Abstract
A t(1;19)(q23;p13) is detected cytogenetically in approximately 5% of childhood acute lymphoblastic leukemias (ALLs) and its presence has been associated with an increased risk of relapse in several previously-completed Pediatric Oncology Group (POG) clinical trials. The t(1;19) fuses E2A to PBX1 in more than 95% of cases and this molecular abnormality can be reliably identified by polymerase chain reaction (PCR)-mediated amplification ofE2A-PBX1 chimeric mRNAs. We used a nested PCR assay, which reproducibly detected a 104- to 105-fold dilution of t(1;19)+ into t(1;19)− cells, to evaluate minimal residual disease (MRD) in 48 children with t(1;19)+ ALL enrolled in POG clinical trials for lower (POG 9005) and higher (POG 9006) risk ALL. Peripheral blood (PB) and bone marrow (BM) samples were collected prospectively at the end of consolidation (weeks 25 and 31 after end of induction) and the presence or absence of PCR-detectable MRD was correlated with clinical outcome. Overall, 41 of 148 (28%) samples were PCR+. Of the 65 time points with informative results from both PB and BM, PCR results were concordant for 51 pairs (10 PB+/BM+, 41 PB−/BM−) and discordant for 14 (5 PB+/BM−, 9 PB−/BM+), indicating that assessment of only PB or only BM can inaccurately classify some PCR+cases as PCR−. There were no significant differences in event-free survival between PCR+ and PCR−patients. We conclude that qualitative detection of MRD by amplification of E2A-PBX1 chimeric mRNAs at the end of consolidation was not significantly predictive of outcome for children treated on POG 9005/9006 and that such results should not be used to alter therapy for patients with t(1;19)+ ALL.
THE ADVENT OF the polymerase chain reaction (PCR) has facilitated detection of minimal residual disease (MRD) in patients with leukemia who are in complete clinical remission during and after therapy.1 PCR-based approaches typically can detect 104- to 106-fold dilutions of cells carrying the specific marker into marker-negative cells, which is significantly more sensitive than the 1% to 5% threshold of detection for traditional methodologies.1 Several different molecular markers can be monitored by PCR, chief among them being leukemia-specific Ig or T-cell receptor (TCR) gene rearrangements that are unique to each patient, and chimeric mRNAs produced by chromosomal translocations. There are advantages and disadvantages to each strategy, with amplification of Ig/TCRgene rearrangements being more widely applicable while amplification of chimeric transcripts has the theoretical advantage of measuring a marker directly linked to leukemogenesis rather than a functionally neutral genetic alteration. A major goal of this field of research is to determine if patients can be reliably stratified on the basis of PCR results into subgroups with differential likelihoods of being cured with contemporary chemotherapy regimens. Development and refinement of PCR-based MRD assays potentially offers the opportunity to “tailor” chemotherapy based on molecular assessment of treatment response.
The t(1;19)(q23;p13) occurs in 5% to 6% of all childhood acute lymphoblastic leukemias (ALLs).2 More than 95% of t(1;19)s fuse the chromosome 19 gene E2A to the chromosome 1 genePBX1, producing fusion mRNAs that encode for E2A-PBX1 chimeric proteins.3-5E2A-PBX1+t(1;19)+ ALLs are typically classified as pre-B ALLs as they express cytoplasmic, but not surface, Ig (cIg+/sIg−). Approximately 25% of pre-B ALLs contain a t(1;19) and E2A-PBX1 fusion.6-9Genomic breakpoints in E2A-PBX1+t(1;19)+ ALLs occur in restricted regions of E2Aand PBX1 and join identical portions of E2A andPBX1 in the processed fusion mRNA.3-5,10 A small percentage of cases have 27 identical nucleotides of uncertain origin located at the junction of E2A and PBX1 sequences in the processed fusion mRNAs.11 Despite this minor difference, fusion mRNAs can be amplified with a single set of primers from all cases of E2A-PBX1+ t(1;19)+ALL molecularly analyzed to date except for a single case with variantE2A-PBX1 fusion mRNAs.5,11-15 We have previously shown that PCR can detect MRD in t(1;19)+ ALL patients in remission who subsequently experienced relapse.5 Others have also anecdotally described PCR detection of MRD in small cohorts of t(1;19)+ ALL patients, but the clinical value of this approach has not been defined.14 16-18
Presence of the t(1;19) has been associated with an inferior treatment outcome on several Pediatric Oncology Group (POG) and St Jude Children's Research Hospital (Memphis, TN) clinical trials and has been shown to largely account for the inferior prognosis observed in pre-B as compared to early pre-B ALLs in these studies.7-9 Published results from other groups have generally, but not invariably, demonstrated similar adverse outcomes for t(1;19)+ ALLs.19,20 Recent trials have shown that this adverse outcome for t(1;19)+ patients can be overcome by more intensive therapies.9 20 In 1991, the POG began a pair of clinical trials for lower (POG 9005) and higher (POG 9006) risk children with ALL. After a standard 3 (9005) or 4 (9006) drug induction, these trials allocated all t(1;19)+patients to a single chemotherapy regimen that was identical between the 9005 and 9006 studies. This study design provided us with the opportunity to evaluate the predictive value of MRD detection by amplification of E2A-PBX1 chimeric mRNAs in a large cohort of similarly treated children with t(1;19)+ ALL.
MATERIALS AND METHODS
Study group and design.
Patients less than 22 years of age enrolled in POG 9005/9006 in whom a t(1;19)(q23;p13) was identified cytogenetically at the time of diagnosis formed the study group. In total, 30 t(1;19)+patients were enrolled in POG 9005 and 44 in POG 9006. After receiving a standard three (9005) or four (9006) drug induction (prednisone, vincristine, L-Asparaginase with or without daunorubicin), t(1;19)+ patients were nonrandomly assigned to an anti-metabolite–based consolidation and continuation treatment arm which was identical between the two studies.21 The protocols called for paired samples of peripheral blood (PB) and bone marrow (BM) to be collected from t(1;19)+ patients at defined times during therapy and shipped by overnight courier to a central laboratory at Stanford University. In the current study, we analyzed all available PB and BM samples collected at two time points at the conclusion of consolidation chemotherapy (weeks 25 and 31 after completion of induction). The 9005/9006 clinical trials and sample collection were approved by the individual Institutional Review Boards of POG member institutions and informed consent was obtained from patients and/or their parents. Patients were enrolled between January 1991 and November 1994. Cutoff for analysis was October 1996.
Molecular analyses.
Mononuclear cells were isolated from PB/BM samples by Ficoll-Hypaque density centrifugation, lysed in 0.5 to 1.0 mL of Solution D22 and frozen at −70°C as cell lysates. Frozen lysates were later transferred to the laboratory of one of the authors (S.P.H.) at the University of Colorado Health Sciences Center (UCHSC). After being stored at −70°C for 1 to 5 years, the solution D lysates were thawed rapidly and total RNA was extracted by the guanidinium acid lysis technique as previously described,5 22 dissolved in diethylpyrocarbonate-treated double-distilled H2O, and frozen at −70°C for later analysis.
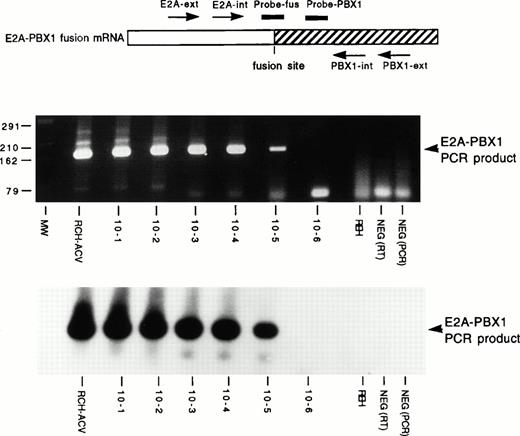
Two micrograms of total RNA was first treated with DNAse I (GIBCO-BRL, Gaithersburg, MD) to eliminate any DNA present in the samples, and then reverse transcribed to cDNA using random hexamer primers (Boehringer Mannheim, Indianapolis, IN).23 One-half of the cDNA was subjected to a two step nested PCR assay using E2A andPBX1 primers flanking the site of mRNA fusion exactly as described previously (Fig1A).23 To insure that amplifiable RNA had been isolated, the remaining one-half of the cDNA was used to amplify a portion of the ubiquitously expressed ABL mRNA using primers and conditions described previously.5 23 After amplification, one-fifth (10 μL) of the second round E2A-PBX1 PCR product and theABL PCR product were size-fractionated by electrophoresis in 3% agarose gel. In a series of dilution experiments, the nestedE2A-PBX1 PCR assay consistently detected a 10−4 dilution of t(1;19)+ RCH-ACV RNA into t(1;19)− REH RNA, detected a 10−5 dilution in 50% to 70% of experiments and never detected a 10−6 dilution. Hybridization of immobilized nested PCR products with a radiolabeled internal oligonucleotide did not increase the sensitivity or consistency of detection; thus we relied on visualization of bands on ethidium bromide-stained gels.
Sensitivity of E2A-PBX1 PCR assay. TheE2A-PBX1 fusion mRNA and the relative location of amplification primers (arrows) and detection oligonucleotides (lines) are schematically depicted at the top. Below this are displayed a photograph of an ethidium bromide-stained agarose gel of nested PCR products and a Southern blot of this gel hybridized with radio-labeled Probe-fus oligonucleotide (identical results were seen with Probe-PBX1). Molecular size (MW) markers are indicated at left in base pairs. Samples include t(1;19)+ RCH-ACV and t(1;19)− REH pre-B ALL cell lines, serial 10-fold dilutions (10−1 to 10−6) of RCH-ACV RNA into REH RNA, and reverse transcriptase (RT) and PCR negative (NEG) controls.
Sensitivity of E2A-PBX1 PCR assay. TheE2A-PBX1 fusion mRNA and the relative location of amplification primers (arrows) and detection oligonucleotides (lines) are schematically depicted at the top. Below this are displayed a photograph of an ethidium bromide-stained agarose gel of nested PCR products and a Southern blot of this gel hybridized with radio-labeled Probe-fus oligonucleotide (identical results were seen with Probe-PBX1). Molecular size (MW) markers are indicated at left in base pairs. Samples include t(1;19)+ RCH-ACV and t(1;19)− REH pre-B ALL cell lines, serial 10-fold dilutions (10−1 to 10−6) of RCH-ACV RNA into REH RNA, and reverse transcriptase (RT) and PCR negative (NEG) controls.
Numerous precautions were taken to decrease the chance of false positive results. All RNAs were isolated before performing anyE2A-PBX1 PCR analyses at UCHSC. To further exclude false positives, RNA was isolated in batches of 10 to 20 samples, which included 1 to 2 samples from patients with t(9;22)+ ALL treated on POG 9005/9006 who were involved in a parallel study of MRD and from whom PB/BM samples had been collected contemporaneously and handled identically in the same central laboratory at Stanford University. All PCR reactions were set up and run in a separate laboratory 2 miles away from that which had been used to isolate the RNAs. After amplification, samples were transported back to the UCHSC campus and PCR products were run on agarose gels and analyzed as described above. Because of these precautions, the initial sample handling, RNA isolation, PCR set up and amplification and gel analyses were completely physically isolated from one another. If any contamination was introduced during sample processing and/or RNA isolation, one would anticipate that false positive results would be seen in the t(9;22)+ samples.
Each run of PCR reactions included the following controls: RNAs from RCH-ACV and REH pre-B ALL cell lines, a 10−4 dilution of RCH-ACV RNA in REH RNA, 1 or 2 RNAs from t(9;22)+ ALL patients treated on 9005/9006, a reverse transcription (RT) negative control in which a sample containing water rather than input RNA was carried through all steps beginning with the RT step, and a PCR negative control in which water rather than input cDNA was subjected to amplification. To consider a result valid, each of the following requirements had to be met: a readily visible ABL PCR product had to be present in the control PCR reaction, readily visibleE2A-PBX1–specific PCR products of the appropriate size had to be amplified from RCH-ACV and the 10−4 dilution of RCH-ACV, and each of the negative controls had to be negative. To further validate positive samples, we required that each positive sample be independently confirmed. Specifically, whenever available, matched PB and BM samples from weeks 25 and/or 31 from an individual patient were analyzed at the same time. If more than one sample was available from a patient and all were concordant (all positive or all negative), results were considered valid. If there was any discordance, all samples were repeated 1 to 2 times. We required two positive results to consider a sample positive. All PCR reactions were performed without knowledge of treatment outcome and PCR results were not available to the treating physicians.
Statistical methods.
To assess the prognostic significance of week 25/31 PCR results, we compared the event-free survival (EFS) two ways. The first compared patients with negative PCR results on all available assays (never positive) against those with positive or mixed results (positive at some times or sample sources but not at others). The second compared patients who were positive on all available assays (never negative) against those with negative or mixed results. By “never” we refer to weeks 25 and 31 only. EFS was the dependent variable and the logrank test24 was used. EFS curves were constructed by the method of Kaplan-Meier.25 The only other formal statistical comparison was to assess the concordance of week 25 and week 31 PCR results, using the exact conditional chi-square.26 It is important to realize that one cannot preclude the possibility of selection bias in terms of the prognostic significance of PCR results, because of the relatively large number of patients with missing data. Further, given the small number of treatment failures, the power to detect moderate-sized differences is relatively low.
RESULTS
Seventy-four patients with t(1;19)+ ALL were enrolled on POG 9005 or 9006, 71 of whom were potentially available for analysis (1 was taken off study at week 4, 1 was lost to follow-up, and 1 relapsed before week 31). Samples were received at week 25 and/or 31 from 68% of these patients (Table 1). By study, samples collected at week 25 and/or 31 were received from 17 of 30 t(1;19)+ patients enrolled in POG 9005 and 31 of 44 enrolled in POG 9006. In total, samples from 54% of potential time points were available for analysis and matched PB and BM analyses were available for 81% of these times. The outcome of patients from whom samples were received is similar to that of all t(1;19)+ patients enrolled in POG 9005/9006 (data not shown).
Samples Available for PCR Analysis of MRD
| Time Point . | Patients From Whom Samples Were Received . |
|---|---|
| Week 25 only | 8 (11%) |
| Week 31 only | 9 (13%) |
| Week 25 and 31 | 31 (44%) |
| Total | 48 (68%) |
| Source of Samples | No. Available |
| Matched PB and BM | 68 (48%) |
| PB only | 2 (1%) |
| BM only | 9 (6%) |
| Total time points | 79 (56%) |
| Total samples | 147 (52%) |
| Time Point . | Patients From Whom Samples Were Received . |
|---|---|
| Week 25 only | 8 (11%) |
| Week 31 only | 9 (13%) |
| Week 25 and 31 | 31 (44%) |
| Total | 48 (68%) |
| Source of Samples | No. Available |
| Matched PB and BM | 68 (48%) |
| PB only | 2 (1%) |
| BM only | 9 (6%) |
| Total time points | 79 (56%) |
| Total samples | 147 (52%) |
A nested PCR assay that reproducibly detected a 10−4to 10−5 dilution of t(1;19)+ into t(1;19)− RNA in pilot experiments was employed for our studies (Fig 1). A 10−4 dilution of t(1;19)+ RNA was included in each individual PCR experiment. The PCR assay consistently detected a standard level of MRD because we did not observe experiments in which the undiluted sample was positive and the 10−4 dilution was negative.
E2A-PBX1 chimeric transcripts were detected in 41 of 148 (28%) week 25 or 31 samples from t(1;19)+ ALL patients, 104 were PCR− and 3 were uninformative because an ABLband was not amplified in the control PCR reaction (representative results are shown in Fig 2). Nineteen of 20 samples from t(9;22)+ ALL patients analyzed were negative for E2A-PBX1 chimeric transcripts and one was uninformative because a control ABL band was not amplified. False positives were not observed in the negative control cell line (REH), RT or PCR negative controls included in each PCR run. There were no major differences in PCR results in terms of clinical parameters including patient age, presenting white blood count or NCI consensus risk status (Table 2). Cryopreservation for up to 5 years did not materially affect the integrity of RNA isolated because a correct-sized ABL PCR product was amplified from 179 of 183 (98%) frozen samples analyzed. There was no correlation between length of sample storage and incidence of PCR positivity (data not shown).
Representative results of E2A-PBX1 PCR assays. Photograph of an ethidium bromide-stained agarose gel containing size-fractionated products of nested PCR forE2A-PBX1 fusion mRNA. Molecular size (MW) markers are indicated at left in base pairs. The sample identity is given at the top of the gel. Samples include bone marrow (B) and/or peripheral blood (P) from weeks 25 and/or 31 for patients 1 to 6, t(1;19)+ RCH-ACV and t(1;19)− REH pre-B ALL cell lines, a 10−4 dilution of RCH-ACV RNA into REH RNA, and RT and PCR negative controls. The migration of the E2A-PBX1PCR product is indicated at the right of the gel. For patient 2, two independently isolated BM samples [Ba and Bb] were analyzed.
Representative results of E2A-PBX1 PCR assays. Photograph of an ethidium bromide-stained agarose gel containing size-fractionated products of nested PCR forE2A-PBX1 fusion mRNA. Molecular size (MW) markers are indicated at left in base pairs. The sample identity is given at the top of the gel. Samples include bone marrow (B) and/or peripheral blood (P) from weeks 25 and/or 31 for patients 1 to 6, t(1;19)+ RCH-ACV and t(1;19)− REH pre-B ALL cell lines, a 10−4 dilution of RCH-ACV RNA into REH RNA, and RT and PCR negative controls. The migration of the E2A-PBX1PCR product is indicated at the right of the gel. For patient 2, two independently isolated BM samples [Ba and Bb] were analyzed.
Demographics
| PCR Status (n) . | Median(quartiles) . | NCI Consensus Risk Status (n) . | ||
|---|---|---|---|---|
| WBC (x 109L) . | Age (Years) . | Standard . | Poor . | |
| Never negative (14) | 27 (8-54) | 4 (3-9) | 7 | 7 |
| At least one negative (34) | 19 (7-40) | 7 (3-11) | 18 | 16 |
| Never positive (20) | 21 (10-41) | 7 (3-12) | 11 | 9 |
| At least one positive (28) | 18 (7-53) | 5 (3-9) | 14 | 14 |
| PCR Status (n) . | Median(quartiles) . | NCI Consensus Risk Status (n) . | ||
|---|---|---|---|---|
| WBC (x 109L) . | Age (Years) . | Standard . | Poor . | |
| Never negative (14) | 27 (8-54) | 4 (3-9) | 7 | 7 |
| At least one negative (34) | 19 (7-40) | 7 (3-11) | 18 | 16 |
| Never positive (20) | 21 (10-41) | 7 (3-12) | 11 | 9 |
| At least one positive (28) | 18 (7-53) | 5 (3-9) | 14 | 14 |
PCR status refers to E2A-PBX1 chimeric transcripts and patients were classified as either never negative (no negative PCR assays from PB/BM samples available from weeks 25 and 31) or at least one negative and never positive (no positive PCR assays from PB/BM samples available from weeks 25 and 31) or at least one positive.
We compared PCR results for the 65 pairs of PB and BM that were collected at the same time point. Results were concordant for 51 pairs (10 PB+/BM+, 41 PB−/BM−) and discordant for 14 (5 PB+/BM−, 9 PB−/BM+). The discordant sample pairs were reproducibly discordant in all cases. Informative PCR results were available from both weeks 25 and 31 for 32 of the patients. There was no apparent association of PCR status at weeks 25 and 31 (Table 3). Approximately one-third of patients who were PCR− at week 25 were PCR+ at week 31, quite similar to the percentage who were PCR+ at week 25 and again positive at week 31 (Pvalue for the exact conditional chi-square test of association is > .99).
Comparison of Week 25 and 31 PCR Results
| . | . | 25 . | |||
|---|---|---|---|---|---|
| NA . | Negative . | Positive . | Total . | ||
| NA | 4 | 3 | 7 | ||
| 31 | Negative | 2 | 14 | 6 | 22 |
| Positive | 7 | 9 | 3 | 19 | |
| Total | 9 | 27 | 12 | 48 | |
| . | . | 25 . | |||
|---|---|---|---|---|---|
| NA . | Negative . | Positive . | Total . | ||
| NA | 4 | 3 | 7 | ||
| 31 | Negative | 2 | 14 | 6 | 22 |
| Positive | 7 | 9 | 3 | 19 | |
| Total | 9 | 27 | 12 | 48 | |
Abbreviation: NA, not available.
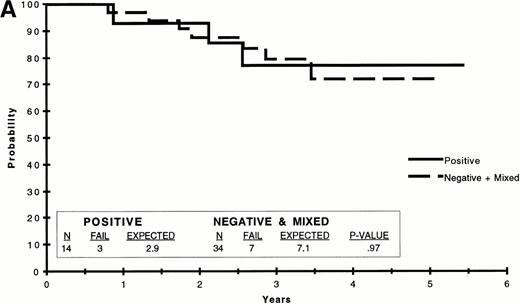
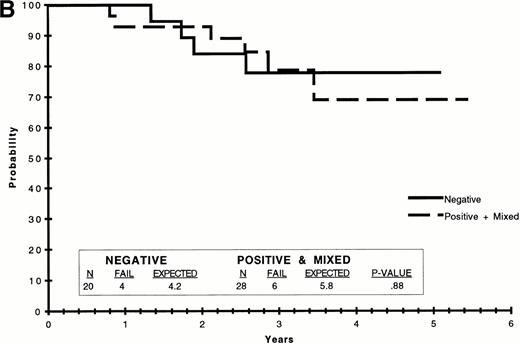
We analyzed EFS to determine if the presence or absence of amplifiableE2A-PBX1 chimeric transcripts was predictive of treatment outcome. Because of the variability of PCR results, we compared several different groupings to determine if any predictive value was apparent. These included comparisons of patients for whom all available PCR results were negative (n = 20) versus those in whom all results were positive (n = 13) versus those with both negative and positive results (n = 15); those who were PCR+ at week 25 and PCR− at week 31 (n = 6) versus those who were PCR− at week 25 and PCR+ at week 31 (n = 9); those who were PCR+ on all available assays (n = 13) versus those who were always PCR− plus those with some positive and some negative results (n = 35); and those who were PCR− on all available assays (n = 20) versus those who were always PCR+ plus those with some positive and some negative results (n = 28). There were no significant differences in treatment outcome between any of these comparisons. Selected EFS curves are shown in Fig 3.
EFS on POG 9005/9006 by t(1;19) PCR. (A) EFS of patients for whom all available PCR results were positive versus that of patients for whom all available results were negative plus those with some positive and some negative PCR results (negative + mixed). (B) EFS of patients for whom all available PCR results were negative versus that of patients for whom all available results were positive plus those with some positive and some negative PCR results (positive + mixed). The data were analyzed by the Kaplan-Meier method and the difference compared by the logrank test, the results of which are included in tabular form within each graph.
EFS on POG 9005/9006 by t(1;19) PCR. (A) EFS of patients for whom all available PCR results were positive versus that of patients for whom all available results were negative plus those with some positive and some negative PCR results (negative + mixed). (B) EFS of patients for whom all available PCR results were negative versus that of patients for whom all available results were positive plus those with some positive and some negative PCR results (positive + mixed). The data were analyzed by the Kaplan-Meier method and the difference compared by the logrank test, the results of which are included in tabular form within each graph.
DISCUSSION
PCR analysis of MRD by amplification of E2A-PBX1 chimeric transcripts at the end of consolidation therapy was not predictive of treatment outcome for children with t(1;19)+ ALL treated on POG 9005/9006. The lack of concordance between samples obtained simultaneously from PB and BM and between samples obtained 6 weeks apart (weeks 25 and 31) was surprising. These discrepancies could potentially be due to the presence of levels of MRD at or near the levels of detection of the assay (10−4 to 10−5). Given the elaborate precautions employed in this study and inclusion of multiple negative controls, it is extremely unlikely that our results are confounded by false positives due to PCR contamination. It is difficult to completely exclude false negative PCR results, but we emphasize that all PCR assays included a 10-4 dilution of positive control RNA which consistently gave positive results. Discrepant samples were reproducibly discrepant in independently-performed PCR reactions. Thus, it seems unlikely that the PCR assay employed in this study lacked sensitivity or reproducibility. Most importantly, the lack of correlation between clinical outcome and PCR results was quite consistent across all comparisons. Based on these factors, we conclude that our results are truly representative and not due to technical problems.
There were 6 time points for which a BM sample was PCR−, but a matched PB sample was not available. Based on the discordance observed in some cases between simultaneously obtained BM and PB samples, we considered the possibility that some or all of these six data points might have been false negatives and repeated the analyses with the results from these six data points excluded. This did not alter our conclusions because there were no significant differences in EFS based on PCR status in these analyses.
It is possible that the strategy we used to detect MRD was suboptimal and that other strategies might be more predictive of outcome. We decided to analyze samples obtained at the end of consolidation because we wanted to assess disease burden before most relapses were likely to occur (and only 1 of 74 patients relapsed before week 31) to avoid missing identification of high-risk patients and the later opportunity for early therapeutic intervention. As the intensive phase of treatment had been completed by week 25, we hypothesized that the level of leukemic cells would have decreased substantially by this time and that levels of MRD might separate patients into cohorts at high and low risk of subsequent relapse. This assumption may be false, or the decrease may evolve over a longer period of time after completion of consolidation and samples obtained later in therapy might have been more informative. In this regard, it is important to emphasize that one group found that Ig PCR assays performed at the end of induction had clinical predictive value, but the same assays performed at the time therapy was discontinued did not.27 28
Another potential problem is that we used a qualitative (positivev negative) rather than a quantitative PCR assay. Recently several semiquantitative PCR assays for MRD detection have been developed. One strategy used for chimeric mRNAs such as BCR-ABLand AML1-ETO involves coamplification of known amounts of artificial fusion templates of unique size and has been shown in several studies to be more predictive of outcome than qualitative PCR.29-32 Others have used recombinant phage plaque counting techniques27,28 or limiting dilution assays with multiple replicates33 to quantify MRD assessed by amplifying unique Ig rearrangements. It is possible that such approaches might better identify high-risk patients based on the level of MRD or trends in the this level over time, rather than simply its absence or presence. Finally, it is important to note that the overall outcome for t(1;19)+ patients treated on POG 9005/9006 is improved from prior POG studies and this high success rate combined with limited sample numbers means that our study had a relatively low power to detect differences in outcome. One problem encountered in our study, as in similar longitudinal studies reported by others, is the incompleteness of sample collection at each time point. It is clear that future studies of MRD, particularly those performed in large cooperative groups, must develop methods to insure near complete sample collection. If missing at random assumptions are not satisfied, the validity of the method of choice, the Cox model,34 with time dependent covariates, becomes severely biased without complete data at every time point for nearly every patient.
Several small studies have described the use of E2A-PBX1 PCR to assess MRD in t(1;19)+ ALL patients. Izraeli et al16 analyzed 7 patients at one or two time points 6 to 36 months after attaining complete remission (CR). They found no obvious correlation between PCR results and clinical outcome and 2 patients remained in CR for >1 year after positive PCR assays at 24 and 29 months. There also was no obvious correlation of PCR results with clinical outcome among 9 patients studied by Devaraj et al14 with 2 patients remaining in CR for 20 to 32 months after being PCR+ 5 to 6 months into therapy.14Privitera et al17 assessed MRD in six t(1;19)+patients. Five became PCR− 3 to 6 months into therapy and remained in CR at last follow-up (15 to 60 months after diagnosis). One was persistently PCR+ during therapy, remained positive when treatment was discontinued but continued in CR 5 years later. Lanza et al18 assessed MRD in 3 t(1;19)+patients; two became PCR− early in therapy, while one remained PCR+ 3 years after diagnosis.18Despite this difference, all three patients remained in clinical and hematological complete remission. Thus, none of the data available at this time suggest that detection of MRD by E2A-PBX1 PCR in t(1;19)+ ALL provides information that is sufficiently predictive of treatment outcome to warrant alterations in patient management.
Our study emphasizes several important facts that have relevance to MRD analysis. First, discordant PCR results were observed at 22% of the time points for which results were available from simultaneously obtained PB and BM specimens (9 PB− /BM+and 5 PB+/BM− ), suggesting that conclusions based upon PCR results obtained from PB only or BM only are problematic. Although most studies of MRD in acute leukemia have focused on BM samples, two other studies analyzed MRD detection in matched BM/PB samples from patients with t(9;22)+ ALL. Similar to our findings, Radich et al35 reported concordant PCR results from 23/31 patients, while 5 were BM+/PB− and 3 BM−/PB+.35 In contrast, van Rhee et al36 found that while that the number ofBCR-ABL transcripts in paired PB and BM samples did not differ significantly overall, four patients were PB−/BM+, while none were PB+/BM−.36
Second and most important, our results underscore the danger in extrapolating results from one method of PCR detection of MRD to another. In several settings, experimental evidence suggests that PCR detection of MRD by amplification of chimeric mRNAs produced by chromosomal translocations provides clinically useful information which may have therapeutic implications. Detection of MRD by amplification ofBCR-ABL fusion mRNAs in patients with CML after BMT has been performed in numerous studies and a consensus is emerging that patients who are reproducibly PCR+ after the early post-BMT phase are at increased risk of relapse when compared to those who are reproducibly PCR−.37,38 Similarly, detection of MRD by amplification of PML-RARA fusion transcripts at the end of therapy appears to provide useful clinical information in patients with acute promyelocytic leukemia.39-44 On the other hand, the presence of amplifiable AML1-ETO fusion transcripts in most or all t(8;21)+ AML patients long after completion of chemotherapy or BMT indicates that qualitative PCR results are not predictive of treatment outcome in this subset of AML, although a recent small series provides preliminary data suggesting that quantitative PCR may be more informative.32 45-48 Thus, it is clear that the potential clinical utility of each individual PCR MRD assay needs to be rigorously determined before results are used to modify treatment of individual patients or groups of patients.
APPENDIX
| Institution . | Grant No. . |
|---|---|
| ALBERTA PEDIATRIC ONCOLOGY CONSORTIUM | |
| Alberta Children's Hospital | |
| Alberta Pediatric Oncology Consortium | |
| Cross Cancer Institute | |
| BAYLOR | |
| Baylor | CA-03161 |
| UT/Galveston | CA-03161 |
| BERGAN-PASSIAC CCOP | |
| Bergan-Passiac CCOP | |
| Hackensack Medical Center | |
| BOSTON FLOATING HOSPITAL23 | |
| Boston Floating Hospital | |
| Eastern Main | |
| BOWMAN GRAY | |
| Bowman Gray | CA-53128 |
| CARDINAL GLENNON CHILDREN'S HOSPITAL | |
| Cardinal Glennon Children's Hospital | |
| CAROLINAS CONSORTIUM | |
| Carolinas Consortium | |
| Carolinas Medical Center | CA-69177 |
| Children's Hospital Greenville System | CA-69177 |
| East Carolina University | CA-69177 |
| Medical University of South Carolina | CA-69177 |
| Presbyterian Hospital | CA-69177 |
| CHILDREN'S HOSPITAL MICHIGAN | |
| Children's Hospital Michigan | CA-29691 |
| Hurley Medical Center | CA-29691 |
| St Johns Hospital | CA-29691 |
| CHILDREN'S MEMORIAL HOSPITAL (CHICAGO) | |
| Children's Memorial Hospital (Chicago) | CA-07431 |
| Christ Hospital | CA-07431 |
| Rush-Presbyterian | CA-07431 |
| DANA-FARBER CANCER INSTITUTE | |
| Dana-Farber Cancer Institute | CA-41573 |
| Maine Children's | CA-41573 |
| DUKE UNIVERSITY | |
| Duke University | CA-15525 |
| West Virginia University, Charleston | CA-15525 |
| West Virginia University, Morgantown | CA-15525 |
| EASTERN PEDIATRIC ONCOLOGY CONSORTIUM | |
| Eastern Pediatric Oncology Consortium | |
| Mount Sinai Medical School (NY) | CA-69428 |
| University of Maryland | CA-69428 |
| Yale University | CA-69428 |
| EMORY UNIVERSITY | |
| Emory University | CA-20549 |
| FLORIDA CCOP | |
| All Children's Hospital | |
| Florida CCOP | |
| Joe DiMaggio Children's | |
| M.D. Anderson Cancer Center Orlando | |
| Sacred Heart Hospital | |
| Tampa Children's Hospital | |
| HAWAII (NP) CCOP | |
| Cancer Center of Hawaii | |
| Hawaii (NP) CCOP | |
| HOSPITAL FOR SICK CHILDREN | |
| Hospital for Sick Children | |
| JOHNS HOPKINS UNIVERSITY | |
| Fairfax Hospital | CA-28476 |
| Johns Hopkins University | CA-28476 |
| LSU CCOP | |
| Children's Hospital New Orleans/LSU CCOP | |
| LSU CCOP | |
| McGILL UNIVERSITY | |
| Children's East Ontario | CA-33587 |
| McGill University | CA-33587 |
| MEDICAL COLLEGE VIRGINIA | |
| Medical College Virginia | |
| MIAMI CHILDREN'S HOSPITAL | |
| Miami Children's Hospital | |
| MIDWEST CHILDREN'S CANCER CENTER | |
| Midwest Children's Cancer Center | CA-32053 |
| NEW ENGLAND CONSORTIUM | |
| Dartmouth Hitchcock | CA-29293 |
| Massachusetts General Hospital | CA-29293 |
| New England Consortium | |
| Rhode Island Hospital | CA-29293 |
| SUNY Stony Brook | CA-29293 |
| University of Vermont | CA-29293 |
| OCHSNER CCOP | |
| Ochsner CCOP | |
| Ochsner Clinic | |
| OKLAHOMA UNIVERSITY | |
| Oklahoma University | CA-11233 |
| Warren Clinics | CA-11233 |
| OPERATIONS OFFICE | |
| Operations Office | CA-30969 |
| ROSWELL PARK CANCER INSTITUTE | |
| Roswell Park Cancer Institute | CA-28383 |
| SPOG CONSORTIUM | |
| SPOG Bern | |
| SPOG Consortium | |
| SPOG Geneva | |
| SPOG Lausanne | |
| SUNY SYRACUSE | |
| SUNY Syracuse | |
| SWMSC | |
| Cook-Ft. Worth Children's Medical Center | CA-33625 |
| SWMSC | |
| Scott & White | CA-33625 |
| Southwestern Medical School | CA-33625 |
| ST CHRISTOPHER'S HOSPITAL | |
| St Christopher's Hospital | |
| ST JUDE CHILDREN'S | |
| East Tennessee State University | CA-31566 |
| St Jude Children's | CA-31566 |
| STANFORD UNIVERSITY | |
| Kaiser/Santa Clara | CA-33603 |
| Stanford University | CA-33603 |
| University of Arizona | CA-33603 |
| STATISTICAL OFFICE | |
| Statistical Officer | CA-29139 |
| UNIVERSITY OF ALABAMA | |
| University of Alabama | CA-25408 |
| UNIVERSITY OF ARKANSAS | |
| University of Arkansas | |
| UNIVERSITY OF FLORIDA | |
| Nemours Children's Clinic | |
| University of Florida | |
| UNIVERSITY OF KANSAS | |
| University of Kansas | |
| UNIVERSITY OF MIAMI | |
| St Mary's Hospital | |
| University of Miami | |
| UNIVERSITY OF MISSISSIPPI MEDICAL CENTER | |
| Keesler AFB Hospital | CA-15898 |
| University of Mississippi Medical Center | CA-15989 |
| UNIVERSITY OF ROCHESTER | |
| University of Rochester | |
| UNIVERSITY OF VIRGINIA | |
| University of Virginia | |
| UC/DAVIS | |
| Sutter Community Hospital | |
| UC/Davis | |
| UCSD CONSORTIUM | |
| Children's Hospital (San Diego) | CA-28439 |
| Kaiser Permanente/San Diego | CA-28439 |
| UC/San Diego | CA-28439 |
| UCSD Consortium | |
| US TEX PED CCOP | |
| San Antonio MPC & BDC | |
| US TEX PED CCOP | |
| UT/San Antonio | |
| USA CCOP | |
| University of South Alabama | |
| USA CCOP | |
| UNIF SERV ONC. CONS | |
| Madigan Army Medical Center | |
| Naval Medical Center, Portsmouth | |
| Tripler Army Medical Center | |
| Unif Serv. Onc. Cons. | |
| Walter Reed Army Medical | |
| WASHINGTON UNIVERSITY | |
| University of Missouri | CA-05587 |
| Washington University | CA-05587 |
| WICHITA CCOP | |
| St Francis Regional | |
| Wichita CCOP |
| Institution . | Grant No. . |
|---|---|
| ALBERTA PEDIATRIC ONCOLOGY CONSORTIUM | |
| Alberta Children's Hospital | |
| Alberta Pediatric Oncology Consortium | |
| Cross Cancer Institute | |
| BAYLOR | |
| Baylor | CA-03161 |
| UT/Galveston | CA-03161 |
| BERGAN-PASSIAC CCOP | |
| Bergan-Passiac CCOP | |
| Hackensack Medical Center | |
| BOSTON FLOATING HOSPITAL23 | |
| Boston Floating Hospital | |
| Eastern Main | |
| BOWMAN GRAY | |
| Bowman Gray | CA-53128 |
| CARDINAL GLENNON CHILDREN'S HOSPITAL | |
| Cardinal Glennon Children's Hospital | |
| CAROLINAS CONSORTIUM | |
| Carolinas Consortium | |
| Carolinas Medical Center | CA-69177 |
| Children's Hospital Greenville System | CA-69177 |
| East Carolina University | CA-69177 |
| Medical University of South Carolina | CA-69177 |
| Presbyterian Hospital | CA-69177 |
| CHILDREN'S HOSPITAL MICHIGAN | |
| Children's Hospital Michigan | CA-29691 |
| Hurley Medical Center | CA-29691 |
| St Johns Hospital | CA-29691 |
| CHILDREN'S MEMORIAL HOSPITAL (CHICAGO) | |
| Children's Memorial Hospital (Chicago) | CA-07431 |
| Christ Hospital | CA-07431 |
| Rush-Presbyterian | CA-07431 |
| DANA-FARBER CANCER INSTITUTE | |
| Dana-Farber Cancer Institute | CA-41573 |
| Maine Children's | CA-41573 |
| DUKE UNIVERSITY | |
| Duke University | CA-15525 |
| West Virginia University, Charleston | CA-15525 |
| West Virginia University, Morgantown | CA-15525 |
| EASTERN PEDIATRIC ONCOLOGY CONSORTIUM | |
| Eastern Pediatric Oncology Consortium | |
| Mount Sinai Medical School (NY) | CA-69428 |
| University of Maryland | CA-69428 |
| Yale University | CA-69428 |
| EMORY UNIVERSITY | |
| Emory University | CA-20549 |
| FLORIDA CCOP | |
| All Children's Hospital | |
| Florida CCOP | |
| Joe DiMaggio Children's | |
| M.D. Anderson Cancer Center Orlando | |
| Sacred Heart Hospital | |
| Tampa Children's Hospital | |
| HAWAII (NP) CCOP | |
| Cancer Center of Hawaii | |
| Hawaii (NP) CCOP | |
| HOSPITAL FOR SICK CHILDREN | |
| Hospital for Sick Children | |
| JOHNS HOPKINS UNIVERSITY | |
| Fairfax Hospital | CA-28476 |
| Johns Hopkins University | CA-28476 |
| LSU CCOP | |
| Children's Hospital New Orleans/LSU CCOP | |
| LSU CCOP | |
| McGILL UNIVERSITY | |
| Children's East Ontario | CA-33587 |
| McGill University | CA-33587 |
| MEDICAL COLLEGE VIRGINIA | |
| Medical College Virginia | |
| MIAMI CHILDREN'S HOSPITAL | |
| Miami Children's Hospital | |
| MIDWEST CHILDREN'S CANCER CENTER | |
| Midwest Children's Cancer Center | CA-32053 |
| NEW ENGLAND CONSORTIUM | |
| Dartmouth Hitchcock | CA-29293 |
| Massachusetts General Hospital | CA-29293 |
| New England Consortium | |
| Rhode Island Hospital | CA-29293 |
| SUNY Stony Brook | CA-29293 |
| University of Vermont | CA-29293 |
| OCHSNER CCOP | |
| Ochsner CCOP | |
| Ochsner Clinic | |
| OKLAHOMA UNIVERSITY | |
| Oklahoma University | CA-11233 |
| Warren Clinics | CA-11233 |
| OPERATIONS OFFICE | |
| Operations Office | CA-30969 |
| ROSWELL PARK CANCER INSTITUTE | |
| Roswell Park Cancer Institute | CA-28383 |
| SPOG CONSORTIUM | |
| SPOG Bern | |
| SPOG Consortium | |
| SPOG Geneva | |
| SPOG Lausanne | |
| SUNY SYRACUSE | |
| SUNY Syracuse | |
| SWMSC | |
| Cook-Ft. Worth Children's Medical Center | CA-33625 |
| SWMSC | |
| Scott & White | CA-33625 |
| Southwestern Medical School | CA-33625 |
| ST CHRISTOPHER'S HOSPITAL | |
| St Christopher's Hospital | |
| ST JUDE CHILDREN'S | |
| East Tennessee State University | CA-31566 |
| St Jude Children's | CA-31566 |
| STANFORD UNIVERSITY | |
| Kaiser/Santa Clara | CA-33603 |
| Stanford University | CA-33603 |
| University of Arizona | CA-33603 |
| STATISTICAL OFFICE | |
| Statistical Officer | CA-29139 |
| UNIVERSITY OF ALABAMA | |
| University of Alabama | CA-25408 |
| UNIVERSITY OF ARKANSAS | |
| University of Arkansas | |
| UNIVERSITY OF FLORIDA | |
| Nemours Children's Clinic | |
| University of Florida | |
| UNIVERSITY OF KANSAS | |
| University of Kansas | |
| UNIVERSITY OF MIAMI | |
| St Mary's Hospital | |
| University of Miami | |
| UNIVERSITY OF MISSISSIPPI MEDICAL CENTER | |
| Keesler AFB Hospital | CA-15898 |
| University of Mississippi Medical Center | CA-15989 |
| UNIVERSITY OF ROCHESTER | |
| University of Rochester | |
| UNIVERSITY OF VIRGINIA | |
| University of Virginia | |
| UC/DAVIS | |
| Sutter Community Hospital | |
| UC/Davis | |
| UCSD CONSORTIUM | |
| Children's Hospital (San Diego) | CA-28439 |
| Kaiser Permanente/San Diego | CA-28439 |
| UC/San Diego | CA-28439 |
| UCSD Consortium | |
| US TEX PED CCOP | |
| San Antonio MPC & BDC | |
| US TEX PED CCOP | |
| UT/San Antonio | |
| USA CCOP | |
| University of South Alabama | |
| USA CCOP | |
| UNIF SERV ONC. CONS | |
| Madigan Army Medical Center | |
| Naval Medical Center, Portsmouth | |
| Tripler Army Medical Center | |
| Unif Serv. Onc. Cons. | |
| Walter Reed Army Medical | |
| WASHINGTON UNIVERSITY | |
| University of Missouri | CA-05587 |
| Washington University | CA-05587 |
| WICHITA CCOP | |
| St Francis Regional | |
| Wichita CCOP |
ACKNOWLEDGMENT
We gratefully acknowledge the invaluable assistance of the physicians who treated these patients and, most importantly, the patients and their parents, without whom these studies would not have been possible. We also thank Sharon Murphy for her encouragement and interest in this project. We are particularly indebted to Qi Wei for technical assistance and the use of his laboratory.
Supported by grants from The Southern California Children's Cancer Service and Couples Against Leukemia, Los Angeles, CA (S.P.H.), The Maxfield Foundation, Saratoga, CA (S.P.H.), and the Ladies Auxiliary to the Veterans of Foreign Wars, Denver, CO (S.P.H.). S.P.H. was supported in part by a Physicians Research Training Fellowship from the American Cancer Society and a National Research Service Award from the National Institutes of Health (Bethesda, MD). S.P.H. is the recipient of a BLOOD/ASH Scholar Award. B.M.C. is the Rebecca Jean Slye Professor of Pediatric Oncology and is supported in part by the Midwest Athletes Against Childhood Cancer (MACC) Fund. The National Cancer Institue grant numbers for participating authors: CA-09184, CA-33603, CA-32053, CA-25408, CA-20549, CA-03161, CA-15989, CA-29139, CA-30969, CA-37379, CA-49605. For complete grant information see the Appendix.
Address reprint requests to Stephen P. Hunger, MD (9000/9005/9006), c/o POG Operations Office, 645 N Michigan Ave, Suite 910, Chicago, IL 60610.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 2. Representative results of E2A-PBX1 PCR assays. Photograph of an ethidium bromide-stained agarose gel containing size-fractionated products of nested PCR forE2A-PBX1 fusion mRNA. Molecular size (MW) markers are indicated at left in base pairs. The sample identity is given at the top of the gel. Samples include bone marrow (B) and/or peripheral blood (P) from weeks 25 and/or 31 for patients 1 to 6, t(1;19)+ RCH-ACV and t(1;19)− REH pre-B ALL cell lines, a 10−4 dilution of RCH-ACV RNA into REH RNA, and RT and PCR negative controls. The migration of the E2A-PBX1PCR product is indicated at the right of the gel. For patient 2, two independently isolated BM samples [Ba and Bb] were analyzed.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/3/10.1182_blood.v91.3.1021/3/m_blod4032202.jpeg?Expires=1769082112&Signature=K9lLj8UzkyLU1gJ52mXuLMDccFhFkQCTFkgVVGViaNA3byQADoONp7Wnp5lwUFoR9mseww~HYpvDvmXYuK6VKwEjuI7NL6fDv7pulZj~xs1pnebTKp62Sj-uXNa0K7XSMGwGmEhQiazoCBjGSvCi0RXruSKc63ZcSfgw9azfUaTGEqqYzFGX6gj~M4zdTKDLJEgMfGT8XipHPly5VRTPL37hT6m1bhYH13vD7CVa7RQfX0BTh6jp5NV86Ph9FD0Q3xUdHQ0iz-EjquhVO5DlBsFNiY4pH5c6AHCzIerQ2libJ2Gtb3f6Ae-fzAlBX2T--j7T81pXjv6diA~7SUlacA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal