Abstract
Death-associated protein kinase (DAP-Kinase) is a novel serine/threonine kinase whose expression is required for γ interferon-induced apoptosis. A previous study suggested that DAP-Kinase expression may be lost epigenetically in cancer cell lines, because treatment of several nonexpressing cell lines with 5-aza-2′-deoxycytidine resulted in the expression of DAP-Kinase. Using methylation-specific polymerase chain reaction (MSP), we examined the DAP-Kinase CpG island for hypermethylation in cancer. Normal lymphocytes and lymphoblastoid cell lines are unmethylated in the 5′ CpG island of DAP-Kinase. However, in primary tumor samples, all Burkitt’s lymphomas and 84% of the B-cell non-Hodgkin’s lymphomas were hypermethylated in the DAP-Kinase CpG island. In contrast, none of the T-cell non-Hodgkin’s lymphoma samples and 15% or less of leukemia samples examined had hypermethylated DAP-Kinase alleles. U937, an unmethylated, DAP-Kinase–expressing leukemia cell line, was treated with γ interferon and underwent apoptosis; however, Raji, a fully methylated, DAP-Kinase nonexpressing Burkitt’s lymphoma cell line, only did so when treated with 5-aza-2′-deoxycytidine followed by γ interferon. Our findings in cell lines and primary tumors suggest that hypermethylation of the DAP-Kinase gene and loss of γ interferon-mediated apoptosis may be important in the development of B-cell malignancies and may provide a promising biomarker for B-cell–lineage lymphomas.
ONE OF THE HALLMARKS of cancer is an imbalance between cell growth and death. Whereas normal cells undergo apoptosis after specific regulatory signals, transformed cells can lose this response and survive, accumulating genetic and phenotypic defects leading to tumor progression. In particular, DNA damage-induced apoptosis in response to genotoxic drug treatment or ionizing radiation may be diminished. Critical genes involved in this response include p53,1,2 Bcl-2,3 and Bax.3,4Alterations in each of these genes have been described in cancers,5 including hematologic malignancies.6,7 For example, rearrangements including Bcl-2 are a prominent feature of follicular lymphomas.8
Apoptosis is also critical to normal B-cell function in the immune system.9 These cells both differentiate and proliferate by clonal selection controlled by T cells or undergo programmed cell death when not positively selected.9 The precise signals responsible for T-cell–induced B-cell apoptosis are not completely characterized. However, in addition to direct cell-cell interactions with B cells, T cells also secrete cytokines,10 which may be important in this regard. Such soluble signals may initiate programmed cell death in B cells, which, if blocked, might contribute to B-cell tumorigenesis.
Death-associated protein kinase (DAP-Kinase) is a 160-kD serine/threonine, microfilament-bound kinase recently shown to be involved in γ interferon-induced apoptosis.11 It was isolated by Deiss et al12 in 1995 by technical knock-out (TKO) studies in which cellular resistance to γ interferon-induced apoptosis was selected for after transfection with an antisense cDNA library. DAP-Kinase was identified as an γ interferon-induced gene and found to phosphorylate itself and exogenous substrates in a process regulated by a Ca2+-calmodulin binding domain.11 DAP-Kinase contains a death domain, and its expression is required for induction of programmed cell death in cells treated with γ interferon.11 Overexpression of DAP-Kinase induces programmed cell death, whereas a catalytically inactive mutant protects cells from γ interferon-induced apoptosis.11
Several B-cell lymphoma cell lines have decreased or absent expression of DAP-Kinase RNA or protein.13 Some of these cell lines are able to re-express DAP-Kinase after treatment with 5-aza-2′-deoxycytidine, a DNA demethylating drug.13This finding suggests that abnormal loss of DAP-Kinase expression could be associated with aberrant promoter region methylation. Such epigenetic loss of tumor-suppressor gene expression is associated with the aberrant methylation of CpG island gene-promoter regions, serving as an alternative to the genetic loss of a tumor-suppressor gene function by deletion or mutation.14-16 However, the role of inactivation of DAP-Kinase in primary malignancies has not been examined. Therefore, we investigated the methylation status of DAP-Kinase in normal lymphocytes, hematopoietic cancer cell lines, and primary hematopoietic tumor samples and studied its link to DAP-Kinase expression and γ interferon-induced apoptosis.
MATERIALS AND METHODS
Samples and DNA preparation.
High molecular weight DNA was obtained from leukemic samples, as described.17 The classification of acute lymphoblastic leukemia (ALL) samples was determined by surface markers, as described.17 Lymphoma DNA was kindly provided by Dr Constance Griffin (Johns Hopkins University, Baltimore, MD), and the clinical diagnosis served as the subtype classification.
Methylation-specific polymerase chain reaction (MSP).
DNA methylation patterns in the CpG island of DAP-Kinase were determined by chemical treatment with sodium bisulfite and subsequent use of the previously described polymerase chain reaction (PCR) procedure.18 MSP distinguishes the methylation status of a given region based on sequence changes produced by sodium bisulfite treatment18 and has been previously validated for genes, including p16,18,19p15, E-cadherin, VHL,18hMLH1,20 the estrogen receptor,21GSTπ,22 and MGMT,23 and for the diagnosis of the imprinting disorder of Prader-Willi and Angelman’s syndromes.24-27 The primer sequences designed for DAP-Kinase spanned 6 CpGs in total within the 5′ region of the gene. Primer sequences for unmethylated reaction were 5′-GGA GGA TAG TTG GAT TGA GTT AAT GTT-3′ (sense) and 5′-CAA ATC CCT CCC AAA CAC CAA-3′ (antisense). Primer sequences for methylated reaction were 5′-GGA TAG TCG GAT CGA GTT AAC GTC-3′ (sense) and 5′-CCC TCC CAA ACG CCG A-3′ (antisense). The 5′ position of the sense unmethylated and methylated primers corresponds to bp 2 and 5 of Genbank sequence no.X76104, respectively. Both antisense primers originate from bp 87 of this sequence. The annealing temperature for both the unmethylated and methylated reactions was 60°C. All MSP reactions were performed with positive and negative controls for both unmethylated and methylated alleles and a no DNA control.
Restriction analysis.
Aberrant methylation of CpG dinucleotides between the primer sequences for DAP-Kinase was determined as previously described.18MSP products were digested by Aci I (New England BioLabs, Beverley, MA), which cuts DNA at the sequence 5′ C ^CGC 3′/3′ GGC ^G 5′, were ethanol precipitated, and were run on 8% polyacrylamide gels.
Cell culture.
Raji, a Burkitt’s Lymphoma cell line, and U937, a leukemia cell line, were grown in RPMI 1640 with glutamine, 10% fetal bovine serum (Sigma, St Louis, MO), 100 U/mL penicillin, and 10 μg/mL streptomycin and maintained in log phase growth. Raji cells were treated with 5-aza-2′-deoxycytidine (Sigma) in 4 different dosage and time courses. Cells were treated with 1 μmolar on days 0 and 2, with media changes on days 2 and 5. Cells were treated with 0.5 μmol/L either on day 0 with media change on day 2 or were treated on days 0 and 2 with media changes on days 2 and 5. Cells were treated with 0.3 μmol/L on days 0, 2, 6, and 8 with media changes on days 2, 5, 6, 8, and 9 and then were carried for 7 days in normal media. Raji cells and U937 cells were treated with 1,000 U/mL γ-interferon (R&D Systems, Minneapolis, MN) or a carrier control of 0.1% bovine serum albumin (BSA).
DNA preparation.
Cells were pelleted and DNA was isolated using 1 mL/107cells of TE9, 1% sodium dodecyl sulfate (SDS), and 0.4 mg/mL proteinase K at 55°C for 1 to 2 hours and then 37°C overnight. DNA was then purified using phenol-chloroform extraction and ethanol precipitated.
Protein preparation and Western analysis.
Cells were counted using a Coulter Counter (Coulter, Hialeah, FL) and lysed (107 cells/mL) with 10 mmol/L phosphate buffer, pH 7.5, 100 mmol/L NaCl, 1% Trition X-100, 0.5% sodium deoxycholate, 0.1% SDS, 5 mmol/L EDTA, 1 μg/mL phenylmethyl sulfonyl fluoride (PMSF), and 1× complete protease inhibitor (Boehringer Mannheim, Indianapolis, IN). Cellular debris was pelleted and protein from 106 cells were run on a 7.5% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel. The protein was transferred to an immobilon-P transfer membrane (Millipore, Burlington, MA), which was blocked with 5% BSA/TTBS for 2 hours and then incubated with DAP-Kinase antibody (1:1,000 dilution in 1% BSA/TTBS; Sigma) for 2 hours and then with antimouse antibody-horseradish peroxidase (HRP; 1:2,000 dilution in 1% BSA/TTBS; Santa Cruz Laboratories, Santa Cruz, CA) for 1 to 2 hours. The blot was then washed in serial TTBS washes for 1 hour, developed with the ECL detection kit (Pierce, Rockford, IL), and exposed to film (Eastman Kodak, Rochester, NY).
Reverse transcriptase-PCR (RT-PCR).
Total cellular RNA was obtained and reverse-transcribed as previously described.28 Controls consisted of RNA treated identically but without the addition of reverse transcriptase and were labeled. RT-PCR was performed with the following primers for DAP-Kinase (5′-GAT AGA AAT GTC CCC AAA CCT CG-3′ and 5′-TCT TCT TG GAT CCT TGA CCA GAA-3′, which amplify a 343-bp product spanning sequence 781 to 1124 from Genbank X76104) and forGAPDH (5′-CGG AGT CAA CGG ATT TGG TCG TAT-3′ and 5′-AGC CTT CTC CAT GGT GGT GAA GAC-3′). Initial hot start was followed by 35 cycles of amplification (95°C for 30 seconds, 58°C for 60 seconds, and 72°C for 60 seconds), and 10 μL of product was electrophoresed on 6% nondenaturing polyacrylamide gels and directly visualized with ethidium bromide staining under UV illumination. GAPDH controls were amplified for 30 cycles (95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds), and 5 μL of product was electrophoresed.
Apoptosis analysis.
Cells were fixed in 80% methanol (106 cells/150 μL) and stored at 4°C. To stain cells for counting, cells were pelleted, rehydrated with 150 μL phosphate-buffered saline (PBS), pelleted again to remove the PBS, and incubated with Hoechst dye (1:500 dilution of 1 mg/mL bisbenzimide H 33342 fluorochrome trihydrochloride [Calbiochem, La Jolla, CA] in PBS). Apoptotic nuclear bodies versus nonapoptotic nuclei were counted under 40× UV microscopy in units of approximately 200 cells, counting up to 1,000 cells total. Standard deviation was calculated when sufficient cell number allowed 2 or more units to be counted.
RESULTS
DAP-Kinase has a region in the first 525 bp of its cDNA sequence that qualifies as a CpG island. Within the first 525 bp of the DAP-Kinase transcription start site, there are 46 CpGs (8.8%) and 49 GpCs, and the sequence is 64% G+C rich. The CG:GC ratio is 0.9. Each of these is a characteristic of a CpG island. To address the normal methylation pattern of this region, we examined lymphocytes from 7 healthy controls and found no evidence for methylation of the gene (Fig 1A). We also examined Epstein-Barr virus (EBV)-immortalized lymphoblastoid cell lines to determine whether growth in cell culture led to methylation of DAP-Kinase. None of the 12 EBV-immortalized cell lines was methylated at the DAP-Kinase CpG island, suggesting that cell culture or immortalization is not related to DAP-Kinase methylation (Fig 1B).
MSP of DAP-Kinase. Primer sets used for amplification are designated as unmethylated (U) or methylated (M). The size of the MSP product for the unmethylated reaction is 106 bp. The size of the MSP product for the methylated reaction is 98 bp. In all MSP reactions, 10λ of product was run on a 6% polyacrylamide gel, stained with ethidium bromide, and visualized under UV illumination. (A) MSP analysis of DNA from lymphocytes of 6 normal individuals shows amplification of only unmethylated alleles in the DAP-Kinase CpG island. (B) MSP of DAP-Kinase in lymphoblastoid cell lines (cell lines immortalized by EBV-transformation) shows amplification of only unmethylated alleles in the DAP-Kinase CpG island. (C) MSP of DAP-Kinase in cell lines. Raji shows amplification of only methylated alleles for the DAP-Kinase CpG island. U937, an expressing cell line, shows amplification of only unmethylated alleles for the DAP-Kinase CpG island. (D) MSP of DAP-Kinase in Burkitt’s lymphoma samples. Each sample shows aberrant methylation of the DAP-Kinase CpG island. None of these samples was microdissected, so the unmethylated alleles amplified in each sample represent either normal tissue found within the tumor samples or heterogeneity of aberrant methylation within the tumor sample itself. (E) MSP of DAP-Kinase in lymphoma samples. Samples: B, B-cell non-Hodgkin’s lymphoma; T, T-cell non-Hodgkin’s lymphoma; H, Hodgkin’s lymphoma. The majority of the B-cell non-Hodgkin’s lymphoma samples show aberrant methylation of the DAP-Kinase CpG island, and none of the T-cell non-Hodgkin’s lymphoma samples do.
MSP of DAP-Kinase. Primer sets used for amplification are designated as unmethylated (U) or methylated (M). The size of the MSP product for the unmethylated reaction is 106 bp. The size of the MSP product for the methylated reaction is 98 bp. In all MSP reactions, 10λ of product was run on a 6% polyacrylamide gel, stained with ethidium bromide, and visualized under UV illumination. (A) MSP analysis of DNA from lymphocytes of 6 normal individuals shows amplification of only unmethylated alleles in the DAP-Kinase CpG island. (B) MSP of DAP-Kinase in lymphoblastoid cell lines (cell lines immortalized by EBV-transformation) shows amplification of only unmethylated alleles in the DAP-Kinase CpG island. (C) MSP of DAP-Kinase in cell lines. Raji shows amplification of only methylated alleles for the DAP-Kinase CpG island. U937, an expressing cell line, shows amplification of only unmethylated alleles for the DAP-Kinase CpG island. (D) MSP of DAP-Kinase in Burkitt’s lymphoma samples. Each sample shows aberrant methylation of the DAP-Kinase CpG island. None of these samples was microdissected, so the unmethylated alleles amplified in each sample represent either normal tissue found within the tumor samples or heterogeneity of aberrant methylation within the tumor sample itself. (E) MSP of DAP-Kinase in lymphoma samples. Samples: B, B-cell non-Hodgkin’s lymphoma; T, T-cell non-Hodgkin’s lymphoma; H, Hodgkin’s lymphoma. The majority of the B-cell non-Hodgkin’s lymphoma samples show aberrant methylation of the DAP-Kinase CpG island, and none of the T-cell non-Hodgkin’s lymphoma samples do.
Next, we investigated by MSP whether the methylation status of the DAP-Kinase gene was indeed responsible for the loss of transcription previously reported in several hematopoietic cell lines. We found that Raji, a Burkitt’s lymphoma cell line that does not express DAP-Kinase,13 has only methylated copies of the DAP-Kinase gene. However, U937, a leukemia cell line that expresses DAP-Kinase,13 has only unmethylated copies of DAP-Kinase (Fig 1C). Thus, our analysis suggests that the presence of hypermethylation of the CpG island correlates with the loss of expression of DAP-Kinase. We studied other cancer cell lines and found several with partial methylation of the DAP-Kinase CpG island. This mix of methylated and unmethylated copies of DAP-Kinase represents either intra-allelic heterogeneity of the CpG island methylation or methylation of the island that varies from allele to allele.
Because DAP-Kinase is aberrantly methylated in hematopoietic cancer cell lines and such methylation is linked to the transcriptional repression of the gene, we analyzed primary hematopoietic cancer samples to see if aberrant methylation of DAP-Kinase occurred in vivo. In 100% of the Burkitt’s lymphoma samples studied (9 of 9), we observed methylation of the DAP-Kinase CpG island (Fig 1D). In 84% of the B-cell lymphoma samples studied (21 of 25), we found methylation of DAP-Kinase as well (Fig 1E). Methylation of the DAP-Kinase gene did not vary markedly between low-grade or high-grade non-Hodgkin’s lymphomas. In contrast, none of the 4 T-cell lymphoma samples we examined had aberrant methylation of the DAP-Kinase CpG island.
In primary leukemia, there was hypermethylation in 15% or less of the samples in each subclass (Table 1). The involved leukemias were most often of the B-cell type. In chronic lymphocytic leukemia, 1 of the 10 samples demonstrated methylation of DAP-Kinase; this case was notable in presenting as a lymphocytic lymphoma. A single chronic myeloid leukemia sample also demonstrated DAP-Kinase hypermethylation; this occurred in a patient in a basophilic blast crisis. In acute leukemia, 1of 26 pediatric acute myeloid leukemias and 3 of 20 B-cell acute lymphoblastic leukemias, but none of 10 T-cell acute lymphoblastic leukemias, showed methylation of the CpG island (data not shown). The single acute myeloid leukemia (AML) sample with methylated DAP-Kinase CpG islands had biphenotypic markers with a B-cell phenotype.
Methylation of DAP-Kinase in Hematologic Malignancies and Normal Cells
| Cell Type . | No. With Aberrant Methylation . | No. of Total Samples . |
|---|---|---|
| Non-cancer cell types | ||
| Normal lymphocytes | 0 | 7 |
| Lymphoblastoid cell lines | 0 | 12 |
| Lymphomas | ||
| Burkitt’s lymphoma cell line (Raji) | 1 | 1 |
| Burkitt’s lymphoma samples | 9 | 9 |
| B-cell lymphoma samples | 21 | 25 |
| T-cell lymphoma samples | 0 | 4 |
| Hodgkin’s lymphoma samples | 2 | 3 |
| Lymphoma samples (unknown cell type) | 9 | 12 |
| Leukemias | ||
| Leukemia cell lines (HL60, KG1a, U937, Jurkat, A3.01) | 3 | 5 |
| B-cell ALL samples | 3 | 20 |
| T-cell ALL samples | 0 | 10 |
| CLL samples | 1 | 11 |
| Pediatric AML samples | 1 | 26 |
| Adult AML samples | 2 | 6 |
| CML samples | 1 | 13 |
| Cell Type . | No. With Aberrant Methylation . | No. of Total Samples . |
|---|---|---|
| Non-cancer cell types | ||
| Normal lymphocytes | 0 | 7 |
| Lymphoblastoid cell lines | 0 | 12 |
| Lymphomas | ||
| Burkitt’s lymphoma cell line (Raji) | 1 | 1 |
| Burkitt’s lymphoma samples | 9 | 9 |
| B-cell lymphoma samples | 21 | 25 |
| T-cell lymphoma samples | 0 | 4 |
| Hodgkin’s lymphoma samples | 2 | 3 |
| Lymphoma samples (unknown cell type) | 9 | 12 |
| Leukemias | ||
| Leukemia cell lines (HL60, KG1a, U937, Jurkat, A3.01) | 3 | 5 |
| B-cell ALL samples | 3 | 20 |
| T-cell ALL samples | 0 | 10 |
| CLL samples | 1 | 11 |
| Pediatric AML samples | 1 | 26 |
| Adult AML samples | 2 | 6 |
| CML samples | 1 | 13 |
To address the density of methylation of the DAP-Kinase CpG island, we also studied the methylation of selected CpG dinucleotides in this region that did not fall under our DAP-Kinase MSP primers, as previously described.18 This technique takes advantage of the divergence of unmethylated and methylated sodium bisulfite-modified CpG dinucleotides in restriction enzyme sites. The restriction enzymeAci I recognizes the sequence 5′-C ^CGC-3′ and 3′-GGC ^G-5′, which is found in the DAP-Kinase gene between the MSP primers. If the CpGs are methylated in this sequence, the sodium bisulfite-modified 3′ to 5′ sequence is unaltered by the bisulfite and Aci I cuts the DNA. If the CpGs are unmethylated, the sodium bisulfite modifies the 3′ to 5′ sequence to 3′-GGTG-5′ and the DNA will not be cut. We analyzed DNA amplified by MSP for methylated and unmethylated sequences from representative MSP product samples: 2 B-cell ALL, 1 T-cell ALL, 2 chronic lymphocytic leukemia (CLL), 2 B-cell lymphomas, 1 T-cell lymphoma, and 2 Burkitt’s lymphomas as well as Raji and normal lymphocytes. In all of the samples analyzed except 1, the unmethylated MSP product was not cut by Aci I, whereas the methylated MSP product was (data not shown); therefore, methylation of the DAP-Kinase CpG island was dense over this region of the CpG island. The single exception to these findings was an unmethylated CLL sample that was partially cleaved by the restriction enzyme. This sample may have had heterogeneous methylation of its DAP-Kinase CpG island, such that several copies of the DAP-Kinase gene in this sample were methylated at this restriction site sequence but were not methylated in the region of the MSP primer sequences.
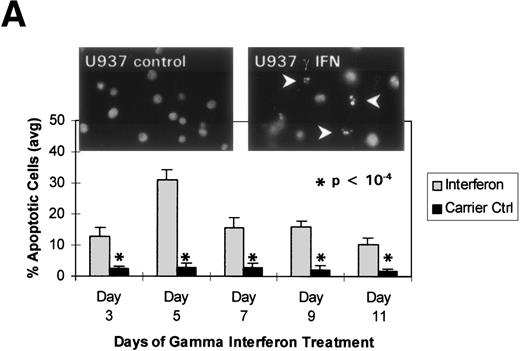
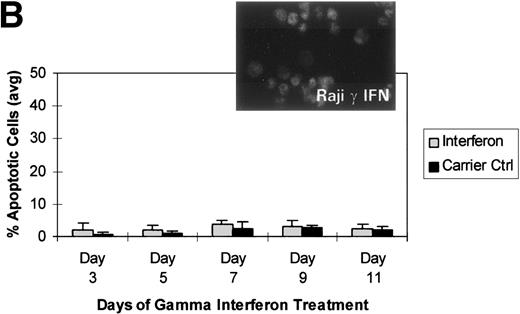
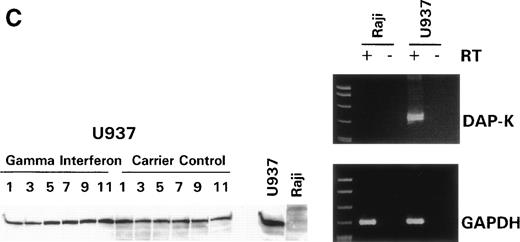
We turned our attention back to established cell lines to study the consequences of loss of expression of DAP-Kinase in association with hypermethylation of the gene. U937 and Raji cell lines were treated with γ interferon for a total of 11 days. After γ interferon exposure, U937, which expresses DAP-Kinase, underwent apoptosis, with maximal cell death occurring at days 3 through 7 of cytokine treatment (Fig 2A). Associated with the induction of this cell death was an upregulation of DAP-Kinase protein expression (Fig 2C). Raji cells, on the other hand, did not express DAP-Kinase at either the RNA or protein level (Fig 2C) and did not undergo apoptosis after γ interferon treatment during this time period (Fig 2B).
γ Interferon treatment of cell lines. U937, a leukemia cell line, and Raji, a Burkitt’s lymphoma cell line, were treated with 1,000 U/mL of γ interferon or carrier control for 11 days. (A) U937 cell apoptosis. U937 cells were analyzed for apoptosis on days 3, 5, 7, 9, and 11. U937 cells underwent apoptosis when treated with γ interferon, with significant differences in cell death between treated cells and carrier control cells on each day (P < 10−4). The inset demonstrates U937 cells treated for 5 days with γ interferon or the carrier control; several of the γ interferon-treated cells are undergoing nuclear apoptotic changes (arrowheads point to apoptotic cells), whereas none of the carrier control-treated cells is. (B) Raji cell apoptosis. Raji cells were analyzed for apoptosis on days 3, 5, 7, 9, and 11. Raji cells did not undergo apoptosis when treated with γ interferon or the carrier control. Raji cells treated for 5 days with γ interferon show no cells undergoing apoptosis, and there are no apoptotic nuclei. (C) DAP kinase expression. Cells (106) were lysed on days 1, 3, 5, 7, 9, and 11 of treatment with γ interferon or carrier control before Western analysis. U937 cells exposed to γ interferon upregulated the expression of DAP-Kinase from days 5 through 11. Carrier control cells expressed DAP-Kinase endogenously but did not upregulate the protein expression. Western analysis of U937 and Raji cells and RT-PCR analysis (far right) with GAPDH as controls demonstrate the lack of endogenous expression of DAP-Kinase in Raji at both the RNA and protein levels. DAP-Kinase RT-PCR amplifies a 343-bp product, whereas the GAPDH controls are 310 bp. + and − indicate the addition or the absence of reverse transcriptase (RT), respectively.
γ Interferon treatment of cell lines. U937, a leukemia cell line, and Raji, a Burkitt’s lymphoma cell line, were treated with 1,000 U/mL of γ interferon or carrier control for 11 days. (A) U937 cell apoptosis. U937 cells were analyzed for apoptosis on days 3, 5, 7, 9, and 11. U937 cells underwent apoptosis when treated with γ interferon, with significant differences in cell death between treated cells and carrier control cells on each day (P < 10−4). The inset demonstrates U937 cells treated for 5 days with γ interferon or the carrier control; several of the γ interferon-treated cells are undergoing nuclear apoptotic changes (arrowheads point to apoptotic cells), whereas none of the carrier control-treated cells is. (B) Raji cell apoptosis. Raji cells were analyzed for apoptosis on days 3, 5, 7, 9, and 11. Raji cells did not undergo apoptosis when treated with γ interferon or the carrier control. Raji cells treated for 5 days with γ interferon show no cells undergoing apoptosis, and there are no apoptotic nuclei. (C) DAP kinase expression. Cells (106) were lysed on days 1, 3, 5, 7, 9, and 11 of treatment with γ interferon or carrier control before Western analysis. U937 cells exposed to γ interferon upregulated the expression of DAP-Kinase from days 5 through 11. Carrier control cells expressed DAP-Kinase endogenously but did not upregulate the protein expression. Western analysis of U937 and Raji cells and RT-PCR analysis (far right) with GAPDH as controls demonstrate the lack of endogenous expression of DAP-Kinase in Raji at both the RNA and protein levels. DAP-Kinase RT-PCR amplifies a 343-bp product, whereas the GAPDH controls are 310 bp. + and − indicate the addition or the absence of reverse transcriptase (RT), respectively.
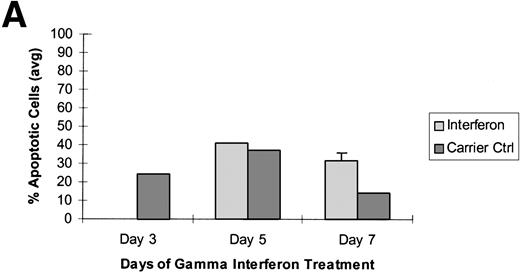
To further determine whether aberrant methylation of the DAP-Kinase CpG island was directly mediating the resistance of Raji cells to γ interferon-induced apoptosis, we treated these cells in three different time course experiments with the demethylating agent, 5-aza-2′-deoxycytidine, followed directly by γ interferon exposure. In each experiment, the DAP-Kinase CpG island was partially demethylated by 5-aza-2′-deoxycytidine within the first 2 days of drug exposure (Fig 3A). Although the acute toxic effects of 5-aza-2′-deoxycytidine caused some apoptotic cell death, Raji cells in which the DAP-Kinase CpG island had been partially demethylated underwent a higher percentage of programmed cell death in each experiment when treated with γ interferon compared with the level of apoptosis in those cells exposed only to carrier control (Fig 3). This γ interferon-induced apoptosis occurred at days 5 and 7 of cytokine treatment, a time similar to that observed for γ interferon-responsive U937 cells.
Raji cells were treated with 5-aza-2′-deoxycytidine in 3 experiments to demethylate the DAP-Kinase CpG island and then exposed to γ interferon. (A) Raji cell apoptosis. Raji cells were treated with 1 μmol/L 5-aza-2′-deoxycytidine for 5 days and then with 1,000 U/mL γ interferon for 7 days. Cells were analyzed for apoptosis on days 3, 5, and 7 of γ inteferon or carrier control treatment. By day 7, Raji cells exposed to γ interferon were undergoing more apoptosis than Raji cells exposed only to carrier control. (B) Raji cell apoptosis. Raji cells were treated with 0.5 μmol/L 5-aza-2′-deoxycytidine for 5 days and then with 1,000 U/mL γ interferon for 7 days. Cells were analyzed for apoptosis on days 3, 5, and 7 of γ inteferon or carrier control treatment. By day 7, Raji cells exposed to γ interferon were undergoing more apoptosis than Raji cells exposed only to carrier control. (C) Raji cell apoptosis. Raji cells were treated with 0.5 μmol/L 5-aza-2′-deoxycytidine for 2 days and then with 1,000 U/mL γ interferon for 7 days. Cells were analyzed for apoptosis on days 3, 5, and 7 of γ inteferon or carrier control treatment. By day 5, a trend had developed in which Raji cells exposed to γ interferon underwent more apoptosis than those exposed with carrier control alone.
Raji cells were treated with 5-aza-2′-deoxycytidine in 3 experiments to demethylate the DAP-Kinase CpG island and then exposed to γ interferon. (A) Raji cell apoptosis. Raji cells were treated with 1 μmol/L 5-aza-2′-deoxycytidine for 5 days and then with 1,000 U/mL γ interferon for 7 days. Cells were analyzed for apoptosis on days 3, 5, and 7 of γ inteferon or carrier control treatment. By day 7, Raji cells exposed to γ interferon were undergoing more apoptosis than Raji cells exposed only to carrier control. (B) Raji cell apoptosis. Raji cells were treated with 0.5 μmol/L 5-aza-2′-deoxycytidine for 5 days and then with 1,000 U/mL γ interferon for 7 days. Cells were analyzed for apoptosis on days 3, 5, and 7 of γ inteferon or carrier control treatment. By day 7, Raji cells exposed to γ interferon were undergoing more apoptosis than Raji cells exposed only to carrier control. (C) Raji cell apoptosis. Raji cells were treated with 0.5 μmol/L 5-aza-2′-deoxycytidine for 2 days and then with 1,000 U/mL γ interferon for 7 days. Cells were analyzed for apoptosis on days 3, 5, and 7 of γ inteferon or carrier control treatment. By day 5, a trend had developed in which Raji cells exposed to γ interferon underwent more apoptosis than those exposed with carrier control alone.
To minimize the toxic effects of 5-aza-2′-deoxycytidine and the apoptosis related to acute 5-aza-2′-deoxycytidine treatment, we conducted a fourth experiment in which Raji cells were exposed to a lower concentration of 5-aza-2′-deoxycytidine in a more chronic treatment, were allowed to recover for 7 days in normal media, and were then treated with γ interferon for 11 days. In this experiment, we again were able to partially demethylate the DAP-Kinase gene (Fig 4A). This treatment protocol now produced a statistically significant increase in γ interferon-induced apoptosis over the carrier control (Fig 4B). During the time course of γ interferon treatment, we observed a loss of sensitivity to γ interferon exposure in this experiment, which coincided with an increase in methylation of the DAP-Kinase CpG island (Fig 4A). This may represent remethylation of the gene by DNA methyltransferase or the outgrowth of cells that had not demethylated the DAP-Kinase gene and thus were protected from γ interferon-induced apoptosis. All of these data suggest that Raji undergoes γ interferon-induced cell death only after demethylation of the DAP-Kinase gene.
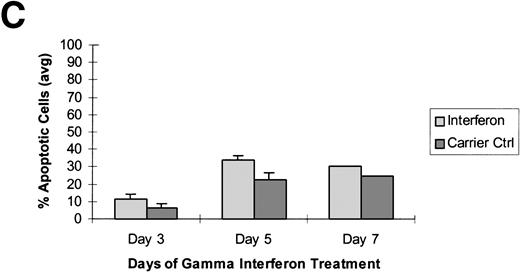
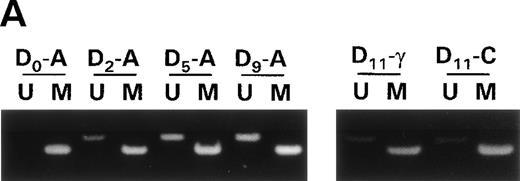
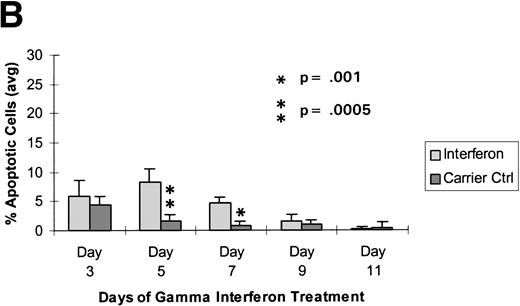
Raji cells were treated with 0.3 μmol/L 5-aza-2′-deoxycytidine for 9 days to demethylate the DAP-Kinase CpG island, placed in normal media for 7 days, and then treated with 1,000 U/mL γ interferon for 11 days. (A) MSP of DAP-Kinase in Raji cells. The methylation status of the DAP-Kinase CpG island was checked on days 0, 2, 5, and 9 of 5-aza-2′-deoxycytidine treatment (D0-9-A) and then after the combined 7 days of normal media and either 11 days of γ interferon (D11-γ) or carrier control (D11-C) media. (B) Raji cell apoptosis. By days 5 and 7, Raji cells exposed to γ interferon underwent significantly more apoptosis than those exposed with carrier control alone (P= .0005 and P = .001, respectively). By days 9 and 11, Raji cells were losing their unmethylated copies of DAP-Kinase and also their sensitivity to γ interferon treatment.
Raji cells were treated with 0.3 μmol/L 5-aza-2′-deoxycytidine for 9 days to demethylate the DAP-Kinase CpG island, placed in normal media for 7 days, and then treated with 1,000 U/mL γ interferon for 11 days. (A) MSP of DAP-Kinase in Raji cells. The methylation status of the DAP-Kinase CpG island was checked on days 0, 2, 5, and 9 of 5-aza-2′-deoxycytidine treatment (D0-9-A) and then after the combined 7 days of normal media and either 11 days of γ interferon (D11-γ) or carrier control (D11-C) media. (B) Raji cell apoptosis. By days 5 and 7, Raji cells exposed to γ interferon underwent significantly more apoptosis than those exposed with carrier control alone (P= .0005 and P = .001, respectively). By days 9 and 11, Raji cells were losing their unmethylated copies of DAP-Kinase and also their sensitivity to γ interferon treatment.
DISCUSSION
DAP-Kinase is a novel protein whose expression is required in γ interferon-induced apoptosis of cells. The gene may be important biologically in normal cell differentiation and selection in the immune system. Recent reports demonstrate expression of DAP-Kinase to be decreased or absent in several cancer cell lines.13 A decrease in expression of this gene imparts resistance to γ interferon-induced apoptosis in cells,11,12 and a link between the loss of DAP-Kinase expression and cellular apoptosis may facilitate metastasis in an experimental system.29 However, despite these compelling data, no involvement of DAP-Kinase in primary malignancy has been investigated.
Our results suggest that the loss of expression of DAP-Kinase observed in cell lines is indeed due to promoter region hypermethylation. This pattern of hypermethylation was also very frequent in primary B-cell malignancies, although not observed in T-cell malignancies or in myelogenous leukemia. Although we examined a smaller number of T-cell malignancies (ALL and non-Hodgkin’s lymphoma) than those with a B-cell phenotype, the incidence of hypermethylation of DAP-Kinase present in B-cell malignancies suggests that we would have easily detected a similar incidence in T-cell malignancies if present. However, DAP-Kinase methylation is not part of the normal differentiation of B cells, because normal peripheral lymphocytes (predominantly B cell) and immortalized lymphoblasts (B cells) do not have hypermethylation of the CpG island of the gene. Thus, hypermethylation of DAP-Kinase is characteristic of the transformed B cell, but not of normal B cells or those immortalized in vitro.
We have previously examined methylation of other tumor-suppressor genes and DNA repair enzymes in many of these same samples. p15 is frequently methylated in ALL and AML samples, p15 and/or p16 are frequently methylated in the Burkitt’s samples, and high-grade non-Hodgkin’s lymphoma frequently has hypermethylation of p16.17 We have also recently examined DNA repair enzymes and find that only rarely is the GSTπ gene methylated in these samples,22 whereas MGMT is often methylated in the high-grade lymphomas.23 In this latter case, whereas DAP-Kinase methylation was associated with only the B-cell phenotype (being present in both high-grade and low-grade non-Hodgkin’s lymphoma), MGMT was very highly associated with the diffuse large-cell lymphoma phenotype.23 Thus, multiple epigenetic events, ie, the hypermethylation and silencing of many genes, along with genetic alterations, such as translocation, deletion, and point mutations, coexist in the constellation of changes associated with the transformed phenotype.
The functional consequences of hypermethylation associated loss of DAP-Kinase expression are demonstrated in transformed cell lines. Raji, a nonexpressing and hypermethylated cell line, did not respond to γ interferon treatment, unlike U937, an expressing and unmethylated cell line. When we treated Raji cells with 5-aza-2′-deoxycytidine and partially demethylated the DAP-Kinase CpG island, these cells became sensitive to γ interferon treatment. This sensitivity was lost as passaged Raji cells remethylated the DAP-Kinase CpG island. Further emphasizing the role of DAP-Kinase in transformation rather than immortalization is the finding that, in vivo, re-expression of DAP-Kinase in cancer cells lowers tumorigenicity but has no effect on cell growth in culture.13 29
DAP-Kinase is a novel protein required for programmed cell death in the presence of γ interferon,11,12 and its lack of expression and concordant hypermethylation protects cells from γ interferon-induced apoptosis. The high frequency of abnormal methylation of DAP-Kinase in primary B-cell cancers suggests not only an important role for this pathway in the development of B-cell malignancies, but also that this DNA modification may serve as a biomarker for these cancers. Because in an experimental model, the loss of DAP-Kinase expression aids in metastatic potential of lung cancer cells,29 loss of expression of this gene may be important in the progression of other cancer types as well.
ACKNOWLEDGMENT
The authors thank Dr Christof Lengauer for assistance in the apoptosis analysis, Drs Connie Griffen and Mike Grever for primary tumor samples, and Dr Paul Corn for cDNA samples.
Supported in part by grants from the Howard Hughes Medical Institute and the V Foundation and National Institutes of Health Grants No. CA43318 and P30CA06973. R.A.K. was a Howard Hughes Medical Institute Medical Student Research Training Fellow. S.B.B. and J.G.H. receive research funding and are entitled to sales royalties from ONCOR, which is developing products related to research described in this report. The terms of this arrangement have been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to James G. Herman, MD, Johns Hopkins Medical Institutions, Department of Oncology, Tumor Biology, 424 N Bond St, Baltimore, MD 21231.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal