ACUTE LYMPHOBLASTIC LEUKEMIA (ALL) is the most prevalent type of cancer, as well as the most common form of leukemia in children.1 This lymphoid malignancy, manifested by the proliferation of lymphopoietic blast cells, represents a heterogeneous group of diseases that vary with respect to morphological, cytogenetic, and immunologic features of the transformed cells. Technical improvements in immunofluorescence staining and flow cytometry together with the availability of numerous monoclonal antibodies (MoAbs) that recognize lineage-associated membrane molecules have illuminated the immunophenotypic heterogeneity in ALL. We now know that leukemia cells from patients with ALL may express various combinations of surface antigens that are found normally on lymphocyte precursors at discrete stages of maturation.2,3 Thus, the malignant clones in patients with ALL are thought to originate from normal lymphoid progenitor cells arrested at early stages of B- or T-lymphocyte ontogeny. Although cells from the majority (≈85%) of pediatric patients express B-lineage–associated antigens, those from approximately 15% of patients express the T-lineage–associated antigens CD1, CD2, CD3, CD4, CD5, CD7, or CD8.4-6 T-lineage ALL in children is associated with numerous unfavorable presenting features, thus it is not surprising that children with T-lineage ALL frequently have been reported to have a worse prognosis than children with B-lineage ALL.4,5,7-10 However, a number of encouraging reports from recent clinical studies using contemporary risk-adjusted multiagent chemotherapy programs have documented remarkably improved outcomes for patients with T-lineage ALL.6 10-14 Moreover, advanced preclinical studies have triggered much optimism that new agent discovery programs may lead to further improvements in outcome in the near future. In this review, we discuss current concepts regarding the etiology, biological characteristics, clinical features, and treatment of pediatric T-lineage ALL.
ETIOLOGY
The role of numerous epidemiological factors, including maternal and paternal exposure to radiation, history of maternal fetal loss or fertility problems, higher birthweight at diagnosis, and use of exogenous growth hormone, remains controversial in the cause of pediatric ALL.15-17 A recent comprehensive review found no relationship between exposure to electromagnetic field (EMF) radiation and incidence of childhood ALL.18 The reported space-time clustering of ALL cases, which might suggest an etiologic agent such as a virus, is also controversial.19-23 Human T-cell leukemia virus-I and II may be associated with adult, but not pediatric T-lineage leukemia or lymphoma,24,25 and Epstein-Barr virus infection has been linked to a limited number of cases of T-cell lymphoma, but not T-lineage ALL, in children.26
The autosomal recessive disorder ataxia telangiectasia (AT) appears to be a true etiologic factor because patients with AT have an increased risk of developing lymphoid malignancies, including T-lineage ALL.27 Translocations involving the T-cell receptor (TCR) loci are reported in approximately 10% of the T cells from patients with AT,28 but interestingly, the most frequent of these translocations appear to involve different regions within the TCR loci compared with those observed in patients with T-lineage ALL without AT.29-31 The molecular basis for these effects as well as other genetic abnormalities that may play a role in T-lineage leukemia will be discussed below. Taken together, these data suggest that multiple factors may be involved in the origin of T-lineage ALL.
BIOLOGICAL FEATURES OF T-LINEAGE ALL
Because leukemic cells are thought to originate from normal T-lymphocyte precursors arrested at early stages of ontogeny,2 32 every pathway that ensures homeostasis of a functional immune system is a potential target for disruption. Still, the fundamental issue of how many different mutations are required for malignant transformation to the leukemic state remains to be delineated. Nevertheless, clear associations have been identified between the occurrence of nonrandom translocations or other gene mutations and the development of T-lineage ALL. Below, we describe the specific molecular defects found in T-lineage leukemias and discuss altered signal transduction pathways that may contribute to the malignancy.
Chromosomal translocations.
An array of nonrandom translocations that are specific to T-lineage ALL have been identified; all appear to occur preferentially in the TCR loci on chromosomes 14 and 7.33 The breakpoints in many cases resemble TCR recombination signals, implying that the aberration arose during TCR rearrangement.34-39 Translocations involving chromosomes 1 and 14, such as t(1;14)(p33;q11) and t(1;14)(p32;q11), have been estimated to occur in approximately 3% of T-lineage ALL cases.40 In such rearrangements, the SCL/TCL5/TAL-1 gene from chromosome 1 and the TCRδ gene on chromosome 1436,41,42 are juxtaposed, resulting in deregulation of normal TAL-1 expression.41,43 TAL-1 was predicted to encode a protein containing a helix-loop-helix DNA binding motif,42 43 suggesting that the t(1;14) translocations could contribute to leukemogenesis by inducing aberrant expression of novel or TAL-1–regulated genes.
A distinct TAL-1 disruption occurs via an interstitial deletion between a locus called SIL (SCL interrupting locus) and the 5′UTR of SCL, resulting in a fusion transcript SIL/SCL, and is estimated to occur with a frequency of 16% to 26% in T-lineage ALL.44-46Presence of a TAL-1 disruption was correlated with high white blood cell (WBC) count, high hemoglobin level, and CD2+/CD10− immunophenotypes, and interestingly, 4-year event-free survival (EFS) was higher for patients with TAL-1 disruption compared to those without TAL-1 alterations (59% ± 11% v 44% ± 7%, respectively), although this difference did not reach conventional significance.46Although TAL-1 is required for development of all hematopoietic lineages in mice,47 the gene is not expressed in B- or T-lineage cells,41,48 and interestingly, SCL-transfected, v-ABL–transformed cells appear to be oncogenic in mice.49Taken together, these data suggest that disruption of normal TAL-1 expression may contribute to the transformation of T-cell precursors into leukemic blasts.
The t(10;14)(q24;q11) translocation, first identified in T-cell neoplasms including T-lineage ALL, involves the TCRα/TCRδ locus on chromosome 1434,35,50 and the TCL3 locus on chromosome 10.34,35 An open reading frame within TCL3 encodes a novel homeobox protein, HOX-11, whose expression is deregulated as a result of the translocation.51-53 Moreover, like TAL-1, HOX-11 is capable of DNA binding and transcriptional activation of reporter genes, suggesting a role for this gene in leukemic transformation.54 Additional studies showed that whereas HOX-11 was expressed in leukemic cell lines and leukemic blasts, it was not expressed in normal T lymphocytes,52,53,55 but was required for normal spleen development.56 Reverse transcriptase-polymerase chain reaction (RT-PCR) assays have suggested that HOX-11 alterations may occur with high frequency in patients with T-lineage ALL.57 Thus, deregulation of HOX-11 is likely to be a biologically significant factor in development of T-lineage ALL.
Translocations t(11;14)(p13;q11) and t(11;14)(p15;q11) also are observed frequently in T-lineage ALL58-60; both involve breakpoints within diversity or J segments the region the TCRα or TCRδ genes on chromosome 14.37,61,62 McGuire et al61 described multiple open reading frames near the chromosome 11 breakpoints and identified one at 11p15 as the open reading frame of the TTG-1 gene. Similarly, Boehm et al63 identified the involved region of 11p15 as the rhombotin gene. Both genes encode proteins characterized by duplicate cysteine-rich zinc-finger protein binding homology domains.61,63 A related gene, rhombotin-2/TTG-2, was shown to be deregulated in cases involving 11p13.63,64 Consistent with the predicted structure of the rhombotins, a recent report described the identification of an ets family transcription factor, ELF-2, that contains rhombotin-2 binding domains, suggesting a transcriptional regulatory role for rhombotin-2.65Clinically, several investigators have associated t(11;14) translocations with an immature stage of thymocyte development,37,59 60 but the overall prognostic significance of this translocation remains unclear.
Although translocations involving chromosome 7 occur in both B-precursor and T-lineage ALL, those involving the TCR-β locus at 7q32-36 are specific for T-lineage ALL.66 One such translocation, t(7;19), truncates the lyl-1 gene on chromosome 19,67 presumably resulting in altered DNA-binding ability for lyl-1.68 Another case, t(7;9), results in truncation of the TAN-1 gene on chromosome 9.69 The mouse homologue of TAN-1 is expressed ubiquitously, but is most abundant in lymphoid tissues, suggesting that normal expression of TAN-1 is disrupted in t(7;9)+ ALL.69
The distinct translocation t(1;7)(p34;q34) was shown to juxtapose the TCR-β constant region enhancer upstream of the LCK gene, which encodes an SRC family protein tyrosine kinase that is involved in signal transduction through CD4.70,71 Notably, overexpression of LCK in transgenic mice causes thymomas or both thymomas and peripheral lymphoid malignancies,72,73suggesting a role for deregulated LCK expression in leukemogenesis. The c-myc locus on chromosome 8 defines yet another class of translocations associated with T-lineage ALL. In t(8;14)(q24;q11), c-myc is translocated with the TCRα loci on chromosome 14, resulting in deregulation of myc expression.74,75 In t(2;8), a fusion protein is produced that consists of c-myc and the product of an unidentified locus on chromosome 2.76 The frequency and significance of these translocations are unclear at present.
We have recently determined the frequency and clinical significance of chromosomal abnormalities in a large cohort of patients with T-lineage ALL enrolled on contemporary CCG studies (Heerema N., et al, submitted for publication). Translocations involving 14q11 and 7q32-q36 were among the most frequent abnormalities, but non-TCR loci, including 9p, 6q, 11q23, and 14q32, also were frequently altered. Notably, none of these abnormalities had prognostic significance in the context of the intensive therapies used in contemporary CCG studies. Nevertheless, the array of chromosomal rearrangements described above are a hallmark of the biological diversity of T-lineage ALL and are likely to result from alterations in underlying cellular control mechanisms. Indeed, recent advances in our understanding of cell signaling and cell cycle control suggest that defective cell surveillance mechanisms are likely to be the major factors leading both to unrestrained proliferation of leukemic cells and to the development of chromosomal abnormalities, including translocations, pseudodiploidy, and hyperdiploidy, that are associated with leukemic cells.77-80 Alterations in such control mechanisms are discussed below.
Mutation or loss of cell cycle control genes.
Mutations present in malignant cells allow them to circumnavigate regulators that control proliferation and differentiation. The retinoblastoma (Rb) gene was originally identified as a tumor suppressor gene because of its inactivation in cases of retinoblastoma; prostate, breast, and lung cancers; and leukemias.81Notably, the telomeric Rb1 gene is located on the long arm of chromosome 13 (13q14), which is inactivated or deleted in approximately 6% of T-lineage ALL cases.82 83
In addition to Rb, other proteins that affect cell cycle progression include the cyclin-dependent kinase inhibitors p21, p27, and p57, as well as the inhibitors of Cdk4 (Ink4): p15Ink4b, p16Ink4a, p18Ink4c, and p19Ink4d.84-89 Among the Ink4 family of inhibitors, p15Ink4b and p16Ink4a have been implicated for a role in the biology of T-lineage ALL.90-95Both genes map to 9p21, a region on the short arm of chromosome 9 previously shown to be deleted frequently in T-lineage ALL.33,96-98 In addition, Batova et al95recently reported that the 5′ promoter region of the p15 gene is preferentially hypermethylated, presumably resulting in loss of transcriptional expression in 38% of newly diagnosed T-lineage ALL.
Another critical regulator of cell cycle progression, the p53 gene, is the most frequently mutated gene in human cancers.81 The major function of p53 is to ensure that cells arrest and attempt to repair genotoxic damage before replicating DNA and entering mitosis.99 In p53-deficient mice, the most common tumor that arises is a T-lineage lymphoid malignancy.100 Although p53 mutations are infrequently observed at diagnosis, they are associated with relapse in pediatric T-lineage ALL.101 102
Another sensor for cell damage appears to be the ATM gene product, which is mutated in patients with AT.103 After insult with agents that induce sublethal DNA damage, cells from patients with AT fail to block DNA synthesis and thereby fail to repair the damaged DNA.104 These effects are apparently caused by a failure of the mutated ATM gene to regulate p53.105 ATM-deficient mice develop an aggressive form of T-lineage leukemia/lymphoma,106,107 and, as described above, children with AT frequently develop T-lineage ALL,27,108 109implicating ATM in leukemogenesis.
Other genes implicated in the malignant transformation of leukemic cells are Ets-1 and IKAROS. The Ets-1 T-lymphocyte transcription factor is thought to be important for normal thymic development and for prevention of cell death in normal mature T cells. A mutation in the DNA binding domain of the Ets-1 was reported in a case of T-lineage ALL,110 but the clinical significance of this finding remains to be proven. The IKAROS gene encodes a zinc finger DNA binding protein that is required for lymphoid cell differentiation.111 Heterozygous transgenic mice harboring a defective IKAROS gene develop a very aggressive form of T-cell leukemia, suggesting that IKAROS may serve as a suppressor of leukemic transformation.112
Leukemic cells also appear to be altered in their responses to various stimuli that induce apoptosis. Debatin et al113,114reported that primary leukemic cells and cell lines from adult patients with T-cell leukemia were sensitive to FasL-induced cell killing in vitro, whereas leukemic cells from pediatric patients with T-lineage ALL were resistant. Resistance was unrelated to the quantity of Fas on the cell surface, but was reversed by treatment with the protein synthesis inhibitor cycloheximide, suggesting that short-lived proteins were required for maintenance of the resistant phenotype. In vivo treatment of a human T-lineage ALL-engrafted severe combined immunodeficiency (SCID) mouse with an anti-Fas antibody resulted in prolonged survival, but did not eradicate the disease, supporting the existence of Fas sensitive and insensitive leukemic cells.115 These data suggest that altered responses to apoptotic stimuli or regulatory factors may contribute to the ability of leukemic cells to escape killing by either immune surveillance or cytotoxic agents.
Bcl-2, which protects cells from non–Fas-mediated apoptosis,116,117 is expressed in both T-lineage and B-lineage leukemias, but it is not yet known how this affects their ability to survive cytotoxic treatments. A related protein, Bax,118 acts as an antagonist to Bcl-2 and may confer radiation sensitivity to cells.119 In a recent CCG study, we found a marked variation in Bcl-2 expression by primary leukemic cells from 238 children with newly diagnosed ALL, including 52 patients with T-lineage ALL.120 High-risk features, such as high WBC count, organomegaly, presence of MLL-AF4 or BCR-ABL fusion transcripts, or leukemic cell growth in SCID mice, were not associated with Bcl-2 expression in these patients. For patients with T-lineage ALL, high Bcl-2 expression was predictive of slow early response (ie, M3 day 14 marrow status). However, with limited follow-up and overall excellent outcome for patients, this correlation did not extend to EFS.
CLINICAL FEATURES AND TREATMENT OF T-LINEAGE ALL
T-lineage ALL is distinct from B-lineage ALL not only biologically, but also clinically. Although the basis for these differences is not well understood, clinical characteristics have been useful prognostic factors for guiding the use of experimental treatments. Below, we describe common presenting features, prognostic variables, and treatment outcome of patients with T-lineage ALL based on data accumulated over the last decade. We then focus on causes for treatment failure and discuss new strategies for improving outcome among subgroups of patients who remain at risk for relapse despite intensive therapy.
Presenting features.
The relationship between T-lineage markers and unfavorable presenting characteristics was first noted by Borella, Sen, and others,121-124 and numerous studies have now confirmed that compared to patients with B-lineage ALL, those with T-lineage ALL more frequently show the highest WBC range (≥50,000/μL), are nonwhite, older, exhibit marked enlargement of the spleen, liver, and lymph nodes, and have a mediastinal mass.5,7,9,10 125
Modal chromosome number is often abnormal among patients with ALL, with hyperdiploidy (>50 chromosomes) correlated with favorable outcome and pseudodiploidy associated with poor outcome.58,98,126-129 The hyperdiploid karyotype is more often associated with pre-B or early pre-B immunophenotypes,129 whereas the pseudodiploid karyotype is more often associated with the T-lineage immunophenotype.33Also, “near tetraploid” chromosome number (>65) is more often associated with T-lineage ALL and poor outcome.130 As described above, nonrandom translocations in T-lineage ALL preferentially occur in the TCR loci on chromosomes 7 and 14,33 and those involving the TCRβ locus at 7q32-36 and the TCRαδ 14q11 collectively occur in approximately 20% of all T-lineage ALL cases.33
Risk classification of T-lineage ALL.
In general, treatment protocols for childhood leukemias have relied on the known prognostic factors of age and WBC count, as well as organomegaly rather than immunophenotype for risk assessment. As a result, even though many patients with T-lineage ALL were previously misclassified or not immunophenotyped, they were likely to receive treatment for high-risk ALL based on their other presenting features. In contemporary trials, various groups have used somewhat different criteria for classification, which has complicated comparisons of results between groups, but nevertheless has generally resulted in similar assignment of patients with T-lineage ALL to more intensive treatment protocols, such as Berlin-Frankfurt-Munster (BFM),131,132 modified BFM,12 and the New York (NY) regimen,13 as well as those of the St Jude Children's Research Hospital133 and Dana Farber Cancer Institute.11
From 1983 through 1993, children daignosed with ALL who exhibited National Cancer Institute (NCI) standard risk features134were classified by the CCG as either low risk (ages 2 through 9 years and WBC <10,000/μL) or intermediate risk (ages 2 through 9 years and WBC <10,000 to 49,999/μL, or age 1 year and WBC <50,000/μL), whereas patients exhibiting NCI poor-risk characteristics were classified as follows: high risk, ages 1 through 9 years with WBC ≥50,000/μL or age >10 years; infants, age <1 year; lymphomatous, patients with specific high-risk features, as described.135 As shown in Table1, patients with T-lineage ALL more frequently were assigned to the higher risk than to the lower risk protocols, which is consistent with their clinical features described above.
CCG and NCI Risk Group Classification of Children With B-Lineage and T-Lineage Acute Lymphoblastic Leukemia
| Risk Group . | B-Lineage ALL N = 3,668 . | T-Lineage ALL N = 730 . | ||
|---|---|---|---|---|
| N . | (%)* . | N . | (%)-151 . | |
| CCG-Low | 705 | (19.2) | 58 | (8.0) |
| CCG-Intermediate | 1,575 | (42.9) | 71 | (9.7) |
| CCG-High | 1,059 | (28.9) | 169 | (23.2) |
| CCG-Lymphomatous | 216 | (5.9) | 425 | (58.2) |
| CCG-Infant | 113 | (3.1) | 7 | (1.0) |
| Total | 3,668 | (100.0) | 730 | (100.0) |
| NCI-Standard | 2,213 | (60.3) | 211 | (28.9) |
| NCI-Poor | 1,455 | (39.7) | 519 | (71.1) |
| Total | 3,668 | (100.0) | 730 | (100.0) |
| Risk Group . | B-Lineage ALL N = 3,668 . | T-Lineage ALL N = 730 . | ||
|---|---|---|---|---|
| N . | (%)* . | N . | (%)-151 . | |
| CCG-Low | 705 | (19.2) | 58 | (8.0) |
| CCG-Intermediate | 1,575 | (42.9) | 71 | (9.7) |
| CCG-High | 1,059 | (28.9) | 169 | (23.2) |
| CCG-Lymphomatous | 216 | (5.9) | 425 | (58.2) |
| CCG-Infant | 113 | (3.1) | 7 | (1.0) |
| Total | 3,668 | (100.0) | 730 | (100.0) |
| NCI-Standard | 2,213 | (60.3) | 211 | (28.9) |
| NCI-Poor | 1,455 | (39.7) | 519 | (71.1) |
| Total | 3,668 | (100.0) | 730 | (100.0) |
*Percentage of patients with B-lineage ALL classified into each risk group.
Percentage of patients with T-lineage ALL classified into each risk group.
Treatment outcome in T-lineage ALL.
As noted above, previous studies showed poorer outcomes for patients with T-lineage ALL compared with patients with B-lineage ALL. For example, in the BFM group, Henze et al136 reported poor outcome for patients with T-lineage ALL who were treated on DAL (adapted from St Jude protocol VII), with 9-year probabilities of continuous complete remission (CCR) of 9% ± 9% and 41% ± 5%, for T-lineage and non–T-lineage, respectively. In contrast, patients treated on BFM achieved CCR of 52% ± 13% and 65% ± 5%, respectively, suggesting that BFM provided superior treatment for T-lineage ALL.
Investigators of the Pediatric Oncology Group7 treated 53 patients with T-lineage ALL with a modified LSA2L2 regimen that had been shown to be efficacious for treatment of T-cell non-Hodgkin's lymphoma.
Although complete remission was achieved for 88% of the patients, the projected overall 3-year EFS was only 40% (SE = 8.3%). Moreover, for patients with WBC count <50,000, the projected 3-year EFS was 67%, whereas for patients with WBC count >50,000, 3-year EFS was only 19%. In a follow-up study, 253 children with T-lineage ALL treated by a modified LSA2L2 regimen together with cranial radiation therapy and triple intrathecal therapy for presymptomatic treatment of central nervous system (CNS) disease achieved an overall 4-year EFS of 43% (SE = 4%).8 Thus, although outcomes improved, the LSA2L2 regimen remained ineffective for the majority of patients with T-lineage ALL. Similarly, in an analysis of data from St Jude studies X and XI, conducted from 1979 to 1983, 120 children with T-lineage ALL had a 5-year EFS of 46% (SE = 18%).137 In a French trial, Garand et al10 treated 88 pediatric patients with T-lineage ALL by protocols such as BFM or FRALLE,138 and an EFS of approximately 58% was reported for a median follow-up of 30 months, suggesting that such therapy could improve outcome for these patients.
Although the studies described above generally found unfavorable outcomes for patients with T-lineage ALL, other recent studies have reported improved outcomes through the use of highly intensive treatment protocols. For example, using an intensive four-drug induction and multidrug continuation, including doxorubicin and prednisone together with prophylaxis for CNS disease and high-dose L-asparaginase, Clavell et al11 reported improved outcome (4-year EFS of 71%) for high-risk patients, including those who had T-lineage ALL. More recently, in a study by Schorin et al1420 patients with T-lineage ALL treated with multiagent chemotherapy together with cranial irradiation and intrathecal methotrexate for 2 years also had favorable outcomes (7-year EFS of 70%, SE = 10%).14 The favorable outcome was attributed to the inclusion of L-asparaginase and doxorubicin in the treatment regimen.
Studies by the CCG also have shown improvements in EFS outcome for high-risk patients with ALL including those with the T-lineage immunophenotype. Steinherz et al13 used an intensive multidrug chemotherapy (NY regimen) to treat 100 patients with characteristics previously correlated with a high risk for relapse. This patient population included 13 patients with T-lineage ALL (defined as E-rosette–+). Four-year EFS for the entire cohort was 69% (SE = 5%), whereas 4-year EFS for patients with T-lineage ALL was 75%. Gaynon et al139 used a modified BFM therapy involving four-drug induction and aggressive continuation therapy to treat high-risk children, including 60 who were E-rosette–+. Overall 3-year EFS was 65% (SD = 3.5%); patients with WBC count >50,000 who were E-rosette–+ had a 3-year EFS of 75% (SD = 6.9%), whereas those who were E-rosette–− had an EFS of 51% (SD = 6.3%).
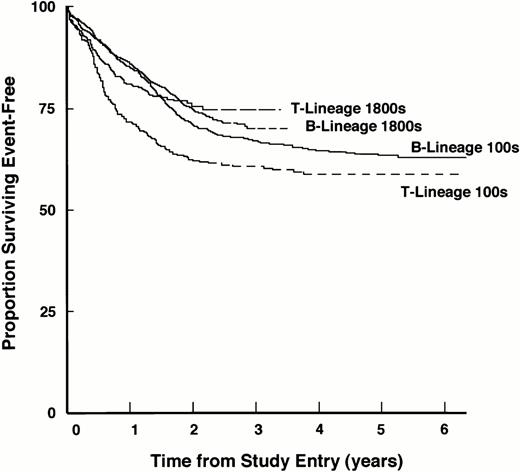
To investigate the outcome of patients with T-lineage ALL on these regimens more thoroughly, we recently analyzed data from the large cohort of patients enrolled on CCG studies conducted between 1983 and 1993.134 Notably, we observed a significant improvement in outcome of patients with T-lineage ALL compared with those on earlier studies because of marked decreases in the incidences of induction failures, early bone marrow relapses, and CNS relapses when more aggressive therapy was given (Fig 1). The probability of 3-year survival for patients with T-lineage ALL increased from 56% in studies conducted between 1978 and 1983, to 65% in studies conducted between 1983 and 1989, and to 78.8% in studies conducted between 1989 and 1993 (Table 2).Taken together, these various studies suggest that current risk for patients with T-lineage ALL treated by intensive therapeutic regimens is similar to that of patients with B-lineage ALL. Thus, a major improvement in treatment of T-lineage ALL has been achieved.
Improved EFS of patients with T-lineage ALL in the context of contemporary intensive chemotherapy programs. EFS for the entire cohort of patients with T-lineage and B-lineage ALL treated on the 1800 series and 100 series of CCG studies are shown. EFS values at designated points in follow-up are given in the text.
Improved EFS of patients with T-lineage ALL in the context of contemporary intensive chemotherapy programs. EFS for the entire cohort of patients with T-lineage and B-lineage ALL treated on the 1800 series and 100 series of CCG studies are shown. EFS values at designated points in follow-up are given in the text.
Outcome for Patients With T-Lineage ALL Treated During Three Consecutive CCG Treatment Eras
| CCG Study Era . | Years . | Event-Free Survival (%) . | |
|---|---|---|---|
| 3-Year . | 5-Year . | ||
| CCG-160s | 1978-1983 | 56.4 | 52.5 |
| CCG-100s | 1983-1989 | 65.8 | 61.0 |
| CCG-1800s | 1989-1993 | 78.2 | 75.2 |
| CCG Study Era . | Years . | Event-Free Survival (%) . | |
|---|---|---|---|
| 3-Year . | 5-Year . | ||
| CCG-160s | 1978-1983 | 56.4 | 52.5 |
| CCG-100s | 1983-1989 | 65.8 | 61.0 |
| CCG-1800s | 1989-1993 | 78.2 | 75.2 |
Prognostic factors in T-lineage ALL.
A number of risk factors for T-lineage ALL were identified in the studies described above. For example, Dowell et al9 and Shuster et al8 reported that compared with patients with T-lineage ALL whose leukemic cells were CD10−, those whose cells were CD10+ were more likely to achieve remission and have significantly improved EFS outcomes. In another study, Pui et al137 reported that CD3 positivity in association with an abnormal karyotype was a significant adverse risk factor; 5-year EFS for patients with both of these characteristics was 35%. In contrast, Shuster et al8 found no prognostic significance for CD3 expression; rather, the most important favorable prognostic factors for patients with low WBC count or high WBC count at diagnosis were CD5 positivity or expression of the THY antigen, respectively.
The findings that many patients with T-lineage ALL now can achieve a much improved outcome has motivated attempts to identify subgroups of patients within T-lineage ALL that may exhibit improved or reduced probabilities of survival. Two previous CCG studies described above noted a favorable association between outcome and E-rosette (CD2) positivity among high-risk patients.13,139To determine comprehensively the clinical significance of CD2 expression in T-lineage ALL, we prospectively immunophenotyped leukemic cells from the large cohort of children enrolled on CCG studies between 1983 and 1993.140 We noted a statistically significant correlation (P = .0006) between the CD2 antigen expression frequency (ie, the average percentage of blasts that were positive for CD2) and EFS. Compared with patients with the highest CD2 expression level, patients with intermediate and low CD2 expression frequencies had relative hazard rates (RHR) of 1.27 and 2.01, indicating an increased risk of treatment failure. After 6 years of follow-up, the EFS estimates for the three CD2 expression groups (low expression frequency to high expression frequency) were 49.3%, 63.5%, and 72.2%, respectively. CD2 expression remained a significant predictor of EFS after adjustment for the effects of other covariates by multivariate regression. Expression of other antigens (CD3, CD5, CD10, or CD34) by leukemic cells was not correlated with EFS. Thus, the expression frequency of CD2 antigen is a powerful predictor of EFS that may be useful for risk classification or assignment to novel therapies aimed at improving patient outcome.
Maturation stage of the predominant leukemic clones also has been suggested as a means for subgrouping patients with T-lineage ALL. Crist et al4 stratified 101 patients with T-lineage ALL into three maturation groups according to expression of T-lineage cell surface antigens, as follows: stage I, CD2+CD7+; stage II, CD2+CD7+CD1+CD4+CD8+; and stage III, CD2+CD7+CD1−(CD4+ or CD8+)CD3+. Although the percentage of patients achieving remission following induction therapy was lower for patients with T-lineage ALL of the earliest maturation stage (79%, 100%, and 94% for stages I, II, and III, respectively), 4-year EFS was equally poor for all three groups (33%, 32%, and 38%, respectively).
Recently, we analyzed data from a large cohort of patients with T-lineage ALL treated on contemporary protocols of the CCG to further investigate the prognostic role of the apparent maturation stage of leukemic T-cell precursors.141 Patients were immunophenotypically classified as follows: pro-thymocyte leukemia (pro-TL), CD7+CD2−CD5−; immature TL, CD7+(CD2+ or CD5+)CD3−; and mature TL, CD7+CD2+CD5+CD3+. No group had a preponderance of favorable or unfavorable presenting characteristics. Four-year EFS was lower for patients with pro-TL (57.1%; SD = 8.4%) compared with patients with immature and mature TL (68.5%, SD = 3.5%; and 77.1%, SD = 4.0%; respectively) with an overall significance of P = .05. Highly significant differences were found for overall survival (P = .005) as a result of the deaths of all patients with pro-TL who relapsed. Although CD2 also was a significant prognostic factor (P = .03), RHRs of 2.11, 1.51, and 1.17 for patients with pro-TL, CD2−immature TL, and CD2+ immature TL, respectively, suggested that the pro-TL maturation stage had added prognostic significance (Fig2). Indeed, multivariate analysis indicated that the influence of ontogeny group was greater than that of CD2. Thus, leukemic cells of the pro-TL maturation stage identified a subgroup of patients with T-lineage ALL who have a significantly worse EFS outcome than patients whose leukemic cells correspond to a more mature stage of development.
EFS of patients with T-lineage ALL according to the apparent maturational stage of bone marrow leukemic blasts. EFS for (A) mature TL, (B) CD2+ immature TL, (C) CD2−immature TL, and (D) pro-TL patients treated on the 1800 series of CCG protocols are shown. EFS values at designated points in follow-up are given in the text.
EFS of patients with T-lineage ALL according to the apparent maturational stage of bone marrow leukemic blasts. EFS for (A) mature TL, (B) CD2+ immature TL, (C) CD2−immature TL, and (D) pro-TL patients treated on the 1800 series of CCG protocols are shown. EFS values at designated points in follow-up are given in the text.
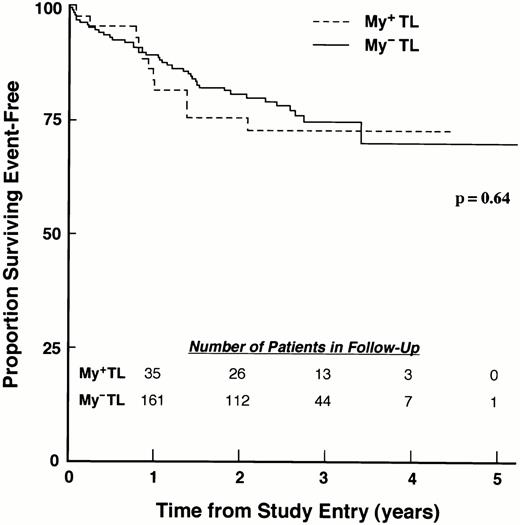
The variant immunophenotype in which leukemic cells coexpress T-lineage– and myeloid–associated antigens represents a controversial prognostic factor. Although numerous investigators have reported that coexpression of myeloid antigens predicted an adverse risk for patients with T-lineage ALL,142,143 others have found similar outcomes for myeloid antigen negative (My−) and myeloid antigen positive (My+) T-lineage ALL.144,145 We recently evaluated the influence of myeloid antigen expression on treatment outcome in a large cohort of children with newly diagnosed ALL enrolled on risk-adjusted CCG studies.146 Patients were classified as My−or My+ T-lineage, according to expression of CD7, CD13, and CD33. Patients with My+ T-lineage ALL were more likely than patients with My− T-lineage ALL to show favorable presenting features, but induction outcome and EFS outcome were similar for patients with My+ and My− T-lineage ALL, with 4-year EFS of 72.7% (SD = 7.1%) and 70.1% (SD = 5.7%), respectively (P = .49; Fig 3).These results show that regardless of treatment intensity, mixed myeloid-lymphoid phenotype was not an adverse prognostic factor for childhood T-lineage ALL.
Myeloid antigen expression in T-lineage ALL is not associated with poor EFS. EFS for My+ TL and MY− TL patients treated on the 1800 series of CCG protocols are shown. EFS values at designated points in follow-up are given in the text.
Myeloid antigen expression in T-lineage ALL is not associated with poor EFS. EFS for My+ TL and MY− TL patients treated on the 1800 series of CCG protocols are shown. EFS values at designated points in follow-up are given in the text.
IMPEDIMENTS TO EFFECTIVE TREATMENT
Drug resistance.
Despite improvements in overall survival, relapse in the bone marrow, CNS, and other sites remains a significant problem for high-risk patients. Pieters et al147 showed that patients with T-lineage ALL were particularly resistant to prednisone (PRED), daunorubicin, cytarabine, mafosfamide, and L-asparaginase, but wide ranges of resistance levels were observed within each immunophenotypic group. For all patients, the probability of continuous complete remission decreased with increasing resistance to PRED. In a later study, these investigators reported that patients with T-lineage ALL were more resistant to a host of drugs including those mentioned above as well as teniposide, ifosfamide, vincristine, vindesine, and dexamethasone.148 Lauer et al149 found that a regimen of intensive rotating drug pairs was effective for prevention of drug resistance in high-risk patients with B-lineage, but not T-lineage ALL, again suggesting that immunophenotype plays a role in drug sensitivity. Others have attributed methotrexate (MTX) resistance in patients with T-lineage ALL to a decreased formation of MTX-polyglutamates, which is a determinant of toxicity.150,151 Resistance to glucocorticoids is thought to be caused by low glucocorticoid receptor (GR) levels. However, the relationship between GR and outcome within the T-lineage immunophenotype is unclear. Quddus et al152 reported that leukemic cell GR level did not predict outcome within the T-lineage group, whereas Costlow et al153 reported that lower GR levels were correlated with unfavorable presenting features including T-lineage. Finally, although multidrug resistance is thought to be mediated by overexpression of P-glycoprotein, the product of the multidrug resistance gene MDR-1,154 the specific significance of this phenomenon in T-lineage ALL has not been determined.
INNOVATIVE TREATMENT STRATEGIES FOR T-LINEAGE ALL
Current strategies for improving treatment of children with ALL have been aimed at maximizing efficacy of treatment according to risk. Reliable and accurate methods for predicting prognosis are required to achieve adequate treatment with the least intensive regimens. Identification of biological and clinical prognostic factors, as discussed above, has aided in stratifying patients according to risk. However, additional methods are required for identifying and more effectively treating subgroups of high-risk patients who are most likely to relapse despite intensive therapy.
Seventy-five percent of children with T-lineage ALL on CCG protocols fit within the NCI high-risk category based on presenting age and WBC count.134 Patients with T-lineage ALL with standard risk represent less than 4% of patients with ALL and less than 6% of all standard-risk patients. Treatment of patients who have relapsed generally has consisted of intensive chemotherapy to achieve a second remission and subsequent use of either nonablative chemotherapy or ablative radiochemotherapy followed by bone marrow transplantation (BMT), and recurrence of leukemia is the major obstacle to the success of either approach. Intensification of cytotoxic therapy using conventional drugs will likely cause overlapping toxicities and may result in delays which may erode the intensity of therapy. Overall, the outcome for patients with relapsed T-lineage ALL is dismal because only a very small fraction can be saved with high-dose radiochemotherapy followed by BMT. Consequently, the development of new potent antileukemia drugs and the design of combinative treatment protocols using these new agents have emerged as exceptional focal points for research in modern therapy of relapsed T-lineage ALL.
Immunotoxins and other targeted biotherapeutics.
Immunotoxins (MoAb-toxin conjugates) are a new class of immunopharmacologic agents that shows considerable promise for more effective treatment of T-lineage ALL. A vast number of MoAbs have been developed with the intent of specifically targeting cytotoxic agents to leukemia cells while limiting the deleterious effects on normal tissues. Immunoconjugates containing toxins such as pokeweed antiviral protein, ricin, Pseudomonas endotoxin, and diphtheria toxin directed against T-lineage–specific surface antigens have been developed for use as systemic therapy of T-lineage ALL.155-158
Murphy et al159 as well as Kreitman et al160 have pioneered the use of genetic engineering to redirect the lethal action of diphtheria toxin towards effective targeting of growth factor receptors on leukemic cells. In one example, researchers have developed a recombinant fusion toxin, DAB486IL-2, in which the native receptor binding domain of diphtheria toxin has been replaced with interleukin-2.161
Deoxyguanosine analogs.
Another new and promising treatment program for T-lineage ALL is based on the potent antileukemia activity of deoxyguanosine analogs. The accumulation and the resulting toxicity of dGTP in T lymphocytes was first described in patients with a genetic deficiency for the enzyme purine nucleoside phophorylase (PNP).162,163 This observation lead to the search for means by which cytotoxic levels of dGTP could be achieved in T-lineage leukemias. An analog of deoxyguanosine, Ara-G (9-β-D-arabinofuranosylguanine) accumulates in T cells and acts as a poor substrate for endogenous PNP, but is efficiently phosphorylated by deoxycytidine kinase164,165; in vitro studies have shown that Ara-G is selectively cytotoxic for T-cell lines and T-lineage leukemic cells.166 167
Recently, a water soluble pro-drug derivative of Ara-G, known as compound 506U/C-506 (2-amino-6-methoxypurine arabinoside), was developed for in vivo therapeutic applications.168Preliminary results of a Phase I trial of C-506 in adult T-cell malignancies suggested that daily infusion of C-506 could achieve and maintain cytotoxic levels of Ara-GTP.169 These data indicate that C-506 warrants investigation as a new therapeutic drug for treatment of pediatric T-lineage ALL.
CONCLUSIONS
The adverse risk previously associated with T-lineage ALL in children has progressively been surmounted by intensive chemotherapeutic regimens. Still, approximately 20% to 25% of children with T-lineage ALL continue to fail therapy. Further augmentation of the currently used intensive chemotherapeutic regimens may not be warranted because of the likelihood of significant adverse effects. Thus, the current challenge is to apply our expanding knowledge of biological regulation in leukemic cells to the development of novel biologic therapeutics, particularly those that specifically target leukemic cells. Such agents could theoretically be used either to trigger cell killing directly or to alter the leukemic cell's response to radiation or chemotherapeutics. Finally, the identification of prognostically distinct patient subgroups may lead to tailored and risk-adjusted therapies for children with T-lineage ALL. Use of these various strategies, singly and in combination, should allow further improvements in outcome for patients with ALL who remain at risk for treatment failure.
Supported in part by research grants including CCG Chairman's Grants No. CA-13539, CA-51425, CA-42633, CA-42111, CA-60437, and CA-27137 from the National Cancer Institute, National Institutes of Health. F.M.U. is a Stohlman Scholar of the Leukemia Society of America, New York, NY.
Address reprint requests to Fatih M. Uckun, MD, PhD, Children's Cancer Group ALL Biology Reference Laboratory and Wayne Hughes Institute, 2665 Long Lake Rd, St Paul, MN 55113.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal