Abstract
Using fluorescence in situ hybridization (FISH) and probes located on 12p12.1 to 13.3, we studied the breakpoints of 23 patients who had various hematologic malignant diseases and who had 12p13-balanced translocations (21 patients), inversion (1 patient), or insertion (1 patient). Among them, 14 patients had breakpoints within YAC964c10, which contains the TEL (ETV6 ) gene and in 12 of these with balanced translocations or insertion, the FISH results suggested that TEL was involved. Two of the 14 patients, patients no. 13 and 14, had breakpoints in YAC 964C10 that were centromeric to TEL but telomeric to KIP1. In the other 9 patients whose breakpoints did not fall within the YAC, the breakpoints were found telomeric to the YAC in at least three different locations on distal 12p. These results indicated that TEL was involved in only half (12 of 23) of the patients with balanced 12p13 rearrangements and that there probably were several other breakpoint cluster regions on 12p13, suggesting that genes other than TEL were involved in these rearrangements.
REARRANGEMENTS of the short arm of chromosome 12 (p11 to p13) are one of the most common recurring abnormalities found in a broad spectrum of hematologic malignancies. They include balanced or unbalanced translocations, insertions, inversions, and deletions, which most frequently involve band 12p13. We have previously reported that the breakpoints of most balanced 12p13 translocations were clustered in the 1.39 Mb YAC964c10 and that this YAC borders the smallest commonly deleted region of most 12p13 deletions,1 suggesting that the gene that is involved in 12p13 rearrangements (including the deletions) should be contained in this YAC. At the time of our initial studies we did not know that YAC 964C10 contained an internal deletion. The exact size of the deletion is unknown but it includes the CDKN1B (KIP1) locus.2 Thus, the smallest region of deletion on 12p13, as previously defined by Sato et al,3 is flanked by sequences from this YAC.
Golub et al4 cloned the TEL (Translocation ETS Leukemia) (ETV6 ) gene at the 12p13 breakpoint of a patient with t(5; 12)(q33; p13) and chronic myelomonocytic leukemia (CMML). This resulted in a fusion of TEL located at 12p13.1 with platelet-derived growth factor receptor β (PDGFRB ) at 5q33. TEL is a member of the ETS family of transcription factors and has an ETS DNA-binding domain near its carboxyterminus and a helix-loop-helix domain near the aminoterminus. We showed that TEL is contained in YAC 964c10.1 We and others have shown that TEL is involved in other balanced 12p13 translocations that result in various chimeric genes; TEL-STL in t(6; 12)(q23; p13),5,TEL-ABL in t(9; 12),6,7,TEL-AML1 in t(12; 21),8,9 and TEL-MN1 in t(12; 22).10 Using fluorescence in situ hybridization (FISH) to analyze five patients with a t(3; 12)(q26; p13), Raynaud et al11 showed that TEL and the MDS1/EVI1 genes were likely involved in three of them. More recently, Wlodarska et al12 reported that TEL is involved in three additional patients with t(5; 12)(q33; p13) and CMML and in one patient with t(10; 12)(q24; p13) and progressive myelodysplastic syndrome (MDS). Höglund et al13 also showed TEL rearrangement on Southern blot analysis in one MDS patient with a 5q33 break and in one acute myeloid leukemia (AML) patient with a 22q12 break.
In a series of recent papers, the TEL-AML1 fusion has been found in about 25% of white children14-16 and about 12% of Japanese children with early B-cell acute lymphoblastic leukemia (ALL)17 but only in about 2% of adult B-cell ALL patients.18
The fact that some 12p13 rearrangements do not involve TEL has been reported on by others.11,13 The breakpoint in a different 5; 12 translocation has been mapped centromeric to TEL in 12p12 using FISH.19 It should be noted that the location of probes may vary by one band depending on whether the mapping is done with Giemsa (G-) or reverse (R-) banding techniques. Thus, it is unclear at present (1) how frequently TEL is involved in balanced 12p13 rearrangements and (2) whether there are other breakpoint cluster regions in 12p13 that would suggest that other genes may be involved in these rearrangements. To answer these questions, we analyzed the location of 12p13 breakpoints of 23 patients who had balanced 12p13 rearrangements using FISH probes located between 12p12.1 and 12p13.3.
MATERIALS AND METHODS
Patients. Patients with hematologic malignant diseases and balanced rearrangements involving band 12p13 studied at the University of Chicago were selected for the present report. Patient samples were obtained with informed consent. Cytogenetic material from 22 patients and one cell line that had balanced translocations (21), insertion, or inversion (one each) were available for FISH analysis. Specimens from 14 patients were obtained before treatment. Clinical and cytogenetic data on these patients are summarized in Table 1. Some of the data of 11 patients and the cell line (patients no. 1-10, 21, and 22) were previously reported by Kobayashi et al,1 and only the new information is presented here. Molecular analysis of patient no. 12 has been reported on by Golub et al,7 and we have described the cytogenetic analysis of patient no. 23 in more detail elsewhere.20
Clinical and Cytogenetic Data of the 23 Cases With Balanced Translocations, Insertions, or Inversions Involving Band 12p13
| . | Age/Sex . | Diagnosis . | Stage . | Source . | Karyotype . |
|---|---|---|---|---|---|
| 1* | 65M | AML | RL | BM | 46,XY,t(5; 12)(q13; p13)[25]/46,XY[5] |
| 2* | 5M | ALL, SUPB2 | DX | cell line | 44,XY,del(2)(q13q37),t(3; 7)(q25; p15),der(4)t(2; 4)(q13; q25),inv(5)(q13q33),t(6; 12)(q23; p13), t(11; 17)(p11.2; p11.2),−14,del(17)(p11p13),dic(21; 21)(q21; q21) |
| 3* | 53M | MDS | CP | BCo | 47,XY,+8,t(8; 12)(q1?1; p13)[16]/unrelated clones[5]/46,XY[3] |
| 4* | 27F | t-AML | DX | BM | 46,XX,t(11; 12; 17)(q24; p13; p13)[29]/46,XX[2] |
| 5 | 3F | ALL-L1 | DX | BM | 46,XX,t(1; 12; 21)(p22; p13; q22.1)[8]/related clones[2]/46,XX[6] |
| 6* | 62M | AML-M1 or M2 | RL | BM | 46,XY,t(12; 22)(p13.3; q13.1),t(9; 13)(q32; q12)[10]/related clone[6]/unrelated clones[4]/46,XY[3] |
| 7* | 24M | CML | BC | LN | 46,XY,t(9; 22)(q34; q11),ins(19; 12)(q13; p13p11)[26] |
| 8* | 62M | ALL | DX | BM | 46,XY,t(6; 1; 12)(p2?5; q2?3; p13),t(4; 11)(q21; q23)[5]/46,XY[9] |
| 9* | 30M | NHL-LB | DX | BM | 46,XY,t(2; 12)(q31; p13),del(11)(q21)[9] |
| 10 | 53F | AML-M6 | DX | BCo | original karyotype: 44,XX,del(5)(q22q35),−7,add(9)(p2?2),del(12)(p11p13),−17,−18, der(19)t(7; 19)(p12; p13),+r[7]/45,idem,+mar1[4]/46,idem,+mar1,+mar2[3]/46,XX[6] |
| revised karyotype: 44,XX,del(5)(q22q35),−7,t(12; 9; mar1)(p13; p2?2; q?),−17,−18, der(19)t(7; 19)(p12; p13),+r[7]/45,idem,+mar2[4]/46,idem,+mar2,+mar3[3]/46,XX[6] | |||||
| 11 | 69M | MDS | RL | BM | original karyotype: 46,XY,t(3; 5)(q25; q34)[16]/46,idem,del(12)(p12p13),add(16)(q24)[4] |
| revised karyotype: 46,XY,t(3; 5)(q25; q34)[16]/46,idem,t(12; 16)(p13; q24)[4] | |||||
| 12* | 81M | AUL | DX | BM | 45,add(X)(p11),Y,−3,der(3)t(3; 15)(q12; q15),t(4; 7)(q21; q22),−5,t(9; 12; 14)(q34; p13; q22), del(12)(p11p13),der(15)del(15)(q11q15)t(3; 15)(q12; q15),der(18)t(3; 18)(q11; q1?2),+mar |
| 13 | 41F | AML-M4 | RL | BM | 46,XX,t(5; 12)(q33; p13),t(11; 19)(q23; p13.1)[7]/46,XX[14] |
| 14* | 12F | ALL-L1 | DX | BM | 55,XX,+X,+X,+X,+4,t(5; 12)(q13; p13),+6,+14,+17,+21,+21[18]/related clones[13] |
| 15 | 38M | CML | CP | BM | 46,XY,t(9; 22),(q34;q11),t(12; 18)(p13; q12)[10]/46,XY,t(9; 22),add(10)(p15)[9] |
| 16 | 55M | CML? | DX | BM | 46,XY,t(8; 12)(?q11; p13),t(9; 22)(q34; q11)[6]/46,XY,t(9; 22)[13] |
| 17 | 8F | ALL | RL | BM | 45,X,−X,t(1; 12)(p22; p13),del(6)(q1?3q2?1)[4]/46,XX[16] |
| 18 | 76F | NHL(B) | DX | LN | 49,XX,+7,+7,+12,−13,−22,del(1)(p31.3p34),del(4)(p14p15),del(6)(q23q27),del(15)(q13q15), t(3; 12)(p21; p13),t(14; 18)(q32; q21),+2der(18)t(14; 18)[10]/related clones[8] |
| 19 | 60F | ALL | DX | BM | 46,XX,t(5; 8; 14)(q11; q24; q32),t(12; 22)(p13; q11),(18)(q23)[21] |
| 20 | 74M | NHL | DX | LN | 49,XY,+Y,t(2; 12)(p11; p13),+12,add(14)(q32),t(1; 14)(q21; q32),+18[8]/ unrelated clones[2]/46,XY[8] |
| 21* | 21F | ALL-L3 | DX | BM | 47,XX,del(12)(p11p13),+del(12)(p11p13),t(12; 22)(p13; q13)[17]/46,XX[7] |
| 22* | 0.5F | AML | DX | BM | 46,XX,t(1; 12)(p32; p13.1),del(6)(q15q23),t(17; 20)(q23; p11.2)[7]/related clones[4]/46,XX[9] |
| 23 | 49F | AML-M6 | DX | BM | 46,XX,add(5)(q1?3),inv(12)(p13.1q11),−15,−17,del(20)(q11q13.1orq13.1q13.3),+der(?) t(?; 15)(?; q14),+mar1[16]/46,idem,del(7)(p11p22)[2]/92,XXXX, add(5)×2,inv(12)×2, −15,−15,−17,−17,del(20)×2,+der(?)×2,+mar1×2[6]/46,XX[4] |
| . | Age/Sex . | Diagnosis . | Stage . | Source . | Karyotype . |
|---|---|---|---|---|---|
| 1* | 65M | AML | RL | BM | 46,XY,t(5; 12)(q13; p13)[25]/46,XY[5] |
| 2* | 5M | ALL, SUPB2 | DX | cell line | 44,XY,del(2)(q13q37),t(3; 7)(q25; p15),der(4)t(2; 4)(q13; q25),inv(5)(q13q33),t(6; 12)(q23; p13), t(11; 17)(p11.2; p11.2),−14,del(17)(p11p13),dic(21; 21)(q21; q21) |
| 3* | 53M | MDS | CP | BCo | 47,XY,+8,t(8; 12)(q1?1; p13)[16]/unrelated clones[5]/46,XY[3] |
| 4* | 27F | t-AML | DX | BM | 46,XX,t(11; 12; 17)(q24; p13; p13)[29]/46,XX[2] |
| 5 | 3F | ALL-L1 | DX | BM | 46,XX,t(1; 12; 21)(p22; p13; q22.1)[8]/related clones[2]/46,XX[6] |
| 6* | 62M | AML-M1 or M2 | RL | BM | 46,XY,t(12; 22)(p13.3; q13.1),t(9; 13)(q32; q12)[10]/related clone[6]/unrelated clones[4]/46,XY[3] |
| 7* | 24M | CML | BC | LN | 46,XY,t(9; 22)(q34; q11),ins(19; 12)(q13; p13p11)[26] |
| 8* | 62M | ALL | DX | BM | 46,XY,t(6; 1; 12)(p2?5; q2?3; p13),t(4; 11)(q21; q23)[5]/46,XY[9] |
| 9* | 30M | NHL-LB | DX | BM | 46,XY,t(2; 12)(q31; p13),del(11)(q21)[9] |
| 10 | 53F | AML-M6 | DX | BCo | original karyotype: 44,XX,del(5)(q22q35),−7,add(9)(p2?2),del(12)(p11p13),−17,−18, der(19)t(7; 19)(p12; p13),+r[7]/45,idem,+mar1[4]/46,idem,+mar1,+mar2[3]/46,XX[6] |
| revised karyotype: 44,XX,del(5)(q22q35),−7,t(12; 9; mar1)(p13; p2?2; q?),−17,−18, der(19)t(7; 19)(p12; p13),+r[7]/45,idem,+mar2[4]/46,idem,+mar2,+mar3[3]/46,XX[6] | |||||
| 11 | 69M | MDS | RL | BM | original karyotype: 46,XY,t(3; 5)(q25; q34)[16]/46,idem,del(12)(p12p13),add(16)(q24)[4] |
| revised karyotype: 46,XY,t(3; 5)(q25; q34)[16]/46,idem,t(12; 16)(p13; q24)[4] | |||||
| 12* | 81M | AUL | DX | BM | 45,add(X)(p11),Y,−3,der(3)t(3; 15)(q12; q15),t(4; 7)(q21; q22),−5,t(9; 12; 14)(q34; p13; q22), del(12)(p11p13),der(15)del(15)(q11q15)t(3; 15)(q12; q15),der(18)t(3; 18)(q11; q1?2),+mar |
| 13 | 41F | AML-M4 | RL | BM | 46,XX,t(5; 12)(q33; p13),t(11; 19)(q23; p13.1)[7]/46,XX[14] |
| 14* | 12F | ALL-L1 | DX | BM | 55,XX,+X,+X,+X,+4,t(5; 12)(q13; p13),+6,+14,+17,+21,+21[18]/related clones[13] |
| 15 | 38M | CML | CP | BM | 46,XY,t(9; 22),(q34;q11),t(12; 18)(p13; q12)[10]/46,XY,t(9; 22),add(10)(p15)[9] |
| 16 | 55M | CML? | DX | BM | 46,XY,t(8; 12)(?q11; p13),t(9; 22)(q34; q11)[6]/46,XY,t(9; 22)[13] |
| 17 | 8F | ALL | RL | BM | 45,X,−X,t(1; 12)(p22; p13),del(6)(q1?3q2?1)[4]/46,XX[16] |
| 18 | 76F | NHL(B) | DX | LN | 49,XX,+7,+7,+12,−13,−22,del(1)(p31.3p34),del(4)(p14p15),del(6)(q23q27),del(15)(q13q15), t(3; 12)(p21; p13),t(14; 18)(q32; q21),+2der(18)t(14; 18)[10]/related clones[8] |
| 19 | 60F | ALL | DX | BM | 46,XX,t(5; 8; 14)(q11; q24; q32),t(12; 22)(p13; q11),(18)(q23)[21] |
| 20 | 74M | NHL | DX | LN | 49,XY,+Y,t(2; 12)(p11; p13),+12,add(14)(q32),t(1; 14)(q21; q32),+18[8]/ unrelated clones[2]/46,XY[8] |
| 21* | 21F | ALL-L3 | DX | BM | 47,XX,del(12)(p11p13),+del(12)(p11p13),t(12; 22)(p13; q13)[17]/46,XX[7] |
| 22* | 0.5F | AML | DX | BM | 46,XX,t(1; 12)(p32; p13.1),del(6)(q15q23),t(17; 20)(q23; p11.2)[7]/related clones[4]/46,XX[9] |
| 23 | 49F | AML-M6 | DX | BM | 46,XX,add(5)(q1?3),inv(12)(p13.1q11),−15,−17,del(20)(q11q13.1orq13.1q13.3),+der(?) t(?; 15)(?; q14),+mar1[16]/46,idem,del(7)(p11p22)[2]/92,XXXX, add(5)×2,inv(12)×2, −15,−15,−17,−17,del(20)×2,+der(?)×2,+mar1×2[6]/46,XX[4] |
Abbreviations: AML, acute myeloid leukemia; RL, relapse; BM, bone marrow; ALL, acute lymphoblastic leukemia; Dx, diagnosis; MDS, myelodysplastic syndrome; CP, chronic phase; BC, blast crisis; t-AML, treatment related AML; CML, chronic myelogenous leukemia; LN, lymph node; NHL, non-Hodgkin lymphoma; AUL, acute undifferentiated leukemia; BCo, bone core.
Cytogenetic analysis. Metaphase cells were prepared from bone marrow cells as previously described.1,21 The karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN 1995).22 Standard cytogenetic analysis suggested that each patient had a balanced rearrangement involving 12p13.
FISH probes. We used 13 cosmid probes (D12S235, D12S237, D12S134, D12S229, HTY3049c1-7, D12S133, 15a4 (TEL exon 1), 95h4 (TEL exon 2), D12S142, D12S119, D12S54, D12S140, and D12S20), two phage probes (D12S54 and GDI.D4), and a plasmid probe (D12S20) that were mapped to 12p12.1 to 13.3 and were ordered as previously described.23 We also used YAC964c10 that contained the TEL gene,1 a cosmid contig that contained about 60 kb of genomic sequences surrounding exon 3 of TEL (TEL ex 3),5 a P1 phage clone that contained a 90-kb insert from the 3′ part of TEL including exons 3 to 8 (P1. TEL), and two P1 phage clones of 90 kb that contained the CDKNIB gene (p27KIP1)24 as well as two more telomeric YACs, 961a6 and 771h4. The order of these probes was telomere - D12S235 - D12S237 - D12S134 - YAC961a6 - YAC771h4 - D12S229 - HTY3049c1 - 7 - D12S133- YAC964 - c10 (15a4; 95h4 ; TEL ex 3 and P1 TEL are contained in this YAC) - p27 KIP1 - D12S142 - D12S119 - GDI. D4 - D12S54 - D12S140 - D12S20 - centromere (Fig 1).1,2 23
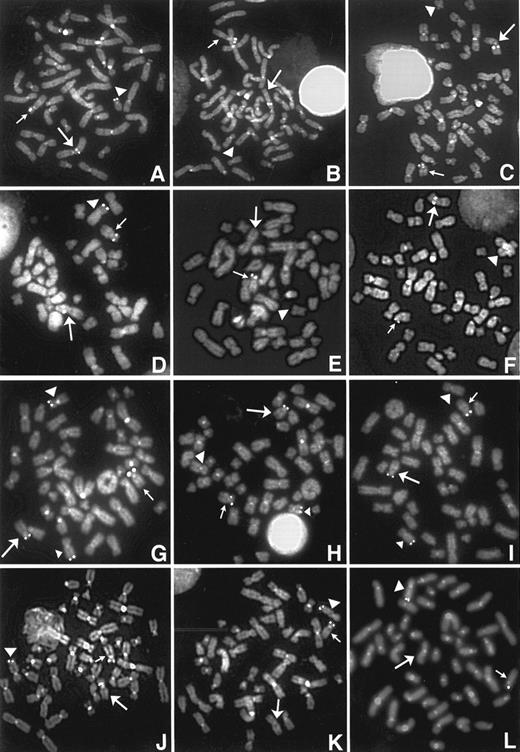
Representative photograph of FISH signals for four patients (patients no. 3, 8, 10, and 13) in whom the FISH signals of the YAC 964c10 were observed to be split between the der(12) and the partner derivative chromosome. The left column (A, D, G, and J) shows the FISH signals of the YAC964c10, the middle column (B, E, H, and K) shows the signals of TEL exon 3 probe, and the right column (C, F, I, and L) shows the signals of P1 TEL probe in each patient. The small arrow indicates the normal chromosome 12, the large arrow indicates the derivative chromosome 12, and the arrowhead identifies the partner derivative chromosome. (A) FISH signals of the YAC964c10 were observed on the der(12), the der(8), and normal chromosome 12 in patient no. 3. (B and C) Signals of the TEL exon 3 and P1 TEL probes were observed only on the der(12) and normal chromosome 12. In patient no. 8, (D) FISH signals of the YAC964c10 were observed on the der(12), the der(6), and normal chromosome 12. (E) The TEL exon 3 probe was deleted from the der(12) and der(6). (F ) The signal of the P1 TEL was observed on the der(12) and normal chromosome 12. In patient no. 10, (G) FISH signals of the YAC964c10 were observed on the der(12), the der(9) (large arrowhead), mar1 (small arrowhead), and normal chromosome 12. (H and I) The signals of the TEL exon 3 and P1 TEL probes were observed only on the der(12), the mar1, and normal chromosome 12. In patient no. 13, (J) FISH signals of the YAC964c10 were observed on the der(12), the der(5), and normal chromosome 12. (K and L) Signals of the TEL exon 3 and P1 TEL probes were observed only on the der(5) and normal chromosome 12.
Representative photograph of FISH signals for four patients (patients no. 3, 8, 10, and 13) in whom the FISH signals of the YAC 964c10 were observed to be split between the der(12) and the partner derivative chromosome. The left column (A, D, G, and J) shows the FISH signals of the YAC964c10, the middle column (B, E, H, and K) shows the signals of TEL exon 3 probe, and the right column (C, F, I, and L) shows the signals of P1 TEL probe in each patient. The small arrow indicates the normal chromosome 12, the large arrow indicates the derivative chromosome 12, and the arrowhead identifies the partner derivative chromosome. (A) FISH signals of the YAC964c10 were observed on the der(12), the der(8), and normal chromosome 12 in patient no. 3. (B and C) Signals of the TEL exon 3 and P1 TEL probes were observed only on the der(12) and normal chromosome 12. In patient no. 8, (D) FISH signals of the YAC964c10 were observed on the der(12), the der(6), and normal chromosome 12. (E) The TEL exon 3 probe was deleted from the der(12) and der(6). (F ) The signal of the P1 TEL was observed on the der(12) and normal chromosome 12. In patient no. 10, (G) FISH signals of the YAC964c10 were observed on the der(12), the der(9) (large arrowhead), mar1 (small arrowhead), and normal chromosome 12. (H and I) The signals of the TEL exon 3 and P1 TEL probes were observed only on the der(12), the mar1, and normal chromosome 12. In patient no. 13, (J) FISH signals of the YAC964c10 were observed on the der(12), the der(5), and normal chromosome 12. (K and L) Signals of the TEL exon 3 and P1 TEL probes were observed only on the der(5) and normal chromosome 12.
FISH. The probes were labeled with biotin-11-dUTP or digoxigenin-11-dUTP using nick translation or polymerase chain reaction (PCR) labeling after sequence-independent amplification and were hybridized to the patient slides as previously described.25 26 Chromosomes were identified using counterstaining with 4′ 6-diamidino-2-phenylindole dihydrochloride (DAPI). The presence or absence of the FISH signals was scored on an average of 11 abnormal metaphase cells (range, 2-18) per probe per patient by two persons blinded to the identity of probes and patients. Images of the hybridized cells were captured with a liquid-cooled, charge-coupled device camera (Photometrics, Tucson, AZ). Separate gray-scale images for the DAPI and the fluorescein isothiocyanate fluorescence were acquired. After adjusting the gray levels with the National Institutes of Health (NIH; Bethesda, MD) image 1.52 software, the images were merged using Adobe Photoshop (Adobe Systems Inc, San Jose, CA) on a Macintosh computer (Apple Computers, Cupertino, CA).
RESULTS
FISH analysis of the patient material. The FISH results using the 12p probes on the 23 patients with balanced 12p13 rearrangements are summarized in Tables 2 and 3. In 14 patients (patients no. 1-14; Table 2), the FISH signals of YAC964c10 were observed on the derivative partner chromosome and the normal 12p [der(12)] (Fig 1A, D, G, and J), indicating that the breakpoint on the der(12) was located within the YAC probe. In patients no. 1 to 6, the FISH signals of the more telomeric probes, D12S133 through D12S235, were found exclusively on the partner derivative chromosomes, and the signals of the more centromeric probes, CDKN1B through D12S20 were found exclusively on the der(12) (Fig 1B and C and Table 2). In patient no. 7, we found the opposite pattern, namely, the signals of the more telomeric probes were on the der(12) and the signals of the more centromeric probes were on the derivative partner chromosome, the der(19), indicating that this region of 12p was inserted into 19p (Table 2). In these 7 patients, the breakpoint in the der(12) should occur telomeric (5′) to the TEL exon 3 probe, because it and P1 TEL probes hybridize to the der(12), whereas the YAC hybridizes to both the der(12) and the other derivative chromosomes (in patient no. 7, again, the reverse was found). In patients no. 2 and 12, the precise position of the breakpoint was determined: In patient no. 2, we showed that the TEL gene was split at nucleotide (nt) 187/188 and fused with a new gene, STL (six-twelve leukemia) on the der(6) using the cell line material.5 In patient no. 12 the breakpoint was determined to be between exons 5 and 6 (nt 1033/1034) of TEL and the fusion partner is ABL at exon 2.7 In patient no. 6, TEL could be split at nt 187/188 and fused to MN1 on the der(22), as has been reported for patients with a t(12; 22)10; however, the breakpoint in our patient was identified as 22q13 not 22q11. In patients no. 8 and 9, the signal of the TEL exon 3 probe was found only on the normal 12p and not on the der(12) or the partner derivative chromosome (Fig 1E), whereas the P1 TEL probe hybridized only to the der(12) and the normal 12p (Fig 1F ). This suggests that there has been an unbalanced rearrangement with deletion of TEL exon 3.
Results of FISH Analysis Using 12p12 to 13 Probes in 14 Patients With Balanced 12p13 Rearrangements in Whom the YAC Signals Were Split Between the der(12) and the Partner Derivative Chromosomes
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | t(5; 12) . | t(6; 12) . | t(8; 12) . | t(11; 12; 17) . | t(1; 12; 21) . | t(12; 22) . | ins(19; 12) . | t(6; 1; 12) . | t(2; 12) . | t(12; 9; mar1) . | t(12; 16) . | t(9; 12; 14) . | t(5; 12) . | t(5; 12) . |
| . | (q13; p13) . | (q23; p13) . | (q11; p13) . | (q24; p13; p13) . | (p22; p13; q22.1) . | (p13.3; q13.1) . | (q13; p11p13) . | (p2?5; q2?3,p13) . | (q31; p13) . | (p13; p2?2; q?) . | (q13; q24) . | (q34; p13; q22) . | (q33; p13) . | (q13; p13) . |
| D12S235 | der(17) | der(21) | der(22) | der(6) | der(14) | |||||||||
| D12S237 | der(22) | der(14) | ||||||||||||
| D12S134 | der(5) | der(6) | der(8) | der(17) | der(12) | der(6) | der(2) | der(14) | der(5) | der(5) | ||||
| D12S229 | der(5) | der(17) | der(22) | der(6) | der(2) | der(14) | ||||||||
| HTY3049 c1-7 | der(8) | der(17) | der(5) | |||||||||||
| D12S133 | der(5) | der(6) | der(8) | der(17) | der(21) | der(22) | der(12) | der(6) | der(2) | der(16) | der(14) | der(5) | der(5) | |
| YAC964c10 | der(5) | |||||||||||||
| → | der(6) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(8) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(17) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(21) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(22) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(12) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(6) | |||||||||||||
| der(19) | ||||||||||||||
| → | der(2) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(9)/mar1 | |||||||||||||
| der(12) | ||||||||||||||
| → | der(16) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(14) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(5) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(5) | |||||||||||||
| der(12) | ||||||||||||||
| → | ||||||||||||||
| der(12) | ||||||||||||||
| 15a4 (TEL exon 1) | mar1 | der(16) | U | N | U | |||||||||
| 95h4 (TEL exon 2) | mar1 | der(16) | U | N | U | |||||||||
| TEL exon 3 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | deleted | deleted | mar1 | ||||
| → | der(16) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(12) | der(5) | deleted | |||||||||||
| der(12) | ||||||||||||||
| P1 TEL exons 3-8 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | mar1 | ||||
| → | der(14) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(5) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(5) → | |||||||||||||
| p27KiP1 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | deleted | der(12) | der(12) | der(12) | der(12) |
| D12S142 | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | der(12) | der(12) | der(12) | |||
| D12S119 | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | der(12) | der(12) | ||||
| GDI.D4 | der(12) | der(12) | ||||||||||||
| D12S54 | der(12) | der(12) | ||||||||||||
| D12S140 | der(12) | |||||||||||||
| D12S20 | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | der(12) | der(12) |
| . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | t(5; 12) . | t(6; 12) . | t(8; 12) . | t(11; 12; 17) . | t(1; 12; 21) . | t(12; 22) . | ins(19; 12) . | t(6; 1; 12) . | t(2; 12) . | t(12; 9; mar1) . | t(12; 16) . | t(9; 12; 14) . | t(5; 12) . | t(5; 12) . |
| . | (q13; p13) . | (q23; p13) . | (q11; p13) . | (q24; p13; p13) . | (p22; p13; q22.1) . | (p13.3; q13.1) . | (q13; p11p13) . | (p2?5; q2?3,p13) . | (q31; p13) . | (p13; p2?2; q?) . | (q13; q24) . | (q34; p13; q22) . | (q33; p13) . | (q13; p13) . |
| D12S235 | der(17) | der(21) | der(22) | der(6) | der(14) | |||||||||
| D12S237 | der(22) | der(14) | ||||||||||||
| D12S134 | der(5) | der(6) | der(8) | der(17) | der(12) | der(6) | der(2) | der(14) | der(5) | der(5) | ||||
| D12S229 | der(5) | der(17) | der(22) | der(6) | der(2) | der(14) | ||||||||
| HTY3049 c1-7 | der(8) | der(17) | der(5) | |||||||||||
| D12S133 | der(5) | der(6) | der(8) | der(17) | der(21) | der(22) | der(12) | der(6) | der(2) | der(16) | der(14) | der(5) | der(5) | |
| YAC964c10 | der(5) | |||||||||||||
| → | der(6) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(8) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(17) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(21) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(22) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(12) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(6) | |||||||||||||
| der(19) | ||||||||||||||
| → | der(2) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(9)/mar1 | |||||||||||||
| der(12) | ||||||||||||||
| → | der(16) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(14) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(5) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(5) | |||||||||||||
| der(12) | ||||||||||||||
| → | ||||||||||||||
| der(12) | ||||||||||||||
| 15a4 (TEL exon 1) | mar1 | der(16) | U | N | U | |||||||||
| 95h4 (TEL exon 2) | mar1 | der(16) | U | N | U | |||||||||
| TEL exon 3 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | deleted | deleted | mar1 | ||||
| → | der(16) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(12) | der(5) | deleted | |||||||||||
| der(12) | ||||||||||||||
| P1 TEL exons 3-8 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | mar1 | ||||
| → | der(14) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(5) | |||||||||||||
| der(12) | ||||||||||||||
| → | der(5) → | |||||||||||||
| p27KiP1 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | deleted | der(12) | der(12) | der(12) | der(12) |
| D12S142 | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | der(12) | der(12) | der(12) | |||
| D12S119 | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | der(12) | der(12) | ||||
| GDI.D4 | der(12) | der(12) | ||||||||||||
| D12S54 | der(12) | der(12) | ||||||||||||
| D12S140 | der(12) | |||||||||||||
| D12S20 | der(12) | der(12) | der(12) | der(12) | der(12) | der(19) | der(12) | der(12) | der(12) | der(12) |
Abbreviations: N, all cells analyzed were normal; U, hybridization unsuccessful.
→ Indicates the breakpoint in chromosome 12 identified by the DNA probe.
Results of FISH Analysis Using 12p12 to p13 Probes in Nine Patients With Balanced 12p13 Rearrangements in Whom the YAC Signals Remained on the der(12)
| Probe order . | 15 . | 16 . | 17 . | 18 . | 193-150 . | 203-150 . | 21 . | 22 . | 23 . |
|---|---|---|---|---|---|---|---|---|---|
| . | t(12; 18) . | t(8; 12) . | t(1; 12) . | t(3; 12) . | t(12; 22) . | t(2; 12) . | t(12; 22) . | t(1; 12) . | inv(12) . |
| . | (p13; q12) . | (q1?1; p13) . | (p22; p13) . | (p21; p13),+12 . | (p13; q11) . | (p11; p13),+12 . | (p13; q13) . | (p32; p13) . | (p13.1q11) . |
| tel | → | clone 1 | clone 1 | ||||||
| D12S235 | der(12) | I | der(1) | der(22) | |||||
| → | |||||||||
| D12S237 | der(12) | der(12) | der(3) | der(22) | der(2) | der(22) | |||
| D12S134 | der(12) | der(1) | der(3) | der(22) | der(2) | der(22) | der(1) | 12p | |
| YAC961a6 | U | I | → | → | |||||
| der(12) | der(12) | ||||||||
| YAC771h4 | U → | I → | der(12) | der(12) | |||||
| D12S229 | der(12) | der(12) | der(12) | der(12) | der(22) | der(1) | 12p | ||
| HTY3049 c1-7 | der(12) | der(12) | der(12) | der(12) | der(22) | ||||
| → | der(1) | ||||||||
| → | 12p | ||||||||
| → | |||||||||
| D12S133 | der(12) | der(12) | der(12) | der(12)3-151 | der(12)3-152 | der(12) | der(12) | 12q | |
| YAC964 c10 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | 12q |
| 15a4 | der(12) | der(12) | N | I | der(12) | U | der(12) | der(12) | 12q |
| 95h4 | der(12) | der(12) | N | I | der(12) | U | ∥ → | der(12) | 12q |
| TEL (exon 3) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(22) | ||
| → | der(12) | 12q | |||||||
| der(12)ρ | |||||||||
| p27KIP2 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | 12q |
| D12S142 | der(12) | der(12) | |||||||
| D12S119 | der(12) | der(12) | der(12) | der(12) | 12q | ||||
| GDI.D4 | der(12) | 12q | |||||||
| D12S54 | der(12) | ||||||||
| D12S140 | der(12) | ||||||||
| D12S20 | der(12) | der(12) | der(12) | 12q | |||||
| cen |
| Probe order . | 15 . | 16 . | 17 . | 18 . | 193-150 . | 203-150 . | 21 . | 22 . | 23 . |
|---|---|---|---|---|---|---|---|---|---|
| . | t(12; 18) . | t(8; 12) . | t(1; 12) . | t(3; 12) . | t(12; 22) . | t(2; 12) . | t(12; 22) . | t(1; 12) . | inv(12) . |
| . | (p13; q12) . | (q1?1; p13) . | (p22; p13) . | (p21; p13),+12 . | (p13; q11) . | (p11; p13),+12 . | (p13; q13) . | (p32; p13) . | (p13.1q11) . |
| tel | → | clone 1 | clone 1 | ||||||
| D12S235 | der(12) | I | der(1) | der(22) | |||||
| → | |||||||||
| D12S237 | der(12) | der(12) | der(3) | der(22) | der(2) | der(22) | |||
| D12S134 | der(12) | der(1) | der(3) | der(22) | der(2) | der(22) | der(1) | 12p | |
| YAC961a6 | U | I | → | → | |||||
| der(12) | der(12) | ||||||||
| YAC771h4 | U → | I → | der(12) | der(12) | |||||
| D12S229 | der(12) | der(12) | der(12) | der(12) | der(22) | der(1) | 12p | ||
| HTY3049 c1-7 | der(12) | der(12) | der(12) | der(12) | der(22) | ||||
| → | der(1) | ||||||||
| → | 12p | ||||||||
| → | |||||||||
| D12S133 | der(12) | der(12) | der(12) | der(12)3-151 | der(12)3-152 | der(12) | der(12) | 12q | |
| YAC964 c10 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | 12q |
| 15a4 | der(12) | der(12) | N | I | der(12) | U | der(12) | der(12) | 12q |
| 95h4 | der(12) | der(12) | N | I | der(12) | U | ∥ → | der(12) | 12q |
| TEL (exon 3) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(22) | ||
| → | der(12) | 12q | |||||||
| der(12)ρ | |||||||||
| p27KIP2 | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | der(12) | 12q |
| D12S142 | der(12) | der(12) | |||||||
| D12S119 | der(12) | der(12) | der(12) | der(12) | 12q | ||||
| GDI.D4 | der(12) | 12q | |||||||
| D12S54 | der(12) | ||||||||
| D12S140 | der(12) | ||||||||
| D12S20 | der(12) | der(12) | der(12) | 12q | |||||
| cen |
Abbreviations: I, inadequate sample with no metaphase cells; U, unsuccessful hybridization; N, only normal cells seen.
These patients could have two clones.
Of 12 cells, six had signal on der(12) and six had signal on der(22).
In two of 14 cells, probe labeled der(2).
ρ Signal on der(12) and der(22) in eight of nine cells.
Only one metaphase seen; signal on normal 12 but not on der(12) or der(22).
In patient no. 10, the breakpoint appears to have occurred within the TEL exon 3 probe, because the signals of the TEL exon 3 and P1 TEL probes were split between the der(12) and the derivative partner chromosomes, namely, the short arm of chromosome 9 and an unidentified marker chromosome. To define the breakpoint in TEL more precisely, we used two other probes 15a4 (exon 1) and 95h4 (exon 2) and showed that both of them localized to the marker chromosome; therefore, the breakpoint is likely to be in TEL intron 2, which is 82 kb.23 The unidentified marker chromosome (the size of a C group chromosome, mar 1; Fig 1H) was not chromosome X or 7 based on FISH painting probes. On standard cytogenetic analysis, this patient was thought to have a simple 12p deletion, [del(12)(p11p13),add(9)(p2?2) and mar 1] (Table 1). However, the YAC964c10 probe labeled not only the del(12p) but also the short arm of the add(9) and the end of the long arm of mar 1 (Fig 1G). Moreover, the chromosome 12 painting probe labeled the terminal region of the add(9) and the marker. Therefore, this patient has a three-way rearrangement involving 12p, 9p, and the long arm of the mar. In this patient, the centromeric region of 12p including the p27KIP1 probe was deleted, although we could not determine the extent of this deletion because of lack of material for additional FISH analysis.
In patient no. 11, signals for the YAC and TEL ex 3 were found on both the der(12) and der(16), whereas probes for TEL exons 1 and 2 hybridized to the der(16). These data indicate that the translocation occurred within TEL intron 2 or exon 3. Using a 16q24-qter probe, we could show that 16q24-qter was translocated to 12p. The standard cytogenetic study showed a del(12)(p12p13) and add(16) (q24) (Table 1).
In patient no. 12, the breakpoint was thought to occur telomeric of the TEL ex 3 probe because its signal remained on the der(12), but that of the P1TEL probe was split between the der(12) and the partner derivative chromosome, der(14). The precise position of the breakpoint was shown to be at nt 1033/1034 of TEL between exons 5 and 6, and TEL was fused with ABL.7 To reconcile the FISH findings with the molecular evidence we have to postulate a complicated rearrangement in which a small portion of chromosome 9 band q34 containing part of the ABL gene is inserted into 12p13 replacing the 3′ portion of TEL.
In patients no. 13 and 14, the breakpoint was thought to occur centromeric to the P1 TEL probe and, thus, 3′ of the TEL-coding region because the signals of the TEL exon 3 and P1 TEL probes were found only on the partner derivative chromosomes, although the TEL exon 3 probe was deleted in patient no. 14 (Fig 1K and L). These results indicate that the breaks occurred centromeric (3′) to the TEL-coding region.
In the remaining nine patients (patients no. 15-23), the YAC signals were observed on the der(12) and normal 12p suggesting that the breakpoints in the der(12) occurred telomeric to the YAC. We used additional telomeric probes to define the position of the breakpoint more precisely. The results are shown in Table 3. In patient no. 15, the breakpoint occurred telomeric to D12S235, which was the most telomeric probe we used. In patient no. 16, the breakpoint occurred telomeric to D12S237; our analysis with D12S235 was unsuccessful. In four patients (patients no. 17-20), the breakpoints were found to cluster between D12S134 and D12S229. In an attempt to define the breakpoint in these four patients more precisely we used two YAC probes (961a8 and 771h4); we were successful in only two of them. For patients no. 19 and 20, the breakpoint was between YAC 961a6 and D12S134; it is possible that the break in patients no. 17 and 18 could be in the same interval. Patients no. 19 and 20 had suggestive evidence of two clones detected only on FISH analysis because probe D12S133 appeared on the der(12) in some cells and on the other derivative chromosomes in other cells (see Table 3 for details). In three patients (patients no. 21-23), the breakpoints were located between HTY3049c1-7 and D12S133. In patient no. 21, the TEL exon 3 probe was split between the der(12) and the der(22) in eight of nine cells, raising the possibility that TEL could be involved in this patient. Lack of material for DNA analysis or for additional FISH studies precludes a definitive answer.
DISCUSSION
We have shown that the breakpoints were found within YAC964c10 in 14 of 23 patients and were telomeric to the YAC in the 9 remaining patients. Of the 14 patients who had breakpoints in the YAC, FISH results in 12 were consistent with a break in the TEL gene. In 2 of the 12 patients, the precise position of the breakpoint was determined: at nt 187/188 of the TEL gene in patient no. 25 and at nt 1033/1034 in patient no. 12.7 The patients with t(5; 12)(q33; p13) and CMML reported by Golub et al4 had the breakpoint at nt 487/488 of TEL. At least four breakpoints have been defined for TEL: at nt 187/188 in TEL/MN1 and TEL/STL; at nt 352/353 in TEL/MN1; at nt 487/488 in TEL/PDGFRB and TEL/ABL (one patient); and at nt 1033/1034 in TEL/AML1 (CBFA2 ) and TEL/ABL (one patient). Höglund et al,13 analyzing the t(3; 12), mapped two breakpoints in intron 1 of TEL (3′ of nt 187) and a third in intron 2 (5′ of nt 352). Our 12 patients who had breakpoints in TEL also showed variable breakpoints based on our FISH results. In patients no. 1 to 7 the YAC was split by the translocation, whereas the third exon beginning at nt 188 of TEL and the 3′ portion of TEL remained on the der(12) chromosomes or der(19) in the case of patient no. 7. This hybridization pattern could be best explained by assuming a TEL/MN1 or TEL/STL type break in intron 2, 5′ of nt 187/188.10 It is possible that TEL is not affected in some of these patients and that a break occurs 5′ or telomeric to TEL. Patients no. 8 and 9 could have the same breakpoint in TEL as the first seven patients with the added complication that the reciprocal fusion gene (other/TEL ) either would not exist or would have a partial deletion of TEL, as has been described in other translocations. Patients no. 10 and 11 also had a breakpoint in TEL intron 2, which was confirmed because TEL exons 1 and 2 were translocated to the partner chromosomes.
In addition to the five translocation partners (not including MDS/EVI1 ) previously reported for TEL, we have identified eight new partner bands (2q31, 5q13, 6p25, 8q11, 9p22, 16q24, 17p13, and 19q13) and, thus potentially, the location of eight new partner genes. Moreover, the phenotype of the hematologic malignancy in these cases is quite diverse including ALL in three patients; AML in five; and MDS, chronic myelogenous leukemia, acute undifferentiated leukemia, and non-Hodgkin's lymphoma-lymphoblastic in one patient each. Patient no. 13 apparently had the same translocation, t(5; 12)(q33; p13), as the patients reported previously,4,12 although the diagnosis was not CMML, but AML-M4; the lack of DNA precluded confirmation that PDGFRB was involved. However, the breakpoint position in TEL was obviously different because it was centromeric to TEL, suggesting that a different gene may be involved. In addition, in patient no. 14, there was also a small interstitial deletion. Raynaud et al11 also reported that one of their t(3; 12) patients (case 4) had a breakpoint 3′ of TEL exon 8.11
In the remaining nine patients (Table 3), two possible breakpoint cluster regions were found telomeric to TEL, one between D12S134 and D12S229 (a distance of about 6 megabases) in four patients and the other between HTY3049c1-7 and D12S133 in three patients. With the use of additional probes between D12S134 and D12S229 we could narrow the breakpoint region in two patients to between D12S134 and YAC 961a6. These four patients all had lymphoid neoplasms (patients 17 through 20). These regions likely contain as yet unknown oncogenes that play a role in leukemogenesis of 12p13 rearrangements and that could contribute to the wide variety of phenotypes resulting from 12p rearrangements.
FISH analysis clarified the 12p rearrangement in a number of patients. A small deleted region was found in three patients (patients no. 8, 9, and 14) in the area of the third TEL exon and in one patient (patient no. 10) in the area of the p27KIP1 probe. A 12p13-balanced translocation rather than a simple del(12) was found in patients no. 10 and 11, and a complex translocation and insertion involving chromosomes 12 and 22 occurred in patient no. 21. Additional rearrangements occurred within the leukemic clone in patients no. 19 and 20. Similar results related to 12p abnormalities have been reported on by others. Thus, Höglund et al13 noted that the standard G-banded karyotypes were modified in 10 of the 20 cases they described based on FISH analysis. Their cases included many with unbalanced rearrangements of 12p and, thus, only two of their patients had TEL rearrangements confirmed on Southern blot analysis. Taken together, it is likely that the region around TEL and D12S133 is prone to chromosomal breakage and that this results in more complex rearrangements. We have already observed the same findings in studying the patients with unbalanced 12p13 rearrangements or del(12p) that included 12p13.1 Whether this genomic instability is unique to this area, or whether this instability occurs in other chromosomal regions, especially those in which specific chromosomal breakpoints are clustered, is unknown. Cloning more of these breakpoints should clarify the mechanism involved in these rearrangements as well as provide some understanding of the genomic mechanisms associated with leukemogenesis.
In summary, the FISH findings reported here show that (1) TEL is involved in about half of the balanced 12p13 rearrangements, (2) there are at least two other regions both telomeric to TEL in which several breakpoints are located suggesting the existence of other critical genes, and (3) the chromosomal region around TEL and D12S133 is genomically unstable, resulting in complex chromosomal rearrangements.
Supported by Grant-in-Aid for International Exchange from Japan Clinical Pathology Foundation (Y.S.); National Institutes of Health Grants No. CA42557 (J.D.R.), CA40046 (M.M.L.); Department of Energy Grant No. DE-FG0286ER60408 (J.D.R.); and The G. Harold and Leila Y. Mathers Charitable Foundation (J.D.R.).
Address reprint requests to Janet D. Rowley, MD, DSc, Blum-Riese Distinguished Service Professor, Departments of Medicine and of Molecular Genetics and Cell Biology, University of Chicago, Medical Center, Section of Hematology/Oncology, 5841 S Maryland Ave, MC 2115, Chicago, IL 60637-1470.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal