Abstract
The E2A-HLF fusion gene, formed by the t(17;19)(q22;p13) chromosomal translocation, is thought to drive the leukemic transformation of early B-cell precursors by repressing an evolutionarily conserved apoptotic pathway. To test this hypothesis, we sought to identify downstream targets of E2A-HLF in t(17;19)+ pro-B leukemia cells (UOC-B1) that had been transfected with a zinc-inducible vector encoding a dominant-negative suppressor (E2A-HLF[dn]) of the oncoprotein. Representational difference analysis of mRNAs from E2A-HLF(dn)+ UOC-B1 cells grown with (E2A-HLF inactive) or without (E2A-HLF active) the addition of zinc yielded several differentially expressed cDNA fragments that were individually subcloned. Two of the clones, designated F-5 and G-4, hybridized with mRNAs that were upregulated by E2A-HLF. Levels of both transcripts declined sharply within 8 to 12 hours after suppression of E2A-HLF DNA-binding activity, becoming undetectable after 96 hours. The F-5 cDNA was identified as a portion of ANNEXIN VIII, whose product was expressed in promyelocytic leukemia cells and UOC-B1 cells, but not in other leukemic cell lines. A novel full-length cDNA cloned with the G-4 fragment encoded a protein that we have named SRPUL (sushi-repeat protein upregulated in leukemia). It is normally expressed in heart, ovary, and placenta, but could not be detected in leukemic cell lines other than UOC-B1. Neither protein prevented apoptosis in interleukin-3–dependent murine pro-B cells, suggesting that they have paraneoplastic roles in leukemias that express E2A-HLF, perhaps in the disseminated intravascular coagulopathy and hypercalcemia that characterize these cases.
THE E2A-HLF FUSION gene is generated by the t(17;19) (q23;p13) in cases of pro-B acute leukemia that occur in older children and adolescents.1,2 The leukemias associated with this genetic alteration do not respond well to standard chemotherapy and typically present with an intravascular coagulopathy and hypercalcemia—unusual findings in children with B-cell precursor acute lymphoblastic leukemia (ALL).3,4 The encoded E2A-HLF protein contains the AD1 and AD2 transactivation domains of E2A,5-8 linked to the bZIP DNA binding/protein dimerization region of HLF. HLF is a member of the PAR subfamily of leucine zipper-type transcription factors that is normally expressed in liver, kidney cells, and neurons within the central nervous system.1,2,9 Overexpression of a dominant-negative inhibitor of the chimera in leukemic cells transformed by E2A-HLF results in programmed cell death.10Similarly, introduction of wild-type E2A-HLF into Baf-3 interleukin-3 (IL-3)-dependent murine pro-B lymphocytes allows the cells to survive in G1 phase of the cell cycle for 2 weeks or longer after IL-3 withdrawal, and to resume exponential growth when IL-3 is reintroduced.10 These findings suggest that E2A-HLF contributes to leukemogenesis by altering the transcriptional control of genes responsible for lineage-specific cell suicide in pro-B cells, leading to the survival of defective cells that normally would be eliminated by apoptosis.10
A critical first step in understanding malignant transformation mediated by E2A-HLF is to identify downstream responder genes targeted by this oncoprotein. Thus, we are exploiting a matched cell system based on the presence or absence of E2A-HLF transcriptional activity. It uses a t(17;19)+ pro-B leukemia cell line (UOC-B1) transduced with a zinc-inducible vector encoding a dominant-negative suppressor of E2A-HLF, which sequesters the chimeric transcription factor within nonfunctional complexes, thereby blocking its ability to bind to the HLF DNA consensus sequence.10 Of several possible approaches to identify potential targets of E2A-HLF action, we have chosen the subtractive process of representational difference analysis (RDA), a polymerase chain reaction (PCR)-based technique that has proved useful for selecting and cloning cDNAs that correspond to differentially expressed mRNAs.11-15
Here we describe the use of RDA to identify two genes whose expression depends on the activity of E2A-HLF. One gene was identified asANNEXIN VIII, which is also expressed in acute promyelocytic leukemia.16,17 The second is a novel gene that encodes a protein we have named SRPUL (sushi-repeat protein upregulated in leukemia), which contains consensus repeat motifs found in the extracellular domain of members of the selectin family.18,19SRPUL is most closely related to a gene called SRPX (sushi repeat-containing protein), orETX1, which may be involved in X-linked retinitis pigmentosa.20 21 The ANNEXIN VIII and SRPUL proteins could not substitute for E2A-HLF in promoting the survival of murine pro-B cells deprived of growth factor, suggesting that they are more likely to be involved in paraneoplastic syndromes characteristic of t(17;19)+ leukemias than in processes leading to malignant transformation.
MATERIALS AND METHODS
Construction of eukaryotic expression vectors.
Expression plasmids containing wild-type E2A-HLF, a dominant-negative suppressor of E2A-HLF binding (E2A-HLF[dn]), ANNEXIN VIII, and SRPUL-CFwere constructed in the pMT-CB6+ eukaryotic expression vector (a gift from Dr F. Rauscher III, Wistar Institute, Philadelphia, PA). This vector contains the inserted cDNA under control of the sheep metallothionine promoter, as well as the neomycin-resistance gene (neoR) driven by the SV40 early promoter.
Cell culture and cell survival assay.
UOC-B1 human pro-B-cell leukemia cells that express E2A-HLF and HL-60 human myeloid leukemia cells were cultured in RPMI 1640 medium containing 10% fetal calf serum. UOC-B1(dn) clone 3 and control UOC-B1 (pMT) cells transfected with either E2A-HLF(dn) or the pMT empty vector were prepared as described previously.10E2A-HLF(dn) expression was induced in the UOC-B1(dn)3 cells by treating them with 100 μmol/L ZnSO4 for 24 hours. Viable cell counts were determined by trypan-blue dye exclusion in triplicate assays. FL5.12 pro-B lymphocytes22 were cultured in RPMI-1640 medium containing 10% fetal calf serum and 10% WEHI-3B–conditioned medium (as a source of IL-3). Transfectants were generated by electroporation using 2 × 107 cells and 80 μg of plasmid DNA with a gene pulser (Bio-Rad, Hercules, CA) set to deliver 320 V and 960 μF. The cells were then cultured in 24-well dishes and selected in the presence of the G418 neomycin analogue (0.6 mg/mL) for 2 to 4 weeks. The levels of protein expression were determined by Western blotting, and cell clones with regulated expression were selected for further experimentation. For cell survival assays, protein expression was induced by treating cells with 100 μmol/L ZnSO4 for 16 hours before growth factor deprivation. IL-3 was removed by repeated centrifugation in fresh media, and the cells were adjusted to 5 × 105 per mL on day 0 and cultured without IL-3. Viable cell counts were determined by trypan-blue dye exclusion.
Northern blot analysis.
One microgram of messenger RNA, extracted with Fast Track (Invitrogen, Carlsbad, CA), was separated by electrophoresis in 1% agarose gels containing 2.2 mol/L formaldehyde and transferred to nylon membranes. Signals were visualized by autoradiography after hybridization to a human HLF, ANNEXIN VIII, SRPUL, or β-ACTIN32P-labeled cDNA. A multiple-tissue Northern blot (Clontech, Palo Alto, CA) contained 2 μg each of poly(A) RNAs isolated from various normal human tissues.
Reverse transcriptase (RT)-PCR.
Total RNA was extracted from leukemia cells using the Trizol reagent (GIBCO-BRL, Gaithersburg, MD), following the manufacturer’s directions. Reverse transcription was performed with 2 μg of total RNA. RNA was resuspended in a final volume of 11 μL with 50 ng of random hexamer primer (Pharmacia Biotech, Uppsala, Sweden), incubated at 80°C for 5 minutes and cooled on ice. Reverse transcription was performed using 200 U SuperScript II reverse transcriptase (GIBCO-BRL) in the manufacturer’s buffer in the presence of 0.5 mmol/L dNTP and 1.0 mmol/L dithiothreitol in a final volume of 20 μL, and incubated at 42°C for 1 hour. Each sample was then incubated with 1 μL of RNaseH (GIBCO-BRL) at 37°C for 20 minutes. As a negative control, the reaction was also performed for each sample without reverse transcriptase. cDNA samples were stored at −20°C. PCR was performed using a DNA Thermal cycler (Perkin Elmer, Applied Biosystems, Inc, Foster City, CA). PCR reactions contained 1 μL of the cDNA product, 1× reaction buffer, 1.5 mmol/L MgC12, 0.2 mmol/L dNTP, 10 pmol of each primer, and 2.5 U of Taq polymerase (Perkin Elmer) in a total volume of 50 μL. Thermocycling parameters for the first PCR and the sequences of the specific primers are as follows: E2A-HLF, 30 cycles at 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 1 minute; primers described previously1; ANNEXIN VIII, 40 cycles at 94°C for 30 seconds, 52°C for 30 seconds, and 72°C for 1 minute; sense primer 5′-ATGCAGAGACCCTCTACAAA-3′ and antisense primer 5′-CATAGTCTTCCTCATACGCC-3′; SRPUL, 40 cycles at 94°C for 30 seconds, 52°C for 30 seconds, and 72°C for 1 minute; sense primer 5′-GAGGAAATTATCACAGCAGC-3′ and antisense primer 5′-CAGTGTAACGAATCACATGC-3′; c-ABL, 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute; sense primer 5′-GTATCATCTGACTTTGAGCC-3′ and antisense primer 5′-GTACCAGGAGTGTTTCTCCA-3′. A second PCR was performed for SRPUL using the same primers and 3 μL of the first PCR products as template for 30 cycles. The amplification of c-ABL mRNA was performed as a control to assess the quality of each RNA sample. These sets of the primers amplify a 342-bp ANNEXIN VIII product, a 501-bp SRPUL product, and a 288-bp c-ABL product. Ten microliters of the PCR products were electrophoresed in a 2.0% agarose gel and transferred to a nylon membrane (NEN Life Science, Boston, MA) with 0.4 N NaOH. The membranes were hybridized with an internal probe end-labeled with 32P using polynucleotide kinase, and visualized by autoradiography. The sequences of the internal probes were as follows: E2A-HLF, 5′-ACCCTCCCTGACCTGTCTCG-3′; ANNEXIN VIII, 5′-CTCATTGTGGCCCTTATGTA-3′; SRPUL, 5′-GAGAAATTGACTGCTCGAGT-3′; c-ABL, 5′-TAACTAAAGGTGAAAAGCTCC-3′.
Immunoblot analysis.
Cells were solubilized in Nonidet P-40 (NP-40) lysis buffer (150 mmol/L NaCl, 1.0% NP-40, 50 mmol/L Tris [pH 8.0]), and total cellular proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After wet electrotransfer of the products onto nitrocellulose membranes, immunoblotting was performed withANNEXIN VIII17 and HLF(C)23 specific rabbit antiserum, preimmune rabbit serum, and FLAG monoclonal antibody (MoAb) (M2; Eastman Kodak, New Haven, CT). The blots were then stained with primary antibodies and studied by horseradish peroxidase–conjugated donkey anti-rabbit– and sheep anti-mouse–enhanced chemiluminescence (Amersham Life Sciences Inc, Arlington Heights, IL).
RDA.
RDA was performed essentially as described by Hubank and Schatz.11 Briefly, mRNAs were isolated from UOC-B1(dn)3 cells harboring the E2A-HLF(dn) construct and grown in the presence or absence of 100 μmol/L zinc in the culture medium. A 10-μg sample of mRNA from each cell population was used for RDA analysis. cDNAs were synthesized from the mRNAs and digested withDpnII. Adapters, 5′-AGCACTCTCCAGCCTCTCACCGCA-3′ (RBgl24) and 5′-GATCTGCGGTGA-3′ (RBgl 12), were ligated to the DpnII-digested cDNA. This mixture was amplified by PCR with RBgl 24 oligonucleotides, and the adapters were excised withDpnII. A second pair of adapters, 5′-ACCGACGTCGACTATCCATGAACA-3′ (JBgl 24) and 5′-GATCTGTTCATG-3′ (JBgl 12), were ligated to the amplified fragments from UOC-B1(dn) cells grown without zinc (tester) and hybridized with the RBgl 24-amplified cDNA fragments (RBgl adapters removed) from zinc-induced UOC-B1(dn) cells (driver) at a ratio of 1:80 for 20 hours. The hybridization mix was used as template for amplification by PCR. A second round of subtraction was performed by removing the JBgl adapters from an aliquot of the first-round PCR product, ligating a third pair of oligonucleotides adapters, 5′-AGGCAACTGTGCTATCCGAGGGAA-3′ (NBgl 24) and 5′-GATCTTCCCTCG-3′ (NBgl 12), and hybridizing with driver amplicons at a ratio of 1:800. A third round of subtraction was performed by removing the NBgl adapters from an aliquot of the second-round PCR product, relegating the JBgl 24 and JBgl 12 adapters, and hybridizing with driver amplicons at a ratio of 1:8000. After PCR amplification with JBgl 24 primers, several bands were evident in 2.0% agarose gels stained ethidium bromide; these were individually subcloned and tested for differential expression by Northern blotting of RNAs from UOC-B1(dn)3 cells grown in the absence or presence of zinc.
Cloning of ANNEXIN VIII full-length cDNA by RT-PCR.
The ANNEXIN VIII cDNA was cloned by RT-PCR using upstream and downstream primers with flanking EcoRI sites (5’-CGTGTGGAATTCCAGCAGAGGCCAACC-3’ and 5’-CTCTGAATTC-ATGGTCTTTGCTCTTG-3’). RT-PCR was performed with a cDNA Cycle Kit (Invitogen). The nucleotide sequence of the insert, confirmed by DNA sequencing, was identical to that of the VACβcDNA.24
Metabolic labeling and immunoprecipitation.
Cells were metabolically labeled by incubation in methionine-free medium for 30 minutes. [35S]methionine (New England Nuclear, Boston, MA) was then added (0.5 mCi/mL of culture medium), and the cells were incubated for an additional 60 to 180 minutes. After removal of the medium, the cells were lysed for 60 minutes on ice in radioimmunoprecipitation assay buffer (150 mmol/L NaCl, 1.0% NP-40, 0.5% SDS, 50 mmol/L Tris-HCl [pH 8.0]), plus 50 μg/mL leupeptin, 0.5% aprotinin, 1 mmol/L Na vanadate, 1 mmol/L phenylmethylsulfonyl fluoride, and 2 mmol/L EDTA. The cell lysates from 2 × 107 cells were clarified by centrifugation at 20,000gfor 30 minutes. These lysates were then mixed with the designated rabbit antiserum or α-FLAG MoAb for 60 to 120 minutes at 4°C, and immune complexes were collected by incubation for 60 minutes with protein A-Sepharose (Amersham Pharmacia Biotech, Uppsala, Sweden). To detect secreted proteins, we labeled FL5.12 cells expressing SRPUL in 4 mL of methionine-free medium, which also was immunoprecipitated with the α-FLAG MoAb and protein A-Sepharose. The proteins were analyzed by SDS-PAGE followed by autoradiography. The level of protein expression was quantified with use of a phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Isolation, sequence analysis, and in vitro transcription-translation of the SRPUL cDNA.
A λ-Zap II human heart cDNA library (Stratagene, La Jolla, CA) and a λ-Zap II UOC-B1(dn) cDNA library (Stratagene) were hybridized with the 340-nucleotide G-4 probe. The pBluescript phagemid was purified from positive clones by in vivo excision from the λ-Zap II vector, using ExAssist helper phage (Stratagene). The full-length SRPULclone hybridized to the same E2A-HLF–regulated transcript in UOC-B1(dn) cells as did the G-4 probe. Sequence analysis of theSRPUL cDNA was performed by the Center for Biotechnology at St Jude Children’s Research Hospital using dye-terminator cycle sequencing ready reaction kits with AmpliTaq DNA polymerase FS (Perkin-Elmer [PE], Applied Biosystems, Inc [ABI]) and synthetic oligonucleotides. Samples were electrophoresed, detected, and analyzed on PE/ABI model 373, model 373 Stretch, or model 377 DNA sequencers. In vitro transcription-translation analysis of phagemid DNA (1.0 μg) was performed with [35S]methionine (Amersham, translation grade) in a coupled transcription-translation assay system (TNT Kit; Promega, Madison, WI). Products were analyzed by electrophoresis in SDS-PAGE, after which the gels were dried and subjected to autoradiography.
Construction of plasmids expressing SRPUL with a C-terminal FLAG-tag (SRPUL-CF).
We added the FLAG epitope, with the amino acid sequence DYKDDDDK, to the C-terminus of the SRPUL protein. To generate pMT-CB6+/SRPUL-CF, we amplified a sequence by PCR with full-length SRPUL as a template that included the translation initiation signal and FLAG coding sequence, using and an upstream primer with a flanking HindIII site (5′-CCCAAGCTTGCCATGGCCAGTCAGC-TAACTCA-3′) and a downstream primer with a flanking BamHI site (5′-CGCGGATCCTCACTTGTCATCGTCGTCCTTGTAGTCCTCGCATATGTCCCTTTGCT-3′). PCR was performed using Pfu DNA polymerase (Stratagene). The PCR product was double-digested with HindIII and BamHI and cloned into HindIII/BamHI-digested pMT-CB6+. The nucleotide sequence of the insert was confirmed by DNA sequencing.
Indirect immunofluorescence.
Indirect immunofluorescence analysis was performed as described by Murti et al,25 with minor modifications. Cells were fixed with 3.7% paraformaldehyde for 20 minutes, washed three times with PBS, and permeabilized in 0.2% Triton-X (Sigma, St Louis, MO)/PBS for 10 minutes at room temperature. After three washes with PBS, the slides were preincubated with 5% skim milk powder, and 1% normal horse serum in PBS for 30 minutes at room temperature, and then incubated with α-FLAG MoAb M2 (10 μg/mL) for 1 hour at room temperature in a humidified chamber. The cells were washed four times with PBS, and then incubated with fluorescein (FITC)-conjugated goat-mouse IgG (1/2,000) (Jackson Immuno Research Laboratories, West Grove, PA) for 30 minutes at room temperature. The samples were examined with a Nikon Labophot-2 microscope equipped with epifluorescence optics for FITC detection and 4.6-diamidino-2-phenylindole (DAPI).
RESULTS
Identification of genes regulated by E2A-HLF.
The effects of blocking E2A-HLF activity in UOC-B1 leukemia cells were studied with a conditional dominant-negative mutant,10E2A-HLF(dn), which lacks the AD1 transactivation domain of E2A6,7 and contains a mutated HLF DNA-binding domain with an intact leucine-zipper domain. Thus, E2A-HLF(dn) will heterodimerize with E2A-HLF and sequester the endogenous oncogenic form of the protein within inactive complexes that are unable to bind to DNA.10
In clones transfected with the E2A-HLF(dn) cDNA in a zinc-inducible vector, expression of the mutant protein was highly dependent on addition of zinc to the culture medium, as shown in Fig 1 for UOC-B1(dn)3 cells. By contrast, the endogenous E2A-HLF oncoprotein was expressed at constant levels in both UOC-B1(dn)3 and control cells, regardless of the addition of zinc. Immunoblot analysis showed that E2A-HLF(dn) levels were 10-fold higher than those of the wild-type protein at 4 to 8 hours after the addition of zinc, even though expression of the mutant mRNA was at lower levels than that of the wild-type E2A-HLF transcript (Fig 2C). This likely reflected either higher translation efficiency or a prolonged half-life of the mutant compared with the wild-type protein. The mutant protein was relatively stable, in that its levels did not decrease appreciably until 2 days after zinc was removed from the cultures (Fig 1). Binding of E2A-HLF by the E2A-HLF(dn) mutant, leading to protein heterocomplexes unable to interact with the HLF consensus sequence, was shown in a previous publication.10 In that study, the E2A-HLF(dn) protein induced apoptosis of UOC-B1(dn) cells, beginning approximately 72 hours after induction of its expression and 48 hours after the loss of E2A-HLF DNA-binding activity. The rate of apoptotic death exceeded the rate of mitotic division, resulting in a progressive loss of viable cells over a 7-day period, with a gradual return to normal survival after removal of zinc from the culture medium.10 Thus, our inducible system provided matched cell populations with active and suppressed E2A-HLF functions, allowing the analysis of differentially expressed genes.
Immunoblot analysis of E2A-HLF and E2A-HLF(dn) proteins. Cell lysates were prepared from UOC-B1(dn)3 cells grown in the absence of zinc (lane 1) or in its presence (100 μmol/L) for the indicated intervals (lanes 2 through 7). After 24 hours, a portion of the cells were washed to remove zinc and cultured for the indicated intervals in the absence of the metal (lanes 8 through 11). Control UOC-B1 cells electroporated with the pMT empty vector (UOC-B1[pMT]) and grown for 24 hours in medium without (lane 12) or with 100 μmol/L zinc (lane 13).
Immunoblot analysis of E2A-HLF and E2A-HLF(dn) proteins. Cell lysates were prepared from UOC-B1(dn)3 cells grown in the absence of zinc (lane 1) or in its presence (100 μmol/L) for the indicated intervals (lanes 2 through 7). After 24 hours, a portion of the cells were washed to remove zinc and cultured for the indicated intervals in the absence of the metal (lanes 8 through 11). Control UOC-B1 cells electroporated with the pMT empty vector (UOC-B1[pMT]) and grown for 24 hours in medium without (lane 12) or with 100 μmol/L zinc (lane 13).
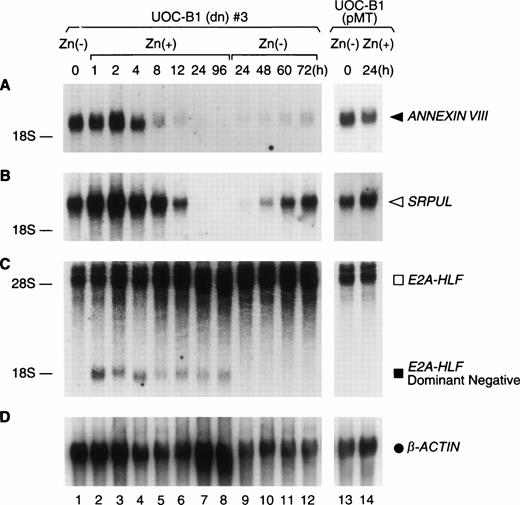
Upregulation of ANNEXIN VIII and SRPULcDNAs by E2A-HLF. Northern blot analysis of poly(A) RNA (1 μg per lane) prepared from UOC-B1(dn)3 cells grown in zinc-free medium (lane 1) or in medium supplemented with zinc for the indicated times (lanes 2 through 8), followed by zinc depletion (lanes 9 through 12). Control experiments were performed with UOC-B1 cells transfected with an empty vector and grown in zinc-free medium (lane 13) or for 24 hours in the presence of zinc (lane 14). (A) The blot was hybridized with ANNEXIN VIII (F-5) (◂), (B)SRPUL (G-4) (◃), or (C) HLF cDNA fragments, and then stripped and rehybridized with a β-ACTIN probe (D) (•).
Upregulation of ANNEXIN VIII and SRPULcDNAs by E2A-HLF. Northern blot analysis of poly(A) RNA (1 μg per lane) prepared from UOC-B1(dn)3 cells grown in zinc-free medium (lane 1) or in medium supplemented with zinc for the indicated times (lanes 2 through 8), followed by zinc depletion (lanes 9 through 12). Control experiments were performed with UOC-B1 cells transfected with an empty vector and grown in zinc-free medium (lane 13) or for 24 hours in the presence of zinc (lane 14). (A) The blot was hybridized with ANNEXIN VIII (F-5) (◂), (B)SRPUL (G-4) (◃), or (C) HLF cDNA fragments, and then stripped and rehybridized with a β-ACTIN probe (D) (•).
Using mRNAs from UOC-B1(dn)3 cells grown in the absence of zinc (E2A-HLF transcriptionally active) or for 24 hours in its presence (E2A-HLF transcriptionally inactive), we prepared cDNAs and subjected them to three rounds of sequential hybridization and PCR amplification according to the RDA protocol of Hubank and Schatz.11Several differentially amplified cDNA fragments were individually subcloned and tested for differential gene expression by Northern blotting of UOC-B1(dn) RNAs. cDNA fragments representing two differentially regulated genes were identified, and designated F-5 and G-4. Sequence comparisons showed that the 351-nucleotide F-5 fragment corresponded to part of the coding region and the 3′ nontranslated region of ANNEXIN VIII, also known as human vascular anticoagulant beta (VAC-β).24 By contrast, the G-4 cDNA fragment represented a novel gene, because it did not correspond to any previously known cDNA identified with the BLAST search utility (National Center for Biotechnology Information). The predicted amino acid sequence of the protein contained so-called sushi repeats,20 26 so we have named this gene SRPUL, for sushi-repeat protein upregulated in leukemia.
Northern blot analysis showed high levels of ANNEXIN VIII andSRPUL mRNA expression in UOCB1(dn)3 cells, which decreased rapidly, coincident with expression of the E2A-HLF(dn) mRNA (Fig 2). The ANNEXIN VIII mRNA level declined fivefold by 8 hours after the addition of zinc and 25-fold by 24 hours, becoming undetectable by 96 hours (Fig 2A). Similarly, there was a threefold decrease in concentrations of SRPUL mRNA within 12 hours and a 60-fold deficit by 24 hours; SRPUL transcripts were also not detectable after 96 hours (Fig 2B). Removal of zinc from the growth medium restored both SRPUL and to a lesser extent ANNEXIN VIIImRNA expression by 72 hours, coincident with a decline in the E2A-HLF(dn) protein level (Fig 1). ANNEXIN VIII andSRPUL were unaffected by zinc in control UOC-B1 cells (UOC-B1[pMT]) lacking the dominant-negative construct, confirming that the observed changes in gene expression were stimulated by E2A-HLF (Fig 2, lanes 13 and 14).
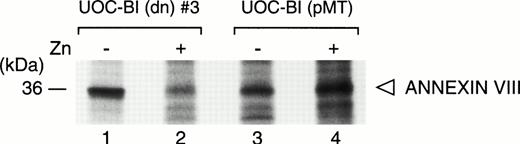
To show ANNEXIN VIII regulation at the protein level, we used specific antiserum to immunoprecipitate metabolically labeled lysates from UOC-B1(dn) and UOC-B1(pMT) cells grown with or without zinc. ANNEXIN VIII protein synthesis rates were threefold lower in UOC-B1(dn) cells grown in the presence of zinc, and remained unchanged in control UOC-B1(pMT) cells, regardless of the zinc concentration (Fig 3).
Regulation of ANNEXIN VIII protein synthesis. UOC-B1(dn)3 (lanes 1 and 2) or control UOC-B1(pMT) (lanes 3 and 4) cells were grown without exogenous zinc (lanes 1 and 3) or for 24 hours in the presence of zinc (100 μmol/L; lanes 2 and 4). Metabolically labeled lysates of these cells were immunoprecipitated with an ANNEXIN VIII antiserum.
Regulation of ANNEXIN VIII protein synthesis. UOC-B1(dn)3 (lanes 1 and 2) or control UOC-B1(pMT) (lanes 3 and 4) cells were grown without exogenous zinc (lanes 1 and 3) or for 24 hours in the presence of zinc (100 μmol/L; lanes 2 and 4). Metabolically labeled lysates of these cells were immunoprecipitated with an ANNEXIN VIII antiserum.
Expression of ANNEXIN VIII and SRPUL is associated with E2A-HLF expression in early B-lineage leukemias.
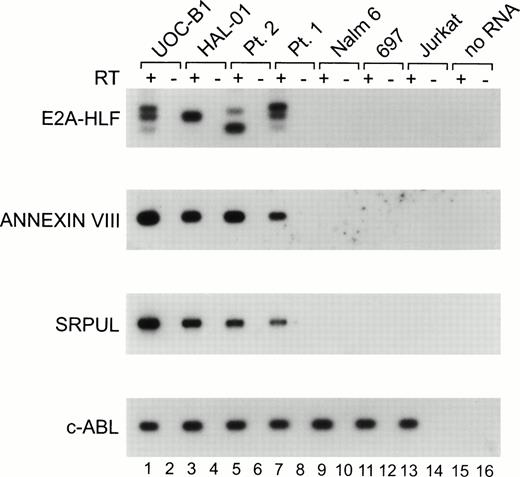
To assess the linkage between expression of the E2A-HLFchimeric oncogene and the expression of ANNEXIN VIII andSRPUL, we performed RT-PCR analysis of leukemic cell lines and cryopreserved primary leukemia patient samples with and without the t(17;19). As shown in Fig 4, the UOC-B1 and HAL-01 t(17;19)-positive cell lines, as well as two primary patient leukemia samples (patients #1 and #21), each express theE2A-HLF fusion transcript as well as mRNAs encoding ANNEXIN VIII and SRPUL. The E2A-HLF fusion transcripts migrate differently for each of the patients shown in Fig 4, because the joining region at the fusion junction contains unique numbers of inserted nucleotides, as described previously.1,2 The UOCB1 cell line was derived from cells of patient 1,1 which explains the identical pattern of E2A-HLF for these two samples. Examples of RNAs from three other leukemia cell lines lacking the t(17;19) are shown in Fig 4, including the Nalm6 and 697 early B-lineage and the Jurkat T-cell cell lines. These cell lines did not express either E2A-HLF or transcripts encoding ANNEXIN VIII or SRPUL, although they did express the c-ABLcontrol, indicating that the RNA was intact for these samples. In all, a total of 10 leukemic cell lines were tested using the RT-PCR assay shown in Fig 4. ANNEXIN VIII expression was detected in the REH early B-lineage and the U937 monocytic cell lines lacking E2A-HLF, in addition to the four samples expressing E2A-HLFshown in Fig 4. SRPUL was not expressed by any of the cell lines evaluated except those shown in Fig 4 that expressedE2A-HLF. Thus, our analysis suggests that expression of ANNEXIN VIII and SRPUL is consistently associated with expression of the E2A-HLF fusion gene, and that expression of these responder genes is relatively rare in leukemias transformed through the auspices of other genetic alterations.
RT-PCR analysis. Each RNA was transcribed with (+; odd lanes) or without (−; even lanes) RT. The PCR products were transferred to a nylon membrane and analyzed by hybridization with end-labeled internal oligonucleotide probes specific for each gene. RNAs were evaluated for the UOC-B1 (lanes 1 and 2) and HAL-01 (lanes 3 and 4) t(17;19)-positive human leukemic cell lines, patient samples with t(17;19)-positive ALL (lanes 5 through 8), the t(17;19)-negative Nalm6 early B-lineage leukemic cell line (lanes 9 and 10), 697 pre-B leukemic cell line (lanes 11 and 12), and Jurkat T-cell leukemic cell line (lanes 13 and 14), as well as a control PCR reaction lacking RNA template (lanes 15 and 16).
RT-PCR analysis. Each RNA was transcribed with (+; odd lanes) or without (−; even lanes) RT. The PCR products were transferred to a nylon membrane and analyzed by hybridization with end-labeled internal oligonucleotide probes specific for each gene. RNAs were evaluated for the UOC-B1 (lanes 1 and 2) and HAL-01 (lanes 3 and 4) t(17;19)-positive human leukemic cell lines, patient samples with t(17;19)-positive ALL (lanes 5 through 8), the t(17;19)-negative Nalm6 early B-lineage leukemic cell line (lanes 9 and 10), 697 pre-B leukemic cell line (lanes 11 and 12), and Jurkat T-cell leukemic cell line (lanes 13 and 14), as well as a control PCR reaction lacking RNA template (lanes 15 and 16).
Overexpression of E2A-HLF induces ANNEXIN VIII but not SRPUL expression in HL-60 cells.
Next, we tested whether expression of the intact E2A-HLF chimeric protein can induce expression of ANNEXIN VIII and SRPUL mRNAs. For these experiments, HL-60 cells were transfected with a pMT-CB6+/E2A-HLF construct to generate cell clones in which the oncogenic form of E2A-HLF is regulated by the sheep metallothionein promoter (eg, HL-60[E2A-HLF]24 in Fig 5). By 4 hours after addition of zinc to the culture medium, E2A-HLF protein levels had increased fivefold (Fig 5A, lane 3) by comparison with background levels mediated by the sheep metallothionine promoter in the absence of zinc (lane 1). ANNEXIN VIII mRNA expression lagged several hours behind the expression of E2A-HLF, becoming detectable 8 hours after zinc addition and reaching its highest level by 24 hours (Fig 5B). E2A-HLF did not induce SRPUL mRNA expression in HL-60 cells (Fig 5C).
ANNEXIN VIII expression in HL-60 cells engineered to express E2A-HLF. (A) Immunoblot analysis of HL-60 cells transfected with zinc-regulated pMT-CB6+/E2A-HLF (HL-60[E2A-HLF]24 cells). Cell lysates were prepared from cells grown in medium lacking exogenous zinc (lane 1) and at serial intervals after the addition of zinc (100 μmol/L) to the medium (lanes 2 through 8). Control HL-60 cells transfected with the empty pMT vector (HL-60 [pMT]) did not express E2A-HLF, whether cells were grown in the absence (lane 9) or presence (lane 10) of zinc. Northern blot analysis of poly(A) RNA (1 μg per lane) prepared from HL-60(E2A-HLF)24 or HL-60(pMT) cells grown under the same conditions hybridized with either (B) ANNEXIN VIII or (C) SRPUL cDNA probes. (D) The blot was stripped and rehybridized with a control β-ACTIN probe.
ANNEXIN VIII expression in HL-60 cells engineered to express E2A-HLF. (A) Immunoblot analysis of HL-60 cells transfected with zinc-regulated pMT-CB6+/E2A-HLF (HL-60[E2A-HLF]24 cells). Cell lysates were prepared from cells grown in medium lacking exogenous zinc (lane 1) and at serial intervals after the addition of zinc (100 μmol/L) to the medium (lanes 2 through 8). Control HL-60 cells transfected with the empty pMT vector (HL-60 [pMT]) did not express E2A-HLF, whether cells were grown in the absence (lane 9) or presence (lane 10) of zinc. Northern blot analysis of poly(A) RNA (1 μg per lane) prepared from HL-60(E2A-HLF)24 or HL-60(pMT) cells grown under the same conditions hybridized with either (B) ANNEXIN VIII or (C) SRPUL cDNA probes. (D) The blot was stripped and rehybridized with a control β-ACTIN probe.
Cloning of full-length SRPUL.
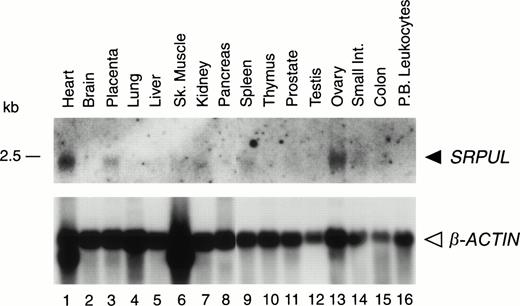
Northern blotting with the 340-bp G-4 probe showed a SRPUL mRNA of 2.5 kb, which was expressed at highest levels in heart, ovary, and placenta (Fig 6). Using the G-4 probe to screen cDNA libraries from mRNA of human heart library and UOC-B1(dn) cells, we obtained positive clones with approximately 2-kb cDNA inserts. The longest cDNA consisted of 2,133 bp, and had one long open-reading frame that extended from nucleotide 419 to 1813, potentially encoding a 465-amino acid protein (GenBank accession no.AF060567). The initial ATG codon was preceded by an in-frame termination codon, and hence constitutes the bona fide initiation codon for this protein. The molecular weight of the protein was predicted to be 53 kD, consistent with results of an in vitro transcription-translation experiment showing a 50-kD product (data not shown). Comparison of the deduced SRPUL amino acid sequence with protein databases revealed three consensus (sushi) repeats of approximately 60 amino acids, starting with amino acid 59 (Fig 7). These repeats contain six conserved cysteines, and are characteristic motifs in members of the selectin family of cell membrane proteins with functional roles in cell adhesion.18,19 The highest homology scores (identity 47%, similarity 67%) were obtained with a sushi repeat–containing protein designated SRPX or ETX1,20,21 which may be involved in X-linked retinitis pigmentosa, and with its rat homolog, whose expression is downregulated by v-src.27
Expression of SRPUL mRNA in normal tissues. Northern blot analysis of poly(A) RNA (2 μg per lane) isolated from various human tissues, hybridized with a SRPUL cDNA probe, and then stripped and rehybridized with a β-ACTIN probe. The mobility of a 2.5-kb RNA marker is indicated.
Expression of SRPUL mRNA in normal tissues. Northern blot analysis of poly(A) RNA (2 μg per lane) isolated from various human tissues, hybridized with a SRPUL cDNA probe, and then stripped and rehybridized with a β-ACTIN probe. The mobility of a 2.5-kb RNA marker is indicated.
Alignment of the SRPUL sushi repeat domains. The numbers to the left of each sequence indicate the first and last residue in each repeat, according to the SRPUL cDNA sequence deposited in Genbank (accession no. AF060567). The conserved cysteine residues are numbered from one to six. A consensus sequence derived from the P-selectin (GMP-140) consensus repeat domains is also shown.18
Alignment of the SRPUL sushi repeat domains. The numbers to the left of each sequence indicate the first and last residue in each repeat, according to the SRPUL cDNA sequence deposited in Genbank (accession no. AF060567). The conserved cysteine residues are numbered from one to six. A consensus sequence derived from the P-selectin (GMP-140) consensus repeat domains is also shown.18
The predicted amino acid sequence of SRPUL also contains 30 hydrophobic amino-terminal amino acids with features typical of a leader peptide, including a signal cleavage site behind the serine residue at position 30.28 SRPUL lacks a hydrophobic amino acid region distal to the leader peptide that could function as a transmembrane domain, suggesting that it is a secreted protein. A precedent for alternative splicing occurs in the mRNA of a related protein, P-selectin (GMP-140), resulting in a secreted protein that lacks a transmembrane domain.17 18 However, of six nearly full-lengthSRPUL cDNAs from heart or UOC-B1(dn) cDNA libraries that we analyzed, none corresponded to an alternatively spliced form with a potential transmembrane domain.
Cellular localization of SRPUL-CF in FL5.12 cells.
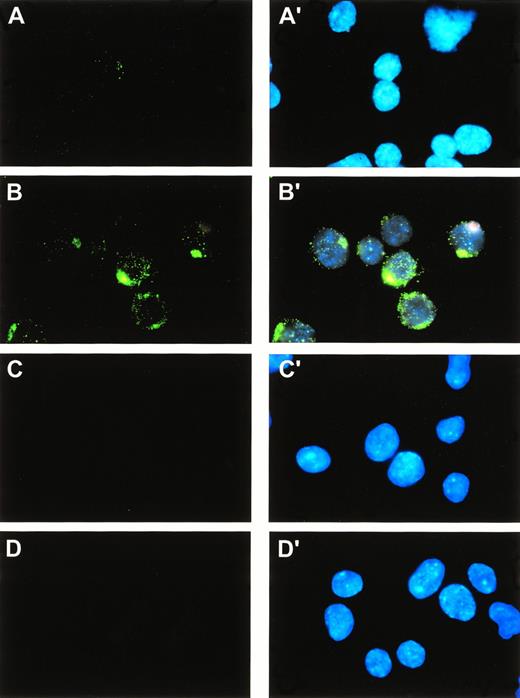
Cellular localization of the SRPUL protein was determined by immunofluorescence staining of FL5.12 cells transfected with a zinc-inducible, C-terminal FLAG-tagged SRPUL cDNA clone. Sixteen hours after the addition of zinc, these cells showed a cytoplasmic distribution of SRPUL-CF expression, suggesting localization to secretory vesicles in a pattern typical of secreted proteins (Fig 8B and B′). FL5.12 (SRPUL-CF) cells grown without zinc showed faint cytoplasmic staining because of low (base-line) levels of expression programmed by the pMT vector in the absence of zinc induction (Fig 8A); mock-transfected control cells were not stained by the antibody (Fig 8C and D).
Subcellular localization of SRPUL-CF proteins. FL5.12 cells expressing SRPUL-CF were immunostained with an -FLAG MoAb (M2). Simultaneous staining with DAPI permitted visualization of cell nuclei (panels A′, B′, C′, D′). The FL5.12 cells were stably transfected with a zinc-regulated pMT vector containing SRPUL-CF (panels A, A′, B, B′) or the empty vector (panels C, C′, D, D′). Cells were grown in the absence of zinc (panels A, A′, C, C′) or 16 hours after its addition to the medium (100 μmol/L; panels B, B′, D, D′).
Subcellular localization of SRPUL-CF proteins. FL5.12 cells expressing SRPUL-CF were immunostained with an -FLAG MoAb (M2). Simultaneous staining with DAPI permitted visualization of cell nuclei (panels A′, B′, C′, D′). The FL5.12 cells were stably transfected with a zinc-regulated pMT vector containing SRPUL-CF (panels A, A′, B, B′) or the empty vector (panels C, C′, D, D′). Cells were grown in the absence of zinc (panels A, A′, C, C′) or 16 hours after its addition to the medium (100 μmol/L; panels B, B′, D, D′).
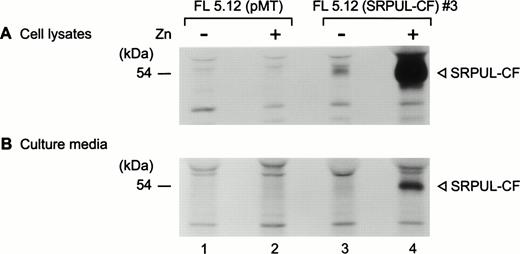
Synthesis and secretion of SRPUL-CF in FL5.12 cells.
FL5.12 (SRPUL-CF) cells and FL5.12 cells transfected with empty vector (pMT) were metabolically labeled after culture for 24 hours in the presence and absence of zinc. SRPUL-CF was immunoprecipitated with a murine α-FLAG MoAb (M2) from cell lysates and the culture media after continuous labeling for 3 hours. In the presence of zinc, FL5.12 (SRPUL-CF) cells produced high levels of the labeled SRPUL-CF protein (Fig 9A). During the 3-hour incubation, a substantial quantity of radiolabeled SRPUL-CF was recovered from the culture medium (Fig 9B), as would be predicted for a secreted protein.
(A) Synthesis and secretion of SRPUL-CF. Metabolically labeled lysates of FL5.12 (SRPUL-CF)3 cells were immunoprecipitated with the -FLAG MoAb (M2) (lanes 3 and 4) and compared with control FL5.12 (pMT) cells (lanes 1 and 2). (B) Secretion of SRPUL-CF by FL5.12 (SRPUL-CF)3 cells (lane 4), as shown by immunoprecipitation from the culture medium 16 hours after the addition of zinc (100 μmol/L).
(A) Synthesis and secretion of SRPUL-CF. Metabolically labeled lysates of FL5.12 (SRPUL-CF)3 cells were immunoprecipitated with the -FLAG MoAb (M2) (lanes 3 and 4) and compared with control FL5.12 (pMT) cells (lanes 1 and 2). (B) Secretion of SRPUL-CF by FL5.12 (SRPUL-CF)3 cells (lane 4), as shown by immunoprecipitation from the culture medium 16 hours after the addition of zinc (100 μmol/L).
Lack of antiapoptotic activity of ANNEXIN VIII and SRPUL-CF.
Because E2A-HLF protects IL-3–dependent murine pro-B cells from apoptosis caused by IL-3 deprivation,10 we asked if either ANNEXIN VIII or SRPUL-CF could perform this function in the absence of the oncoprotein. Thus, we transfected FL5.12 cells with the zinc-regulated vector encoding ANNEXIN VIII or SRPUL-CF. Independent clones were isolated after G-418 selection that expressed either of the proteins in the presence of 100 μmol/L ZnSO4, at levels that were approximately 10-fold higher than the background level expressed by cells grown in medium lacking the metal (Fig 10A). As shown in Fig 10B, FL5.12 cells expressing E2A-HLF survived with little loss of viability, whereas those expressing ANNEXIN VIII or SRPUL-CF rapidly underwent apoptosis after removal of IL-3 from the growth medium. These observations indicate that neither protein blocks apoptosis in our test cell system. Thus, their dysregulation by E2A-HLF in UOC-B1 human leukemia cells probably contributes to properties of t(17;19)+ leukemias other than prolonged cell survival.
Effects of E2A-HLF, ANNEXIN VIII, and SRPUL-CF on the survival of FL5.12 cells deprived of growth factor. (A) Immunoblot analysis with antisera to HLF(C) and ANNEXIN VIII and with the -FLAG MoAb (M2). Results are shown for three independently derived clones of G418-resistant FL5.12 cells transfected with each of the full-length cDNAs (FL5.12 [E2A-HLF], FL5.12 [ANNEXIN VIII] and FL5.12 [SRPUL-CF]) under control of the zinc-regulated pMT promoter as well as controls transfected with the empty vector (FL5.12 [pMT]). (B) Survival of FL5.12 cells expressing E2A-HLF, ANNEXIN VIII, or SRPUL-CF, in the absence of IL-3. Cells growing exponentially in IL-3–supplemented media for 16 hours in the presence or absence of zinc were adjusted to 5 × 105 cells/mL on day 0, and cultured after removal of IL-3. The cell number of viable cells is shown for E2A-HLF-, ANNEXIN VIII-, and SRPUL-CF–expressing transfected FL5.12 clones grown in the presence (open symbols) or absence (closed symbols) of zinc (100 μmol/L).
Effects of E2A-HLF, ANNEXIN VIII, and SRPUL-CF on the survival of FL5.12 cells deprived of growth factor. (A) Immunoblot analysis with antisera to HLF(C) and ANNEXIN VIII and with the -FLAG MoAb (M2). Results are shown for three independently derived clones of G418-resistant FL5.12 cells transfected with each of the full-length cDNAs (FL5.12 [E2A-HLF], FL5.12 [ANNEXIN VIII] and FL5.12 [SRPUL-CF]) under control of the zinc-regulated pMT promoter as well as controls transfected with the empty vector (FL5.12 [pMT]). (B) Survival of FL5.12 cells expressing E2A-HLF, ANNEXIN VIII, or SRPUL-CF, in the absence of IL-3. Cells growing exponentially in IL-3–supplemented media for 16 hours in the presence or absence of zinc were adjusted to 5 × 105 cells/mL on day 0, and cultured after removal of IL-3. The cell number of viable cells is shown for E2A-HLF-, ANNEXIN VIII-, and SRPUL-CF–expressing transfected FL5.12 clones grown in the presence (open symbols) or absence (closed symbols) of zinc (100 μmol/L).
DISCUSSION
A large number of chimeric or otherwise dysregulated transcription factors are produced by the diverse chromosomal translocations found in specific types of acute leukemia and sarcomas.29-31Research into mechanisms by which these aberrant proteins subvert normal programs of cell proliferation, differentiation, and survival is now focused on the downstream genes transcriptionally regulated in transformed cells. Our goal is to identify the genetic programs responsible for the transformed phenotype of ALLs with the t(17;19), which express the chimeric transcription factor E2A-HLF and carry a poor prognosis.3 4 Two classes of genes are relevant in this regard—those mediating the antiapoptotic properties of E2A-HLF and those contributing to the unusual clinical features of patients with t(17;19)+ acute leukemias, such as coagulopathy and hypercalcemia.
Using the RDA procedure we identified two genes, ANNEXIN VIIIand SRPUL, whose expression clearly depended on the DNA-binding activity of E2A-HLF. We also were able to show upregulation ofANNEXIN VIII in HL-60 cells by conditional expression of the oncogenic form of E2A-HLF, providing further evidence of ANNEXIN VIII regulation by the fusion protein. Although SRPUL mRNA levels showed the same pattern of regulation as those at ANNEXIN VIII in response to the dominant-negative form of E2A-HLF in UOC-B1 cells, the two genes were not coordinately upregulated in HL-60 cells. This discrepancy likely reflects cell-type–specific differences that determine whether individual genes are E2A-HLF responsive, such as the requirement for other transcriptional regulatory proteins or local epigenetic factors such as chromatin configuration or DNA methylation status.
Our studies do not establish whether the promoter of either ANNEXIN VIII or SRPUL is a direct target of E2A-HLF, but rather implicate these genes as elements of biochemical cascades responsive to modulation by the chimeric transcription factor. The fact that we were able to isolate only two differentially regulated RDA PCR products suggests the presence of other, undetected transcripts that were upregulated by E2A-HLF. Braun et al,12 in their RDA analysis of genes induced by the EWS-FLI fusion protein, found that at least a 10-fold difference in transcript levels between the tester and driver populations was needed to ensure detection of differentially expressed mRNA species.12 Our results favor the interpretation that RDA preferentially identifies highly regulated genes, in that both ANNEXIN VIII and SRPUL mRNA levels declined more than 25-fold to undetectable levels as E2A-HLF transcriptional activity was progressively suppressed through dominant-negative interference with DNA binding.
ANNEXIN VIII belongs to a family of Ca2+-dependent phospholipid-binding proteins that includes 13 members with similar structures, characterized by the presence of four to eight repeats of a 70-amino acid domain and a variable region at the N-terminus.32-34 Annexins are cytosolic proteins, and their ability to associate with negatively charged phospholipids of the inner membrane bilayer in the presence of Ca2+ seems to be required for their antiphospholipase, anticoagulant, antiinflammatory, and antiprotein kinase C activities. Annexin VIII was initially identified as VAC-β, a protein whose anticoagulant activity depends on its phospholipid-binding properties and that also possesses phospholipase A2 activity.24 Moreover, the annexins drive Ca2+-dependent aggregation of secretory granules and are postulated to have roles in endosomal vesicle trafficking and cell-matrix interactions.32-34
Of greatest relevance to the present study, ANNEXIN VIII is specifically upregulated in cases of acute promyelocytic leukemia (APL), but not in most other forms of chronic or acute leukemia.16 Treatment of APL cells with all-trans-retinoic acid induces downregulation of ANNEXIN VIII at the transcriptional level, suggesting that it is a target of either normal retinoic acid receptors or the PML-RARα chimeric protein in these cells.16,17 Interestingly, patients with acute leukemias characterized by either the E2A-HLF or PML-RARα chimeric protein share a defining presenting feature—a bleeding diathesis that resolves after effective antileukemic therapy.3,4,35 The anticoagulant activity of ANNEXIN VIII may depend on its ability to inhibit phospholipid-dependent activation of prothrombin by factor Xa.24 The localization of ANNEXIN VIII at the cytosolic face of the plasma membrane has been interpreted to argue against a role for this protein in the coagulopathy that affects patients with leukemias harboring PML-RARα fusion proteins.17 However, a high rate of spontaneous leukemic cell death is a prominent feature in newly diagnosed patients, providing a means by which ANNEXIN VIII could be released from cells and contribute to the bleeding problems associated with these two forms of acute leukemia.
The ability of ANNEXIN VIII to inhibit phospholipase A2(PLA2)24,36 would predict a contribution of the protein to the aberrant survival of leukemic cells expressing E2A-HLF. PLA2 activation through tumor necrosis factor (TNF)- or Fas-mediated pathways has been linked to the production of second messengers (such as arachidonic acid) that participate in cell death pathways.37 Other inhibitors of cystolic PLA2, including a trifluoromethylketone analog of arachidonic acid,38 have been shown to partially inhibit TNF-induced apoptosis. However, ANNEXIN VIII did not produce an antiapoptotic effect when it was overexpressed in murine pro-B lymphocytes deprived of growth factor (Fig 10). This suggests that any antiapoptotic activity by ANNEXIN VIII in transformed cells would likely be restricted to situations that involve TNF or FAS signaling, in which PLA2 activity would be activated through specific caspase-induced cleavage.39
The SRPUL gene product possesses substantial homology with a sushi repeat–containing protein (SRPX or ETX1), which was independently isolated as a rat gene downregulated through the activity of v-src.20,21,27SRPX (ETX1) is abundantly expressed in the retina and has been implicated as a candidate gene for one form of X-linked retinitis pigmentosa,20,21 a heterogeneous group of human genetic disorders that together constitute the most common cause of inherited blindness. Although retinitis pigmentosa is characterized by the progressive loss of retinal cells leading to retinal degeneration and blindness, apparently because of the aberrant programmed death of photoreceptor cells,40 there was no indication in the present study of an antiapoptotic role for SRPUL in pro-B cells deprived of growth factor.
The three sushi repeats (or complement control sequences) that characterize the SRPUL protein were first identified in plasma β2 glycoprotein41 and transglutaminases, such as factor XIIIa.26 They also occur in members of the selectin family of adhesion molecules found on endothelium, platelets, and leukocytes,18,19,42-44 an association with implications for the function of SRPUL in pro-B lymphocytes transformed by E2A-HLF. Sequence analysis of each of the three members of the selectin family predict type-1 transmembrane proteins with N-terminal lectin-like domains, epidermal growth factor (EGF) repeats, and variable numbers of modules—60 amino acids each—that comprise the sushi or complement consensus repeats.19 The lectin and EGF domains seem to have major roles in selectin-mediated adhesion,45,46 with the sushi repeats preventing direct contact of the lectin and EGF regions with the cell surface, thereby enhancing adhesive function.19 Importantly, inactivation of sushi modules in L-selectin and E-selectin inhibits the adhesive function of both molecules.47
The selectins are thought to contribute to the adhesive properties of migrating tumor cells,48,49 and hence to play a role in tumor metastasis, particularly in gastrointestinal cancers and lymphomas.50,51 Although expression of cell adhesion molecules is known to be upregulated in leukemic B-cell precursors,52-56 relatively little attention has been paid to be correlation between these observations and the dissemination of leukemic cells or prognosis. Because E2A-HLF–positive leukemias are characterized by bone invasion and hypercalcemia, rare observations in early B-lineage leukemias lacking the t(17;19),4 57 we suggest that aberrant secretion of SRPUL may contribute to this unique feature of the disease.
We considered it likely that many of the genes stimulated directly or indirectly by E2A-HLF do not participate in the antiapoptotic activity of the oncoprotein, but rather contribute to the leukemic phenotype in other ways. Thus, identifying the primary leukemogenic target of E2A-HLF remains a crucial step in elucidating the mechanism of action of this fusion protein. Representational difference analysis by PCR may well lack the sensitivity to detect genes that are regulated at only modest levels,12 so that modifications of this procedure or perhaps completely different techniques will be needed to advance results of the present study. One promising approach is analysis of cDNA microarrays on glass slides,58-61 which would permit one to identify genes whose levels are more modestly induced or repressed as part of the complete gene program regulated by the activity of the E2A-HLF oncoprotein. By identifying each of the genes that are transcriptionally regulated in response to the expression of E2A-HLF, we should ultimately be able to understand the mechanisms through which this fusion protein transforms early lymphoid progenitors.
ACKNOWLEDGMENT
We thank J. Wu for excellent technical assistance. We also thank F. Rauscher III for providing the pMT-CB6+ expression vector, R. Hauptmann and G.R. Adolf for VAC-β–specific antiserum and MoAb, K. Ohyashiki and K. Toyama for the HAL-01 cell line, the St Jude Tumor Processing Laboratory for primary ALL samples with the (17;19) chromosomal translocation, and J. Gilbert for scientific editing and helpful discussions.
Supported in part by grants from the National Cancer Institute (CA 59571 and CA 21765) and by the American Lebanese Syrian Associated Charities (ALSAC), St Jude Children’s Research Hospital.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to A. Thomas Look, MD, Department of Experimental Oncology, St Jude Children’s Research Hospital, 332 N Lauderdale, Memphis, TN 38105-2794; e-mail: thomas.look@stjude.org.

![Fig. 1. Immunoblot analysis of E2A-HLF and E2A-HLF(dn) proteins. Cell lysates were prepared from UOC-B1(dn)3 cells grown in the absence of zinc (lane 1) or in its presence (100 μmol/L) for the indicated intervals (lanes 2 through 7). After 24 hours, a portion of the cells were washed to remove zinc and cultured for the indicated intervals in the absence of the metal (lanes 8 through 11). Control UOC-B1 cells electroporated with the pMT empty vector (UOC-B1[pMT]) and grown for 24 hours in medium without (lane 12) or with 100 μmol/L zinc (lane 13).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/1/10.1182_blood.v93.1.321/5/m_blod40105001w.jpeg?Expires=1769205446&Signature=XwmpvAZhIQRbVl1t9YUDu4Mi9LIvweamJDU0ZfJuk0pby5E~oXdABgQFdZGxCaRC5uPvrYOaqyKRzZB7Ukg9Uhvsock2hP19weIcMcSqJmnBCfT0hPW-LkNQFq3DFeY2GdJHGz~TBI-zCgbFj7EPv76wLmQuOV7fBlPdvbJqpAv0uTqxQ9RgB9fTUgszJBklqJYqoQrYs~i8jisfzWltomQW1Khwf-zNVmCmR6plPN8hKqebBbc2K-I0QzTISaSSfI5Qq3Db2dgpEOpM7u-3MSlxebt3TbPMuNZsRzbtnoSehsGaDzdyFTyHasIp-gD4LgsbQIz6EUa8s1TUE5GS5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. ANNEXIN VIII expression in HL-60 cells engineered to express E2A-HLF. (A) Immunoblot analysis of HL-60 cells transfected with zinc-regulated pMT-CB6+/E2A-HLF (HL-60[E2A-HLF]24 cells). Cell lysates were prepared from cells grown in medium lacking exogenous zinc (lane 1) and at serial intervals after the addition of zinc (100 μmol/L) to the medium (lanes 2 through 8). Control HL-60 cells transfected with the empty pMT vector (HL-60 [pMT]) did not express E2A-HLF, whether cells were grown in the absence (lane 9) or presence (lane 10) of zinc. Northern blot analysis of poly(A) RNA (1 μg per lane) prepared from HL-60(E2A-HLF)24 or HL-60(pMT) cells grown under the same conditions hybridized with either (B) ANNEXIN VIII or (C) SRPUL cDNA probes. (D) The blot was stripped and rehybridized with a control β-ACTIN probe.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/1/10.1182_blood.v93.1.321/5/m_blod40105005w.jpeg?Expires=1769205446&Signature=hPhPyeEgkpMpZrDT8qwesl1s7lKcX-yhlnBC3tqGd9rSebT6b6LWYzIyCAY5pGSCNvW3z48akCy8KS6sOXIGCOxi16KWgWtqR2sA-NFPSXjE7PhVwK7SUh2SsCh3CO41~vkelliOdLIFnpvjR0vnoZ2kNUIvxJ6VRYcKuM~Vo8cl05oQp6jzxtjLLNHdwGNMUO0riPrn7OSDNoPonJ0BHeeFZ9EyvmcuZZTUFqpxOo8IGSrZ3mUNbH1pX~Ae4cWIpa-~WngBdWLPoMjWmdnksg85fCGdJFEzDlQlISsNZjOXPky1HSmiOO9yuzlCyO8tXdboTRLfip4u-l~~-XUCcw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 10. Effects of E2A-HLF, ANNEXIN VIII, and SRPUL-CF on the survival of FL5.12 cells deprived of growth factor. (A) Immunoblot analysis with antisera to HLF(C) and ANNEXIN VIII and with the -FLAG MoAb (M2). Results are shown for three independently derived clones of G418-resistant FL5.12 cells transfected with each of the full-length cDNAs (FL5.12 [E2A-HLF], FL5.12 [ANNEXIN VIII] and FL5.12 [SRPUL-CF]) under control of the zinc-regulated pMT promoter as well as controls transfected with the empty vector (FL5.12 [pMT]). (B) Survival of FL5.12 cells expressing E2A-HLF, ANNEXIN VIII, or SRPUL-CF, in the absence of IL-3. Cells growing exponentially in IL-3–supplemented media for 16 hours in the presence or absence of zinc were adjusted to 5 × 105 cells/mL on day 0, and cultured after removal of IL-3. The cell number of viable cells is shown for E2A-HLF-, ANNEXIN VIII-, and SRPUL-CF–expressing transfected FL5.12 clones grown in the presence (open symbols) or absence (closed symbols) of zinc (100 μmol/L).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/1/10.1182_blood.v93.1.321/5/m_blod40105010w.jpeg?Expires=1769205446&Signature=OSS9RngqEcmWO9Wo86p7JI-9HtFt7RuRLWl9BUnkxJvqFonkFNXrppgqP-Ce0EXRxq7ILO10VkMQ73xD530JNDTdlz09aWV0MXbCs5P9lhLCY7x07jv9XyXZ4oUqUI2QyLItXxGt2a9qy3reWZhPWNmxnwWfGFc9RfYkeCW9VTH39qwfN89viC53XfsNjY53j4h5H11nrrzfKJI~u6fV6M1KWt1dRe50UdrCmK0wWgKDLJqVWzR6Ab-53y5ztm7HkW0yeNZ15ZQUri7M6-gVkuXkZnVEmcW~W870OGWH8sB61AISPmJr8KwI7QVd1wutIa7Cm2HsElrmdp6sWqNeVg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal