Abstract
Endothelial cell adhesion to von Willebrand Factor is mainly mediated through an interaction between the αvβ3 integrin and the RGD sequence of von Willebrand factor (vWF ). To define the potential involvement of glycoprotein Ibα (GPIbα) as an endothelial vWF receptor, we compared cell adhesion to three recombinant vWF, the wild-type (WT-rvWF ) and two mutants, RGGS-rvWF (D1746G), defective for binding to platelet αIIbβ3, and ΔA1-rvWF with a deletion between amino-acids 478 and 716, which does not bind to platelet GPIbα. Adhesion of human umbilical vein endothelial cells to purified vWF recombinants was measured by automatized cell counting using an image analyzer. Whereas cell adhesion to ΔA1-rvWF was unchanged compared with WT-rvWF, reaching a plateau of 40% total cells at a concentration of 2.5 μg/mL rvWF, adhesion to RGGS-rvWF was only 10% of total cells. Cell stimulation by tumor necrosis factor-α (TNFα), reported to upregulate the expression of the putative endothelial GPIbα, did not modify adhesion to these rvWF. Monoclonal antibodies to vWF or GPIbα, blocking vWF interaction with platelet GPIbα, were unable to inhibit endothelial cell adhesion to rvWF. In contrast, antibody 9 to vWF, blocking the αvβ3-dependent endothelial cell adhesion to plasma vWF, inhibited adhesion to WT-rvWF as efficiently as to ΔA1-rvWF (50% inhibition at a concentration of 11 and 15 μg/mL, respectively). In agreement with the fact that endothelial cell adhesion to vWF appeared independent of the GPIbα-binding domain, we were unable to detect endothelial surface expression of GPIbα by flow cytometry or in cell lysates by immunoprecipitation followed by immunoblotting. Moreover, expression of GPIbα mRNA was undetectable in endothelial cells, even after stimulation by TNFα. These studies indicate that GPIbα is not expressed in human cultured endothelial cells and is not involved in adhesion to vWF-containing surfaces. Thus, in static conditions, cultured endothelial cells adhere to vWF through an αvβ3-dependent, GPIbα-independent mechanism.
THE ADHESIVE PROTEIN von Willebrand factor (vWF ) is synthesized by endothelial cells. The biosynthesis involves the formation of pro-vWF containing a 741 amino-acid (aa) propeptide and a subunit of 2,050 aa, the formation of dimers with a molecular mass of 500 kD, the assembly into multimers of greater than 15,000 kD, and the proteolytic cleavage of the propeptide necessary for the release in plasma of the fully processed vWF.1 Under hemodynamic conditions of high shear rates, vWF is the main effector of the platelet response to vascular damage, consisting of platelet adhesion and aggregation that leads to the formation of thrombi.2 Two platelet receptors are involved in the interaction with vWF, the glycoprotein (GP) Ib-IX complex and the αIIbβ3 integrin (GPIIb/IIIa). Binding site for vWF resides on GPIbα, the largest subunit of the complex, which also contains GPIbβ and GPIX.3 The interaction with platelet GPIbα is highly susceptible to the conformation of vWF, as evidenced by the absence of interaction between soluble (fluid-phase) vWF and platelet GPIbα. Nonphysiologic substances, such as ristocetin or botrocetin, induce the binding of soluble vWF to GPIbα through an interaction with different regions of vWF.4,5 In addition, the conformation of solid-phase vWF differs from that of vWF in the fluid phase and allows the binding of GPIbα in the absence of any stimulus.6 Binding to GPIbα involves discrete sequences localized within the first type A repeat (A1 domain, aa 497-716) that contains a disulfide bond between Cys 509 and Cys 695 required for binding to GPIbα.7-9 The binding to αIIbβ3 exposed on the surface of activated platelets involves the RGD sequence (aa 1744-1746) of vWF and mediates platelet aggregation.10
In contrast to the platelet, the nature of endothelial cell receptors for vWF is not entirely established. The endothelial αvβ3 integrin has been identified as a common receptor for a number of RGD-containing ligands, including vWF.11,12 A second endothelial vWF receptor was found to be related to platelet GPIbα by immunologic as well as functional studies in the presence of ristocetin.13,14 However, evidence is missing that endothelial cells can bind vWF in the presence of botrocetin, a more specific inducer of vWF interaction with platelet GPIbα than ristocetin. More recently, it was reported that the expression of GPIbα mRNA or protein was very low in unstimulated cells, but could be increased by cytokines such as tumor necrosis factor-α (TNFα).15,16 Site-directed mutagenesis studies of vWF have shown that the RGD sequence is an absolute requirement for endothelial cell adhesion.17 However, this approach has also suggested the involvement of an additional functional region of vWF, the A1 domain, interacting with an endothelial GPIbα-related receptor.17 These studies were extended by using monoclonal antibodies (MoAbs) to platelet GPIbα and a vWF recombinant fragment overlapping the A1 domain.18 However, the latter results were obtained in the absence of cell stimulation and are therefore difficult to reconcile with earlier findings on the cytokine-induced transcription of GPIbα.15 16
Using proteolytic fragments of vWF, we have previously found that the RGD-containing SpII fragment of vWF (aa 1366-2050) is able to promote endothelial cell adhesion and spreading, whereas the SpIII fragment (aa 1-1365), overlapping the A1 domain and containing the platelet GPIbα-binding region, is not. Our results clearly indicated that endothelial cell adhesion to vWF was mediated by αvβ3, whereas it did not involve GPIbα.19
Thus, ambiguities remain concerning the expression, or at least the function, of this GPIbα-related molecule in endothelial cells. The aim of the present work is to assess the importance of vWF domains in the interaction with endothelial cells, in particular to determine the influence of cell stimulation by TNFα on the putative GPIbα-dependent adhesion to vWF. We compared as adhesion-promoting ligands two recombinant mutated vWF (rvWF ), RGGS-rvWF with an Asp 1746 to Gly mutation, previously shown to be unable to bind to platelet αIIbβ3, and ΔA1-rvWF containing a deletion of the A1 domain and completely defective for its interaction with platelet GPIbα.20 In addition, we address the unresolved issue of GPIbα protein and mRNA expression in endothelial cells.
MATERIALS AND METHODS
Expression of recombinant vWF mutants.Recombinant vWF mutants were obtained by stable expression in Baby Hamster Kidney (BHK) cells, as previously described.21 Briefly, the full-length cDNA for human vWF was cloned into the pNUT vector and the construct was named pNUT-vWFWT. Site-directed mutagenesis changing the AC to the GA nucleotides at position 7527-7528 resulted in the substitution of Asp 1746 by Gly. An EagI-EcoRV fragment, containing the cDNA mutation, was subcloned into the pNUT-vWF expression vector, obtaining pNUT-vWFRGGS. A second construct containing the deletion of the cDNA for aa 478 to 716 was cloned into pNUT-vWF obtaining pNUT-vWFΔA1.21 A stable BHK cell line overexpressing furin, the enzyme responsible for the cleavage of the propeptide, was used for coexpression of vWF mutants, using the calcium phosphate precipitation method.22 Selection of stable cell lines was performed in the presence of G418 (GIBCO, Paisley, UK). BHK cells overexpressing furin were transfected with pNUT-vWFWT, pNUT-vWFRGGS, or pNUT-vWFΔA1 and stable transformants were selected by the addition of 100 μmol/L methotrexate (Sigma, St Louis, MO). Cultures were grown in Dulbecco's modified Eagle medium MEM/Ham's F-12 (DMEM/F-12) containing 1% Ultroser (GIBCO).
Purification and characterization of rvWF mutants.The cell lines produced high levels (∼10 μg/mL) of WT-rvWF, RGGS-rvWF, or ΔA1-rvWF. These rvWF were purified by immunoaffinity chromatography on CNBr-activated Sepharose-4B (Pharmacia, Uppsala, Sweden) coupled to a MoAb 453 directed against an epitope on the C-terminal part of the vWF subunit. Characterization of purified rvWF was performed as follows. The amount of vWF:Ag was estimated by enzyme-linked immunosorbent assay. vWF ristocetin cofactor activity was determined using freeze-dried platelets (Organon Teknika, Fresnes, France). Fractions with the highest [vWF:RCo]/[vWF:Ag] ratio were pooled, except for ΔA1-rvWF, which had no detectable ristocetin cofactor activity. Binding to purified αIIbβ3 indicated that WT-rvWF or ΔA1-rvWF bound to the same extent, whereas the binding of RGGS-rvWF was completely abolished. Analysis of rvWF subunit showed a single band indicating complete processing into the mature subunit, and multimeric analysis of rvWF showed the whole range of multimers in all rvWF, except ΔA1-rvWF lacking the highest molecular weight multimers, as previously described.21 22
Antibodies.Different murine MoAbs to vWF produced in our laboratory were used as purified IgG. The epitopes of some MoAbs have been previously localized on the vWF subunit23: MoAb 9 has an epitope localized between aa 1704 and 1746, thus overlaps the RGD sequence, and is known to inhibit vWF binding to platelet αIIbβ324; MoAb 713 (aa 593-678) blocks vWF binding to GPIbα in the presence of ristocetin; and MoAb 724 (aa 565-587) blocks the interaction of vWF with platelet GPIbα in the presence of botrocetin but not ristocetin.25 26 MoAb 453, directed against the C-terminal part of vWF (aa 1366-2050), and MoAb 723 (aa 523-588) do not interfere with any known function of vWF.
We also used several murine MoAbs directed against the components of the platelet GPIb-IX complex: 6D127 (a kind gift of Dr B.S. Coller, SUNY, Stony Brook, NY), Ib-2328 (kindly provided by Dr B. Steiner, Hoffmann-La Roche Ltd, Basel, Switzerland), AS-729 (kindly provided by Dr J. Miller, SUNY, Syracuse, NY), and SZ2 (Immunotech, Marseille, France), which all recognize the amino-terminal 45-kD domain of GPIbα and block its binding to vWF. In addition, we used SZ1 reactive with GPIb-IX complex30 (kindly provided by Dr C. Ruan, Suzhou Medical College, Suzhou, People's Republic of China). Rabbit polyclonal antibody raised against GPIbα was a kind gift of Dr K.J. Clemetson (Theodor Kocher Institut, Bern, Switzerland).31 The MoAb AP3 directed against the β3 integrin subunit32 was a gift of Dr P.J. Newman (The Blood Center of Southeastern Wisconsin, Milwaukee, WI). The polyclonal rabbit antiserum raised against β3 was previously characterized.31 The MoAb 23C6 directed against the αvβ3 integrin complex33 and a polyclonal rabbit antiserum raised against the αv subunit were gifts of Dr M.A. Horton (University College London, London, UK). The MoAb against ICAM-1 (clone 84H10) and isotypic controls were from Immunotech. The murine myeloma monoclonal IgG1 MOPC21 was from Sigma. These antibodies were used as purified IgG at 10 to 20 μg/mL, except for AP3 used at a 100-fold dilution of ascitic fluid.
Cell culture and stimulation.Endothelial cells were isolated from human umbilical veins and grown to confluency in Opti-MEM culture medium (GIBCO) supplemented with 20% fetal calf serum (Boehringer, Meylan, France).19 Cells of a second passage were used unless otherwise specified. In some experiments, endothelial cells were stimulated by 10 ng/mL of recombinant human TNFα (Genzyme, Cambridge, MA) for 24 hours as described.16 This concentration was determined from dose-response studies based on the expression of endothelial activation markers (VCAM-1 and ICAM-1) as well as morphologic changes of the cells that exhibited an elongated phenotype upon stimulation. HEL 5J20 cells of a subclone, selected for increased GPIbα expression compared with the parental human leukemic cell line HEL, were a kind gift of Dr N. Kieffer (French-Luxembourg Biomedical Research Laboratory, Luxembourg, Grand Duchy of Luxembourg) and were cultured as suspension in RPMI and 10% fetal calf serum as reported.34
Adhesion assay.Attachment of endothelial cells was performed in 96-multiwell plastic wells (Dutscher, Strasbourg, France) precoated overnight at 4°C with serial dilutions of purified rvWF. Heat-denatured bovine serum albumin (BSA; Calbiochem, La Jolla, CA) was used as control. Confluent cells were detached by 10 minutes of exposure to EDTA (0.5 mmol/L) and washed twice in Opti-MEM by centrifugation at 250g for 10 minutes. The cells were resuspended at a concentration of 60,000 cells/mL in adhesion buffer (10 mmol/L HEPES, pH 7.4, 140 mmol/L NaCl, 5.4 mmol/L KCl, 5.56 mmol/L glucose, 2 mmol/L CaCl2 , 1 mmol/L MgCl2 , 1 mmol/L MnCl2 ) containing 3% BSA, a procedure that led to single-cell suspensions without cell clumps or aggregates. For inhibition studies with antibodies, the cells were preincubated with 10% human heat-denatured AB serum (Institut Jacques Boy, Reims, France) for 10 minutes at 4°C to quench potential Fcγ binding sites, washed, and incubated with the appropriate dilution of antibody for 30 minutes at 4°C. One hundred microliters of cell suspensions was then added to the wells. After 2 hours of incubation at 37°C, adherent cells were fixed with 100 μL of 2% paraformaldehyde for 30 minutes at 20°C, rinsed twice with water, and stained with haematoxylin for 10 minutes, followed by erythrosin B (4 mg/mL) and Orange G (20 mg/mL) for 10 minutes. Adherent cells were observed by light microscopy in an inverted microscope (Axiovert 135; Carl Zeiss, Göttingen, Germany).
Quantitation of adhesion was performed with a real time digital imaging processing system (Samba 2005; Unilog, Meylan, France). The system consisted of a CCD video camera (Sony, Tokyo, Japan) connected to a fast and intelligent image processing and acquisition board (MVP/AT Matrox, Montreal, Quebec, Canada) located in a personal computer (Deskpro XE433S; Compaq, Les Ulis, France). A specific digital imaging software (IPS/ITB 2005) was developed to control automatic focusing, positioning in the center of the well, and displacement along the x and y axis by a motorized stage (Märzhauser, Wetzlar, Germany). A minimum of 9 fields was studied at a 10-fold magnification. Acquisition of the data was interactive to separate contiguous cells from each other and to quantitate every single cell. The number of adherent cells per well was calculated from the average number of counted cells (usually ∼25 to 30 per field) and normalized for the total surface of the well. Adhesion to BSA used as negative control varied between 2% and 4% and was subtracted from the data.
Results were expressed as the percentage of adherent cells relative to the total number of cells. Mean ± SEM were calculated for three experiments performed in duplicate, unless specified otherwise. Statistical significance of differences between means was evaluated using the Student's t-test for paired samples.
Flow cytometry.Cells were detached with EDTA (0.5 mmol/L), washed with Opti-MEM, and incubated for 30 minutes at 4°C with an appropriate dilution of the primary antibody. After washing in cold phosphate-buffered saline, antibody binding was assessed by flow cytometry (FACScan; Becton Dickinson, Le-Pont-de-Claix, France) by incubating the cells for 30 minutes at 4°C with a 100-fold dilution of fluorescein isothiocyanate (FITC)-conjugated goat F(ab′)2 fragment directed against a mouse IgG (Caltag Laboratories, South San Francisco, CA).
Immunoprecipitation.Endothelial cells were detached with EDTA; centrifuged at 280g for 10 minutes at 4°C; resuspended at 107/mL in 150 mmol/L NaCl, 10 mmol/L Tris, 3 mmol/L EDTA lysis buffer, pH 7.4 containing proteinase inhibitors (1 mmol/L phenylmethyl sulfonyl fluoride [PMSF], 5 mmol/L benzamidine, 0.1 mmol/L aprotinin, 0.1 mmol/L leupeptin, and 5 mmol/L N-ethylmaleimide [NEM]); and lyzed with 1% (vol:vol) Triton X-100 for 30 minutes at 4°C with constant shaking. Insoluble cell debris and nucleus were removed by centrifugation at 12,000g for 10 minutes at 4°C. In some experiments, cells were incubated overnight with 5 μg/mL cytochalasin B (Sigma) before lysis. Human platelets were isolated from plasma proteins as previously described,31 resuspended at 5 × 109/mL in the lysis buffer with proteinase inhibitors, and lysed with Triton X-100 as described above. For immunoprecipitation, cell lysates were adjusted at 2 mg/mL total protein in immunoprecipitation buffer (IP buffer; lysis buffer containing 1 mmol/L PMSF, 5 mmol/L benzamidine, and 0.5% Triton X-100) and processed at 4°C. For each immunoprecipitation, 1 mg of endothelial cell proteins and 0.1 mg of platelet proteins were incubated with 5 μg of MOPC 21 IgG for 60 minutes with gentle stirring, then with 50 μg of rabbit polyclonal IgG against mouse IgG (Nordic Immunology, Tilburg, The Netherlands) for a further 60 minutes, and finally for another 60 minutes with 25 μL of protein A Sepharose CL-4B beads (Pharmacia) and were then washed and diluted one-fifth in IP buffer. The beads were sedimented at 14,000g for 1 minute and the cleared supernatant was cautiously aspirated and kept for further specific immunoprecipitation, whereas the beads were washed three times with IP buffer, before the nonspecific immune complexes bound to the beads were extracted by adding 50 μL of solubilization buffer (150 mmol/L NaCl,10 mmol/L Tris, 3 mmol/L EDTA, 5 mmol/L NEM, 2% [wt:vol] SDS, 5% [vol:vol] 2-mercaptoethanol, pH 6.8) and heating for 10 minutes at 100°C. Beads were sedimented, and the supernatant was aspirated and kept at −20°C until sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis. The cleared Triton X-100 supernatants were then identically immunoprecipitated using specific MoAbs, ie, SZ2 for GPIbα and 23C6 for the αvβ3 integrin. Final specific immune complexes were extracted from protein A Sepharose beads and solubilized with SDS as described above.
SDS-PAGE and immunoblot analysis.Proteins in the immune complexes, together with total endothelial cell or platelet proteins (50 μg and 10 μg per well, respectively) from the initial, unprecipitated Triton extracts further solubilized with 2% (wt:vol) SDS and 5% (vol:vol) 2-mercaptoethanol were separated under reducing conditions by SDS-PAGE on 7% to 15% gradient acrylamide gels. Gels were calibrated for relative molecular mass (Mr) using calibration standard proteins from Bio-Rad (Richmond, CA). Proteins were electrotransferred from unstained gels on nitrocellulose membranes (0.45-μm pores; Schleicher and Schuell, Dassel, Germany) and probed with the polyclonal rabbit antiserum to GPIbα diluted 1/500 or with a mixture of the polyclonal rabbit antisera to αv (diluted 1/50) and to β3 (diluted 1/500), as previously described.31 Bound antibodies were shown by incubation of membranes with 1/1,000 diluted [125I]-Protein A (affinity purified, 1,600 MBq/mg; Amersham International plc, Little Chalfont, UK) and were exposed to Kodak X-Omat MA or AR films (Kodak-Pathé, Marne-la-Vallée, France) for 1 to 2 days.
mRNA analysis.Confluent endothelial cells from primary cultures obtained by a pool of four umbilical cords (4 × 106 cells) or subcultured for two passages (7 × 106 cells) were detached by EDTA and used after washing as well as HEL 5J20 cells (7 × 106 cells). Poly (A)+ RNA was directly isolated by using the mini-message maker kit (R & D Systems, Abingdon, UK). RNA was analyzed by Northern blotting, fixed to a positively charged nylon membrane (Ambion, Austin, TX), and prehybridized for 1.5 hours at 68°C in 0.1% SDS, 6× SSC, and 2× Denhardt's reagent containing 100 μg/mL heat-denatured salmon sperm DNA.35 Hybridization was performed overnight at 68°C in the prehybridization solution containing the following heat-denatured radiolabeled cDNA probes: HEL-derived GPIbα cDNA GPIb2.4 (kind gift of Dr J. Lopez, VA Medical Center, Baylor College of Medicine, Houston, TX36), human ICAM-1 probe cocktail (R & D Systems), and human vWF cDNA probe of 1,760 bp (from clone pvWFIPC8) corresponding to nucleotides 1209 to 2967 of the full-length cDNA.37 Probes were labeled with α32P-dCTP by random-priming with the kit ready-to-go (Pharmacia) or with γ32P-dATP and polynucleotide kinase (Boehringer) at 37°C. Blots were washed for 20 minutes in 2× SSC, 0.1% SDS and twice for 10 minutes in 0.2× SSC, 0.1% SDS at 68°C and analyzed by autoradiography.
RESULTS
Relative importance of the RGD sequence and the A1 domain of rvWF in supporting endothelial cell adhesion.To define the involvement of functional domains of vWF in its interaction with endothelial cells, we compared adhesion to full-length WT-rvWF and two mutants. These rvWF have been previously characterized for their defective interaction with platelet receptors, and we confirmed that RGGS-rvWF was unable to bind to αIIbβ3 and ΔA1-rvWF (deleted for aa 478-716) to platelet GPIbα in the presence of ristocetin or botrocetin.20 21 In contrast, RGGS-rvWF binding to platelet GPIbα and ΔA1-rvWF binding to platelet αIIbβ3 were unchanged compared with WT-rvWF (data not shown). Increasing concentrations of purified rvWF were immobilized onto microtiter plates. Comparison of binding isotherms of 125I-rvWF to plastic wells indicated that rvWF had similar affinities and that saturation was not reached in the range of concentrations tested (data not shown).
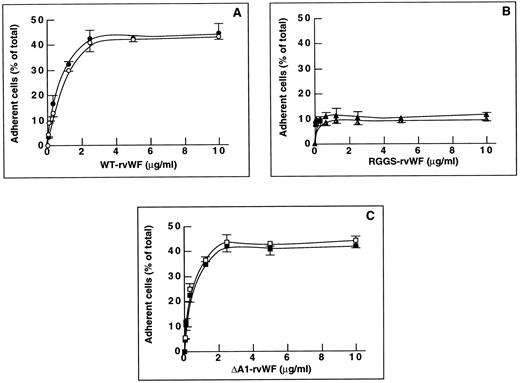
Figure 1A shows that endothelial cell adhesion to solid-phase WT-rvWF increased as a function of the rvWF concentration added to the well, up to 2.5 μg/mL of WT-rvWF where a plateau of adhesion was reached (41.1% ± 4.5% total cells). In contrast, adhesion to RGGS-rvWF did not exceed 11.1% ± 2.9% of total cells, which is significantly lower than adhesion to WT-rvWF (P < .05; Fig 1B). Increasing the concentration of RGGS-rvWF up to 10 μg/mL did not improve adhesion, indicating that this mutant had a defective interaction with endothelial cells. Interestingly, adhesion to ΔA1-rvWF was not decreased compared with WT-rvWF, because it reached a maximum of 42.4% ± 4.2% at 2.5 μg/mL of ΔA1-rvWF (Fig 1C). The same adhesion and spreading pattern was seen on WT-rvWF and on ΔA1-rvWF. After 2 hours, a large proportion of adherent cells were fully spread (∼70%). In contrast, most of the sparse cells that adhered to RGGS-rvWF remained in a round shape and none appeared fully spread. Occasionally, some cells were seen at an early stage of spreading, starting to extend some pseudopodia.
Endothelial cell adhesion to purified WT-rvWF, RGGS-rvWF, or ΔA1-rvWF. Endothelial cells were added to microtiter wells precoated with rvWF. After 2 hours of incubation at 37°C, adherent cells were fixed with paraformaldehyde and stained. Quantitation of adhesion was performed with a real time digital imaging processing system. Adhesion to BSA (20 μg/mL) was subtracted from the data. Results were expressed as the percentage of adherent cells relative to the total number of cells. The mean ± SEM were calculated for three experiments performed in duplicate. Adhesion of nonstimulated cells is shown using solid symbols and adhesion of TNFα-stimulated cells with open symbols. Adhesion to increasing concentrations of purified rvWF: (A) WT-rvWF (•, ○), (B) RGGS-rvWF (▴, ▵); and (C) ΔA1-rvWF (▪, □).
Endothelial cell adhesion to purified WT-rvWF, RGGS-rvWF, or ΔA1-rvWF. Endothelial cells were added to microtiter wells precoated with rvWF. After 2 hours of incubation at 37°C, adherent cells were fixed with paraformaldehyde and stained. Quantitation of adhesion was performed with a real time digital imaging processing system. Adhesion to BSA (20 μg/mL) was subtracted from the data. Results were expressed as the percentage of adherent cells relative to the total number of cells. The mean ± SEM were calculated for three experiments performed in duplicate. Adhesion of nonstimulated cells is shown using solid symbols and adhesion of TNFα-stimulated cells with open symbols. Adhesion to increasing concentrations of purified rvWF: (A) WT-rvWF (•, ○), (B) RGGS-rvWF (▴, ▵); and (C) ΔA1-rvWF (▪, □).
To determine whether cytokines could upregulate the activity of a putative endothelial GPIbα receptor, we compared the adhesion of nonstimulated cells and cells stimulated for 24 hours by TNFα. Adhesion of stimulated cells increased in a dose-dependent manner and reached a plateau at 2.5 μg/mL rvWF. Cell stimulation did not increase the percentage of adherent cells to WT-rvWF (37% ± 2.2%) or to RGGS-rvWF (11.1% ± 1.4%) compared with nonstimulated cells (Fig 1A and B). In addition, adhesion to ΔA1-rvWF reached a plateau of 44.2% ± 2.9% total cells, which is not significantly different from that of nonstimulated cells (Fig 1C). When cells were stimulated with TNFα, the morphology of adherent cells to WT-rvWF, ΔA1-rvWF, and RGGS-rvWF was superimposable to nonstimulated cells and was thus not dependent on stimulation. Therefore, most data of further experiments will be presented without cell stimulation.
These results suggest that the absence of the A1 domain containing the platelet GPIbα-binding site does not impair the ability of vWF to support endothelial cell adhesion and that adhesion to ΔA1-rvWF may be mediated by an interaction of the αvβ3 integrin with its RGD sequence. In contrast, a mutation of the RGD sequence results in a strongly decreased adhesion.
Effect of MoAbs on endothelial cell adhesion.To further assess the importance of the GPIbα-binding site of vWF, we used MoAbs blocking vWF interaction with platelet GPIbα. When comparing adhesion to WT-rvWF and RGGS-rvWF in the presence of these antibodies, none of the anti-vWF (713 and 724) or anti-GPIbα (6D1, AS-7, Ib-23, and SZ2) had any effect on adhesion to WT-rvWF or RGGS-rvWF (Table 1). Furthermore, the MoAb SZ1 to platelet GPIX complexed to GPIb could not block endothelial cell adhesion to any significant extent (Table 1).
Effect of MoAbs Blocking vWF Binding to Platelet GPIbα
| MoAb . | Adherent Cells (% of total) . | ||
|---|---|---|---|
| Target . | Clone . | WT-rvWF . | RGGS-rvWF . |
| None | — | 45.4 ± 2.4 (n = 4) | 16.6 ± 1.1 (n = 4) |
| vWF | 723 | 45.2 ± 1.9 (n = 4) | 17.3 ± 1 (n = 4) |
| vWF | 713 | 43.7 ± 1.7 (n = 4) | 17.9 ± 1.3 (n = 4) |
| vWF | 724 | 39.6 ± 4.6 (n = 4) | 17.1 ± 1.3 (n = 3) |
| GPIbα | 6D1 | 42.6 ± 3.9 (n = 4) | 16.8 ± 1.2 (n = 3) |
| GPIbα | AS-7 | 39.7 ± 7.1 (n = 4) | 15.6 ± 1 (n = 3) |
| GPIbα | Ib-23 | 46.6 (n = 2) | 17.7 (n = 2) |
| GPIbα | SZ2 | 43.7 (n = 2) | 19.1 (n = 2) |
| GPIX | SZ1 | 45.5 (n = 2) | 16.6 (n = 2) |
| MoAb . | Adherent Cells (% of total) . | ||
|---|---|---|---|
| Target . | Clone . | WT-rvWF . | RGGS-rvWF . |
| None | — | 45.4 ± 2.4 (n = 4) | 16.6 ± 1.1 (n = 4) |
| vWF | 723 | 45.2 ± 1.9 (n = 4) | 17.3 ± 1 (n = 4) |
| vWF | 713 | 43.7 ± 1.7 (n = 4) | 17.9 ± 1.3 (n = 4) |
| vWF | 724 | 39.6 ± 4.6 (n = 4) | 17.1 ± 1.3 (n = 3) |
| GPIbα | 6D1 | 42.6 ± 3.9 (n = 4) | 16.8 ± 1.2 (n = 3) |
| GPIbα | AS-7 | 39.7 ± 7.1 (n = 4) | 15.6 ± 1 (n = 3) |
| GPIbα | Ib-23 | 46.6 (n = 2) | 17.7 (n = 2) |
| GPIbα | SZ2 | 43.7 (n = 2) | 19.1 (n = 2) |
| GPIX | SZ1 | 45.5 (n = 2) | 16.6 (n = 2) |
Nonstimulated endothelial cells were preincubated for 30 minutes at 4°C with 20 μg/mL of purified IgG corresponding to the different clones indicated in the table. The percentage of cell adhesion was measured as described in the Materials and Methods. The number of experiments in duplicate is indicated into parenthesis. None of the anti-vWF or anti-GPIbα antibodies had any effect on endothelial cell adhesion to WT-rvWF or RGGS-rvWF.
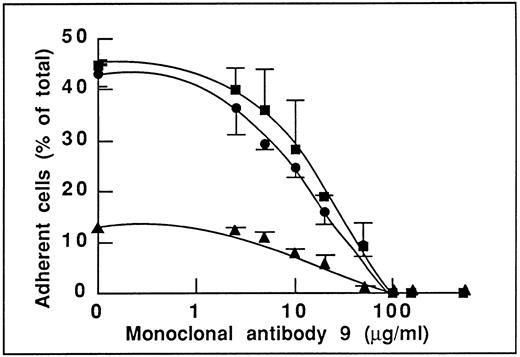
This result is indeed not in favor of an involvement of the GPIbα-binding site of vWF in endothelial cell adhesion. To determine the domain(s) of vWF involved in adhesion, we studied the effect of MoAb 9 to vWF, which we have previously reported as inhibiting the αvβ3-dependent endothelial cell adhesion to plasma vWF.19 As expected, we found that this antibody inhibited adhesion to WT-rvWF (Fig 2). More interestingly, MoAb 9 also inhibited endothelial cell adhesion to ΔA1-rvWF in a dose-dependent manner that was very similar to its effect on WT-rvWF. Adhesion to WT-rvWF and ΔA1-rvWF was inhibited by 50% in the presence of 11 and 15 μg/mL MoAb 9, respectively. Adhesion to either substrate was completely abolished in the presence of 100 μg/mL MoAb 9 (Fig 2). In addition, we found that the low residual adhesion to RGGS-rvWF was also inhibited in a dose-dependent manner by MoAb 9 (50% inhibition at 17 μg/mL), suggesting the involvement of additional sequences besides the RGD motif and located within the carboxy-terminal part of vWF or distantly affected by the binding of the antibody to its epitope.
Effect of MoAb 9 to vWF blocking the αvβ3-dependent endothelial adhesion to vWF. Nonstimulated endothelial cells were incubated with increasing concentrations of MoAb 9 before adhesion to WT-rvWF (•), RGGS-rvWF (▴), or to ΔA1-rvWF (▪). Adhesion was measured as described in the legend to Fig 1. MoAb 9 inhibits adhesion to WT-rvWF to the same extent as adhesion to ΔA1-rvWF. Of note is that adhesion to RGGS-rvWF is also inhibited by MoAb 9.
Effect of MoAb 9 to vWF blocking the αvβ3-dependent endothelial adhesion to vWF. Nonstimulated endothelial cells were incubated with increasing concentrations of MoAb 9 before adhesion to WT-rvWF (•), RGGS-rvWF (▴), or to ΔA1-rvWF (▪). Adhesion was measured as described in the legend to Fig 1. MoAb 9 inhibits adhesion to WT-rvWF to the same extent as adhesion to ΔA1-rvWF. Of note is that adhesion to RGGS-rvWF is also inhibited by MoAb 9.
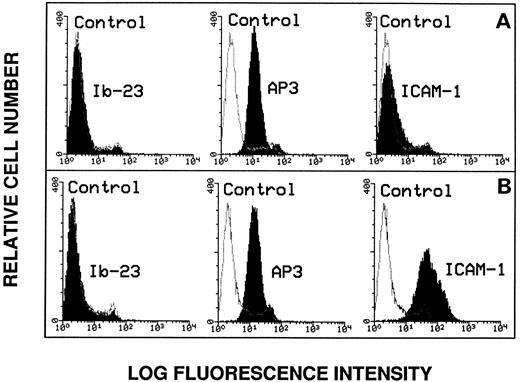
Lack of evidence for endothelial GPIbα expression.Because we found no evidence that endothelial cells interact with vWF through its GPIb-binding domain, it was of importance to directly assess endothelial GPIbα surface expression by flow cytometry. As shown in Fig 3A, there was no significant change in fluorescence intensity of the cells using MoAb Ib-23 directed against the amino-terminal portion of the GPIbα subunit. As positive control, the expression of the β3 integrin subunit was assessed in the presence of MoAb AP3.
Flow cytometry analysis of endothelial cell surface markers. The anti-GPIbα MoAb Ib-23 binding to nonstimulated cells (A) was compared with its binding to TNFα-stimulated cells (B). Cells were incubated with 10 μg/mL of the primary antibody followed by a 100-fold dilution of FITC-conjugated secondary antibody and examined by flow cytometry. Ib-23 does not recognize endothelial cells, whereas the AP3 anti-β3 subunit MoAb was used as a positive control of both cell populations and the anti–ICAM-1 MoAb was used as a positive control for cell stimulation. The negative controls are represented by the nonshaded peaks. Shaded areas are representative of fluorescence depicted (from left to right) in the presence of Ib-23, AP-3, and anti–ICAM-1 antibody.
Flow cytometry analysis of endothelial cell surface markers. The anti-GPIbα MoAb Ib-23 binding to nonstimulated cells (A) was compared with its binding to TNFα-stimulated cells (B). Cells were incubated with 10 μg/mL of the primary antibody followed by a 100-fold dilution of FITC-conjugated secondary antibody and examined by flow cytometry. Ib-23 does not recognize endothelial cells, whereas the AP3 anti-β3 subunit MoAb was used as a positive control of both cell populations and the anti–ICAM-1 MoAb was used as a positive control for cell stimulation. The negative controls are represented by the nonshaded peaks. Shaded areas are representative of fluorescence depicted (from left to right) in the presence of Ib-23, AP-3, and anti–ICAM-1 antibody.
To determine whether endothelial GPIbα surface expression may be upregulated by cytokines, we also studied TNFα-stimulated cells (Fig 3B). Cell stimulation was effective because the expression of ICAM-1 was increased compared with nonstimulated cells. However, we did not detect any change in the fluorescence intensity of the stimulated cell population using MoAb Ib-23 (Fig 3A and B). In addition, we could not detect any expression of the epitopes for MoAbs SZ2, A-S7, or 6D1 as well as for AK3 directed against the macroglycopeptide portion of GPIbα regardless of whether the cells were stimulated or not (data not shown).
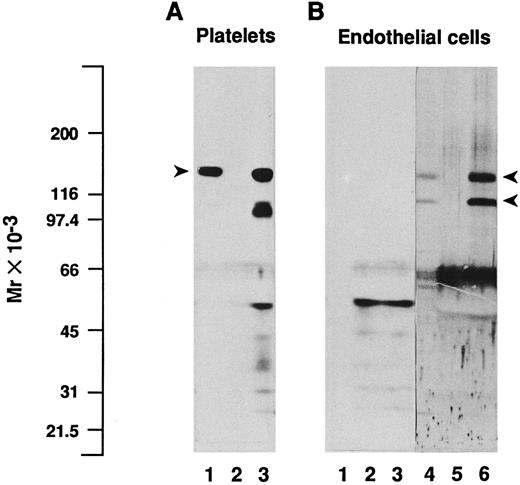
In addition, we performed immunoprecipitations on endothelial cell or platelet lysates using the MoAb SZ2 directed to GPIbα, followed by SDS-PAGE and immunoblotting analysis of the immune complexes and of the unprecipitated lysates, with a polyclonal antiserum to GPIbα (Fig 4). Precipitation of platelet extracts with SZ2 resulted in a major band with Mr = 137,000 (Fig 4A, lane 3) in the position of platelet GPIbα, as detected in the lysate (lane 1). Despite the continuous presence of proteinase inhibitors during immunoprecipitation experiments, partially degraded GPIbα was also clearly visible on the immunoblot of immunoprecipitates, with fragments in the range of Mr = 97,000 to 103,000. In contrast, no band corresponding to GPIbα or any of its fragments could be detected using the same conditions of immunoblotting in endothelial cell lysates or immunoprecipitates (Fig 4B, lanes 1 and 3), even after prolonged exposure on films (up to 6 days). Only labeling of material with Mr less than 66,000 was observed, as well as in platelet immunoprecipitates, whether using SZ2 or the nonspecific MOPC21 antibody (Fig 4A and B, lanes 2 and 5), which corresponded mostly to IgG fragments. Because cytochalasin B has been reported to increase the amount of GPIbα that can be detected by immunoprecipitation and/or immunoblotting of endothelial cell extracts,18 we also immunoprecipitated lysates of cytochalasin B-treated endothelial cells with SZ2. However, although cytochalasin B was effective in abolishing cell spreading, we were unable to detect GPIbα in the immunoprecipitates (data not shown). As a positive control, immunoblotting of the endothelial cell lysates with two polyclonal antibodies to each of the αv and β3 subunits identified two bands with Mr = 138,000 and 113,000, corresponding to the reduced αv heavy chain and β3 polypeptides, respectively (Fig 4B, lane 4).11 The intensity of these bands was markedly increased in immunoprecipitates obtained with the MoAb 23C6 against the αvβ3 integrin complex (Fig 4B, lane 6), whereas they were absent from immunoprecipitates obtained with MOPC21 (Fig 4B, lane 5).
Immunoblotting and immunoprecipitation analysis of platelet and endothelial cell protein extracts. Triton X-100–soluble proteins from platelets (A) or from endothelial cells (B) were analyzed directly by immunoblotting (lanes 1 and 4) or after immunoprecipitation with the MoAb SZ2 to GPIbα (lanes 3), with the irrelevant monoclonal IgG MOPC21 (lanes 2 and 5), or with the MoAb 23C6 to the αvβ3 integrin complex (lane 6). Proteins were separated by SDS-PAGE under reduced conditions, transferred to nitrocellulose membranes, and immunoblotted with a polyclonal antiserum to GPIbα (lanes 1 to 3, 2 days of exposure on film) or with a mixture of two polyclonal antisera to each of the αv and β3 subunits (lanes 4 through 6, 1 day of exposure on film), as detailed in the Materials and Methods. Immunoprecipitation of platelet lysates with SZ2 shows a major band in the position of platelet GPIbα, indicated by an arrowhead (A), whereas no such band is detected in endothelial cell immunoprecipitates. On (B), arrowheads depict the position of endothelial αv heavy chain and β3 subunits. Molecular mass markers are indicated on the left-hand side.
Immunoblotting and immunoprecipitation analysis of platelet and endothelial cell protein extracts. Triton X-100–soluble proteins from platelets (A) or from endothelial cells (B) were analyzed directly by immunoblotting (lanes 1 and 4) or after immunoprecipitation with the MoAb SZ2 to GPIbα (lanes 3), with the irrelevant monoclonal IgG MOPC21 (lanes 2 and 5), or with the MoAb 23C6 to the αvβ3 integrin complex (lane 6). Proteins were separated by SDS-PAGE under reduced conditions, transferred to nitrocellulose membranes, and immunoblotted with a polyclonal antiserum to GPIbα (lanes 1 to 3, 2 days of exposure on film) or with a mixture of two polyclonal antisera to each of the αv and β3 subunits (lanes 4 through 6, 1 day of exposure on film), as detailed in the Materials and Methods. Immunoprecipitation of platelet lysates with SZ2 shows a major band in the position of platelet GPIbα, indicated by an arrowhead (A), whereas no such band is detected in endothelial cell immunoprecipitates. On (B), arrowheads depict the position of endothelial αv heavy chain and β3 subunits. Molecular mass markers are indicated on the left-hand side.
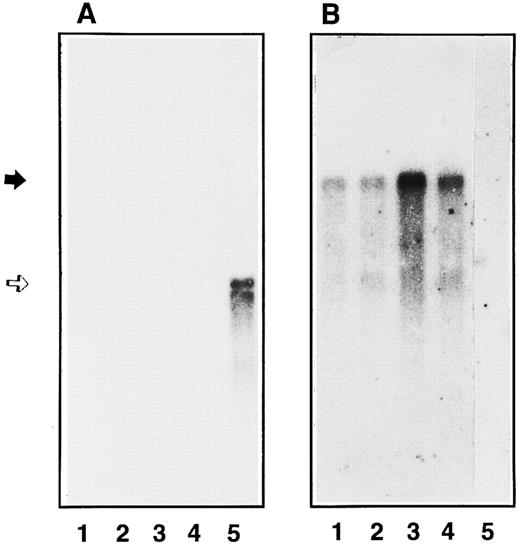
Study of GPIbα mRNA expression in endothelial cells and in HEL 5J20 cells.Conflicting data concerning the expression of GPIbα mRNA in endothelial cells after stimulation with TNFα have been reported.15,16 18 Therefore, we have studied by Northern blot the expression of GPIbα mRNA and compared it with the inducible ICAM-1 mRNA and with the constitutively expressed vWF mRNA. As shown in Fig 5A, a 2.8-kb band was observed in the HEL 5J20 extract corresponding to GPIbα mRNA (lane 5). In contrast, we were unable to detect such a band in endothelial extracts even after 1 week of exposure of the autoradiograph (lanes 1 through 4). The endothelial origin was confirmed by the presence of a 9-kb vWF mRNA in nonstimulated and stimulated cell extracts (Fig 5B). Cell stimulation was assessed by the presence of ICAM-1 mRNA as a 3.3-kb band in the TNFα-stimulated endothelial cells (data not shown).
Northern blot analysis of endothelial GPIbα mRNA. Poly(A+) mRNA were extracted and analyzed by Northern blot using radiolabeled GPIbα cDNA (A) or vWF cDNA (B) as the probe in the different cell extracts: primary endothelial cells (4 × 106 cells, lanes 1 and 2), second passage endothelial cells (7 × 106 cells, lanes 3 and 4), and HEL 5J20 cells (7 × 106 cells, lane 5). mRNA expression was observed in unstimulated (lanes 1 and 3) endothelial cells or after TNFα treatment (lanes 2 and 4). The position of GPIbα mRNA is indicated by an open arrow corresponding to 2.8 kb, whereas vWF mRNA band is shown by a solid arrow (9 kb).
Northern blot analysis of endothelial GPIbα mRNA. Poly(A+) mRNA were extracted and analyzed by Northern blot using radiolabeled GPIbα cDNA (A) or vWF cDNA (B) as the probe in the different cell extracts: primary endothelial cells (4 × 106 cells, lanes 1 and 2), second passage endothelial cells (7 × 106 cells, lanes 3 and 4), and HEL 5J20 cells (7 × 106 cells, lane 5). mRNA expression was observed in unstimulated (lanes 1 and 3) endothelial cells or after TNFα treatment (lanes 2 and 4). The position of GPIbα mRNA is indicated by an open arrow corresponding to 2.8 kb, whereas vWF mRNA band is shown by a solid arrow (9 kb).
To rule out a heterogeneity in the level of GPIbα mRNA due to the number of passage in cultured cells, we compared confluent umbilical endothelial cells before any passage with second-passage cultured cells. As shown on Fig 5, a 9-kb band was found in all endothelial cell extracts (lanes 1 through 4), but no GPIbα mRNA was detectable. TNFα was able to upregulate ICAM-1 expression in primary and second passage cells (data not shown).
DISCUSSION
In the present study, we compared the adhesive properties of two recombinant vWF, each containing a molecular defect impairing its ability to bind to the GPIb-IX or αIIbβ3 platelet receptor, with the aim of localizing the endothelial cell binding sites on the vWF subunit. Our results indicate that endothelial cell adhesion to RGGS-rvWF is strongly impaired, whereas adhesion to ΔA1-rvWF is not different from that to WT-rvWF. In addition, we provide data obtained by immunochemical analysis and functional studies using a large panel of MoAbs showing the lack of expression of an endothelial GPIbα-related receptor on cell surface and in cell lysates. Therefore, we conclude that endothelial cells adhere to vWF through a GPIbα-independent mechanism.
A main indication that the GPIbα-binding domain of vWF is not involved in endothelial cell adhesion is provided by the ability of ΔA1-rvWF to support adhesion and spreading to the same extent as WT-rvWF. This is in contrast with the absolute requirement of the A1 domain for vWF interaction with platelet GPIbα, because platelet adhesion to immobilized ΔA1-rvWF was decreased to the level of the albumin control in either static or flow conditions.20,21 Thus, our result extends our previous observation that endothelial cells are unable to adhere and spread on the SpIII fragment overlapping the GPIb-binding domain, whereas the complementary SpII fragment (aa 1366-2050) supports adhesion to the same extent as vWF.19 Accordingly, we find that endothelial cell adhesion to RGGS-rvWF is strongly impaired and that spreading is completely abolished, because only 10% of cells adhere to RGGS-rvWF, compared with approximately 40% adherent cells to WT-rvWF (Fig 1). Interestingly, this value compares very well with the previously reported 75% decrease of cell attachment to two RGD mutants of vWF relative to WT-rvWF, which was assessed by counting cell-associated radioactivity.17 18 To detect low-affinity interactions, we have developed a sensitive quantitative adhesion assay based on an automated digital imaging processing system. Automatic focusing and displacement of the microscope stage allows accurate determination of cell adhesion with high reproducibility and small interexperimental variations. This method has also the advantage over other quantitative assays for cell adhesion to show the cell morphology.
In addition, we find that the low residual adhesion to RGGS-rvWF can be completely inhibited in a dose-dependent way by MoAb 9 to vWF, suggesting the involvement of additional sequences beside RGD that are located within the carboxyterminal part of vWF and allow optimal interaction with the αvβ3 integrin (Fig 2). Thus, it appears that the presence of RGD and its neighboring sequences is sufficient to support full adhesion and spreading of endothelial cells. Deleting the A1 domain does not result in a more pronounced dependency on the αvβ3-mediated endothelial cell adhesion, as shown by the similar inhibition of adhesion to ΔA1-rvWF and WT-rvWF by anti-vWF MoAb 9 (Fig 2). This finding suggests that no additional functional site in the A1 domain, working in cooperation with the RGD-containing region, can be demonstrated.
However, in contrast to reported data on a MoAb (AS-7) that inhibits endothelial cell adhesion to RGD-mutated-rvWF,18 we have been unable to specifically block a putative GPIb-dependent endothelial cell adhesion to RGGS-rvWF (Table 1). This is shown by using different antibodies reported to block vWF binding to platelet GPIb, including AS-7.25-30 This lack of inhibition of endothelial cell adhesion is correlated with the absence of detection of protein expression on the cell surface by flow cytometry (Fig 3). It could be postulated that heterogeneity of endothelial cells may influence the expression of the GPIbα epitope. Although the use of different culture media and serum batches may account for these differences, we have carefully selected culture conditions for human umbilical vein endothelial cells from a second passage close to those used in previously reported functional studies.16-18 However, the following remark may provide some explanation for the discrepancies between our results and those reported by Beacham et al.18 To prevent proteolysis of GPIb-IX by trypsin, we detach the cells with EDTA in conditions in which endothelial cell adhesion to WT-rvWF is completely inhibitable by the anti-αvβ3 MoAb LM609 (data not shown). However, this group reported a decreased adhesion to WT-rvWF when using EDTA-treated cells, suggesting an inactivation of αvβ3 by EDTA and hence its possible downregulation due to culture conditions.18
TNFα has been reported to both increase GPIbα and decrease αvβ3 expression on endothelial cells.16-18,38 However, cytokine stimulation is not an absolute requirement, because adhesion may involve a GPIbα-dependent mechanism even in the absence of cell stimulation.18 Therefore, we also separately addressed the issue of cell stimulation by comparing adhesion of nonstimulated and TNFα-stimulated endothelial cells. Cell stimulation is effective, as indicated by the increased expression of ICAM-1 (Fig 3). However, we found that cell stimulation by TNFα neither increases cell adhesion to RGGS-rvWF nor unravels an inhibition of adhesion to WT-rvWF or RGGS-rvWF by anti-vWF or anti-GPIbα antibodies. Thus, in our hands, cytokine stimulation does not induce the expression of a functional endothelial GPIbα. This result is confirmed by our failure to identify endothelial GPIbα expression on the surface of stimulated cells by flow cytometry using a wide variety of antibodies to the platelet receptor.
Endothelial GPIbα has been previously reported by its immunoreactivity with antibodies to glycocalicin, the extracellular portion of platelet GPIbα.13,14,18 Immunoprecipitation of endothelial cell lysates indicated two proteins of Mr 145,000 and 90,000 that displayed an increased intensity after pretreatment of cells with cytochalasin B.18 While using the same conditions of immunoblotting in endothelial cell lysates or immunoprecipitates, we have been unable to detect a band in the position of platelet GPIbα or any of its fragments (Fig 4). Because an increase in endothelial GPIbα mRNA expression has been shown after exposure to TNFα, we have also attempted to identify increased gene expression.15,16 This expression has been shown in human tonsilar endothelial cells that may be subjected to high cytokines levels, but very low levels of GPIbα mRNA have also been reported in nonstimulated cells.15,16,39 After hybridization with the same cDNA probe as reported by others,36 we do not detect any significant expression of GPIbα mRNA by Northern blot analysis. In addition, GPIbα mRNA is not detectable after endothelial cell stimulation, whereas ICAM-1 mRNA is clearly enhanced.
Variability in mRNA levels of different proteins has been reported according to the lineage and time in culture of human umbilical vein endothelial cells.40 To address this issue, we have compared the level of expression of GPIbα mRNA as a function of the number of cell passages. As reported, we found a significant expression of vWF mRNA in primary and second passage human umbilical vein cells.40 In contrast, the message for GPIbα could not be shown in primary or secondary endothelial cells even after cytokine stimulation.
A marked heterogeneity of expression of endothelial cells has been reported between different adult tissues antigens, eg, vWF is more expressed in large vessels than in capillaries.41 This heterogeneity may be involved in a specialization of the endothelium to perform different functions. Therefore, one cannot rule out that GPIbα expression may be found in adult endothelial cells of other origin. Because high shear stress induces the binding of plasma vWF to platelet GPIbα, the effect of shear on the expression and the function of the putative endothelial GPIbα is obviously an important question to be addressed in tissues that are exposed to these conditions.
ACKNOWLEDGMENT
The authors are indebted to Dr N. Kieffer for the generous gift of the HEL 5J20 cell line and to Dr J. Lopez for the GPIbα cDNA probe. Drs B.S. Coller, J.L. Miller, C. Ruan, B. Steiner, P.J. Newman, M.A. Horton, and K.J. Clemetson are thanked for providing antibodies. We are grateful to Paulette Legendre for technical assistance in cell culture and to Nathalie Janel for help in performing Northern blots. We are indebted to the nursing staff of the Department of Obstetrics of the International Hospital of the University of Paris for their help in obtaining umbilical cords. We thank Dr L. Coulombel for revision of the manuscript.
Supported by EC Biomed Grant No. CT931685 (D.B.), by a GEHT-Baxter grant (C.P.), and by CNRS (D.P.).
Address reprint requests to Dominique Baruch, MD, PhD, INSERM U143, Hopital de Bicetre, 94275 Bicetre, Cedex, France.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal