMALLY FUNCTIONS AS A CRITICAL REGULATOR OF HEMATOPOIESIS
Given the unusual phenotype of the leukemic cells from which SCL was first identified, it seemed possible that the protein encoded by the SCL gene would also prove an important molecule in normal hematopoietic cells. The first evidence that this was the case came from analysis of the amino acid sequence for the predicted SCL protein.9 Two important domains within the protein were immediately apparent. One domain is a helix-loop-helix (HLH) region that is common to a large family of functionally important transcription factors. This region allows SCL to heterodimerize with E12 and E47, products of the E2A gene and themselves HLH transcription factors. In fact, the E2A gene [which is involved in the t(1;19) translocation in human B-ALL] was a founding member of the HLH family along with C-MYC [involved in the t(8;14) translocation of Burkitt’s lymphoma], MYO-D (an important regulator of muscle differentiation), and daughterless (aDrosophila gene involved in sex determination).83
The second conspicuous domain within the SCL protein is a basic region, which is also present in many other HLH proteins: the SCL/E2A heterodimer binds DNA using the basic domains present within these proteins.84 The site on the DNA to which the SCL/E2A heterodimer binds conforms to an “E-box” motif, which is also common to other proteins of the HLH class.83 85
A role for SCL in the regulation of normal hematopoiesis was suggested by its normal expression pattern; SCL is restricted to hematopoietic cells, endothelial cells, the central nervous system, and embryonic skeleton.23,86,87 Within the hematopoietic compartment, SCL is normally expressed within erythroid cells, mast cells, megakaryocytes, and progenitor/“stem” cells.10,21-24 50
The first clue to a normal function of the SCL protein came from antisense experiments showing that SCL regulates proliferation and self-renewal in a multipotent hematopoietic cell line.51 A positive role for SCL in erythroid differentiation was suggested by the observation that SCL mRNA levels increased during erythroid differentiation induced either by chemicals or erythropoietin,22,23,50,88-90 although a concurrent decrease in levels of SCL protein implies additional levels of control.91 A more direct role for SCL in erythroid differentiation was provided by gene delivery experiments where enforced expression of SCL enhanced erythroid differentiation of hematopoietic cell lines.58,90 Conversely, as cells differentiated along the myeloid/macrophage pathway, levels of SCL mRNA and protein decreased, becoming essentially undetectable.50,53,54,90,92 The decrease in SCL expression not only accompanies macrophage differentiation, but is also necessary because enforced SCL expression prevents macrophage differentiation from proceeding normally.53,54,90 These results in cell lines have recently been confirmed and extended using normal human CD34-positive “stem” cells. In these cells, retrovirally directed SCL expression enforces and hastens erythroid differentiation and also directs megakaryocytic differentiation, with less striking effects on the myeloid compartment.59 60
Dramatic confirmation that the SCL gene encodes a vital stem cell regulator came from studies in which SCL function was destroyed (Fig 2). This was achieved using gene targeting in embryonic stem (ES) cells to derive SCL-null mutant animals (“knock-out” mice). Mice lacking SCL function died at about 8.5 days of embryonic development (E8.5) with a complete failure of yolk sac hematopoiesis,93,94 suggesting that SCL is crucial for the generation of hematopoietic cells in the early embryo (“primitive hematopoiesis”). SCL plays a similar essential role in adult, “definitive” hematopoiesis. After injection of SCL-null ES cells into blastocysts, analysis of the resulting chimeric animals showed the complete failure of the SCL-null ES cells to give rise to hematopoietic cells in the adult animals, although the mutant ES cells did contribute to all other tissues examined.95,96Furthermore, these SCL-null ES cells failed to give rise to hematopoietic cells during in vitro culture and failed to express hematopoietic-specific genes.97 Taken together, these observations show that SCL plays a critical role in the generation and/or subsequent behavior of pluripotent hematopoietic stem cells. This role is “cell-autonomous,” and occurs within the stem cell itself. These results also predict that normal SCL-expressing progenitors should function as multipotent cells, displaying the ability to generate hematopoietic cells committed to multiple lineages. This prediction has recently been confirmed using a gene-targeting approach; a marker gene (Lac-Z) was introduced into the SCL locus, thereby allowing the isolation of SCL-expressing cells for functional analysis.98 These results confirm the status of SCL as a “master regulator” for hematopoiesis.99
SCL is required for development of hematopoietic stem cells. (A) Wild type (+/+) and SCL mutant (−/−) mouse embryos. Note the red-brown coloration of the normal embryo due to blood within the fetal liver and vessels. This is absent in the mutant which also has a runted appearance. (B) Histological section through the mouse yolk sac with blood islands (arrows) present in the wild-type and lacking in the SCL mutant yolk sac. (C) Histological section through the embryo showing hematopoietic cells in the paired dorsal aortae in the wild type (arrows) and the absence of these cells in the dorsal aortae of the SCL mutant (arrows). (Reprinted with permission from Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey RP, Metcalf D, Begley CG: Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. The Proceedings of the National Academy of Sciences, U.S.A., vol 92, p 7075, 1995. Copyright 1995 National Academy of Sciences, U.S.A.93)
SCL is required for development of hematopoietic stem cells. (A) Wild type (+/+) and SCL mutant (−/−) mouse embryos. Note the red-brown coloration of the normal embryo due to blood within the fetal liver and vessels. This is absent in the mutant which also has a runted appearance. (B) Histological section through the mouse yolk sac with blood islands (arrows) present in the wild-type and lacking in the SCL mutant yolk sac. (C) Histological section through the embryo showing hematopoietic cells in the paired dorsal aortae in the wild type (arrows) and the absence of these cells in the dorsal aortae of the SCL mutant (arrows). (Reprinted with permission from Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey RP, Metcalf D, Begley CG: Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. The Proceedings of the National Academy of Sciences, U.S.A., vol 92, p 7075, 1995. Copyright 1995 National Academy of Sciences, U.S.A.93)
The results with SCL are remarkably similar to those seen when the LMO-2 gene is destroyed. LMO-2 null animals also lack any detectable hematopoietic cells within the yolk sac and embryo, and die at about E8.5.100 Similarly, LMO-2 null ES cells fail to contribute to adult hematopoiesis,101 thus reinforcing the parallels between SCL and LMO-2. In addition to interacting in erythroid and some leukemic T cells, it therefore seems likely that SCL and LMO-2 are components of a critical multiprotein complex present in hematopoietic stem cells.
Thus, the phenotype of the initial stem cell leukemia implied SCL was capable of regulating hematopoietic differentiation in the leukemic cells. Pursuit of this early clue has unmasked a master gene which functions as a critical regulator of hematopoiesis.
HIDDEN TALENTS: SCL ALSO CONTROLS HEMANGIOBLAST FORMATION AND ENDOTHELIAL DEVELOPMENT
Given the crucial function of SCL within hematopoietic stem cells, the expression of SCL within endothelial cells raised the possibility that SCL could also function within an even more primitive cell, the putative common hematopoietic and endothelial precursor, the hemangioblast. There is a long-recognized, close relationship between the development of blood and endothelium, suggesting that they may well share a common precursor. Both cell types emerge at a similar time during the formation of the yolk sac blood islands,102-104and early intra-embryonic sites of hematopoiesis and are closely associated with vessels.105-111 In addition, a number of molecules are expressed in both endothelium and in hematopoietic progenitors including CD34.112,113 Furthermore, mice lacking the early endothelial marker Flk-1 display a defect in both hematopoietic cells and vasculature,114 and Flk-1 appears to play an important and cell-autonomous role in the formation of both lineages.115 Consistent with this concept VEGF, the ligand for Flk-1, can stimulate the generation of hematopoietic cells from ES cells116 and single Flk-1 positive avian cells can generate either hematopoietic or endothelial cells in vitro.117Moreover, in the zebrafish mutant cloche, the numbers of both endothelial and hematopoietic cells are severely reduced.118 These studies are all consistent with the notion of a common precursor for hematopoietic and endothelial cells.
Several lines of evidence suggest a functional role for SCL in endothelial cell development. SCL is expressed in normal endothelium86,119 and in SCL-null animals there is a defect in the development of yolk sac blood vessels with the failure of formation of large vitelline vessels.93 This was shown to reflect a primary consequence of SCL loss in elegant transgenic experiments that rescued the hematopoietic, but not the vascular defect, in the SCL-null mice.120 Furthermore, in the zebrafish mutant cloche, both the endothelial and hematopoietic defects are partially rescued by ectopic expression of SCL.121
Direct evidence for a role for SCL in hemangioblast formation has been reported by Gering et al122(Fig 3). SCL was coexpressed with Flk-1 in presumptive hemangioblasts within early postero-lateral mesoderm of zebrafish embryos. Ectopic expression of SCL mRNA during zebrafish development resulted in an expansion of hematopoietic and endothelial precursors at the expense of somitic and pronephric duct tissues. Taken together, these data show that SCL is capable of specifying or directing hemangioblast development from early mesoderm, and underline the striking similarities between the role of SCL in hematopoiesis/vasculogenesis and the function of other HLH proteins in muscle and neural development.123 124
SCL specifies hemangioblast development from early zebrafish mesoderm. Appearance of uninjected zebrafish embryos (a) and embryos injected with SCL mRNA at the 2-4 cell stage (b). SCL injected embryos developed a marked excess of hemoglobinized blood cells (arrowhead), a striking absence of blood circulation despite a beating heart and an abnormally curved axis. In situ hybridization experiments with SCL alone (c) or SCL and Flk-1 (d) showed a marked increase in cells coexpressing both endogenous SCL and Flk-1 in SCL-injected embryos. This increase was often unilateral (as illustrated in this figure), because the injected mRNA was frequently localized to one half of an embryo. NT, neural tube; NC, notochord; SM, somitic mesoderm; LM, lateral mesoderm. (Reprinted from Gering M, Rodaway ARF, Göttgens B, Patient RK, Green AR: The SCL gene specifies haemangioblast development from early mesoderm. The EMBO Journal, vol 17, p 4029, 1998, by permission of Oxford University Press.122)
SCL specifies hemangioblast development from early zebrafish mesoderm. Appearance of uninjected zebrafish embryos (a) and embryos injected with SCL mRNA at the 2-4 cell stage (b). SCL injected embryos developed a marked excess of hemoglobinized blood cells (arrowhead), a striking absence of blood circulation despite a beating heart and an abnormally curved axis. In situ hybridization experiments with SCL alone (c) or SCL and Flk-1 (d) showed a marked increase in cells coexpressing both endogenous SCL and Flk-1 in SCL-injected embryos. This increase was often unilateral (as illustrated in this figure), because the injected mRNA was frequently localized to one half of an embryo. NT, neural tube; NC, notochord; SM, somitic mesoderm; LM, lateral mesoderm. (Reprinted from Gering M, Rodaway ARF, Göttgens B, Patient RK, Green AR: The SCL gene specifies haemangioblast development from early mesoderm. The EMBO Journal, vol 17, p 4029, 1998, by permission of Oxford University Press.122)
It is interesting to contrast the loss of function phenotype, in which endothelial cells develop but fail to contribute normally to vessel formation, with the gain of function experiments described above. These results are consistent with previous studies of HLH proteins in myogenesis and neurogenesis, suggesting a cascade of HLH proteins that may have overlapping functions and expression patterns.123 124 In the case of SCL, functional redundancy with related HLH proteins may explain the relatively normal development of endothelial cells in SCL null mice.
DEUS EX MACHINA: WHAT REGULATES THE REGULATOR?
Multiple tiers of regulation.
Although multiple mechanisms exist and at different levels, to ensure that the critical functions of SCL are tightly controlled, these mechanisms and their significance are poorly understood. However, the importance of these mechanisms is amply demonstrated when they go awry in the T lymphocyte.
At the level of SCL protein, there is a requirement for HLH partner proteins such as E12 and E47. Additional control is provided via a subgroup of HLH proteins, the prototypic example of which is Id. These HLH family members lack a DNA-binding basic domain and thereby isolate other HLH proteins in inactive complexes.125 Moreover, the SCL protein itself exists in several different forms as a result of multiple sites of protein initiation within the SCL gene.17-19,28,126-128 These proteins can occur in a cell-type–specific manner and may differ in their ability to transactivate downstream genes. Regulation of protein turnover91 and phosphorylation on serine127 are probably also important in regulating the function of SCL protein.
Signaling pathways.
Several lines of evidence suggest links between SCL and a number of signal transduction pathways. During early development the formation of blood progenitors is thought to be initiated by growth factors, including bone morphogenetic protein 4 (BMP-4). During hematopoietic development in Xenopus, SCL expression is induced by BMP-4 and its expression is inhibited by a dominant-negative BMP-4 receptor.129 However, the precise position of SCL within the hierarchy of transcription factors modulated by BMP-4 is not known.
In the context of adult hematopoiesis, SCL has been linked to growth factor signaling pathways that suggest a “downstream” role for SCL. During erythroid differentiation, levels of SCL increase in response to stem cell factor130,131 and erythropoietin.88,89 Conversely, levels of SCL mRNA and protein decrease during growth-factor–induced macrophage differentiation.53,54 SCL is also a target for MAP kinases, including ERK-1.132
By contrast, antisense and dominant-negative SCL constructs reduced levels of c-kit (the receptor for stem cell factor) and SCL transactivated the c-kit promoter.134 These data raise the possibility that c-kit is regulated by SCL.
These observations are relevant to current views of hematopoietic stem cell behavior and lineage commitment. Several lines of evidence suggest that multiple components of distinct differentiation programs may be primed and undergo low-level transcription within individual multipotent stem cells.135 Commitment to a particular lineage may result from poorly understood and possibly stochastic processes that result in a single program becoming dominant and reinforced with competitive feedback loops. Transcription factors play a pivotal role in these processes. Thus, upregulation of c-kit or other growth factor receptors by SCL may modulate the behavior of multipotent cells and, as part of a self-sustaining feedback loop, participate in the lineage-commitment process.
Cis-acting elements.
The functions of many transcription factors hinge on their cellular context, the presence or absence of interacting proteins, and the accessibility of potential target genes. Therefore, the biological function of a given transcription factor is likely to depend critically upon its pattern of expression. This reasoning has focused attention on the cis-acting elements that regulate SCL transcription. Characterization of SCL regulatory elements that drive expression in hemangioblasts or multipotent progenitors will illuminate the molecular basis for the formation of these cell types and also provide important tools for experimental or therapeutic manipulation of hematopoietic stem cells.
Two promoters in alternative 5′ exons have been identified in both murine and human SCL genes,17,19,136-138 together with a third promoter within the body of the gene, the normal function of which remains unclear.133 Both 5′ promoters exhibit lineage restricted activity. Promoter 1a is regulated by GATA-1, SP1, and SP3, and is active in transient reporter assays in erythroid and mast cells.58,136,137,139 Promoter 1b is regulated by PU-1, SP1, and SP3, and is active in mast and primitive myeloid cells.138 139 Thus, in committed hematopoietic lineages, SCL appears to be regulated at least in part by a transcription factor with a pivotal role in erythroid cells (GATA-1) and by a critical myeloid transcription factor (PU-1).
However, although these promoters recapitulate aspects of the lineage-restricted pattern of SCL expression in transient reporter assays, they are not sufficient to direct reporter gene expression once integrated in chromatin.138 This observation stimulated a search for distant regulatory elements (“enhancers”) in the vicinity of the SCL gene. Therefore, a systematic survey of chromatin structure surrounding the SCL locus was undertaken140,141and resulted in identification of a number of enhancers active in both transient and stable transfection assays.141 More recently, transgenic assays have been used to study the activities of these elements and have identified spatially distinct enhancers that mimic components of normal SCL expression in endothelium, midbrain, hindbrain/spinal cord, and hematopoietic progenitors (A. Sinclair, M.-J. Sanchez, and A.R. Green, unpublished data, 1998). One element is particularly exciting because it appears to target hematopoietic progenitors throughout ontogeny.
UNRESOLVED ISSUES
SCL is clearly pivotal for normal hematopoiesis and vasculogenesis and is also likely to have a critical role in the nervous system. A plethora of questions remain unanswered. How does SCL induce an undifferentiated mesodermal cell to become a hemangioblast and then a multipotent hematopoietic stem cell? Does SCL perform the same role in different classes of hematopoietic stem cells throughout ontogeny? What is the role of SCL in the nervous system? Why is ectopic SCL expression leukemogenic in T-cell progenitors?
Answers to these and other questions are likely to require advances in at least three areas. First, we need to understand the significance of the multiprotein complexes that include SCL. How do the complexes differ in distinct cell types and what are the functional consequences of those differences? Second, it seems likely that different complexes will activate distinct target genes, and so it will also be essential to identify direct target genes downstream of SCL in different SCL-expressing cell types. Only in this way will we be able to put molecular flesh onto our skeletal ideas of SCL’s various functions. Third, how is SCL itself regulated? What triggers SCL expression in early mesoderm and therefore results in hemangioblast specification? How is SCL downregulated during differentiation along specific hematopoietic lineages, and is the modulation cause or consequence? Is there an SCL enhancer that can be used to target hematopoietic stem cells? At the current rate of progress, we should not have long to wait for some answers. Hopefully this will lead back to the bedside with new approaches to the manipulation of hematopoiesis and the treatment of T-ALL.
Supported by The Wellcome Trust, Leukaemia Research Fund (UK), Kay Kendall Leukaemia Fund (UK), Medical Research Council (UK), National Health and Medical Research Council (Canberra), Anti-Cancer Council of Victoria, and the Bone Marrow Donor Institute, Melbourne.
REFERENCES
Author notes
Address reprint requests to C. Glenn Begley, MD, PhD, the Walter and Eliza Hall Institute of Medical Research and the Rotary Bone Marrow Research Laboratories, PO Royal Melbourne Hospital, Parkville, Victoria, Australia 3050.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal