Abstract
Both genetic and environmental factors contribute to the normal population variability of plasma von Willebrand Factor (vWF) levels, however, regulatory mechanisms at the vWF gene locus itself have not yet been identified. We have investigated the association between polymorphic variation in the 5′-regulatory region of the vWF gene and levels of plasma vWF:Ag in a study of 261 group O blood donors. Three novel single nucleotide polymorphisms (SNPs) were identified in the vWF promoter: C/T at -1234, A/G at -1185, and G/A at -1051. These SNPs had identical allele frequencies of 0.36 for the -1234C, -1185A, and -1051G alleles and 0.64 for the -1234T, -1185G, and -1051A alleles and were in strong linkage disequilibrium. In fact, these polymorphisms segregated as two distinct haplotypes: -1234C/-1185A/-1051G (haplotype 1) and -1234T/-1185G/-1051A (haplotype 2) with 12.6% of subjects homozygous for haplotype 1, 40.6% homozygous for haplotype 2, and 42.5% of subjects heterozygous for both haplotypes. Only 4.3% of individuals had other genotypes. A significant association between promoter genotype and level of plasma vWF:Ag was established (analysis of covariance [ANCOVA], P = .008; Kruskal-Wallis test,P = .006); individuals with the CC/AA/GG genotype had the highest mean vWF:Ag levels (0.962 U/mL), intermediate values of vWF:Ag (0.867 U/mL) were observed for heterozygotes (CT/AG/GA), and those with the TT/GG/AA genotype had the lowest mean plasma vWF:Ag levels (0.776 U/mL). Interestingly, when the sample was subgrouped according to age, the significant association between promoter genotype and plasma vWF:Ag level was accentuated in subjects > 40 years of age (analysis of variance [ANOVA], P = .003; Kruskal-Wallis test, P= .001), but was not maintained for subjects ≤ 40 years of age (ANOVA, P > .4; Kruskal-Wallis test, P > .4). In the former subgroup, mean levels of plasma vWF:Ag for subjects with the CC/AA/GG, CT/AG/GA, and TT/GG/AA genotypes were 1.075, 0.954, and 0.794 U/mL, respectively. By searching a transcription factor binding site profile database, these polymorphic sequences were predicted to interact with several transcription factors expressed in endothelial cells, including Sp1, GATA-2, c-Ets, and NFκB. Furthermore, the binding sites at the -1234 and -1051 SNPs appeared to indicate allelic preferences for some of these proteins. Electrophoretic mobility shift assays (EMSAs) performed with recombinant human NFκB p50 showed preferential binding of the -1234T allele (confirmed by supershift EMSAs), and EMSAs using bovine aortic endothelial cell (BAEC) nuclear extracts produced specific binding of a nuclear protein to the -1051A allele, but not the -1051G allele. These findings suggest that circulating levels of vWF:Ag may be determined, at least in part, by polymorphic variation in the promoter region of the vWF gene, and that this association may be mediated by differential binding of nuclear proteins involved in the regulation of vWF gene expression.
THE FORMATION OF a primary platelet plug at sites of vascular injury is an essential component of hemostasis, which is mediated in the initial stages by the adhesive multimeric glycoprotein von Willebrand factor (vWF). A deficiency or qualitative defect of vWF results in the inherited bleeding disorder von Willebrand disease (vWD), thereby illustrating the physiologic importance of this protein. In contrast to its role in bleeding disorders, the involvement of vWF in the pathogenesis of thrombotic disease is less apparent. Nevertheless, there is growing evidence to implicate vWF in at least the terminal thrombotic complications of processes such as atherosclerosis. Porcine models of vWD, in which there is a marked vWF deficiency, have helped to establish that occlusive coronary thrombosis is a vWF-dependent condition (reviewed in Brinkhous et al1). Elevated levels of vWF have been associated with cardiovascular disorders such as ischemic heart disease, cerebrovascular disease, peripheral and pulmonary vascular disease, and several conventional risk factors for thrombotic disease, such as diabetes, hypertension, hypercholesterolemia, and smoking have also been linked with increased levels of plasma vWF (reviewed in Lip and Blann2). In some instances, the concentration of plasma vWF has been established as a significant and independent predictor of cardiovascular risk in individuals with coronary artery disease.3-5
vWF, synthesized exclusively by endothelial cells and megakaryocytes, performs two critical hemostatic functions, mediating platelet adhesion6 and thrombus formation7 at sites of vascular damage and serving as the carrier for procoagulant factor VIII in circulating blood.8 The gene encoding vWF, approximately 180 kb in length and containing 52 exons, has been localized to chromosome 12.9,10 Characterization of the vWF regulatory sequence has indicated that control of vWF gene expression occurs through the interaction of positive and negative regulatory elements located within this region. A cell-type–specific promoter was identified in bovine aortic endothelial cells (BAECs) from -487 bp upstream of the transcription start site to +247 bp, which contains a ubiquitous core promoter between -90 and +22 bp, a strong negative regulatory element upstream of the core promoter (-313 to -487 bp) and a cell-specific positive regulatory region located downstream of the core promoter in the first exon.11 Endothelial cell-specific gene expression requires the presence of the positive regulatory region with an intact GATA binding site11; the inhibitory effect of the negative regulatory element has been attributed to interactions with an NF1-like protein12 and Oct-1.13 An additional element in the vWF promoter, a polymorphic dinucleotide repeat, (GT)n, which starts at -668 bp has also been identified,14 but the functional significance of this element remains to be determined.
Normal levels of plasma vWF can exhibit a sixfold variability, ranging from approximately 40% to 240% of the mean population level, which is designated a value of 1 U/mL, based on an earlier report describing serial studies of vWF:Ag in normal individuals.15 Several genetic and environmental factors, such as ABO blood group,16 age,17 pregnancy,18 and the acute phase response,19 have been described as influencing levels of plasma vWF, however, knowledge of mechanisms at the vWF locus itself, which might influence its expression, remains elusive. Numerous recent reports have documented an association between variation within regulatory regions of gene loci involved in hemostasis and circulating levels of these proteins. Furthermore, these sequence variations, which include polymorphisms in the β-fibrinogen,20 plasminogen activator inhibitor-1 (PAI-1),21 and protein C22 gene promoters, and infrequent mutations of the thrombomodulin23 gene promoter have also been established as independent risk factors for thrombotic vascular disease. These studies suggest that regulatory sequence variations in “hemostatic” genes may be critical elements in the regulation of gene transcription, protein biosynthesis, and possibly, determination of risk for thrombotic disease. This report has examined the potential association of three novel single nucleotide polymorphisms (SNPs) in the vWF regulatory sequence with the concentration of plasma vWF in vivo; the binding of nuclear proteins involved in vWF transcription at each of these polymorphic sites has also been investigated.
MATERIALS AND METHODS
Subjects.
Blood samples were collected from 261 blood donors attending the Canadian Red Cross donor center in Ottawa, Ontario, Canada. The study protocol had been approved by the local institutional ethics review board, and all samples were given a unique coded identifier. Blood donors at this clinic represent a variety of ethnic groups, but are predominantly Caucasian. Samples were collected exclusively from group O donors to minimize the variability of plasma vWF:Ag levels that is recognized to result from differences in ABO blood type.
Measurement of plasma vWF:Ag.
Plasma vWF:Ag was determined by enzyme-linked immunosorbent assay (ELISA) using a polyclonal goat antihuman vWF antibody (Affinity Biologicals, Hamilton, Ontario, Canada). A “CryoCheck” normal reference plasma (Precision Biologicals, Dartmouth, Nova Scotia, Canada), for which a variety of hemostatic parameters including vWF:Ag have been quantified using World Health Organization standards (WHO Lot 91/666), was used to calculate levels of plasma vWF:Ag for each subject. Each individual assay for vWF:Ag was performed in triplicate.
Genomic amplification and sequencing.
Genomic DNA was extracted from EDTA-anticoagulated whole blood by a previously described salt extraction method.24 A portion of the vWF gene, from -1380 to -534 bp upstream of the transcription start site, was screened for promoter polymorphisms. This 846-bp fragment was amplified by polymerase chain reaction (PCR) from genomic DNA samples under the following conditions: 94°C for 1 minute, 56°C for 30 seconds, and 72°C for 30 seconds (30 cycles in total). The downstream and upstream oligonucleotide primers were 5′-ATAAGAGCTGGAAGTGGAAA-3′ and 5′-GGGAGTGATGGTTTGAGTCT-3′ (Cortec DNA Service Laboratories, Kingston, Ontario, Canada), respectively. Several of these amplified products were inserted into the multiple cloning site of the pCR2.1 vector (Invitrogen, Carlsbad, CA). SNPs were detected in these clones by either manual direct dideoxy sequencing using a T7 Sequenase v. 2.0 kit (United States Biochemical, Cleveland, OH) or by automated sequencing (Mobix, Hamilton, Ontario, Canada).
SNP genotyping by allele-specific oligonucleotide (ASO) hybridization.
vWF -1234C/T, -1185 A/G, and -1051 G/A promoter genotypes were established for each subject by ASO hybridization; the sequence and melting temperature (Tm) for each centrally mismatched ASO is listed in Table 1. Using 15 U of T4 polynucleotide kinase (New England BioLabs, Beverly, MA), 20 pmol of each ASO were end-labelled with 10 μCi [γ-32P] adenosine 5′ triphosphate (ATP) and applied to a Sephadex G-50 (Pharmacia Biotech, Piscataway, NJ) spin column to remove unincorporated isotope. Amplified genomic DNA (5 μL, conditions as previously described) was denatured, applied to Hybond-N+ nylon membrane (Amersham, Arlington Heights, IL) in a Minifold II slot blot apparatus (Schleicher and Schuell, Keene, NH), and baked at 80°C for 2 hours. Membranes were prepared in duplicate and prehybridized in 5X SSPE (0.75 mol/L NaCl, 50 mmol/L NaH2PO4, 5 mmol/L EDTA), 5X Denhardt’s solution (0.1% Ficoll 400, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin), and 0.5% sodium dodecyl sulfate (SDS) for 1.5 hours at 37°C. Each of the end-labelled ASOs was hybridized separately to one of the membranes in the prehybridization solution at 37°C for 16 hours. Membranes were subsequently washed to a stringency of 150 mmol/L NaCl and 15 mmol/L sodium citrate (1X SSC, pH 7.0) at temperatures corresponding to the Tm of each ASO probe (Table 1) and autoradiographed for 2 hours at -70°C with Kodak X-OMAT AR film (Eastman Kodak, Rochester, NY). Genomic DNA samples with known SNP genotypes, confirmed by direct sequence analysis, were included on each membrane as controls for hybridization and washing conditions.
ASOs and Double-Stranded EMSA Oligonucleotides for Each Polymorphic Site
| SNP . | ASO and ds EMSA oligo* . | ASO Tm (°C) . |
|---|---|---|
| 5′-ACGCCATT(C/T)TCCTGCCT-3′ | ||
| −1234C/T | 5′-GGGTTCACGCCATT(C/T)TCCTGCCT-3′ | 54/52 |
| 3′-GTGCGGTAA(G/A)AGGACGGAGTCGG-5′ | ||
| 5′-CCGCCACC(A/G)CGCCTGGC-3′ | ||
| −1185A/G | 5′-TACAGGCGCCCGCCACC(A/G)CGCCTG-3′ | 62/64 |
| 3′-CGCGGGCGGTGG(T/C)GCGGACCGATT-5′ | ||
| 5′-GAACCACC(G/A)TGCCCGGC-3′ | ||
| −1051G/A | 5′-GCGTGAACCACC(G/A)TGCCCGGCCC-3′ | 60/58 |
| 3′-TTGGTGG(C/T)ACGGGCCGGGGGGTT-5′ |
| SNP . | ASO and ds EMSA oligo* . | ASO Tm (°C) . |
|---|---|---|
| 5′-ACGCCATT(C/T)TCCTGCCT-3′ | ||
| −1234C/T | 5′-GGGTTCACGCCATT(C/T)TCCTGCCT-3′ | 54/52 |
| 3′-GTGCGGTAA(G/A)AGGACGGAGTCGG-5′ | ||
| 5′-CCGCCACC(A/G)CGCCTGGC-3′ | ||
| −1185A/G | 5′-TACAGGCGCCCGCCACC(A/G)CGCCTG-3′ | 62/64 |
| 3′-CGCGGGCGGTGG(T/C)GCGGACCGATT-5′ | ||
| 5′-GAACCACC(G/A)TGCCCGGC-3′ | ||
| −1051G/A | 5′-GCGTGAACCACC(G/A)TGCCCGGCCC-3′ | 60/58 |
| 3′-TTGGTGG(C/T)ACGGGCCGGGGGGTT-5′ |
Synthesized by Cortec DNA Service Laboratories.
Electrophoretic mobility shift assays (EMSAs).
The polymorphic promoter sequences at -1234, -1185, and -1051 were assessed for potential transcription factor binding sites with the TFSEARCH program (v. 1.3),25 which searches sequence fragments against the MATRIX table of TRANSFAC,26 a transcription factor binding site profile database.
Nuclear extracts were prepared from BAECs using a previously described rapid extraction method.27 Double-stranded oligonucleotides (200 ng), designed for each type of polymorphic sequence (Table 1), were labelled with 15 μCi [α-32P] dATP and 5 U of the Klenow fragment of DNA polymerase I (Gibco BRL/Life Technologies, Gaithersburg, MD) in a reaction containing (final concentrations) 10 mmol/L Tris (pH 7.5), 50 mmol/L NaCl, 10 mmol/L MgCl2, 1 mmol/L dithiothreitol (DTT), and 0.5 mmol/L deoxynucleotide triphosphates (dNTPs; excluding dATP). After a 15-minute incubation at room temperature (RT), 5 mmol/L nonradiolabelled dATP was added and incubated for an additional 10 minutes. Finally, labelled oligos were purified with chloroform:isoamyl alcohol (24:1) and applied to a Sephadex G-50 spin column to remove unincorporated isotope. For EMSAs, protein-DNA complexes were formed essentially as described,28 but with a few modifications. The EMSA reaction contained 25 mmol/L HEPES (pH 7.6), 34 mmol/L KCl, 5 mmol/L MgCl2, 1 μg of double-stranded poly (dI-dC) competitor DNA, and 10 μg of bovine serum albumin. To the radiolabelled oligo/buffer mixture, 5 μg of BAEC nuclear extract or 0.1 μg of recombinant human NFκB (p50 subunit) (Promega, Madison, WI) was added and incubated at RT for 20 to 30 minutes. For supershift assays, the EMSA protocol was identical, but with a subsequent incubation (15 minutes at RT) with 0.1 to 0.2 μg of polyclonal antibodies directed against NFκB p50 (Santa Cruz Biotechnology, Santa Cruz, CA). Competition EMSAs were performed with 10-fold, 50-fold, and 100-fold excess of unlabelled oligonucleotide for -1234C/T and -1051G/A.
Statistical analysis.
Allele frequencies for each SNP were estimated by gene counting; χ2 analysis was used to evaluate deviation of genotype distributions from Hardy-Weinberg equilibrium and also to test for allelic association between these SNPs. The skewness of the plasma vWF:Ag distribution was normalized by logarithmic (ln) transformation, however, untransformed mean vWF:Ag levels are reported in all tables for convenience. Transformed levels of vWF:Ag were compared between genotype groups using analysis of covariance (ANCOVA), in which age was incorporated as a covariate into a one-way analysis of variance (ANOVA). In addition, subjects were divided into two age groups (≤40 years and >40 years); subgroup analysis was performed using one-way ANOVA. Alternatively, a nonparametric Kruskal-Wallis test was performed to assess differences in untransformed vWF:Ag levels between genotype groups. Student’s unpaired t-tests were used to compare age and transformed plasma vWF:Ag levels between male and female subjects.
RESULTS
Range of plasma vWF:Ag values.
The general characteristics of the subjects in this study are summarized in Table 2. The mean (standard deviation [SD]) plasma vWF:Ag level of the entire sample was 0.831 (0.390) U/mL, which is in a range previously reported for group O individuals.16 The normal range of plasma vWF:Ag levels in these subjects was considered to be 0.332 to 1.994 U/mL (40% to 240% of the sample mean).15 Therefore, according to this criterion, 95% of subjects in this study had normal levels of plasma vWF:Ag, while subnormal levels of vWF:Ag were identified in 3.8% of subjects, and 1.2% of individuals had abnormally high vWF:Ag levels. Mean age and mean level of plasma vWF:Ag did not differ significantly between male and female subjects (P > .1).
Characteristics of Subjects—Age and Level of Plasma VWF:Ag
| Subjects . | n . | Mean Age (SD) (yr) . | Mean VWF:Ag Level (SD) (U/mL) . |
|---|---|---|---|
| Male | 154 | 42.9 (10.9) | 0.814 (0.358) |
| Female | 105 | 41.1 (10.8) | 0.859 (0.433) |
| Total | 261* | 42.2 (10.9) | 0.831 (0.390) |
| Subjects . | n . | Mean Age (SD) (yr) . | Mean VWF:Ag Level (SD) (U/mL) . |
|---|---|---|---|
| Male | 154 | 42.9 (10.9) | 0.814 (0.358) |
| Female | 105 | 41.1 (10.8) | 0.859 (0.433) |
| Total | 261* | 42.2 (10.9) | 0.831 (0.390) |
Unpaired t-tests comparing mean ages and mean transformed levels of plasma VWF:Ag between male and female subjects were not significant (P > .1).
Two samples with unspecified gender.
Characterization of vWF promoter SNPs.
Direct sequence analysis of genomic clones containing 846 bp of vWF promoter sequence, from -1380 to -534, showed the presence of three novel SNPs: C/T at -1234, A/G at -1185, and G/A at -1051. These SNPs had identical allele frequencies of 0.36 for the -1234C, -1185A, and -1051G alleles and 0.64 for the -1234T, -1185G, and -1051A alleles. The distribution of vWF -1234C/T, -1185A/G, and -1051G/A genotypes, shown in Table 3, was not significantly different from the distribution expected from Hardy-Weinberg equilibrium (P > .1). Of the 27 possible genotypes, only three main genotypes were represented (-1234/-1185/-1051): CC/AA/GG (12.6%), CT/AG/GA (42.5%), and TT/GG/AA (40.6%). Only 4.3% of subjects had genotypes different from those listed above. Using contingency tables constructed for each pair of SNPs, it was determined that the vWF -1234C/T, -1185A/G, and -1051G/A polymorphisms were in strong linkage disequilibrium (P= .001). Therefore, these SNPs were segregating as two distinct haplotypes: -1234C/-1185A/-1051G (haplotype 1) and -1234T/-1185G/-1051A (haplotype 2).
Distribution of VWF −1234C/T, −1185A/G, and −1051G/A Genotypes in 261 Normal Subjects
| Genotype3-150,3-151 . | n (%) . | ||
|---|---|---|---|
| −1234 . | −1185 . | −1051 . | |
| C/C | A/A | G/G | 33 (12.6) |
| C/T | A/G | G/G | 7 (2.7) |
| C/T | A/G | G/A | 111 (42.5) |
| C/T | A/G | A/A | 1 (0.4) |
| C/T | G/G | A/A | 1 (0.4) |
| T/T | A/G | A/A | 2 (0.8) |
| T/T | G/G | A/A | 106 (40.6) |
| Genotype3-150,3-151 . | n (%) . | ||
|---|---|---|---|
| −1234 . | −1185 . | −1051 . | |
| C/C | A/A | G/G | 33 (12.6) |
| C/T | A/G | G/G | 7 (2.7) |
| C/T | A/G | G/A | 111 (42.5) |
| C/T | A/G | A/A | 1 (0.4) |
| C/T | G/G | A/A | 1 (0.4) |
| T/T | A/G | A/A | 2 (0.8) |
| T/T | G/G | A/A | 106 (40.6) |
The distribution of genotypes for each SNP was not significantly different from Hardy-Weinberg equilibrium (P > .1).
The −1234C/T, −1185A/G, and −1051G/A SNPs were in strong linkage disequilibrium (P = .001).
Association of vWF promoter genotype with level of plasma vWF:Ag.
Mean levels of plasma vWF:Ag were significantly different between SNP genotype groups (Table 4) such that individuals homozygous for haplotype 1 (ie, CC/AA/GG) had a higher mean vWF:Ag level than subjects homozygous for haplotype 2 (ie, TT/GG/AA) (0.962 U/mL v 0.776 U/mL), with heterozygotes possessing an intermediate level (0.867 U/mL) of vWF:Ag (ANCOVA, P = .008; Kruskal-Wallis test, P = .006). Although age was not significantly different between genotype groups (ANOVA, P > .5), it was determined to be significantly associated with the level of plasma vWF:Ag (ANCOVA, P = .011). Before the ANCOVA analysis, the relationship between age and plasma vWF:Ag within each specific genotype group was examined and compared. This relationship was not significantly different between SNP genotype groups, indicating that the association between age and plasma vWF:Ag was essentially identical, regardless of SNP genotype, which was the assumption of the ANCOVA model. Interestingly, the effect of vWF:Ag variation associated with these SNPs differed dramatically when individuals were grouped according to age (≤40 years and >40 years) in the subgroup analysis. The correlation between SNP genotype and level of plasma vWF:Ag was accentuated in subjects >40 years of age (Table 4), with a difference of 0.28 U/mL between mean plasma vWF:Ag levels in subjects homozygous for haplotypes 1 and 2 (ANOVA, P = .003; Kruskal-Wallis test,P = .001). In contrast, the association between promoter genotype and vWF:Ag level was not maintained in subjects ≤40 years of age (ANOVA, P > .4; Kruskal-Wallis test, P > .4), as shown in Table 4. It should be noted that allele frequencies for these SNPs did not differ between age groups.
Levels of Plasma VWF:Ag for Each −1234C/T, −1185A/G, and −1051G/A Genotype
| Genotype . | n (%) . | Mean VWF:Ag Level (SD) (U/mL) . | P Value4-150 . |
|---|---|---|---|
| All subjects (n = 250) | |||
| 1/1 | 33 (13.2) | 0.962 (0.443) | |
| 1/2 | 111 (44.4) | 0.867 (0.354) | .008/.006 |
| 2/2 | 106 (42.4) | 0.776 (0.407) | |
| Subjects >40 yrs of age (n = 131) | |||
| 1/1 | 17 (13.0) | 1.075 (0.498) | |
| 1/2 | 57 (43.5) | 0.954 (0.355) | .003/.001 |
| 2/2 | 57 (43.5) | 0.794 (0.467) | |
| Subjects ≤40 yrs of age (n = 112) | |||
| 1/1 | 14 (12.5) | 0.875 (0.358) | |
| 1/2 | 52 (46.4) | 0.768 (0.336) | .438/.427 |
| 2/2 | 46 (41.1) | 0.754 (0.338) |
| Genotype . | n (%) . | Mean VWF:Ag Level (SD) (U/mL) . | P Value4-150 . |
|---|---|---|---|
| All subjects (n = 250) | |||
| 1/1 | 33 (13.2) | 0.962 (0.443) | |
| 1/2 | 111 (44.4) | 0.867 (0.354) | .008/.006 |
| 2/2 | 106 (42.4) | 0.776 (0.407) | |
| Subjects >40 yrs of age (n = 131) | |||
| 1/1 | 17 (13.0) | 1.075 (0.498) | |
| 1/2 | 57 (43.5) | 0.954 (0.355) | .003/.001 |
| 2/2 | 57 (43.5) | 0.794 (0.467) | |
| Subjects ≤40 yrs of age (n = 112) | |||
| 1/1 | 14 (12.5) | 0.875 (0.358) | |
| 1/2 | 52 (46.4) | 0.768 (0.336) | .438/.427 |
| 2/2 | 46 (41.1) | 0.754 (0.338) |
Haplotype 1: −1234C/−1185A/−1051G. Haplotype 2: −1234T/−1185G/−1051A.
ANCOVA (ANOVA)/Kruskal-Wallis test.
Binding of endothelial cell nuclear proteins to vWF promoter SNPs.
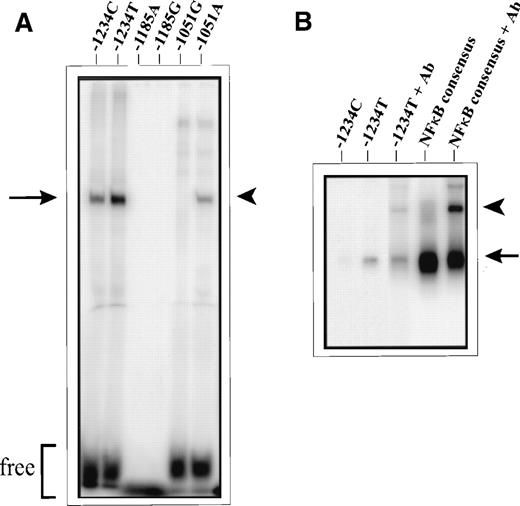
To investigate the possibility that these SNPs alter binding of nuclear proteins, EMSAs were performed using double-stranded oligonucleotides corresponding to the allele sequences at -1234, -1185, and -1051. Incubation of these allele sequences with nuclear extracts from unstimulated (“resting”) BAECs produced a different pattern of protein-DNA complex formation for each polymorphic site (Fig 1A). For the -1234C/T polymorphism, both alleles bound one major factor present in the BAEC nuclear extract, however, the gel-retarded complex is less intense in the assay with the C allele. EMSAs with the -1051G/A SNP showed a nuclear protein bound by the A allele that was not bound by the G allele. Neither of the alleles at -1185 (A or G) bound proteins in the BAEC nuclear extract. The formation of protein-DNA complexes (or lack thereof) in EMSAs as described was verified by five independent assays. Additionally, competition EMSAs, performed with 10-fold, 50-fold, and 100-fold excess of unlabelled oligonucleotide for both -1234C/T and -1051G/A, produced a marked reduction in protein-DNA complex formation in all reactions (data not shown), which indicated specificity of binding. A database search (TFSEARCH/TRANSFAC) with the sequences for these SNPs identified several transcription factors expressed in endothelial cells, which might bind these polymorphic sites, including Sp1, GATA-2, c-Ets, and NFκB. Furthermore, it appeared as though some of these binding sites demonstrated allelic preference; for example, NFκB was predicted to bind preferentially to the T allele at -1234, and at -1051, a binding site for GATA-2 was evident for the A allele, but not the G allele. The preferential binding of NFκB (p50) by the -1234T allele was confirmed by EMSAs with rhNFκB (p50) (Fig 1B). Protein-DNA complex formation was evident only for the -1234T allele, and this complex was supershifted by antibodies directed against NFκB (p50).
Differential binding of nuclear proteins to -1234C/T, -1185A/G, and -1051G/A allele sequences. (A) EMSAs performed with nuclear extracts from unstimulated BAECs. Arrow, -1234C/T-specific factor; arrowhead, -1051A allele-specific factor; free, free (unbound) probe. Allele sequences for -1234C/T and -1051G/A exhibited differential DNA-protein complex formation, while the -1185A/G sequences did not appear to bind factors present in BAEC nuclear extracts. (B) EMSAs performed with recombinant human NFκB (p50). Arrow, oligo/rhNFκB p50 complex formation; arrowhead, supershift with anti-NFκB (p50) antibody (Ab). Markedly enhanced binding of rNFκB p50 occurred to the -1234T allelic probe; this complex was supershifted by antibodies directed against NFκB p50. A NFκB consensus oligonucleotide was included in this EMSA as a positive control.
Differential binding of nuclear proteins to -1234C/T, -1185A/G, and -1051G/A allele sequences. (A) EMSAs performed with nuclear extracts from unstimulated BAECs. Arrow, -1234C/T-specific factor; arrowhead, -1051A allele-specific factor; free, free (unbound) probe. Allele sequences for -1234C/T and -1051G/A exhibited differential DNA-protein complex formation, while the -1185A/G sequences did not appear to bind factors present in BAEC nuclear extracts. (B) EMSAs performed with recombinant human NFκB (p50). Arrow, oligo/rhNFκB p50 complex formation; arrowhead, supershift with anti-NFκB (p50) antibody (Ab). Markedly enhanced binding of rNFκB p50 occurred to the -1234T allelic probe; this complex was supershifted by antibodies directed against NFκB p50. A NFκB consensus oligonucleotide was included in this EMSA as a positive control.
DISCUSSION
In this study, three novel SNPs of the vWF promoter are described, -1234C/T, -1185A/G, and -1051G/A; the strong allelic association of these polymorphisms was expected, given their close physical proximity within the promoter. The objective of this study was to evaluate the effect of genotypic variation at the vWF locus on levels of plasma vWF:Ag in vivo. A clear correlation between vWF promoter SNP genotype and plasma vWF:Ag levels was observed in this sample of 261 group O blood donors; because of the unbiased manner in which subjects were ascertained for this study, it is likely that this sample is representative of a larger population of normal individuals. The mean levels of plasma vWF:Ag differed significantly according to SNP genotype, with higher levels of vWF:Ag found in subjects with the CC/AA/GG genotype, intermediate levels in CT/AG/GA subjects, and the lowest levels in subjects with the TT/GG/AA genotype. ABO blood type and age, factors known to influence plasma levels of vWF, did not account for the differences described above. To our knowledge, this is the first report of an association between genotypic variation at the vWF locus and levels of plasma vWF:Ag. Although this association has not yet been assessed in nongroup O subjects, it is likely that the findings of this study will be applicable to other blood groups. One of these polymorphisms, -1051G/A, and an additional SNP, C/G at -1792, were recently reported in a large population of patients with ischemic heart disease (IHD) and healthy, age-matched controls not to be correlated with circulating levels of vWF.29 However, in that study, which did not control for ABO blood group, any potential association may have been obscured by the variability of plasma vWF levels known to result from this factor. Interestingly, in this preliminary report, both the -1792C/G and -1051G/A polymorphisms were identified as potential predictors of risk for IHD. This finding confirms the observations of previous studies of hemostatic variables, such as β-fibrinogen,20 PAI-1,21 protein C,22 and thrombomodulin,23 which have suggested that some individuals may be at greater risk for manifesting thrombotic disease because of inherited differences in hemostatic protein expression. The relative contributions of genetic and environmental factors to vWF expression have not been clearly defined, however, the significant finding in this study that promoter genotype is related to plasma levels of vWF:Ag would imply a significant role for genetic determinants of vWF:Ag concentration, particularly when combined with the well-documented relationship between ABO blood type and circulating levels of vWF. However, the magnitude of this genotypic effect is likely modulated by several environmental factors, as interactions between genotype and environmental variables have been reported for the determination of plasma levels of other hemostatic proteins such as fibrinogen (reviewed in Humphries et al30), and PAI-1.31 In fact, a genotype-environment interaction might account for the significant correlation between promoter genotype and plasma vWF:Ag levels in subjects >40 years of age and the lack of such an association in subjects ≤40 years of age. Although reports of similar findings are rare, it is not unreasonable to propose that some types of genotypic variation may only be functionally relevant within a specific environmental context, for example, that of the aging circulation. Although the mechanisms mediating expression of vWF in endothelial cells are largely unknown, the biomechanical forces generated by blood flow, such as fluid shear stress, may well play a role in this regulation, and blood flow characteristics are well known to change with advancing age. Interestingly, a contrasting result was recently reported in the study of the relationship between a SNP in the β-fibrinogen promoter (-455G/A) and plasma fibrinogen levels within different age groups. When grouped by decade of age, the association of -455G/A genotype with plasma fibrinogen levels, although significant in the 45 to 55-year old age group, was diminished in the older age groups (55 to 65 years and >65 years).32
Several different mechanisms could account for the association between vWF promoter SNP genotype and plasma vWF:Ag level. The three polymorphisms determining the promoter genotype might affect the binding affinity of nuclear proteins involved in regulating transcription of the vWF gene. Differential binding of nuclear proteins has been demonstrated at the site of the 4G/5G PAI-1 polymorphism; both alleles bound a transcriptional activator, whereas the 5G allele also bound a repressor protein, which was associated with reduced basal levels of PAI-1 transcription.21 Alternatively, these SNPs may be in linkage disequilibrium with functionally important sites elsewhere, either in the coding region of the gene, which might affect posttranscriptional processing of vWF, or in regulatory sequences in close proximity to the vWF gene. Preliminary evidence that the genotype of two of these three SNPs alters the binding affinity of nuclear proteins involved in transcriptional regulation has been presented in this study. Although these sequences are greater than 1 kb upstream of the transcriptional start site, the in vivo study of vWF regulatory sequences has suggested that elements in addition to the region of the promoter from -487 to +250 bp are required for widespread cell-type–specific expression.33 Thus, sequences further upstream of -487 may very well play an important role in transcriptional regulation of this locus in the intact organism. Differential binding of at least two proteins (NFκB and GATA-2) potentially involved in the regulation of vWF transcription was predicted by searching the sequences surrounding these polymorphic sites against a transcription factor profile database. EMSAs with rhNFκB p50 and BAEC nuclear extracts showed significant differential protein binding between the C and T alleles at -1234 and the G and A alleles at -1051, respectively. A supershift EMSA performed with the -1234T allele sequence confirmed the preferential binding of NFκB p50 to this allele. Intriguingly, the differential protein binding at both -1234 and -1051 indicates enhanced binding with the allelic sequences that constitute part of haplotype 2 (TGA), the haplotype associated with lower plasma levels of vWF:Ag. If the influence of these SNP genotypes is mediated through the direct interaction of proteins with these sites, the implication of these findings is that at least one of the proteins binding to these sites possesses the properties of a transcriptional repressor. Transient transfection studies, using constructs containing either haplotype 1 or haplotype 2 SNP allele sequences situated upstream of a luciferase reporter gene, are currently in progress to address this issue. Interestingly, even though NFκB has been, to date, widely regarded as a positive regulator of gene expression, it was recently reported that developmental silencing at the human ζ-globin gene locus was mediated by a NFκB site in the 3′-flanking region.34 Regardless, the differential binding of endothelial cell nuclear proteins to SNP allele sequences suggests that the polymorphisms in the vWF promoter may have a direct functional role in the control of gene expression.
While a significant correlation between SNP genotype in the vWF promoter and circulating levels of vWF:Ag was observed for this group of subjects, it is quite possible that these SNPs may act in concert with other polymorphic sequence elements in the vWF promoter, or within the coding region of the gene, to influence levels of plasma vWF. However, if the recent assessment of sequence diversity in the human lipoprotein lipase gene, which reported 79 single nucleotide substitutions within a 9.7 kb region,35 is any indication of the extent of sequence variation within other genes, then the identification of functionally relevant sites, or combination of sites, could be challenging. Nevertheless, given the apparent clinical significance of elevated levels of plasma vWF, further studies should investigate whether the genotype-specific increase in plasma vWF:Ag levels contributes directly to the increased risk of thrombotic disease in these individuals and if the promoter genotype could predict for therapeutic response.
ACKNOWLEDGMENT
The authors acknowledge Dr Tony Giulivi, of the Canadian Red Cross, for provision of the blood samples.
Supported by Grant No. NA-3661 from the Heart and Stroke Foundation of Ontario, Canada. A.M.K. is the recipient of a Medical Research Council of Canada Studentship and D.L. is a Career Investigator of the Heart and Stroke Foundation of Ontario.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to David Lillicrap, MD, Department of Pathology, Queen’s University, Kingston, Ontario, Canada K7L 3N6; email: lillicrap@cliff.path.queensu.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal