Abstract
Deletions of the long arm of chromosome 11 (11q) are one of the most frequent structural chromosome aberrations in various types of lymphoproliferative disorders. However, in most conventional chromosome banding studies of B-cell chronic lymphocytic leukemia (B-CLL), 11q deletions were not identified as a frequent aberration. The objective of this study was to analyze the frequency and clinical impact of 11q deletions in B-CLL by interphase cytogenetics using fluorescence in situ hybridization (FISH). Mononuclear cells from 214 patients with B-CLL were studied by FISH using the yeast artificial chromosome (YAC) clone 755b11 from chromosome region 11q22.3-q23.1; we previously showed that this clone was contained within a 2- to 3-Mb sized segment of 11q commonly deleted in lymphoproliferative disorders. Forty-three of the 214 (20%) tumors exhibited 11q deletions; 11q deletions were the second most frequent chromosome aberration following 13q14 (RB1 and/or D13S25) deletions (45%); they were more frequent than trisomy 12 (15%) or deletion of 17p (TP53 gene) (10%). Patients with 11q deletions were younger (P = .01) and had more advanced clinical stages (P = .01). 11q deletions were associated with extensive peripheral, abdominal, and mediastinal lymphadenopathy (P < .001). Patients with 11q deletions had a more rapid disease progression as shown by a shorter treatment-free interval (9 months v 43 months; P < .001). The prognostic effect of 11q deletion on survival strongly depended on the age: in patients less than 55 years old, the median survival time was significantly shorter in the deletion group (64 months v 209 months; P < .001), whereas in patients ≥ 55 years old there was no significant difference (94 months v 111 months; P = .82). 11q deletions identify a new clinical subset of B-CLL characterized by extensive lymph node involvement. In younger B-CLL patients, this aberration is an important predictor of survival.
IN B-CELL chronic lymphocytic leukemia (B-CLL) clonal chromosome aberrations are detected in approximately 40% to 50% of cases by conventional chromosome banding analysis. The most common aberrations diagnosed in these studies included trisomy 12 and structural abnormalities of chromosome bands 13q14 and 14q32.1-3
Structural aberrations of the long arm of chromosome 11 (11q) have been identified as a recurring abnormality in various types of lymphoproliferative disorders. Nevertheless, in most studies of conventional banding analysis in B-CLL,1-3 11q aberrations, in particular 11q deletions, have not been reported as a frequent chromosome change. In the First International Working Party on Chromosomes in CLL (IWCCLL), which compiled data from 433 patients,2 37 of 391 (9%) evaluable tumors had structural aberrations of 11q. Twelve of these 37 tumors had aberrations involving band 11q13, of which 10 carried the translocation t(11; 14) that is strongly associated with mantle cell lymphoma (MCL).4 No information on the type of abnormality was provided for the remaining 25 tumors. Evidence for the significance of chromosome loss from 11q came from smaller chromosome banding studies in B-CLL.5-8 In the study by Fegan et al,8 11q deletions were among the most common chromosome aberrations, in particular in patients with disease progression. Hernandez et al9 analyzed 609 patients with various B-cell chronic lymphoproliferative disorders, including 423 cases classified as classical B-CLL, B-CLL mixed cell type, and atypical B-CLL. Although 11q deletions were the most common structural aberration in these tumors, the prevalence was only 6% (25 of 423 cases). Finally, in a compilation of data from the Catalog of Chromosome Aberrations in Cancer,10 one of the most common structural aberrations resulting in loss of chromosome material in the categories “lymphoproliferative disorders” and “non-Hodgkin's lymphomas” were deletions affecting the region 11q21-q25, most frequently chromosome band 11q23.
The recurring loss of material from chromosome bands 11q21-q25 suggests that a novel tumor suppressor gene is localized in this genomic region. To delineate the commonly deleted segment of these 11q deletions, we previously applied fluorescence in situ hybridization (FISH) using a panel of 17 yeast artificial chromosome (YAC) clones that span chromosome bands 11q14.3-q23.3.11 12 In a series of 43 tumors from patients with various lymphoproliferative disorders exhibiting 11q deletions, we identified a 2- to 3-Mb sized genomic region in bands 11q22.3-q23.1 that was deleted in all tumors. In the present study, we report on the incidence of 11q deletions as assessed by interphase cytogenetics using a YAC clone from the commonly deleted segment and on the clinical implication of this aberration in 214 patients with B-CLL.
MATERIALS AND METHODS
Patients
Between October 1990 and March 1996, 214 patients with B-CLL were studied. All patients were seen at our institution that is both a secondary and tertiary care referral center for patients with B-CLL. Cases classified as prolymphocytic leukemia, Waldenström's macroglobulinemia, leukemic follicle center cell, or mantle cell lymphoma were excluded from the analysis. One hundred thirty-one patients were male and 83 female; the age at the time of diagnosis ranged from 33 to 85 years (median, 59 years). The diagnosis of B-CLL required a persistent lymphocytosis of >5.0 × 109/L. Immunophenotypic data were available for 200 of the 214 patients: all leukemias were CD19+, 185 of 195 tested were CD5+, and 186 of 194 were CD23+. At the time of study, 25 patients had Rai stage 0, 21 stage 1, 105 stage 2, 27 stage 3, and 36 stage 4 disease.13
In many patients the diagnosis of B-CLL was made before interphase cytogenetic analysis became a routine investigation in B-CLL at our institution. Therefore, in many patients the analysis was performed several months or even several years after diagnosis (median, 20 months; range, 0 to 247 months). No sequential cytogenetic analyses were done. One hundred thirty-eight patients were previously untreated, 38 patients had received one, and 38 patients two or more chemotherapeutic regimens before interphase cytogenetic analysis.
Chromosome banding data were available from 59 of the 214 tumors. Structural aberrations of chromosome 11 were identified in 7 cases: del(11)(q21q23) (3 cases); del(11)(q21) (2 cases); del(11)(q13q23) (1 case); the remaining tumor exhibited a balanced translocation t(X; 11)(q13; q23).
Interphase Cytogenetic Analysis
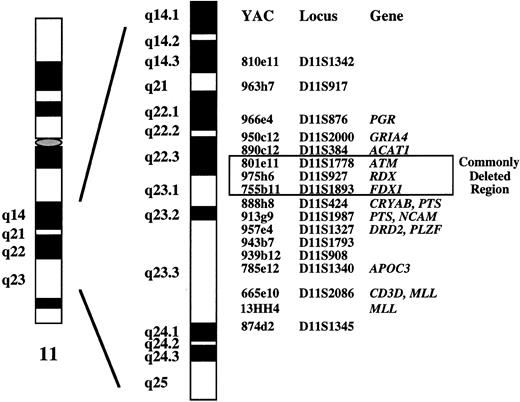
DNA probes.In a previous study, we characterized deletions and translocations affecting chromosome bands 11q21-q23 in 43 tumors classified as B-CLL (n = 40) and mantle cell lymphoma (n = 3).11 Seventeen representative clones from a contig map of YACs encompassing bands 11q14.3-q23.3 were selected as probes (Fig 1). Because overlapping YACs were applied, it was possible to systematically delineate the extent of the deletions at the molecular level. We identified a single critical region of 2 to 3 Mb in bands 11q22.3-q23.1 in which all deletions clustered. This genomic fragment defined by YACs 801e11, 975h6, and 755b11 contains the genes coding for ATM (ataxia telangiectasia mutated), RDX (radixin), and FDX1 (ferredoxin 1) (Fig 1). For the 11q deletion screening in the present study, we used YAC clone 755b11. The 1.8-Mb genomic segment identified by clone 755b11 was not only found to be deleted in all tumors exhibiting an 11q deletion, but it also contained the breakpoint of two balanced translocations, one of them being the t(X; 11)(q13; q23) described above.
Molecular cytogenetic delineation of the critical region of 11q deletions in B-CLL (see ref 11). In a previous study, we identified a 2- to 3-Mb sized commonly deleted region in B-CLL tumors that was defined by YAC clones 801e11, 975h6, and 755b11. Clone 755b11 that we used for deletion screening in the present study also contained the breakpoints of two balanced translocations.
Molecular cytogenetic delineation of the critical region of 11q deletions in B-CLL (see ref 11). In a previous study, we identified a 2- to 3-Mb sized commonly deleted region in B-CLL tumors that was defined by YAC clones 801e11, 975h6, and 755b11. Clone 755b11 that we used for deletion screening in the present study also contained the breakpoints of two balanced translocations.
To distinguish “classical” B-CLL from leukemic MCLs associated with the translocation t(11; 14)(q13; q32), all tumors were studied for the presence of the t(11; 14). For the identification of the t(11; 14) by dual-color FISH we used the following DNA clones: for chromosome 14 a pool of cosmid clones cos-Cα1/2, recognizing the cα1 and cα2 gene segments proximal to the JH-region,14 and of 320-kb YAC clone Y6 (kindly provided by Dr Fumihiko Matsuda, Kyoto, Japan), identifying VH-segments telomeric to the JH-breakpoints in the Ig heavy chain gene15; and for chromosome 11 the differently labeled 540-kb YAC clone 55g7 recognizing DNA sequences spanning the region between the major translocation cluster and the CCND1 gene in the BCL1 locus at 11q13.16
Preparation and labeling of DNA probes.Human sequences from YAC clones 755b11, Y6, and 55g7 were generated by a polymerase chain reaction (PCR) protocol using primers directed against Alu-sequences.17 Amplification was performed in a 100-μL reaction mixture containing approximately 160 ng YAC DNA, 100 mmol/L of the four dNTPs (Boehringer Mannheim, Mannheim, Germany), 10 μL PCR buffer (Boehringer Mannheim), and 2.0 mmol/L MgCl2 (Boehringer Mannheim). Three Alu-PCR reactions were performed using either the primers CL1, CL2, or a combination of both (0.5 μmol/L).17 The products of all three reactions were combined for the FISH experiments. Cos-Cα1/2 cosmid DNA was prepared according to the plasmid MIDI KIT protocol (Qiagen, Hilden, Germany). The probes were labeled by nick translation with biotin-16-dUTP or digoxigenin-11-dUTP (Boehringer Mannheim).
FISH.The hybridization mixture contained approximately 250 ng labeled YAC or cosmid DNA, 10 μg Cot-1 DNA fraction (BRL/Life-Technologies, Gaithersburg, MD), and 10 μg herring sperm DNA (Boehringer Mannheim). Hybridization was performed as described previously.18-20 To prevent false-positive results caused by inadequate hybridization or chromatin loss, analysis was only performed on slides with high hybridization efficiency, indicated by two signals of a control YAC clone in more than 90% of the nuclei. Fluorescence signals were enumerated in 150 to 300 interphase cells. Images were captured using a cooled charge coupled device (CCD) camera (Photometrics Ltd, Tucson, AZ) linked to an Apple Macintosh computer (Ismaning, Germany).
Statistical Analysis
Comparisons of clinical and laboratory parameters between patients with and without 11q deletion were performed using the Wilcoxon rank sum test (quantitative variables), the Fisher's exact test (binary variables), and the Cochran-Armitage trend test (ordinal variables). Survival times and censored waiting times measured from the date of diagnosis were plotted from life tables using Kaplan-Meier estimates. Differences between curves were analyzed by the log-rank test.21 The proportional hazards regression model of Cox was used for the multivariate analysis of survival time data.22 The statistical analyses were performed using the software packages S-Plus (MathSoft, Inc, Seattle, WA) and StatXact (Cytel Software Corp, Cambridge, MA) together with the Design software library.23
RESULTS
Interphase Cytogenetic Analysis
To define the cut-off level for the diagnosis of 11q deletion, hybridization experiments of blood specimens from five probands were performed. By analogy with our previous studies on the deletion screening in B-CLL,19 20 the cut-off level was defined by the mean + 3 SD of the frequency of control cells exhibiting only one 755b11 signal (mean, 3.16%; SD, 1.79%; cut-off level, 8.5%).
Hybridization of clone 755b11 (detected via fluoroscein isothiocyanate) and an internal control (detected via Cy3) to nuclei from a B-CLL tumor exhibiting an 11q deletion. All nuclei show only one hybridization signal with the diagnostic 755b11 FISH probe indicating deletion of the corresponding region.
Hybridization of clone 755b11 (detected via fluoroscein isothiocyanate) and an internal control (detected via Cy3) to nuclei from a B-CLL tumor exhibiting an 11q deletion. All nuclei show only one hybridization signal with the diagnostic 755b11 FISH probe indicating deletion of the corresponding region.
Clinical and Laboratory Data at the Time of Study
| . | No 11q Deletion (N = 171) . | 11q Deletion (N = 43) . | P Value . |
|---|---|---|---|
| Median age (yr) | 63 (37-84) | 58 (36-85) | .01† |
| Male sex | 104 (61%) | 27 (63%) | .86‡ |
| Stage at study | .01ρ | ||
| Rai 0 | 24 (14.0%) | 1 (2.3%) | |
| Rai 1 | 17 (9.9%) | 4 (9.3%) | |
| Rai 2 | 86 (50.3%) | 19 (44.2%) | |
| Rai 3 | 18 (10.5%) | 9 (20.9%) | |
| Rai 4 | 26 (15.2%) | 10 (23.3%) | |
| B symptoms (%) | 37 (21.6%) | 16 (37.2%) | .04‡ |
| White blood cell count (×109/L) | 35.8 | 55.4 | .35† |
| Hemoglobin (g/dL) | 13.1 | 11.7 | .03† |
| Platelet count (×109/L) | 166 | 154 | .20† |
| Lactate dehydrogenase (IU/L) | 174 | 197 | .07† |
| Alkaline phosphatase (IU/L) | 128 | 133 | .30† |
| Albumin (g/L) | 45 | 45 | .58† |
| IgG (g/L) | 9.39 | 8.77 | .42† |
| IgA (g/L) | 1.28 | 0.85 | .04† |
| IgM (g/L) | 0.60 | 0.73 | .60† |
| Splenomegaly (%) | 122 (71.3%) | 35 (81.4%) | .32‡ |
| Peripheral lymphadenopathy (cm2 )* | 4 | 17 | <.001† |
| Mediastinal lymphadenopathy (%) | 5.3 | 23.3 | <.001‡ |
| Abdominal lymphadenopathy (%) | 48.0 | 88.4 | <.001‡ |
| Largest lymph node diameter (cm) | 2.0 | 4.3 | <.001† |
| . | No 11q Deletion (N = 171) . | 11q Deletion (N = 43) . | P Value . |
|---|---|---|---|
| Median age (yr) | 63 (37-84) | 58 (36-85) | .01† |
| Male sex | 104 (61%) | 27 (63%) | .86‡ |
| Stage at study | .01ρ | ||
| Rai 0 | 24 (14.0%) | 1 (2.3%) | |
| Rai 1 | 17 (9.9%) | 4 (9.3%) | |
| Rai 2 | 86 (50.3%) | 19 (44.2%) | |
| Rai 3 | 18 (10.5%) | 9 (20.9%) | |
| Rai 4 | 26 (15.2%) | 10 (23.3%) | |
| B symptoms (%) | 37 (21.6%) | 16 (37.2%) | .04‡ |
| White blood cell count (×109/L) | 35.8 | 55.4 | .35† |
| Hemoglobin (g/dL) | 13.1 | 11.7 | .03† |
| Platelet count (×109/L) | 166 | 154 | .20† |
| Lactate dehydrogenase (IU/L) | 174 | 197 | .07† |
| Alkaline phosphatase (IU/L) | 128 | 133 | .30† |
| Albumin (g/L) | 45 | 45 | .58† |
| IgG (g/L) | 9.39 | 8.77 | .42† |
| IgA (g/L) | 1.28 | 0.85 | .04† |
| IgM (g/L) | 0.60 | 0.73 | .60† |
| Splenomegaly (%) | 122 (71.3%) | 35 (81.4%) | .32‡ |
| Peripheral lymphadenopathy (cm2 )* | 4 | 17 | <.001† |
| Mediastinal lymphadenopathy (%) | 5.3 | 23.3 | <.001‡ |
| Abdominal lymphadenopathy (%) | 48.0 | 88.4 | <.001‡ |
| Largest lymph node diameter (cm) | 2.0 | 4.3 | <.001† |
Median values (and range) are given for quantitative variables.
Median sum of the products of the diameters of the largest cervical, axillar, and inguinal lymph nodes.
Wilcoxon rank sum test.
Fisher's exact test.
ρ Exact trend test (Cochran-Armitage test).
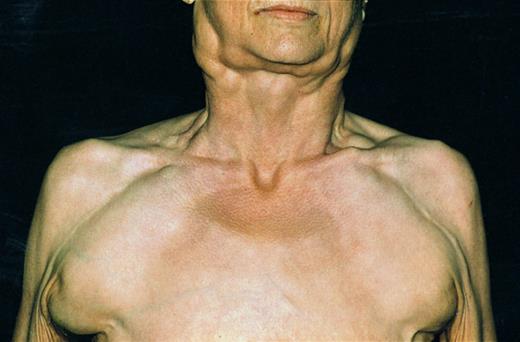
Patient with B-CLL and 11q deletion suffering from massive lymph node enlargement. Extensive lymphadenopathy characterized the clinical course of the disease in almost all patients.
Patient with B-CLL and 11q deletion suffering from massive lymph node enlargement. Extensive lymphadenopathy characterized the clinical course of the disease in almost all patients.
Forty-three of the 214 (20%) B-CLL tumors exhibited a monoallelic deletion of the DNA targeted by clone 755b11 (Fig 2). The percentage of nuclei with one fluorescence signal ranged from 9.3% to 98.0%. Six of the 43 tumors had lower percentages (9.5% to 25.0%) of cells exhibiting the deletion; in 8 tumors the percentages ranged between 48.0% and 70.0%; however, in the remaining 29 tumors more than 80.0% of cells carried the deletion.
All tumors were also analyzed for other recurring chromosome aberrations in B-CLL, such as 13q14 deletions, trisomy 12, and 17p13 deletions. 11q deletions were the second most frequent aberration following 13q14 deletions (45.0%) as assessed by DNA probes recognizing the retinoblastoma (RB1) gene and/or the locus D13S25.19 24 11q deletions were even more frequent than trisomy 12 (15.0%) and 17p (TP53) deletions (10.0%).20 25 In 25 of the 43 tumors with 11q deletions, other chromosome aberrations were detected by our DNA probe set: 18 of the 43 tumors had additional 13q14 deletion; 4 had trisomy 12; 1 had both 13q14 deletion and trisomy 12; and 2 tumors had TP53 gene deletion. None of the tumors carried the translocation t(11; 14).
Treatment-free interval in the 214 patients with B-CLL: The median time from diagnosis to first treatment in the 43 patients with 11q deletion was only 9 months compared to 43 months in the 171 patients without 11q deletion (P < .001).
Treatment-free interval in the 214 patients with B-CLL: The median time from diagnosis to first treatment in the 43 patients with 11q deletion was only 9 months compared to 43 months in the 171 patients without 11q deletion (P < .001).
Correlations With Clinical and Laboratory Data
The clinical characteristics and laboratory data at the time of study of the patients with and without 11q deletion are shown in Table 1. There was no significant difference in sex, white blood cell count, platelet count, lactate dehydrogenase, alkaline phosphatase, IgG, and IgM between the two groups. Patients with 11q deletions were younger (58 years v 63 years; P = .01) and had more advanced clinical stages (P = .01); they had significantly lower hemoglobin levels (11.7 g/dL v 13.1 g/dL; P = .03) and lower serum IgA concentrations (0.85 g/L v 1.28 g/L; P = .04). Furthermore, patients with 11q deletions had significantly more lymph node involvement than patients without 11q deletions as assessed by the median sum of the products of the diameters of the largest cervical, axillar, and inguinal lymph nodes (17 cm2v 4 cm2; P < .001), the presence of mediastinal (23.3% v 5.3%; P < .001) and abdominal (88.4% v 48.0%; P < .001) lymphadenopathy, and by the largest lymph node diameter (4.3 cm v 2.0 cm; P < .001). In most patients with 11q deletions, extensive nodal involvement dominated the clinical course of the disease (Fig 3). Patients with 11q deletions suffered from B symptoms more frequently (P = .04).
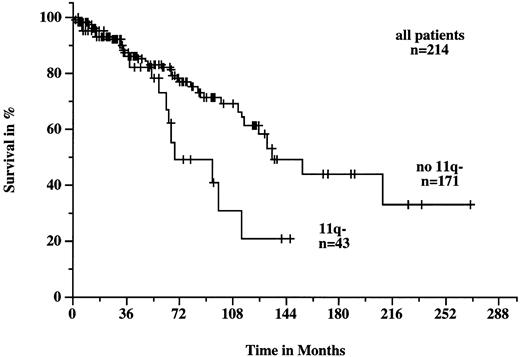
Patients with 11q deletions exhibited a more rapid disease progression and shorter survival times. The treatment-free interval was significantly shorter in patients with 11q deletions than in patients without deletions (9 months v 43 months; P < .001; Fig 4). Figure 5 shows the survival probabilities of the two patient groups measured from the time of diagnosis; the difference between the two curves was statistically significant (P = .02). The median survival time of the patients with 11q deletions from the date of diagnosis was 68 months compared to 134 months for patients without deletions. Patients with 11q deletions had an estimated twofold greater risk of death than patients without a deletion.
Survival probabilities from the time of diagnosis in the 214 patients with B-CLL with (n = 43) and without (n = 171) 11q deletion. The difference between the two curves was significant (P = .002).
Survival probabilities from the time of diagnosis in the 214 patients with B-CLL with (n = 43) and without (n = 171) 11q deletion. The difference between the two curves was significant (P = .002).
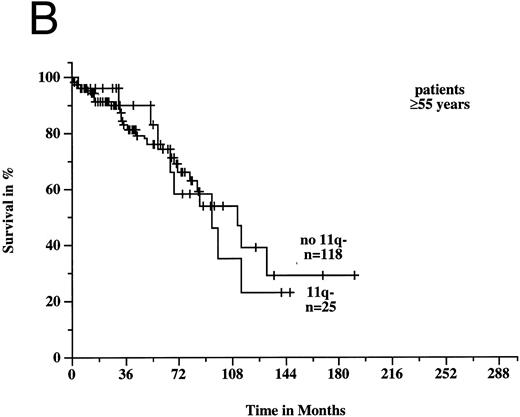
To estimate prognosis, we used a Cox proportional hazards model with survival time from diagnosis as dependent variable, and age, sex, Rai stage, and 11q deletion as possible prognostic factors. At the time of diagnosis, 39 patients had Rai stage 0, 38 stage 1, 99 stage 2, 9 stage 3, and 13 stage 4 disease; the latter two categories were combined to one variable for the survival model. Sixteen patients, for whom there was no information on Rai stage at diagnosis, were excluded from consideration, but we obtained similar results if we used a model with missing values imputation instead. In the survival model, three variables gave significant prognostic information: age (P < .001), 11q deletion (P < .001), and Rai stage (P < .001). There was a significant estimated interaction effect between age and 11q deletion (P = .002): the negative prognostic impact of 11q deletion was only seen in younger patients. Figures 6A and B show the survival probabilities in two different age groups: in patients less than 55 years old, the median survival time was significantly shorter in the 11q deletion group (64 months v 209 months; P < .001). In contrast, in patients ≥55 years old there was no significant difference between patients with and without 11q deletions (94 months v 111 months; P = .82). Estimated relative hazards together with their 95% confidence limits were computed from the model (Table 2). The age effect was measured separately in patients with and without 11q deletions comparing the upper and lower quartile of the age distribution. The effect of 11q deletion was determined for the lower, median, and upper quartile of the age distribution.
Survival probabilities from the time of diagnosis depending on the age: (A) Patients < 55 years: the median survival time of the 18 patients with 11q deletion was 64 months compared to 209 months in the 53 patients without 11q deletion. The difference between the two curves was highly significant (P < .001). (B) Patients ≥ 55 years: the median survival time of the 25 patients with 11q deletion was 94 months compared to 111 months in the 118 patients without 11q deletion (P = .82).
Survival probabilities from the time of diagnosis depending on the age: (A) Patients < 55 years: the median survival time of the 18 patients with 11q deletion was 64 months compared to 209 months in the 53 patients without 11q deletion. The difference between the two curves was highly significant (P < .001). (B) Patients ≥ 55 years: the median survival time of the 25 patients with 11q deletion was 94 months compared to 111 months in the 118 patients without 11q deletion (P = .82).
Estimated Relative Risk (Hazard Ratio, HR) and 95% Confidence Interval (CI)
| Prognostic Factor . | Levels . | HR . | (95% CI) . |
|---|---|---|---|
| Rai stage | Rai 1:0 | 3.14 | (0.6, 16.5) |
| Rai 2:0 | 8.81 | (2.0, 38.8) | |
| Rai 3 or 4:0 | 28.00 | (5.8, 136.0) | |
| Age (yr)* | |||
| No 11q deletion | 66:52 | 4.07 | (2.17, 7.62) |
| 11q deletion | 66:52 | 0.54 | (0.18, 1.57) |
| 11q deletion† | |||
| Age 52 | Yes:no | 3.95 | (1.64, 9.51) |
| Age 59 | Yes:no | 1.47 | (0.68, 3.18) |
| Age 66 | Yes:no | 0.54 | (0.18, 1.65) |
| Prognostic Factor . | Levels . | HR . | (95% CI) . |
|---|---|---|---|
| Rai stage | Rai 1:0 | 3.14 | (0.6, 16.5) |
| Rai 2:0 | 8.81 | (2.0, 38.8) | |
| Rai 3 or 4:0 | 28.00 | (5.8, 136.0) | |
| Age (yr)* | |||
| No 11q deletion | 66:52 | 4.07 | (2.17, 7.62) |
| 11q deletion | 66:52 | 0.54 | (0.18, 1.57) |
| 11q deletion† | |||
| Age 52 | Yes:no | 3.95 | (1.64, 9.51) |
| Age 59 | Yes:no | 1.47 | (0.68, 3.18) |
| Age 66 | Yes:no | 0.54 | (0.18, 1.65) |
The effect of age was measured separately for patients with and without 11q deletion comparing the upper and lower quartile of the age distribution.
The effect of 11q deletion was computed for the lower, median, and upper quartile of the age distribution.
DISCUSSION
In this interphase cytogenetic study, we have identified a new clinical subset of B-CLL that is defined by deletion of a genomic region in chromosome bands 11q22.3-q23.1.
Structural abnormalites of chromosome 11 are recurrent aberrations in various types of lymphoproliferative disorders. However, research has so far focussed on the molecular analysis of balanced translocations. The translocation t(11; 14)(q13; q32) is associated with MCL and results in fusion of the BCL1 locus to the IgH locus.4,26,27 The MLL gene at 11q23 was shown to be rearranged in few cases of non-Hodgkin's lymphoma.28 Other genes on 11q that were identified in rare cases of malignant lymphoma include RCK [cloned from the t(11; 14)(q23; q32) breakpoint of the RC-K8 lymphoma cell line],29LPC [cloned from the t(11; 14)(q32; q32) breakpoint of a sclerosing B-cell lymphoma],30 or the B-cell transcriptional activator BOB1 [fused to the LAZ3/BCL6 gene by the t(3; 11)(q27; q23.1) in the B-cell leukemia line Karpas 231].31BOB1 seems to map proximally to NCAM, its precise genomic location has to our knowledge not yet been determined.31MLL, RCK, and LPC are not contained within the commonly deleted segment that we recently identified in a series of B-CLL tumors exhibiting 11q deletions or translocations.11 This 2- to 3-Mb sized critical genomic region is located in bands 11q22.3-q23.1 and contains the ATM (ataxia telangiectasia mutated), RDX (radixin), and FDX1 (ferredoxin 1) genes (see Fig 1). The 1.8-Mb genomic fragment within the critical region that is recognized by clone 755b11 also contained the breakpoints of two reciprocal translocations. These two breakpoints may point to the location of a novel gene of pathogenic significance in B-CLL, in particular because they map to a deletion cluster region. Therefore, we used clone 755b11 for deletion screening in the present study.
11q deletions were found in 20% of the B-CLL tumors and were the second most frequent aberration following 13q14 deletions. 11q deletions were more frequent than trisomy 12 that in most chromosome banding studies was the most common aberration.1-3 This difference in frequencies is unlikely to result from patient selection, but rather underlines the importance of obtaining cytogenetic data beyond the level of metaphase cells in this disease that is very difficult to study by conventional chromosome banding analysis. In the past, the discrepancy between metaphase and interphase cytogenetic analysis has also been shown for other recurring numerical and structural chromosome abnormalities in B-CLL such as trisomy 12, 13q, and 17p deletions.19,20,25 32-34
Presence of the 11q deletion was associated with a characteristic clinical picture. The patients with 11q deletions were younger and had more advanced clinical stages. Most impressively, 11q deletions were associated with extensive lymphadenopathy as assessed by the extent of peripheral lymph node involvement, the frequency of mediastinal or abdominal lymphadenopathy, and the largest lymph node diameter measured (Fig 3). Along with this marked nodal involvement, the patients suffered from B symptoms more frequently. Our data are in agreement with a recent chromosome banding study that evaluated karyotypic evolution in a series of 45 B-CLL tumors.8 11q deletions were the most common chromosome aberration in the group of tumors exhibiting clonal evolution, and, by analogy to our study, the patients with 11q deletions had advanced or progressive disease. Because in our study not all leukemias were analyzed at the time of diagnosis and because no serial studies were performed, it cannot be assessed whether the 11q deletions occurred as the primary genetic event. The significantly shorter treatment-free interval in the 11q deletion group (Fig 4) could indicate that the deletions occurred early in the course of the disease or even represented the primary event. Serial interphase cytogenetic analyses will be necessary to determine at what timepoint 11q deletions occur in the course of the disease.
11q deletions were predictive of poor survival. Most interestingly, in the multivariate survival model, there was a significant interaction effect between age and 11q deletion: the negative prognostic effect of 11q deletion was only seen in the younger patients (Fig 6). In the age group less than 55 years, the patients with 11q deletions had a median survival time of only 64 months, whereas the patients without the deletion had an excellent outcome. It is important to emphasize that 16 of the 18 (89%) patients with 11q deletions younger than 55 years had Rai stages 0-2 at diagnosis. Thus, 11q deletion was a very important predictor of early disease progression and survival that was independent of stage. Only a few studies have addressed the question of presenting features or prognostic factors in younger B-CLL patients. De Rossi et al35 studied 133 patients with B-CLL younger than 55 years. In univariate analysis, only hemoglobin level, blood, and bone marrow lymphocytosis, but none of the clinical staging systems, had a prognostic value. In contrast, in the study of 117 patients younger than 50 years by Montserrat et al,36 clinical stages, pattern of bone marrow involvement and lymphocyte doubling time were significant prognostic factors, similar to those found in the older age group. Given these conflicting data with regard to clinical risk factors, the 11q deletion adds a new and biologic prognostic factor for younger patients with B-CLL. Our finding is of great clinical relevance because these patients should be considered for experimental treatment approaches such as high-dose chemotherapy and radiotherapy followed by autologous or allogeneic hematopoietic stem cell transplantation.
ACKNOWLEDGMENT
We gratefully acknowledge Daniela Diehl and Kathrin Wildenberger for excellent technical assistance, Drs Stefan Joos and Fumihiko Matsuda for providing us with DNA probes, and Dr Lutz Edler for statistical advice.
Supported by grants from the Tumorzentrum Heidelberg/Mannheim (I/I.1), and the Deutsche Krebshilfe (10-0917-Dö I).
Address reprint requests to Hartmut Döhner, MD, Medizinische Klinik and Poliklinik V, University of Heidelberg, Hospitalstr. 3, 69115 Heidelberg, Germany.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal