Abstract
The helix-loop-helix transcription factor, scl, plays an essential role in hematopoietic development. Embryos in which the gene has been disrupted fail to develop yolk sac erythropoiesis, and scl-null embryonic stem cells do not contribute to hematopoiesis in chimeric mice. To analyze the molecular consequences of scl deficiency, we compared the gene expression profiles of control (wild-type and scl-heterozygous) and scl-null embryonic stem cells differentiated in vitro for up to 12 days. In control and scl-null embryoid bodies the temporal expression pattern of genes associated with the formation of ventral mesoderm, such as Brachyury, bone morphogenetic protein-4, and flk-1, was identical. Similarly, GATA-2, CD34, and c-kit, which are coexpressed in endothelial and hematopoietic lineages, were expressed normally in scl-null embryonic stem cell lines. However, hematopoietic-restricted genes, including the transcription factors GATA-1, EKLF, and PU.1 as well as globin genes and myeloperoxidase, were only expressed in wild-type and scl-heterozygous embryonic stem cells. Indirect immunofluorescence was used to confirm the observations that GATA-1 and globins were only present in control embryoid bodies but that CD34 was found on both control and scl-null embryoid bodies. These data extend the previous gene ablation studies and support a model whereby scl is absolutely required for commitment of a putative hemangioblast to the hematopoietic lineage but that it is dispensable for endothelial differentiation.
DURING MURINE EMBRYOGENESIS, ventral mesoderm migrates into the yolk sac at embryonic day 7.5 (E7.5) and forms aggregates of cells (hemangioblasts) that may represent the putative common precursor for endothelial and hematopoietic cells.1 2 These progenitor cells give rise to blood islands, which consist of an outer layer of endothelium enclosing primitive hematopoietic cells. Insights into the molecular mechanisms important in primitive hematopoiesis have been made through the analysis of gene ablation in the mouse and in vitro studies in Xenopus laevis.
Bone morphogenetic protein-4 (BMP-4), a member of the transforming growth factor (TGF ) β family, which induces ventral mesoderm,3 has been identified as a key component in this cascade. A dominant negative BMP-4 receptor dorsalizes Xenopus embryos that are then deficient in blood formation.4-8 The relevance of this signaling pathway to mammalian hematopoiesis is supported by a recent study by Johansson and Wiles,9 which showed that exogenous BMP-4 was required for mesoderm and globin gene induction in embryoid bodies (EBs) and was a prerequisite for BMP-4 expression from its endogenous locus.
Vertebrate homologues of the Drosophila mothers against decapentaplegic (mad ) gene (Smad1-5) function as homo- and hetero-dimeric signal transduction molecules downstream of TGFβ family receptors.10,11 It has been shown that BMP-4 uses Smad1 or 5 in a heterodimeric complex with Smad4 and that Smad1 can compensate for a dominant negative BMP receptor.12,13 Similar complementation studies have identified a number of homeobox genes including Mix.1,14PV.1,15 Xvent-1,16 Xvent-2/Vox/Xbr-1,17-19 and Xom20 as ventralizing genes downstream of BMP-4. Recently, the identification of a transcriptional partner for Smad2 in Xenopus, a novel winged-helix transcription factor denoted FAST-1 that cooperates to transactivate the Mix.2 gene in response to activin, has provided a definite link between TGFβ signaling and these homeodomain proteins in vertebrates.21 The large number of genes identified thus far reflects the complexity of the process and detailed analysis in the mouse awaits the cloning of their murine homologues.
Gene ablation studies in the mouse have also been instrumental in defining molecules critical for the initiation of hematopoiesis, such as BMP-4, flk-1, scl, and LMO2. Surprisingly, embryos mutant for BMP-4 or its type I receptor display a more severe phenotype than anticipated. Not only do they lack ventral mesoderm and hematopoietic cells, but they die early in development after a failure of epiblast proliferation, a necessary prerequisite for gastrulation.22,23 Ablation of the tyrosine kinase receptor flk-1, expressed on yolk sac hemangioblasts and endothelial cells,24 also results in early embryonic lethality, in this case associated with complete absence of yolk sac blood islands.2
Recent work has shown that the helix-loop-helix transcription factor, scl, the oncogene most frequently activated in childhood T-cell leukemia,25 plays an essential role in hematopoietic development. scl localizes to embryonic and yolk sac mesoderm in E7.5 embryos before the formation of recognizable hematopoietic elements and expression subsequently continues in both endothelial and hematopoietic components of the blood island as well as in several nonhematopoietic sites.26,27 In the adult animal, scl expression is largely restricted to erythroid cells, megakaryocytes, mast cells, early myeloid cells, and committed progenitor cells.28 Embryos in which the scl gene has been disrupted fail to develop yolk sac hematopoiesis, although endothelium appears to form normally.29,30 Furthermore, scl-null embryonic stem (ES) cells do not give rise to colony-forming cells in vitro and fail to contribute to hematopoiesis in chimeric mice, implying that scl is also critical for definitive hematopoiesis.31 32
Oligonucleotide Primers Used for RT-PCR
| Gene . | Size (bp)* . | T (°C)† . | 5′ Primer . | 3′ Primer . | Internal Probe . | Reference . |
|---|---|---|---|---|---|---|
| Mesodermal markers | ||||||
| Brachyury | 947 | 52 | TGCTGCCTGTGAGTCATAAC | TCCAGGTGCTATATATTGCC | GAGCTCAGTTCTTTCGAGGC | 49 |
| BMP RIA | 451 | 55 | CCAGAAGTGCTGGATGAAAGCC | CCTATTCCATGCTAATCCCACC | ACAGCGATGAATGTCTTCGAG | 6 |
| BMP-4 | 570 | 55 | ACTGTGAGGAGTTTCCATCACG | TCTTATTCTTCTTCCTGGACCG | GGACACCAGACTAGTCCATCAC | 9 |
| Smad1 | 336 | 55 | CCAGGCGGCATATTGGAAAAGG | ATCTCAATCCAGCAGGGGGTGC | AGCCGGAACTGCAACTACCACC | 69 |
| Transcription factors | ||||||
| scl | 396 | 55 | TATGAGATGGAGATTTCTGATG | GCTCCTCTGTGTAACTGTCC | GTCCTCACACCAAAGTAGTG | 70 |
| GATA-1 | 404 | 60 | GGAATTCGGGCCCCTTGTGAGG | CGGGGTACCTCACGCTCCAG | GTCTGCAATGCCTGTGGCCTCTA | 71 |
| CCAGAGAG | CCAGATTCGACCC | |||||
| GATA-2 | ∼720 | 60 | CGGAATTCGACACACCACCCGA | CGGAATTCGCCTACGCCATGGC | GTCTGCAATGCCTGTGGCCTCTA | 59 |
| TACCCACCTAT | AGTCACCATGCT | |||||
| EKLF | 359 | 55 | TCGCCGGAGACGCAGGCT | CCCAGTCCTTGTGCAGGA | ACGCAGCCGGCGAACTTTGG | 49 |
| NF-E2 | 391 | 62.5 | GAGCCCTGGCCATGAAGATTCC | CACCATCAGCAGCCTGTTGCAG | GCAAGAACAAGGTGGCAGCCC | 55 |
| c-myb | 681 | 55 | CCTCTAGGAGCTCATTTGTG | TTCAAGGCCAGCATTCTTGC | TACTAAACCATTTCATGAGG | 49 |
| PU.1 | 615 | 55 | TGGAAGGGTTTTCCCTCACC | TGCTGTCCTTCATGTCGCCG | GAGAACTTCCCTGAGAACCAC | 72 |
| Receptors | ||||||
| CD34 | 618 | 55 | GTTACCTCTGGGATCCCTTCAG | CTCCAGAGGTGACCAATGAATA | CAACCACAGACTTCCCCAAC | 45 |
| GCTC | AG | |||||
| flk-1 | 398 | 60 | TAGGTGCCTCCCCATACCCTGG | TGGCCGGCTCTTTCGCTTACTG | GGCAAAGACTATATTGTTCTTC | 73 |
| flk-2 | 300 | 60 | CTTCTGTGCAAATGCTGTGCGC | GTTCTCTCTCGGGAACTCCCAC | TTCTCATTGTGTTGATCTGCC | 45 |
| GTAC | TTAAG | |||||
| c-kit | 765 | 50 | TGTCTCTCCAGTTTCCCTGC | TTCAGGGACTCATGGGCTCA | ACAACACTCTTATCGTAGACC | 49 |
| Epo R | 356 | 62 | TTGACGCTGTCTCTCATTCTGG | CAATACCAAGTAGGTGTCCTGG | CGAGTTTGAGGGTCTCTTCACC | 74 |
| Erythroid and myeloid genes | ||||||
| ζ globin | 371 | 55 | GCTCAGGCCGAGCCCATTGG | TAGCGGTACTTCTCAGTCAG | CGGTCAACTTCAAGCTCCTG | 59 |
| α globin | 334 | 55 | CTCTCTGGGGAAGACAAAAGCA | GGTGGCTAGCCAAGGTCACCAG | GCCCTGGAAAGGATGTTTGCTA | 59 |
| AC | CA | GC | ||||
| βH1 globin | 265 | 55 | CTCAAGGAGACCTTTGCTCA | AGTCCCCATGGAGTCAAAGA | TCCAATCACCAGCTTCTGCC | 49 |
| εy2 globin | 157 | 55 | GGAGAGTCCATTAAGAACCTAG | CTGTGAATTCATTGCCGAA | CCTGAGAACTTCAAACTCTTGG | 59 |
| ACAA | GTAC | GT | ||||
| βmaj globin | 578 | 55 | CACAACCCCAGAAACAGACA | CTGACAGATGCTCTCTTGGG | ACCTTCTGGAAGGCAGCCTGTGC | 49 |
| MPO | 379 | 55 | GGTGCTGAAGAACCTGGAGTTG | CTAGGTCTCCTTCCAGGAAGTC | GATGGTGATCGGTTTTGGTGG | 75 |
| Lysozyme | 344 | 60 | TGCAGGATGACATCACTGCAGC | GCTGTGCTGACTGACAAGGGAG | CATTCGAGCATGGGTGGCATG | 76 |
| vav | 604 | 55 | GTATAACGTGGAGGTCAAGCA | GGAGGACTGGAGAAAATCAGT | GCAGAATTCCCTCAAAGATTGC | 49 |
| LMO2 | 505 | 60 | TCGCGAATTCCACCATGTCCTC | CGAGTTCTAGACTAGATGATCC | GAGGATTGCCTCAGCTGTGACC | 29 |
| GGCCATCGAAAGGA | CATTGATCTTGGT | |||||
| HPRT | 249 | 55 | GCTGGTGAAAAGGACCTCT | CACAGGACTAGAACACCTGC | GGATACAGGCCAGACTTTGT | 49 |
| Gene . | Size (bp)* . | T (°C)† . | 5′ Primer . | 3′ Primer . | Internal Probe . | Reference . |
|---|---|---|---|---|---|---|
| Mesodermal markers | ||||||
| Brachyury | 947 | 52 | TGCTGCCTGTGAGTCATAAC | TCCAGGTGCTATATATTGCC | GAGCTCAGTTCTTTCGAGGC | 49 |
| BMP RIA | 451 | 55 | CCAGAAGTGCTGGATGAAAGCC | CCTATTCCATGCTAATCCCACC | ACAGCGATGAATGTCTTCGAG | 6 |
| BMP-4 | 570 | 55 | ACTGTGAGGAGTTTCCATCACG | TCTTATTCTTCTTCCTGGACCG | GGACACCAGACTAGTCCATCAC | 9 |
| Smad1 | 336 | 55 | CCAGGCGGCATATTGGAAAAGG | ATCTCAATCCAGCAGGGGGTGC | AGCCGGAACTGCAACTACCACC | 69 |
| Transcription factors | ||||||
| scl | 396 | 55 | TATGAGATGGAGATTTCTGATG | GCTCCTCTGTGTAACTGTCC | GTCCTCACACCAAAGTAGTG | 70 |
| GATA-1 | 404 | 60 | GGAATTCGGGCCCCTTGTGAGG | CGGGGTACCTCACGCTCCAG | GTCTGCAATGCCTGTGGCCTCTA | 71 |
| CCAGAGAG | CCAGATTCGACCC | |||||
| GATA-2 | ∼720 | 60 | CGGAATTCGACACACCACCCGA | CGGAATTCGCCTACGCCATGGC | GTCTGCAATGCCTGTGGCCTCTA | 59 |
| TACCCACCTAT | AGTCACCATGCT | |||||
| EKLF | 359 | 55 | TCGCCGGAGACGCAGGCT | CCCAGTCCTTGTGCAGGA | ACGCAGCCGGCGAACTTTGG | 49 |
| NF-E2 | 391 | 62.5 | GAGCCCTGGCCATGAAGATTCC | CACCATCAGCAGCCTGTTGCAG | GCAAGAACAAGGTGGCAGCCC | 55 |
| c-myb | 681 | 55 | CCTCTAGGAGCTCATTTGTG | TTCAAGGCCAGCATTCTTGC | TACTAAACCATTTCATGAGG | 49 |
| PU.1 | 615 | 55 | TGGAAGGGTTTTCCCTCACC | TGCTGTCCTTCATGTCGCCG | GAGAACTTCCCTGAGAACCAC | 72 |
| Receptors | ||||||
| CD34 | 618 | 55 | GTTACCTCTGGGATCCCTTCAG | CTCCAGAGGTGACCAATGAATA | CAACCACAGACTTCCCCAAC | 45 |
| GCTC | AG | |||||
| flk-1 | 398 | 60 | TAGGTGCCTCCCCATACCCTGG | TGGCCGGCTCTTTCGCTTACTG | GGCAAAGACTATATTGTTCTTC | 73 |
| flk-2 | 300 | 60 | CTTCTGTGCAAATGCTGTGCGC | GTTCTCTCTCGGGAACTCCCAC | TTCTCATTGTGTTGATCTGCC | 45 |
| GTAC | TTAAG | |||||
| c-kit | 765 | 50 | TGTCTCTCCAGTTTCCCTGC | TTCAGGGACTCATGGGCTCA | ACAACACTCTTATCGTAGACC | 49 |
| Epo R | 356 | 62 | TTGACGCTGTCTCTCATTCTGG | CAATACCAAGTAGGTGTCCTGG | CGAGTTTGAGGGTCTCTTCACC | 74 |
| Erythroid and myeloid genes | ||||||
| ζ globin | 371 | 55 | GCTCAGGCCGAGCCCATTGG | TAGCGGTACTTCTCAGTCAG | CGGTCAACTTCAAGCTCCTG | 59 |
| α globin | 334 | 55 | CTCTCTGGGGAAGACAAAAGCA | GGTGGCTAGCCAAGGTCACCAG | GCCCTGGAAAGGATGTTTGCTA | 59 |
| AC | CA | GC | ||||
| βH1 globin | 265 | 55 | CTCAAGGAGACCTTTGCTCA | AGTCCCCATGGAGTCAAAGA | TCCAATCACCAGCTTCTGCC | 49 |
| εy2 globin | 157 | 55 | GGAGAGTCCATTAAGAACCTAG | CTGTGAATTCATTGCCGAA | CCTGAGAACTTCAAACTCTTGG | 59 |
| ACAA | GTAC | GT | ||||
| βmaj globin | 578 | 55 | CACAACCCCAGAAACAGACA | CTGACAGATGCTCTCTTGGG | ACCTTCTGGAAGGCAGCCTGTGC | 49 |
| MPO | 379 | 55 | GGTGCTGAAGAACCTGGAGTTG | CTAGGTCTCCTTCCAGGAAGTC | GATGGTGATCGGTTTTGGTGG | 75 |
| Lysozyme | 344 | 60 | TGCAGGATGACATCACTGCAGC | GCTGTGCTGACTGACAAGGGAG | CATTCGAGCATGGGTGGCATG | 76 |
| vav | 604 | 55 | GTATAACGTGGAGGTCAAGCA | GGAGGACTGGAGAAAATCAGT | GCAGAATTCCCTCAAAGATTGC | 49 |
| LMO2 | 505 | 60 | TCGCGAATTCCACCATGTCCTC | CGAGTTCTAGACTAGATGATCC | GAGGATTGCCTCAGCTGTGACC | 29 |
| GGCCATCGAAAGGA | CATTGATCTTGGT | |||||
| HPRT | 249 | 55 | GCTGGTGAAAAGGACCTCT | CACAGGACTAGAACACCTGC | GGATACAGGCCAGACTTTGT | 49 |
Abbreviations: BMP, bone morphogenetic protein; Epo R, erythropoietin receptor; MPO, myeloperoxidase; HPRT, hypoxanthine phosphoribosyl transferase.
Size of the predicted PCR product.
Annealing temperature.
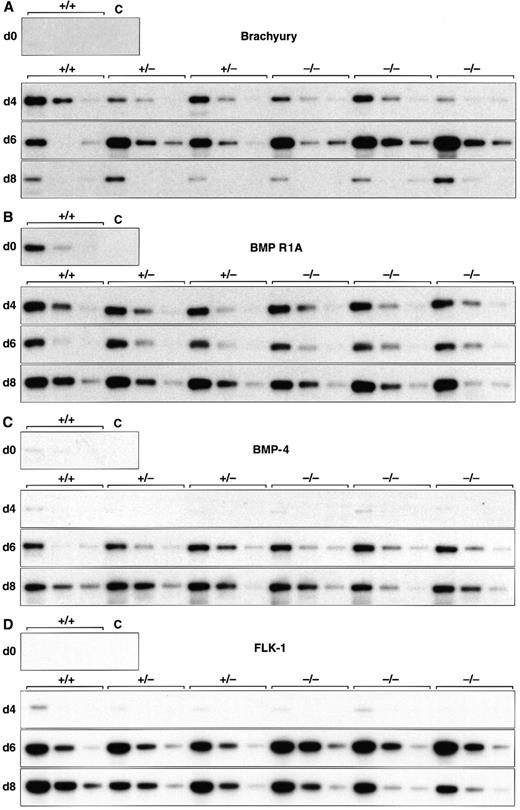
Normal induction of early mesoderm-associated genes in control and scl-null EBs. (A through D) Expression of the early pan-mesodermal marker Brachyury, the ventral mesoderm inducing gene BMP-4, its type I receptor (BMP R1A), and the VEGF receptor tyrosine kinase flk-1 are shown at day 0, 4, 6, and 8 of embryoid body differentiation. At each time point, wild-type (+/+) or scl-heterozygous (+/−) lines are compared with three independent scl-null lines (−/−), except for day 0, when expression is shown for undifferentiated wild-type (+/+) ES cells only. RT-PCR amplification of three fivefold serial dilutions of cDNA is shown for each line. C, control lane with no cDNA added.
Normal induction of early mesoderm-associated genes in control and scl-null EBs. (A through D) Expression of the early pan-mesodermal marker Brachyury, the ventral mesoderm inducing gene BMP-4, its type I receptor (BMP R1A), and the VEGF receptor tyrosine kinase flk-1 are shown at day 0, 4, 6, and 8 of embryoid body differentiation. At each time point, wild-type (+/+) or scl-heterozygous (+/−) lines are compared with three independent scl-null lines (−/−), except for day 0, when expression is shown for undifferentiated wild-type (+/+) ES cells only. RT-PCR amplification of three fivefold serial dilutions of cDNA is shown for each line. C, control lane with no cDNA added.
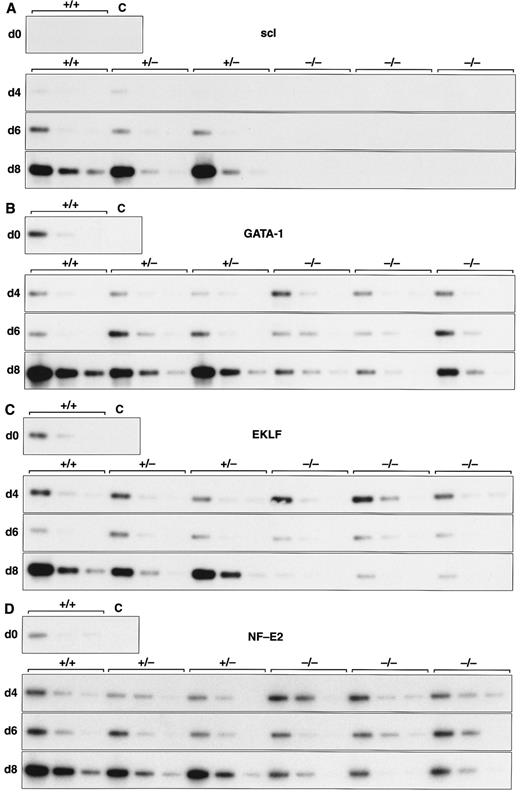
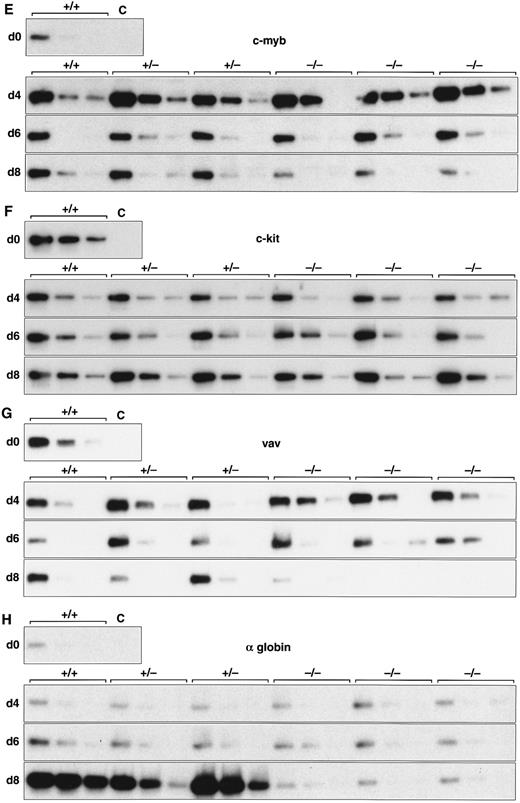
Expression of hematopoietic genes in differentiating control and scl-null EBs. RT-PCR amplification of three fivefold serial dilutions of cDNA for each of the indicated genes is shown for each line at day 0, 4, 6, and 8 of embryoid body differentiation. As in Fig 1, the genotype of the cell line (+/+, +/−, or −/−) is indicated above the panel. Note the discordance in expression between control cells and scl-null cells for GATA-1, EKLF, NF-E2, c-myb, vav, and α globin at the day-8 timepoint.
Expression of hematopoietic genes in differentiating control and scl-null EBs. RT-PCR amplification of three fivefold serial dilutions of cDNA for each of the indicated genes is shown for each line at day 0, 4, 6, and 8 of embryoid body differentiation. As in Fig 1, the genotype of the cell line (+/+, +/−, or −/−) is indicated above the panel. Note the discordance in expression between control cells and scl-null cells for GATA-1, EKLF, NF-E2, c-myb, vav, and α globin at the day-8 timepoint.
LMO2 is a lin-11,is1-1,mec-3 (LIM) domain protein that was also identified by virtue of its involvement in chromosomal translocations in pediatric T-cell leukemia.33 It interacts in vivo with scl28 and displays a similar phenotype in gene-targeted mice, with failure of yolk sac erythropoiesis leading to embryonic lethality around E10.5.33
To analyze the molecular nature of the block in hematopoiesis, which is a consequence of scl gene ablation, we examined the temporal expression pattern of genes involved in early hematopoiesis in differentiating EBs. In contrast with wild-type and scl-heterozygous (scl+/−) EBs, we observed a failure of induction of hematopoietic-specific genes in scl-null EBs, although expression of genes coexpressed in endothelium was normal. This is consistent with the hypothesis that scl is dispensible for endothelial differentiation but is absolutely required for hematopoietic commitment from a primitive hemangioblast. Thus, by applying an extensive analysis of hematopoietic gene expression to control and scl-null EBs, we have extended previous studies and have been able to more clearly define the place of scl within the network of interacting genes that regulate early hematopoiesis.
MATERIALS AND METHODS
Generation and in vitro differentiation of ES cell lines.The three scl-null and the two scl+/− ES cell lines used in this study were generated by electroporation of the scl+/− cell line 93 with the linearized targeting vector 607 and selecting for homologous or nonhomologous integrants as described.32 Parental (W9.5), scl+/− and scl-null cell lines were weaned off feeder layers and maintained in leukemia inhibitory factor (LIF ). Primary EBs were formed in 0.9% methylcellulose cultures (including a selected batch of fetal calf serum at 15%) supplemented with 100 ng/mL stem cell factor (SCF ) and 2 U/mL of recombinant human erythropoietin (Epo) in the absence of LIF. Cultures were established at 7.5 to 10 × 103 cells/mL and 10 to 20 EBs were generated per 1,000 cells plated.
cDNA synthesis and reverse transcription polymerase chain reaction (RT-PCR).Total RNA was isolated from EBs after 4, 6, 8, and 12 days of differentiation and undifferentiated ES cells using Trizol (GIBCO-BRL, Gaithersburg, MD) according to the manufacturer's instructions. Samples were treated with DNase I (Boehringer Mannheim, Mannheim, Germany) and oligo(dT)-primed first strand cDNA synthesis from 3 μg RNA was performed using avian myeloblastosis virus (AMV) reverse transcriptase (Boehringer Mannheim) in a total reaction volume of 60 μL. One microliter of fivefold serial dilutions of each sample was polymerase chain reaction (PCR) amplified with primers for hypoxanthine phosphoribasyl transferase (HPRT; Table 1) using 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 60 seconds. This was performed in a 20 μL reaction mixture containing 1 × PCR buffer (Boehringer Mannheim), 60 μmol/L each dNTP, 1.25 U Taq polymerase (Boehringer Mannheim) preincubated with TaqStart antibody (Clontech, San Diego, CA) at a ratio of 2:1:1 of Taq:TaqStart:TaqStart dilution buffer and 0.5 μmol/L of each specific primer. The product was electrophoresed on a 2% agarose gel, transferred to GeneScreen Plus (Dupont, Wilmington, DE) and the filter hybridized with a 32P-end labeled internal oligonucleotide (Table 1). Signal intensity was quantitated using a Phosphorimager (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software (Molecular Dynamics). This information was used to equilibrate the quantity of serially diluted cDNA in each sample amplified in subsequent reactions with primers for hematopoietic genes. Thirty-cycle PCR reactions were used for all genes and the annealing temperatures are shown in Table 1. Most genes were analyzed on more than one occasion using cDNA from independently derived RNA samples.
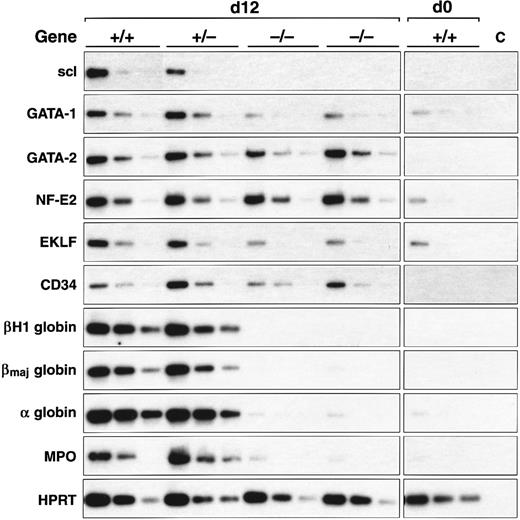
scl-null EBs express endothelial genes but not specific erythroid and myeloid genes. RT-PCR amplification of three fivefold serial dilutions of cDNA for each of the indicated genes is shown for each ES cell line of indicated genotype after day 0 and day 12 of differentiation.
scl-null EBs express endothelial genes but not specific erythroid and myeloid genes. RT-PCR amplification of three fivefold serial dilutions of cDNA for each of the indicated genes is shown for each ES cell line of indicated genotype after day 0 and day 12 of differentiation.
To achieve signals within the linear amplification range for hematopoietic genes, serial dilutions commenced with cDNA synthesized from approximately 10 ng of RNA. For the HPRT control, serial dilutions commenced with cDNA synthesized from approximately 400 pg of DNase I-treated RNA. The hematopoietic gene primers, selected to cross introns where possible, are listed in Table 1. The duration of exposure of the autoradiographs shown in Figs 1-3 was chosen to approximate the intensity of signal on the ethidium bromide stained gel.
Immunofluorescence and confocal microscopy.EBs were harvested after 12 days and treated with trypsin to produce cell suspensions, which were allowed to adhere to poly-(L-lysine) (Sigma Chemical Co, St Louis, MO) coated glass coverslips for 5 minutes. Immunofluorescence was performed as previously described.34 Briefly, cells were fixed in 3.7% paraformaldehyde and stained with 1:25 dilution of biotinylated antimouse CD34 monoclonal antibody (Pharmingen, San Diego, CA) followed by 1:100 dilution of fluorescein isothiocyanate (FITC)-conjugated Avidin D (Calbiochem, San Diego, CA). For detection of globin, cells were permeabilized with 0.5% Triton-X100 after fixation and stained with 1:500 dilution of rabbit antimouse hemoglobin (Cappel Research Products, Durham, NC) followed by 1:100 dilution of FITC-conjugated antirabbit IgG (Silenus, Melbourne, Australia). Cells were briefly (45 seconds) permeabilized with 0.5% Triton-X100 before fixation to detect GATA-1 with a rat anti-mouse GATA-1 (N6) monoclonal antibody35 used at 30 μg/mL followed by 1:100 dilution of FITC-conjugated antirat IgG (Jackson Immunoresearch Laboratories, West Grove, PA).
Images of single optical sections were collected using a Leica Confocal Laser Scanning Microscope and Scanware software (Leica Lasertechnik Gmbh, Heidelberg, Germany).
RESULTS
To study the molecular consequences of scl deletion, gene expression profiles were compared in three control cell lines (two scl-heterozygous (+/−) and one wild-type (+/+) ES cell line) and three scl-null (−/−) ES cell lines that were differentiated in methylcellulose cultures in the presence of SCF and Epo. EBs were harvested after 4, 6, and 8 days of differentiation and cDNA was prepared. Gene expression in the ES cell lines was compared using PCR in a semiquantitative manner. As described in Materials and Methods, this involved careful titration of the quantity of cDNA amplified, the use of serial dilutions of cDNA, and a limitation of PCR cycle numbers to 30. In a further series of experiments, EBs were analyzed after 12 days of differentiation with and without the same growth factors. Four sets of genes were studied: genes involved in mesoderm induction and hematopoietic specification, transcription factors, cell surface molecules, and genes expressed in differentiated erythroid and myeloid cells. The results for all genes are summarized in Table 2, and representative Southern blots of RT-PCR reactions of selected examples are displayed in Figs 1-3.
Hematopoietic Gene Expression During Embryoid Body Development in Control and scl-Null ES Cells
| Gene . | Gene Expression During ES Cell Differentiation (days)* . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 0 . | 4 . | 6 . | 8 . | 12 . | Induction† . | Ratio‡ . | . | . | |||||
| . | c . | c . | null . | c . | null . | c . | null . | c . | null . | c . | null . | c:null . | . | . |
| Mesodermal markers | ||||||||||||||
| Brachyury | − | + | + | ++ | ++ | ± | ± | nd | nd | >500 | >500 | ∼1 | ||
| BMP R1A | + | ++ | ++ | ++ | ++ | +++ | +++ | nd | nd | ∼5 | ∼5 | ∼1 | ||
| BMP-4 | ± | ± | ± | + | + | ++ | ++ | nd | nd | ∼50 | ∼50 | ∼1 | ||
| Smad1 | + | + | + | ++ | ++ | ++ | ++ | ++ | ++ | >5 | >5 | ∼1 | ||
| Transcription factors | ||||||||||||||
| scl | − | ± | − | + | − | ++ | − | + | − | >500 | 0 | >500 | ||
| GATA-1 | ± | ± | ± | ± | ± | ++ | ± | ++ | ± | >5 | ∼1 | >5 | ||
| GATA-2 | − | + | + | ++ | ++ | ++ | ++ | ++ | ++ | >50 | >50 | ∼1 | ||
| EKLF | + | + | + | + | + | ++ | + | ++ | + | >5 | ∼1 | >5 | ||
| NF-E2 | ± | + | + | + | + | ++ | + | ++ | ++ | >5 | >5 | ∼1 | ||
| c-myb | + | ++ | ++ | + | + | + | ± | nd | nd | <5 | ∼1ρ | <5 | ||
| PU.1 | ± | ± | ± | ± | ± | + | ± | + | ± | >5 | ∼1 | >5 | ||
| Receptors | ||||||||||||||
| CD34 | ± | ± | ± | ± | ± | + | ± | ++ | ++ | >50 | >50 | ∼1 | ||
| flk-1 | − | + | + | +++ | +++ | +++ | +++ | nd | nd | >50 | >50 | ∼1 | ||
| flk-2 | ± | ± | ± | ± | ± | ± | ± | ± | ± | >5 | >5 | <5 | ||
| c-kit | ++ | ++ | ++ | ++ | ++ | ++ | ++ | nd | nd | ∼1 | ∼1 | ∼1 | ||
| Epo R | + | ++ | ++ | ++ | ++ | ++ | ++ | nd | nd | >5 | >5 | ∼1 | ||
| Erythroid and myeloid genes | ||||||||||||||
| ζ globin | − | − | − | ± | − | +++ | − | +++ | − | >500 | 0 | >500 | ||
| α globin | ± | ± | ± | ± | ± | +++ | ± | +++ | ± | >500 | ∼1 | >500 | ||
| βH1 globin | − | − | − | ± | − | +++ | − | +++ | − | >500 | 0 | >500 | ||
| ε2 globin | − | − | − | − | − | +++ | − | +++ | − | >500 | 0 | >500 | ||
| βmaj globin | − | − | − | − | − | +++ | − | +++ | − | >500 | 0 | >500 | ||
| MPO | − | ± | ± | ± | ± | + | ± | + | ± | >50 | <5 | >5 | ||
| Lysozyme | − | ± | ± | ± | ± | + | + | nd | nd | >50 | >50 | <5 | ||
| vav | + | + | + | + | + | + | ± | + | ± | ∼1 | 0ρ | >5 | ||
| LMO2 | − | + | + | ++ | ++ | ++ | ++ | nd | nd | >500 | >50 | <5 | ||
| Gene . | Gene Expression During ES Cell Differentiation (days)* . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 0 . | 4 . | 6 . | 8 . | 12 . | Induction† . | Ratio‡ . | . | . | |||||
| . | c . | c . | null . | c . | null . | c . | null . | c . | null . | c . | null . | c:null . | . | . |
| Mesodermal markers | ||||||||||||||
| Brachyury | − | + | + | ++ | ++ | ± | ± | nd | nd | >500 | >500 | ∼1 | ||
| BMP R1A | + | ++ | ++ | ++ | ++ | +++ | +++ | nd | nd | ∼5 | ∼5 | ∼1 | ||
| BMP-4 | ± | ± | ± | + | + | ++ | ++ | nd | nd | ∼50 | ∼50 | ∼1 | ||
| Smad1 | + | + | + | ++ | ++ | ++ | ++ | ++ | ++ | >5 | >5 | ∼1 | ||
| Transcription factors | ||||||||||||||
| scl | − | ± | − | + | − | ++ | − | + | − | >500 | 0 | >500 | ||
| GATA-1 | ± | ± | ± | ± | ± | ++ | ± | ++ | ± | >5 | ∼1 | >5 | ||
| GATA-2 | − | + | + | ++ | ++ | ++ | ++ | ++ | ++ | >50 | >50 | ∼1 | ||
| EKLF | + | + | + | + | + | ++ | + | ++ | + | >5 | ∼1 | >5 | ||
| NF-E2 | ± | + | + | + | + | ++ | + | ++ | ++ | >5 | >5 | ∼1 | ||
| c-myb | + | ++ | ++ | + | + | + | ± | nd | nd | <5 | ∼1ρ | <5 | ||
| PU.1 | ± | ± | ± | ± | ± | + | ± | + | ± | >5 | ∼1 | >5 | ||
| Receptors | ||||||||||||||
| CD34 | ± | ± | ± | ± | ± | + | ± | ++ | ++ | >50 | >50 | ∼1 | ||
| flk-1 | − | + | + | +++ | +++ | +++ | +++ | nd | nd | >50 | >50 | ∼1 | ||
| flk-2 | ± | ± | ± | ± | ± | ± | ± | ± | ± | >5 | >5 | <5 | ||
| c-kit | ++ | ++ | ++ | ++ | ++ | ++ | ++ | nd | nd | ∼1 | ∼1 | ∼1 | ||
| Epo R | + | ++ | ++ | ++ | ++ | ++ | ++ | nd | nd | >5 | >5 | ∼1 | ||
| Erythroid and myeloid genes | ||||||||||||||
| ζ globin | − | − | − | ± | − | +++ | − | +++ | − | >500 | 0 | >500 | ||
| α globin | ± | ± | ± | ± | ± | +++ | ± | +++ | ± | >500 | ∼1 | >500 | ||
| βH1 globin | − | − | − | ± | − | +++ | − | +++ | − | >500 | 0 | >500 | ||
| ε2 globin | − | − | − | − | − | +++ | − | +++ | − | >500 | 0 | >500 | ||
| βmaj globin | − | − | − | − | − | +++ | − | +++ | − | >500 | 0 | >500 | ||
| MPO | − | ± | ± | ± | ± | + | ± | + | ± | >50 | <5 | >5 | ||
| Lysozyme | − | ± | ± | ± | ± | + | + | nd | nd | >50 | >50 | <5 | ||
| vav | + | + | + | + | + | + | ± | + | ± | ∼1 | 0ρ | >5 | ||
| LMO2 | − | + | + | ++ | ++ | ++ | ++ | nd | nd | >500 | >50 | <5 | ||
Abbreviations: ES, embryonic stem; BMP, bone morphogenetic protein; Epo, erythropoietin; MPO, myeloperoxidase.
Gene expression profiles determined by RT-PCR after in vitro differentiation of ES cells. The mean level of expression is compared between two (day 12 samples) or three lines with at least one wild-type scl allele (c) and three lines with both scl alleles deleted (null). Samples were transferred to nylon membranes after electrophoresis, probed with 32P end-labeled specific internal oligonucleotides, and the signals quantitated on a phosphorimager. Scoring was as follows: −, no signal; ±, signal detected after hybridization only; +, ++, +++, signal of increasing intensity visible on ethidium bromide stained gel; nd, not determined.
Gene induction (fold) during differentiation compared with levels in undifferentiated ES cells.
The ratio of mean signal intensity in control versus null lines calculated at day 8 or 12 of differentiation.
ρ After initial induction, expression fell in null cell lines. See also Fig 2E and G.
EBs derived from both sets of cell lines recapitulated the temporal expression pattern of genes required for the formation of ventral mesoderm (Fig 1 and Table 2). The earliest mesoderm marker, Brachyury, expressed in the early primitive streak at E6.5,36 was induced equally in all six cell lines analyzed, peaking after 6 days differentiation and then declining by day 8 (Fig 1A). BMP-4 is detected, first in the E6.5 to E7.5 posterior primitive streak, and later in posterior-ventral mesoderm (which includes yolk sac blood islands among its descendants).22 BMP-4 levels increased from day 4 of differentiation (Fig 1C), whereas its receptor, BMP R1A, and one of the major downstream signaling molecules, Smad1, were present in uninduced ES cells (Fig 1B and Table 2). The VEGF receptor tyrosine kinase, flk-1, was expressed in all lines by day 4 and was present at higher levels by 6 days of differentiation (Fig 1D). This paralleled the induction of BMP-4 and antedated the expression of most hematopoietic genes (discussed later). In keeping with previous observations in scl-null embryos, these results showed that initial molecular events crucial for formation of ventral mesoderm, hemangioblasts, and vasculogenesis were preserved in the scl-mutant lines.
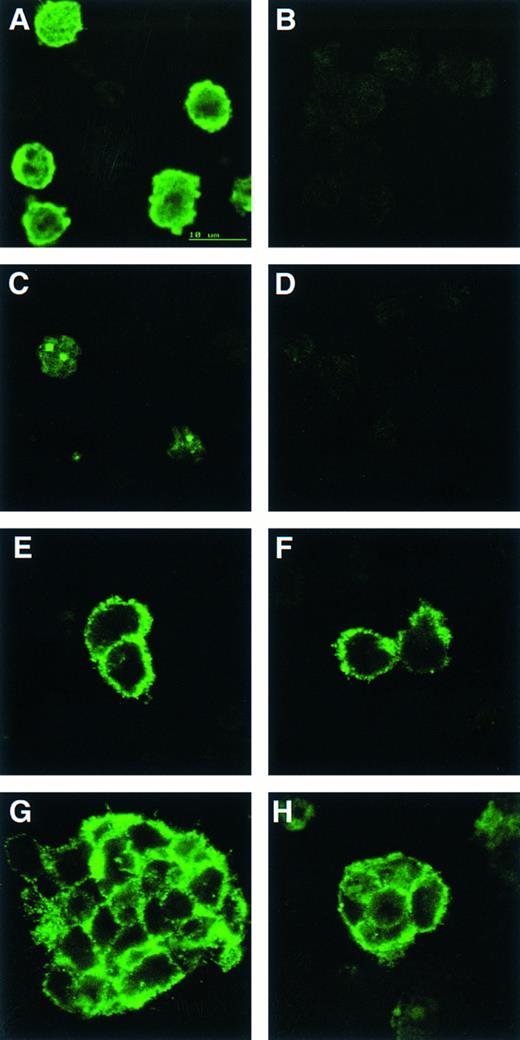
CD34 is detected on cells from both wild-type and scl-null EBs but only wild-type EBs contain cells positive for globin or GATA-1. Indirect immunofluorescence of cells from disaggregated day-12 scl+/+ (A, C, E, and F ) and scl-null (B, D, F, and H) EBs stained with antibodies to globin (A and B), GATA-1 (C and D), and CD34 (E through H).
CD34 is detected on cells from both wild-type and scl-null EBs but only wild-type EBs contain cells positive for globin or GATA-1. Indirect immunofluorescence of cells from disaggregated day-12 scl+/+ (A, C, E, and F ) and scl-null (B, D, F, and H) EBs stained with antibodies to globin (A and B), GATA-1 (C and D), and CD34 (E through H).
Analysis of hematopoietic transcription factors in control ES cells revealed that scl was first detected at day 4, coincident with the onset of flk-1 expression. Expression of scl increased exponentially over the next 4 days (Fig 2A). scl transcripts were absent from the scl-null cell lines. The patterns of expression of GATA-1, EKLF, and NF-E2 were consistent with their roles as transcription factors relevant in later, committed progenitor cells (Fig 2B to D). Low levels of transcription of all three genes were evident in undifferentiated ES cells and persisted unchanged in both scl control and scl-null cell lines until day 6. The significance of this was unclear (Fig 4). However, by day 8, levels of GATA-1, EKLF, and NF-E2 were elevated in scl control lines, consistent with an expansion of hematopoietic cells and their precursors. In contrast, expression remained at basal levels for the scl-null cell lines. Other transcription factors playing a role in hematopoiesis include c-myb, shown to be essential for definitive hematopoiesis,37 and PU.1, a factor important in lymphoid and macrophage differentiation.38 39 Although there was no induction of PU.1 in the scl-null EB (Table 2), c-myb was highly expressed in all lines by day 4, after which time its expression began to wane. In contrast to the stable levels of c-myb transcripts in the day-8 samples from scl control lines, there was a distinct fall in expression in the scl-null cell lines (Fig 2E) suggestive of a failure of formation or expansion of a hematopoietic population.
Two other genes that play pivotal roles in early hematopoiesis are LMO233 and GATA-2.40 LMO2 was first detected at day 4 and expressed at higher levels from day 6 in all cell lines (Table 2). Our analysis of GATA-2 revealed a similar expression profile to that of scl in control EBs. This pattern was unaltered in the scl-null EBs (Fig 3 and Table 2). This accords with the known expression of GATA-2 in endothelial cells, a lineage that appears to develop normally in an scl-null background.29 30
A number of cell surface molecules expressed on primitive hematopoietic cells are coexpressed on other cell types, notably CD34, c-kit, and the Epo receptor, which include endothelial cells in their tissue repertoire.41-43 Furthermore, transcripts for both c-kit and the Epo receptor have been detected in undifferentiated ES cells.44,45 It was not surprising, therefore, that CD34, c-kit, and the Epo receptor were expressed approximately equally in our cohort of differentiating EBs independent of scl-status (Figs 2F and 3 and Table 2). The flk-2 receptor is predominantly expressed in definitive hematopoietic stem cells but is also found in placenta, testis, and brain.46 47 However, because we detected only very low levels of expression, it was not possible to be sure that minor differences between the control and scl-null lines were significant (Table 2).
We investigated genes expressed in hematopoietic cells at later stages of differentiation. The vav gene, which probably functions as a signaling intermediate downstream of receptor tyrosine kinases in hematopoietic cells,48 was detected in all cell lines from days 0 to 6 of differentiation (Fig 2G). However, reminiscent of the expression pattern for c-myb, we noted a marked fall in the amount of vav cDNA from day 8 in the scl-null cell lines. This again suggested the failure to form or expand a hematopoietic population.
Most globin genes were not expressed in scl-null EBs (Figs 2H and 3 and Table 2). However, a consistent exception was a low level of spliced α globin cDNA in all cell lines even in the absence of differentiation induction (Fig 2H). This pattern of expression was similar to that seen with the GATA-1, NF-E2, and EKLF transcription factors but was not seen with other globin genes and was not associated with translation to immunologically detectable protein (Fig 4). The macrophage gene, lysozyme, was barely detectable after 8 days of differentiation in all cell lines (Table 2). On the other hand, the primitive myeloid gene, myeloperoxidase, was expressed from day 8 in scl control EBs but was virtually absent in the scl-null lines (Fig 3 and Table 2).
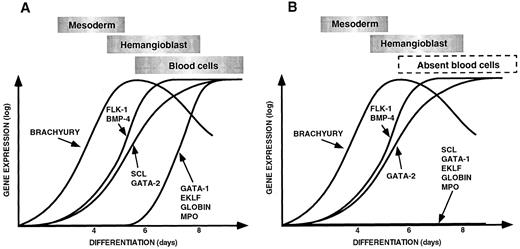
(A) Sequential expression of genes required for ventral mesoderm induction, hemangioblast formation, and hematopoiesis and vasculogenesis during control EB differentiation. (B) Pattern of gene expression observed during differentiation of scl-null EBs showing a failure of expression of hematopoietic-restricted genes. The data for this figure was derived from gene expression in embryoid bodies harvested at 4, 6, and 8 days of differentiation compared with the expression of each gene in undifferentiated ES cells. Expression levels were quantitated on the phosphorimager as described in Materials and Methods. The expression of each gene in undifferentiated ES cells was normalized to the base line and the maximal level of expression given the same arbitrary value for each gene. Gene expression is shown on a logarithmic scale on the Y axis, and the day of differentiation is indicated on the X axis. The points have been linked to form a curve to make the data easier to appreciate. The approximate timing of the major developmental processes of mesoderm induction, hemangioblast formation, and blood cell differentiation are shown in boxes above the curves.
(A) Sequential expression of genes required for ventral mesoderm induction, hemangioblast formation, and hematopoiesis and vasculogenesis during control EB differentiation. (B) Pattern of gene expression observed during differentiation of scl-null EBs showing a failure of expression of hematopoietic-restricted genes. The data for this figure was derived from gene expression in embryoid bodies harvested at 4, 6, and 8 days of differentiation compared with the expression of each gene in undifferentiated ES cells. Expression levels were quantitated on the phosphorimager as described in Materials and Methods. The expression of each gene in undifferentiated ES cells was normalized to the base line and the maximal level of expression given the same arbitrary value for each gene. Gene expression is shown on a logarithmic scale on the Y axis, and the day of differentiation is indicated on the X axis. The points have been linked to form a curve to make the data easier to appreciate. The approximate timing of the major developmental processes of mesoderm induction, hemangioblast formation, and blood cell differentiation are shown in boxes above the curves.
Expression of key genes was reexamined in EBs after 12 days of culture (summarized in Fig 3 and Table 2) to ensure that we did not mistake a delay in gene induction for a failure of expression. These data were consistent with the earlier timepoints examined: globin and myeloperoxidase expression were absent in the scl-null lines, there was persistent failure of induction of GATA-1 and EKLF but normal expression of GATA-2 and CD34. Interestingly, by day 12, NF-E2 levels were equivalent in all cell lines in contrast to our day 8 observations in which expression was clearly diminished in the scl-null EBs (compare Fig 2D and Fig 3).
We also compared gene expression from EBs grown with or without SCF and Epo. The results were not influenced by the addition or omission of these growth factors (data not shown).
To correlate the cDNA expression data with protein production, we immunostained cell suspensions made from day 12 EBs with antibodies to globins, GATA-1, and CD34 (Fig 4). Globin and GATA-1, which displayed its characteristic subnuclear localization to GATA bodies,34 were only detectable in cells derived from control EB (Fig 4A to D). This suggested that the low level, noninducible transcripts observed for both α globin and GATA-1 in the scl-null cell lines did not result in significant quantities of translated product. CD34 gene expression was equivalent in all the cell lines and immunoreactive protein was present in both control and scl-null ES cell lines (Fig 4E to H). Both single cells and larger clumps of CD34+ cells were detected, although the percentage of positive cells was lower in the scl-null EBs (1.3% versus 3.1% of 200 to 700 cells counted per cell line per experiment. Two experiments were performed using 1 or 2 control and 2 or 3 scl-null cell lines in each).
DISCUSSION
We have characterized the expression profiles of ES cells bearing wild-type or mutant scl genes allowed to differentiate in vitro for up to 12 days and established a temporal sequence for the induction of genes important in hematopoiesis. Equilibration of the quantity of cDNA amplified, limitation of cycle numbers, and the use of three independent control and scl-null ES cell lines reasonably permits a comparison of the gene expression between both groups of differentiating ES cells.
Induction of hematopoietic genes in control ES cells.In wild-type or scl+/− ES cells, the first 4 to 6 days of in vitro differentiation was associated with the sequential induction of genes that correlate with the embryonic processes of mesoderm induction, its ventral specification, and formation of mesenchymal aggregates of yolk sac hemangioblasts. It was interesting that the BMP-4 type 1A receptor and Smad1 were expressed in undifferentiatied ES cells in the light of evidence that differentiating EBs are not competent to respond to the inductive effects of BMP-4 for the first 3 to 4 days.9 In control ES cells, expression of flk-1 paralleled BMP-4 and transcripts for scl, and GATA-2 also progressively increased in abundance from day 4. It was only after day 6 that a second cohort of hematopoietic transcription factors appeared (GATA-1, EKLF, NF-E2, and PU.1), associated with more committed progenitors, and globin and myeloperoxidase were expressed. This sequence of events is displayed diagrammatically in Fig 5A.
Our results in control EBs are generally in agreement with other studies of hematopoietic gene expression during in vitro differentiation of ES cells. For example, Keller et al49 also showed that Brachyury expression peaked before the induction of GATA-1 and the globin genes. However, unlike our study in which scl expression preceded GATA-1, Keller et al observed that scl and GATA-1 were expressed contemporaneously. Resolution of the sequence of gene induction may have been more difficult in their study because of the rapid transit from mesoderm formation to hematopoiesis (approximately 2 days). These differences in kinetics may be explicable by differences between the ES cell lines (CCE versus W9.5) and/or by differences in reagents or the culture conditions. The time course of flk-1 induction we observed was similar to that reported in a recent study examining endothelial cell differentiation in EBs.50 In their evaluation of cytokine and receptor gene expression during ES cell differentiation, both Schmitt et al44 and McClanahan et al45 showed significant levels of c-kit and the Epo receptor throughout ES cell differentiation, similar to our findings, and the latter authors also showed increasing CD34 levels during EB differentiation from day 7 onwards.
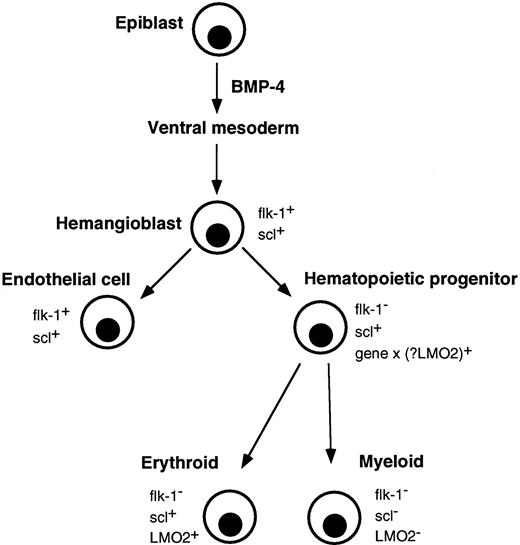
Correlation of major events in hematopoietic development with expression of flk-1, scl, and LMO2. Gastrulating epiblast cells give rise to ventral mesoderm under the influence of BMP-4. Putative hemangioblasts expressing flk-1 and scl differentiate down endothelial and hematopoietic pathways. The molecular event(s) (gene x), which specifies a hematopoietic progenitor and downregulates flk-1, is not known, although LMO2 is a possible candidate. Differentiating erythroid lineage cells continue to express scl and LMO2 but their expression is extinguished in myeloid cells.
Correlation of major events in hematopoietic development with expression of flk-1, scl, and LMO2. Gastrulating epiblast cells give rise to ventral mesoderm under the influence of BMP-4. Putative hemangioblasts expressing flk-1 and scl differentiate down endothelial and hematopoietic pathways. The molecular event(s) (gene x), which specifies a hematopoietic progenitor and downregulates flk-1, is not known, although LMO2 is a possible candidate. Differentiating erythroid lineage cells continue to express scl and LMO2 but their expression is extinguished in myeloid cells.
Gene expression in scl-null embryoid bodies.A major objective of this study was to define the expression patterns of key hematopoietic genes during differentiation of scl-null EBs. As displayed in Fig 5B, the results indicate that whereas the scl-null EBs were competent to express mesoderm-related genes and express genes associated with primitive hemangioblasts and endothelium (for example flk-1, GATA-2, and CD34), they did not undergo differentiation to hematopoietic precursors expressing transcription factors such as GATA-1, EKLF, or PU.1. Consistent with this, markers of more differentiated hematopoietic cells such as globin genes or myeloperoxidase were also absent.
For both c-myb and vav genes, early higher levels of expression could correlate with known nonhematopoietic sites of expression.48 51 The marked differences in levels between control and scl-null EBs from day 8 onwards probably reflect proliferation of hematopoietic cells restricted to control EBs.
LMO2 was induced equally in all ES cell lines with kinetics similar to flk-1 and GATA-2 (Table 2 and data not shown). LMO2 is expressed in nonhematopoietic tissues, such as the brain and kidney,52-54 but the cell type(s) responsible for its early embryonic expression is not known. Also, the leucine zipper transcription factor NF-E255 appeared to be present at normal levels in scl-null ES cells at day 12 of differentiation, arguing for an as yet unidentified nonhematopoietic site of transcription.
Expression of early endothelial genes in scl-null embryoid bodies. flk-1 expression is first detected in the yolk sac at approximately E7 in prospective blood islands, but by E8.5, expression is confined to the endothelium surrounding the hematopoietic cells.2 24 This suggests that hemangioblasts express flk-1 before lineage commitment and that flk-1 expression is retained by the endothelial arm and downregulated in hematopoietic cells. Therefore, our observation that scl-null EBs expressed flk-1 at normal levels argues for the preservation of primitive hemangioblasts in the absence of scl. Experiments to confirm this hypothesis are in progress.
Although scl is also detected in the yolk sac mesoderm, it differs from flk-1 in that expression continues in both endothelial and hematopoietic components of the blood island.27 Thus, it is likely, although not formally proven, that flk-1 and scl are coexpressed in the same cells. Whereas deletion of either gene completely abrogates in vivo hematopoiesis, it is possible that the key role of flk-1 is to provide an endothelially lined microenvironment to foster scl-dependent hematopoiesis. Indeed, the derivation of a small number of hematopoietic colonies from flk-1–null yolk sacs suggests that this receptor may not be absolutely required for hematopoietic commitment.2
Several other genes shared by primitive hematopoietic cells and vascular endothelium, such as GATA-2,56,57 CD34,41 and c-kit,42 were expressed in differentiating scl-null ES cells. This is not entirely surprising because endothelium appears normal in scl-null embryos.29,30 However, as GATA-2 and c-kit transcripts were detected early in differentiation, they could conceivably also mark the common hemangioblast that may be formed in scl-null embryos. As noted for scl, GATA-2, CD34, and c-kit are also dispensable for endothelial development,40,42 58 although gene ablation has hematopoietic consequences of variable severity.
Comparison of the differentiation of scl-null ES cells with other mutant ES cell lines.The in vitro differentiation of doubly targeted ES cells has been studied for a number of hematopoietic regulatory genes. By morphological criteria, LMO2-null ES cells retained macrophage colony forming ability but molecular markers were not used to confirm lineage assignment.33 Because GATA-2–null EBs gave rise to hematopoietic colonies, commitment to hematopoiesis occurred, and other transcription factors were able to partially compensate for GATA-2 deficiency.40 Although GATA-1–null ES cell-derived erythroblasts were blocked in differentiation at the proerythroblast stage, there was little perturbation of gene expression,59 and ES cells in which both copies of GATA-3 were deleted displayed a selective defect in the earliest stages of T-cell differentiation.60 EBs derived from GATA-4–null ES cells did not form visceral yolk sac endoderm61 resulting in disorganization and reduction of vascular and hematopoietic elements.62 However, normal flk-1 and globin gene expression showed that hematopoiesis does not absolutely depend on endodermal signals.
A number of genes specifically affect definitive hematopoiesis. These include c-myb and members of the heterodimeric core binding factor (CBF ) complex, CBFα2 (AML1) and CBFβ. Mice deficient in c-myb had a selective defect in fetal liver hematopoiesis, but analysis of doubly targeted ES cells has not been published.37 Animals homozygous for mutations in either CBFα2 (AML1)63,64 or CBFβ65,66 died between E11.5 and E13.5 with central nervous system hemorrhage and failure of fetal liver hematopoiesis. Vasculogenesis and primitive hematopoiesis were intact in the mutant embryos and CBFα2-null ES cells formed hemoglobinized EBs containing primitive nucleated erythrocytes. Disruption of the PU.1 gene, an ets-family transcription factor, showed a multilineage defect involving monocytes, granulocytes, and lymphocytes, but primitive erythropoiesis was not affected.38,39 Consistent with this late defect, PU.1-null EB cells retained the capacity to express myeloperoxidase and β globin after in vitro differentiation.67 68
Additional molecular events in hematopoietic commitment.The earliest ventral mesoderm genes identified in yolk sac hemangioblasts are flk-1 and scl. Because both genes continue to be expressed in endothelium, as are GATA-2, CD34, and c-kit, it is clear that specification to a hematopoietic phenotype requires additional genetic event(s). Although it is likely that LMO2, which complexes with scl and is induced with similar kinetics, is important, its role remains speculative because its expression pattern in the developing yolk sac or endothelium has not been reported. It may be that as yet unknown gene products are essential, acting either to downregulate flk-1, induce LMO2, or to directly complement genes like scl, GATA-2, CD34, and c-kit (Fig 6). The elucidation of these genetic events remains a major challenge in developmental hematopoiesis.
ACKNOWLEDGMENT
We thank Professor D. Metcalf and Professor T. Graf for helpful discussions and for critically reviewing the manuscript.
Supported in part by the AntiCancer Council of Victoria; the National Health and Medical Research Council, Canberra; and the Rotary Bone Marrow Research Laboratories, the Royal Melbourne Hospital. A.G.E. is a recipient of a Neil Hamilton Fairley Fellowship of the NH&MRC.
Address reprint requests to Andrew G. Elefanty, FRACP, PhD, The Walter and Eliza Hall Institute of Medical Research, PO Royal Melbourne Hospital, Parkville, 3050 Victoria, Australia.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal