Abstract
A large fraction of non-Hodgkin's lymphomas (NHLs) accumulate a wild-type form of the p53 tumor suppressor protein at the nuclear level. In normal cells, p53 induction is associated with a temporary cell growth arrest at the G1-S boundary of the cell cycle. This activity of p53 as a G1 checkpoint molecule is strictly dependent on its ability to induce the transcription of the inhibitor of the cyclin dependent kinase, p21. To verify the functionality of the wild-type p53 protein accumulated in NHL cells, 70 cases were comparatively analyzed for p53 and p21 expression and status of the respective genes. Overexpression of the wt p53 protein was associated with the accumulation of p21, indicating that p53 is functional with respect to p21 induction in these tumors. The coaccumulation of p53 with Ki-67 antigen indicates that wt p53-positive cells and p21-positive cells, as well, are actively proliferative elements, supporting the notion that p53-induced, p21-mediated growth arrest is somehow overridden in NHL cells. No p21 mutation or particular allele variant was shown to correlate with p21 protein accumulation, thus excluding a role for p21 structural abnormalities. Taken together, our data suggest the existence in NHL of a peculiar mechanism of functional inactivation of the p53 G1 checkpoint pathway occurring downstream of the CDK inhibitor p21.
P53 IS A NUCLEAR phosphoprotein that plays a key role in the control of the cell cycle and tissue homeostasis. In normal cells, p53 synthesis is almost restricted to the G1-S boundary and is functional to the evocation of a temporary cell growth arrest during which the cell is allowed to repair possible genomic damages or, in the case of profound DNA alterations or cellular stresses, to undergo apoptosis. These functions, which account for the denomination “guardian of the genome” given to p53, are strictly dependent on the activity of p53 as a transcriptional regulator, the transcriptional defective mutants of p53 being unable to evoke these phenomena.1-3
While the mechanisms by which p53 induces apoptosis are still largely unknown, it is well established that the ability of p53 to induce G1 arrest is related to the synthesis of the CDK inhibitor p21. The synthesis of p53 at the G1-S boundary induces, in turn, the synthesis of p21, which promotes cell growth arrest by interacting with and inhibiting the cyclin-CDK complexes responsible for cell cycle progression.4-6
p53 is frequently mutated in many tumors and most of the biologically significant mutations impair the ability of p53 to function as a transcriptional regulator, thus preventing the induction of p53-dependent p21 synthesis and, consequently, the evocation of the G1 checkpoint.6-8 Unlike functional p53, which is rapidly degraded within a few minutes after its synthesis, mutated versions of the p53 protein tend to accumulate into the cells, thus becoming immunohistochemically detectable. The tendency of the mutated form of p53 to be refractory to normal p53 turnover has led to the conventional paradigm that p53 accumulation in neoplastic cells is synonymous with p53 mutation. Although this holds true for most human and animal tumors, it is far from being a general rule. We and other investigators have recently demonstrated that a large fraction of non-Hodgkin's lymphomas (NHLs) accumulates a wild-type form of p53 (wt-p53) at the nuclear level.9-11 Because the inactivation and consequent stabilization of p53 may also occur through the complexion with viral and cellular proteins, the role of human immunodeficiency virus (HIV) and Epstein-Barr virus (EBV)-encoded antigens, as well as the involvement of the cellular p53-binding protein MDM2, was evaluated. No significant correlation between p53 accumulation and HIV or EBV infection was detected, or the overexpression of MDM2, whose binding to p53 prevents it from acting as a transcriptional activator, was shown to account for p53 stabilization in NHL.9 12 Therefore, up to now, there is no considerable evidence of the functional inactivation of the wt-p53 protein in this type of tumor.
These observations raise the hypothesis that the wild-type form of p53 accumulated in NHL cells may be actually a functional protein, suggesting the existence of mechanisms of inactivation involving p53-downstream targets. To test this hypothesis, 70 NHLs were analyzed for p53 protein expression, p53 gene status, and induction of the CDK inhibitor p21. p21 accumulation significantly correlated with the overexpression of wt-p53 protein, indicating that the wt-p53 protein overexpressed by NHL cells is functional as a transcriptional regulator, in particular as an inducer of p21 synthesis. No p21 mutation or particular allele variant was found to correlate with wt-p53 stabilization, suggesting also that p21 is a potentially functional protein in these tumors. Taken together, our data suggest the existence in NHLs of a specific mechanism of functional inactivation of the p53 pathway involving genetic targets downstream the CDK inhibitor p21.
MATERIALS AND METHODS
Samples.A selected series of 70 NHLs representing most of NHL typologies was analyzed for p53, p21, and Ki-67 expression. The cases were retrieved from the surgical pathology files of the Division of Pathology at the Centro di Riferimento Oncologico, Aviano and the City Hospital, Belluno, Italy. All the cases were defined according to the histologic lymphoma classification of the Working Formulation for NHLs, as shown in Table 1. The Revised European-American Lymphoma (REAL) classification13 was also used (Table 2).
Case Series of NHLs (47 cases, p53-positive group and 23 cases, p53-negative control group) Subdivided According to the Working Formulation Categories, Immunohistochemical Expression of p53 and p21 Proteins
| Working Formulation . | No. of Tested Cases . | p53+ p21+ . | p53+ p21− . | p53− p21+ . | p53− p21− . |
|---|---|---|---|---|---|
| Low-grade | |||||
| Small lymphocytic | 4 | 1 | 3 | — | — |
| Follicular predominantly small cleaved cell | 6 | — | — | — | 6 |
| Follicular, mixed, small cleaved and large cell | 3 | — | 2 | — | 1 |
| Intermediate-grade | |||||
| Follicular, predominantly large cell | 1 | — | — | — | 1 |
| Diffuse mixed, small and large cell | 2 | 1 | — | — | 1 |
| Diffuse, large cell | 24 | 7 | 11 | — | 6 |
| High-grade | |||||
| Large cell, immunoblastic | 8 | 3 | 5 | — | — |
| Small noncleaved | 9 | 2 | 7 | — | — |
| Miscellaneous | |||||
| Anaplastic large cell (Ki-1+) | 6 | 1 | 3 | 2 | — |
| Intestinal T cell | 1 | 1 | — | — | — |
| MALT-type | 5 | — | — | — | 5 |
| Monocytoid | 1 | — | — | 1 | — |
| Total | 70 | 16 | 31 | 3 | 20 |
| Working Formulation . | No. of Tested Cases . | p53+ p21+ . | p53+ p21− . | p53− p21+ . | p53− p21− . |
|---|---|---|---|---|---|
| Low-grade | |||||
| Small lymphocytic | 4 | 1 | 3 | — | — |
| Follicular predominantly small cleaved cell | 6 | — | — | — | 6 |
| Follicular, mixed, small cleaved and large cell | 3 | — | 2 | — | 1 |
| Intermediate-grade | |||||
| Follicular, predominantly large cell | 1 | — | — | — | 1 |
| Diffuse mixed, small and large cell | 2 | 1 | — | — | 1 |
| Diffuse, large cell | 24 | 7 | 11 | — | 6 |
| High-grade | |||||
| Large cell, immunoblastic | 8 | 3 | 5 | — | — |
| Small noncleaved | 9 | 2 | 7 | — | — |
| Miscellaneous | |||||
| Anaplastic large cell (Ki-1+) | 6 | 1 | 3 | 2 | — |
| Intestinal T cell | 1 | 1 | — | — | — |
| MALT-type | 5 | — | — | — | 5 |
| Monocytoid | 1 | — | — | 1 | — |
| Total | 70 | 16 | 31 | 3 | 20 |
Abbreviation: MALT, mucosa-associated lymphoma tissue.
p53, p21, and Ki-67 Immunohistochemical Expression (IHC) and p53 Gene Status in NHLs
| Case No. . | Histologic Diagnosis . | p53 . | p21 . | Ki-67 . | ||
|---|---|---|---|---|---|---|
| . | W.F. Classification . | REAL Classification . | IHC (%) . | Mutation . | IHC (%) . | IHC (%) . |
| N124 | NHL-J | Burkitt's | 65 | cod.286, G→A, Glu→Lys | — | 90 |
| N311 | NHL-J | Burkitt's | 20 | cod.132, A→C, Lys→Gln | — | 90 |
| N312 | NHL-J | Burkitt's | 80 | cod.130, C→G, Leu→Val | — | 90 |
| N313 | NHL-J | Burkitt's | 30 | cod.132, A→C, Lys→Gln | — | NE |
| N315 | NHL-J | Burkitt's | 80 | cod.273, G→T, Arg→Leu | 80 | 90 |
| N519 | NHL-J | Burkitt's | 90 | cod.236, T→C, Tyr→His | 90 | 90 |
| N523 | NHL-J | Burkitt's | 10 | cod.267, G→C, Arg→Pro | — | 70 |
| N521 | NHL-H | Diffuse large B cell | 50 | cod.244, G→A, Gly→Asp | — | 70 |
| L802 | NHL-G | Diffuse large B cell | 50 | cod.141, G→A, Cys→Tyr | — | 50 |
| N611 | NHL-C | Follicular center (grade II) | 20 | cod.161, G→A, Ala→Thr | — | 30 |
| N612 | NHL-C | Follicular center (grade II) | 60 | cod.248, G→A, Arg→Glu | — | 60 |
| L711 | NHL-A | Small lymphocytic | 40 | cod.193, A→G, His→Arg | — | 50 |
| L914 | NHL-A | Small lymphocytic | 60 | cod.258, G→A, Glu→Lys | — | 60 |
| N610 | NHL-A | Small lymphocytic | 60 | cod.275, G→A, Cys→Thr | — | 60 |
| F11 | NHL-H | Diffuse large B cell | 80 | cod.179, A→G, His→Arg | — | 70 |
| F12 | NHL-H | Diffuse large B cell | 80 | cod.282, C→T, Arg→Trp | — | 70 |
| F13 | NHL-G | Diffuse large B cell | 80 | cod.248, C→T, Arg→Trp | — | 60 |
| F15 | NHL-G | Diffuse large B cell | 80 | cod.285, G→A, Glu→Lys | — | 50 |
| N167 | NHL-Misc ALC (Ki-1+) | Diffuse large B cell | 30 | cod.175, G→A, Arg→His | — | 40 |
| N240 | NHL-Misc ALC (Ki-1+) | Anaplastic large cell | 30 | cod.176, T→A, Cys→Ser | 10 | 30 |
| VR | NHL-Misc Intestinal T-cell | Intestinal T cell | 80 | NO | 50 | 50 |
| F33H | NHL-G | Diffuse large B cell | 30 | NO | 40 | 40 |
| N609 | NHL-A | Small lymphocytic | 50 | NO | 10 | 50 |
| N522 | NHL-G | Diffuse large B cell | 60 | NO | 10 | 70 |
| N615 | NHL-G | Diffuse large B cell | 40 | NO | 10 | 80 |
| F8 | NHL-H | Diffuse large B cell | 20 | NO | 25 | 45 |
| F3 | NHL-F | Angioimmunoblastic T cell | 15 | NO | 10 | 25 |
| F14 | NHL-H | Diffuse large B cell | 20 | NO | 15 | 50 |
| F10 | NHL-G | Diffuse large B cell | 15 | NO | 15 | 30 |
| F2 | NHL-G | Diffuse large B cell | 10 | NO | 15 | 45 |
| F4 | NHL-G | Diffuse large B cell | 10 | NO | — | 60 |
| F9 | NHL-G | Diffuse large B cell | 15 | NO | — | 70 |
| N32 | NHL-J | Burkitt's | 50 | NO | — | 80 |
| L985 | NHL-G | Diffuse large B cell | 10 | NO | — | 50 |
| N72 | NHL-H | Diffuse large B cell | 10 | NO | — | 70 |
| N614 | NHL-G | Diffuse large B cell | 10 | NO | — | 30 |
| N226 | NHL-Misc ALC (Ki-1+) | Anaplastic large cell | 10 | NO | — | 20 |
| N227 | NHL-Misc ALC (Ki-1+) | Anaplastic large cell | 10 | NO | — | ND |
| N169 | NHL-J | Burkitt's | 10 | NO | — | 80 |
| N318 | NHL-F | Diffuse large B cell | — | NO | — | 50 |
| N267 | NHL-Misc ALC (Ki-1+) | Diffuse large B cell | — | NO | 10 | 30 |
| N268 | NHL-Misc ALC (Ki-1+) | Diffuse large B cell | — | NO | 10 | 70 |
| N520 | NHL-G | Diffuse large B cell | — | NO | — | 80 |
| N503 | NHL-C | Follicular center (grade II) | — | NO | — | 50 |
| N613 | NHL-Misc MALT-type | Extranodal MZBC | — | NO | — | 20 |
| N319 | NHL-G | Diffuse large B cell | — | NO | — | 70 |
| F5 | NHL-G | Diffuse large B cell | — | NO | — | ND |
| F1 | NHL-Misc Monocytoid | Nodal MZBC | — | NO | 10 | 10 |
| F6 | NHL-B | Follicular center (grade I) | — | NO | — | 10 |
| F7 | NHL-B | Follicular center (grade I) | — | NO | — | ND |
| F33L | NHL-B | Follicular center (grade I) | — | NO | — | 10 |
| F16 | NHL-B | Follicular center (grade I) | — | ND | — | ND |
| F17 | NHL-G | Diffuse large B cell | — | ND | — | 60 |
| F18 | NHL-G | Diffuse large B cell | — | ND | — | 35 |
| F19 | NHL-D | Follicular center (grade III) | — | ND | — | 5 |
| F20 | NHL-B | Follicular center (grade I) | — | ND | — | ND |
| F21 | NHL-B | Follicular center (grade I) | — | ND | — | 15 |
| F22 | NHL-G | Diffuse large B cell | — | ND | — | 35 |
| F23 | NHL-Misc MALT-type | Extranodal MZBC | — | ND | — | 5 |
| F24 | NHL-Misc MALT-type | Extranodal MZBC | — | ND | — | 5 |
| F25 | NHL-Misc MALT-type | Extranodal MZBC | — | ND | — | 5 |
| F26 | NHL-Misc MALT-type | Extranodal MZBC | — | ND | — | 10 |
| F27 | NHL-H | Diffuse large B cell | 50 | ND | 20 | 70 |
| F28 | NHL-G | Diffuse large B cell | 10 | ND | 10 | 40 |
| F29 | NHL-G | Diffuse large B cell | 30 | ND | 10 | 45 |
| F30 | NHL-G | Diffuse large B cell | 10 | ND | — | 60 |
| F31 | NHL-G | Diffuse large B cell | 10 | ND | — | ND |
| F32 | NHL-G | Diffuse large B cell | 15 | ND | — | 55 |
| F34 | NHL-H | Diffuse large B cell | 20 | ND | — | 35 |
| F35 | NHL-G | Diffuse large B cell | 20 | ND | — | 20 |
| Case No. . | Histologic Diagnosis . | p53 . | p21 . | Ki-67 . | ||
|---|---|---|---|---|---|---|
| . | W.F. Classification . | REAL Classification . | IHC (%) . | Mutation . | IHC (%) . | IHC (%) . |
| N124 | NHL-J | Burkitt's | 65 | cod.286, G→A, Glu→Lys | — | 90 |
| N311 | NHL-J | Burkitt's | 20 | cod.132, A→C, Lys→Gln | — | 90 |
| N312 | NHL-J | Burkitt's | 80 | cod.130, C→G, Leu→Val | — | 90 |
| N313 | NHL-J | Burkitt's | 30 | cod.132, A→C, Lys→Gln | — | NE |
| N315 | NHL-J | Burkitt's | 80 | cod.273, G→T, Arg→Leu | 80 | 90 |
| N519 | NHL-J | Burkitt's | 90 | cod.236, T→C, Tyr→His | 90 | 90 |
| N523 | NHL-J | Burkitt's | 10 | cod.267, G→C, Arg→Pro | — | 70 |
| N521 | NHL-H | Diffuse large B cell | 50 | cod.244, G→A, Gly→Asp | — | 70 |
| L802 | NHL-G | Diffuse large B cell | 50 | cod.141, G→A, Cys→Tyr | — | 50 |
| N611 | NHL-C | Follicular center (grade II) | 20 | cod.161, G→A, Ala→Thr | — | 30 |
| N612 | NHL-C | Follicular center (grade II) | 60 | cod.248, G→A, Arg→Glu | — | 60 |
| L711 | NHL-A | Small lymphocytic | 40 | cod.193, A→G, His→Arg | — | 50 |
| L914 | NHL-A | Small lymphocytic | 60 | cod.258, G→A, Glu→Lys | — | 60 |
| N610 | NHL-A | Small lymphocytic | 60 | cod.275, G→A, Cys→Thr | — | 60 |
| F11 | NHL-H | Diffuse large B cell | 80 | cod.179, A→G, His→Arg | — | 70 |
| F12 | NHL-H | Diffuse large B cell | 80 | cod.282, C→T, Arg→Trp | — | 70 |
| F13 | NHL-G | Diffuse large B cell | 80 | cod.248, C→T, Arg→Trp | — | 60 |
| F15 | NHL-G | Diffuse large B cell | 80 | cod.285, G→A, Glu→Lys | — | 50 |
| N167 | NHL-Misc ALC (Ki-1+) | Diffuse large B cell | 30 | cod.175, G→A, Arg→His | — | 40 |
| N240 | NHL-Misc ALC (Ki-1+) | Anaplastic large cell | 30 | cod.176, T→A, Cys→Ser | 10 | 30 |
| VR | NHL-Misc Intestinal T-cell | Intestinal T cell | 80 | NO | 50 | 50 |
| F33H | NHL-G | Diffuse large B cell | 30 | NO | 40 | 40 |
| N609 | NHL-A | Small lymphocytic | 50 | NO | 10 | 50 |
| N522 | NHL-G | Diffuse large B cell | 60 | NO | 10 | 70 |
| N615 | NHL-G | Diffuse large B cell | 40 | NO | 10 | 80 |
| F8 | NHL-H | Diffuse large B cell | 20 | NO | 25 | 45 |
| F3 | NHL-F | Angioimmunoblastic T cell | 15 | NO | 10 | 25 |
| F14 | NHL-H | Diffuse large B cell | 20 | NO | 15 | 50 |
| F10 | NHL-G | Diffuse large B cell | 15 | NO | 15 | 30 |
| F2 | NHL-G | Diffuse large B cell | 10 | NO | 15 | 45 |
| F4 | NHL-G | Diffuse large B cell | 10 | NO | — | 60 |
| F9 | NHL-G | Diffuse large B cell | 15 | NO | — | 70 |
| N32 | NHL-J | Burkitt's | 50 | NO | — | 80 |
| L985 | NHL-G | Diffuse large B cell | 10 | NO | — | 50 |
| N72 | NHL-H | Diffuse large B cell | 10 | NO | — | 70 |
| N614 | NHL-G | Diffuse large B cell | 10 | NO | — | 30 |
| N226 | NHL-Misc ALC (Ki-1+) | Anaplastic large cell | 10 | NO | — | 20 |
| N227 | NHL-Misc ALC (Ki-1+) | Anaplastic large cell | 10 | NO | — | ND |
| N169 | NHL-J | Burkitt's | 10 | NO | — | 80 |
| N318 | NHL-F | Diffuse large B cell | — | NO | — | 50 |
| N267 | NHL-Misc ALC (Ki-1+) | Diffuse large B cell | — | NO | 10 | 30 |
| N268 | NHL-Misc ALC (Ki-1+) | Diffuse large B cell | — | NO | 10 | 70 |
| N520 | NHL-G | Diffuse large B cell | — | NO | — | 80 |
| N503 | NHL-C | Follicular center (grade II) | — | NO | — | 50 |
| N613 | NHL-Misc MALT-type | Extranodal MZBC | — | NO | — | 20 |
| N319 | NHL-G | Diffuse large B cell | — | NO | — | 70 |
| F5 | NHL-G | Diffuse large B cell | — | NO | — | ND |
| F1 | NHL-Misc Monocytoid | Nodal MZBC | — | NO | 10 | 10 |
| F6 | NHL-B | Follicular center (grade I) | — | NO | — | 10 |
| F7 | NHL-B | Follicular center (grade I) | — | NO | — | ND |
| F33L | NHL-B | Follicular center (grade I) | — | NO | — | 10 |
| F16 | NHL-B | Follicular center (grade I) | — | ND | — | ND |
| F17 | NHL-G | Diffuse large B cell | — | ND | — | 60 |
| F18 | NHL-G | Diffuse large B cell | — | ND | — | 35 |
| F19 | NHL-D | Follicular center (grade III) | — | ND | — | 5 |
| F20 | NHL-B | Follicular center (grade I) | — | ND | — | ND |
| F21 | NHL-B | Follicular center (grade I) | — | ND | — | 15 |
| F22 | NHL-G | Diffuse large B cell | — | ND | — | 35 |
| F23 | NHL-Misc MALT-type | Extranodal MZBC | — | ND | — | 5 |
| F24 | NHL-Misc MALT-type | Extranodal MZBC | — | ND | — | 5 |
| F25 | NHL-Misc MALT-type | Extranodal MZBC | — | ND | — | 5 |
| F26 | NHL-Misc MALT-type | Extranodal MZBC | — | ND | — | 10 |
| F27 | NHL-H | Diffuse large B cell | 50 | ND | 20 | 70 |
| F28 | NHL-G | Diffuse large B cell | 10 | ND | 10 | 40 |
| F29 | NHL-G | Diffuse large B cell | 30 | ND | 10 | 45 |
| F30 | NHL-G | Diffuse large B cell | 10 | ND | — | 60 |
| F31 | NHL-G | Diffuse large B cell | 10 | ND | — | ND |
| F32 | NHL-G | Diffuse large B cell | 15 | ND | — | 55 |
| F34 | NHL-H | Diffuse large B cell | 20 | ND | — | 35 |
| F35 | NHL-G | Diffuse large B cell | 20 | ND | — | 20 |
Abbreviations: ALC, anaplastic large cell; MZBC, marginal zone B-cell; −, negative; NO: no mutation; ND, not done; NE, not evaluable.
Immunohistochemical analysis (IHC).Immunohistochemical analysis, as well as result evaluation, was performed independently in two different immunopathology laboratories using the same approach.
Lymphoma samples were tested for p53 protein expression with the monoclonal antibody (MoAb) DO-7 (Novocastra Laboratories Ltd, Newcastle upon Tyne, UK; dilution 1:100), which specifically detects human p53 in routinely processed specimens, and with the MoAb EA10 (Oncogene Science, Manhasset, NY; dilution 1:100), which detects p21 accumulation. Furthermore, the samples were analyzed for Ki-67 expression, which indicates the proliferative activity of the tumor, with MIB1 (Immunotech, Marseille, France; dilution 1:100). Immunohistochemical analysis was performed on formalin or Bouin-fixed, paraffin-embedded serial sections pretreated in a microwave oven (Jet 900W: Philips, Eindhoven, Holland) twice for 5 minutes at 700 W. Immunostaining was performed incubating primary MoAbs overnight at 4°C and then using the avidin biotin peroxidase complex (ABC) method (ABC-Elite kit; Vector, Burlingame, CA).
For control purposes, some randomly selected cases were also immunostained in duplicate by the alkaline-phosphatase antialkaline phosphatase (APAAP) method in frozen tissue sections.
DNA extraction.DNA was purified from 51 cases (39 were p53-IHC–positive and 12 were p53-IHC–negative) by digestion with proteinase K-RNAse and extraction with phenol-chloroform, according to standard methods.14 Frozen material was available in 34 cases. Cases of series F1 to F35 were formalin fixed and paraffin embedded. For these cases, DNA extraction was preceded by a pretreatment consisting of deparaffinization in xylene followed by ethanol rinses and drying, as previously described.15
Hematoxylin and eosin sections were reviewed to confirm the presence of neoplastic cells, which was always greater than 70%.
Oligonucleotide primers.All the oligonucleotides used for polymerase chain reaction (PCR) amplifications were synthesized using an Applied Biosystem synthesizer (Foster City, CA). The oligonucleotides used to amplify the sequences of the p53 gene were the same as described previously.9
The open reading frame of the p21 gene was amplified using the following primers: exon 2, codons 1-89, sense: CATAGTGTCTAATCTCCGCCGT; antisense: CAACTCATCCCGGCCTCGC; exon 2, codons 74-148, sense: CCCAAGCTCTACCTTCCC; antisense: AGCCCTTGGACCATGGATTCTG; exon 3, codons 149-164, sense: GTCTTCCTCAGTTGGGCAGCTC; antisense: AGACACACAAACTGAGACTAAG.
Single-strand conformation polymorphism (SSCP) analysis.SSCP analysis was performed as described previously.9 Two percent formamide was included in the PCR reaction for the amplification of exon 2 (codons 1-89) of the p21 gene. PCR conditions consisted of 30 cycles of denaturation at 94°C for 1 minute, annealing at 50 to 60°C for 1 minute, and strand elongation at 72°C for 1 minute. After amplification, 1/20 of the reaction was mixed 1:1 with 95% formamide, 20 mmol/L EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol, heat denatured and loaded on 20 × 40 × 0.04 cm nondenaturing 0.5× MDE (Mutation Detection Enhancement; AT Biochem, Malvern, PA) 0.6× tris-borate/EDTA gels. Two sets of run conditions were performed: 10 V/cm for 15 hours at room temperature and 20 V/cm for 5 hours at room temperature with fan cooling. After electrophoresis, gels were vacuum dried and autoradiographed.
Sequencing.Sequence analysis was performed as described previously.16 Briefly, five pooled reactions (100 μL) were performed using the same primers as SSCP analysis. PCR products were then loaded on 2% preparative agarose gel and recovered by the QIAEX Gel Extraction Kit (QIAGEN, Chattsworth, CA). DNA sequencing was performed by the dideoxy chain termination method using a Sequenase kit (USB, Cleveland, OH). Both DNA strands were sequenced.
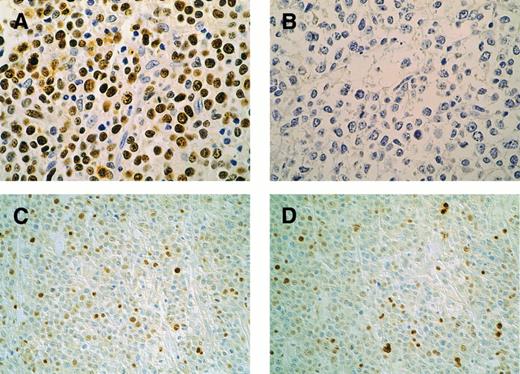
Patterns of p53 and p21 immunohistochemical expression in high-grade lymphomas. Case F12, p53-mutated case: p53 overexpression in the majority of neoplastic cells (A); coupled with absent p21 (B). Original magnification × 400. Case F8, p53-nonmutated case: p53 is present in 25% of lymphomatous cells (C); p21 expression in a similar percentage of cells (D). Original magnification × 200. DO7 (p53) and EA10 (p21) MoAb. Streptavidin-peroxidase method with DAB; light hematoxylin counterstain.
Patterns of p53 and p21 immunohistochemical expression in high-grade lymphomas. Case F12, p53-mutated case: p53 overexpression in the majority of neoplastic cells (A); coupled with absent p21 (B). Original magnification × 400. Case F8, p53-nonmutated case: p53 is present in 25% of lymphomatous cells (C); p21 expression in a similar percentage of cells (D). Original magnification × 200. DO7 (p53) and EA10 (p21) MoAb. Streptavidin-peroxidase method with DAB; light hematoxylin counterstain.
RESULTS
Immunohistochemical analysis.Seventy NHLs, representing most of NHL typologies (Table 1), were analyzed for p53 expression using the MoAb DO-7.
Forty-seven cases showed the presence of a strong nuclear immunoreactivity (68%), with a proportion of p53-positive cells ranging from 10% to 90% (Table 2). No case showed cytoplasmic positivity. The pattern of p53 protein accumulation varied from diffuse to scattered, with almost all of the p53-positive elements dispersed as single cells in a p53-negative context (cases showing 10% of p53-positivity).
Nineteen of the 70 cases analyzed showed the presence of p21-reactive nuclei (27%), with a percentage of reactive cells ranging between 10% and 90%. Sixteen of 19 cases were also p53-IHC-positive, indicating a strict correlation between p53 accumulation and p21 overexpression.
The analysis of serial sections showed that the pattern of p21 immunoreactivity almost overlapped the pattern of p53 accumulation, the p21-positive population being comprised in the p53-positive one (Fig 1).
The expression of Ki-67 (MIB1), a nuclear protein related to cell proliferation, was used to verify the proliferative activity of the tumor mass. Serial section analysis allowed us to verify the relationship between proliferative activity and p53/p21 immunoreactivity. The percentage of Ki-67–reactive cells ranged from 5% to 90%. The p53-positive cell population was included in the Ki-67–reactive one, indicating that p53-positive cells are actively proliferating elements.
Molecular analysis.Molecular analysis was performed to verify the relationship between accumulation of p53/p21 proteins and status of the respective genes.
Molecular analysis was performed on 51 cases (39 were p53 IHC-positive and 12 were p53 IHC-negative) using SSCP analysis and PCR direct sequencing (Fig 2).
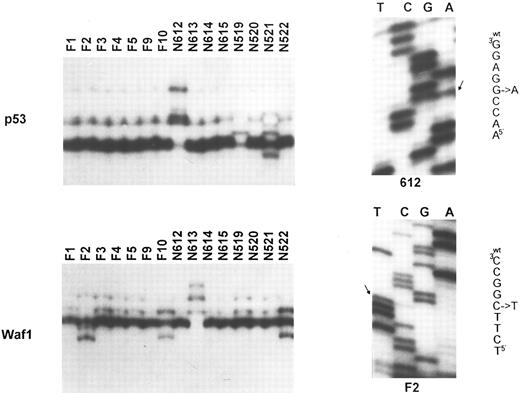
Molecular analysis of the p53 and p21 genes. Top (left): SSCP analysis of the p53 gene (exon 7). Tumor identification numbers are given on the top of each lane. Cases N612, N519, and N521 show an abnormal migration pattern; (right) sequence analysis of case N612, showing the presence of a G to A transition at codon 248. Bottom (left): SSCP analysis of the p21 gene (exon 2). Cases F2, F10, N613, and N522 show a band shift. This was due to a C to T polymorphism at codon 22 for case F2 (sequence analysis on the right), a C to A polymorphism at codon 31 for cases F10 and N522, and to a C to T somatic mutation at codon 20 for case N613.
Molecular analysis of the p53 and p21 genes. Top (left): SSCP analysis of the p53 gene (exon 7). Tumor identification numbers are given on the top of each lane. Cases N612, N519, and N521 show an abnormal migration pattern; (right) sequence analysis of case N612, showing the presence of a G to A transition at codon 248. Bottom (left): SSCP analysis of the p21 gene (exon 2). Cases F2, F10, N613, and N522 show a band shift. This was due to a C to T polymorphism at codon 22 for case F2 (sequence analysis on the right), a C to A polymorphism at codon 31 for cases F10 and N522, and to a C to T somatic mutation at codon 20 for case N613.
Twenty of the 39 p53-IHC–positive cases (51%) analyzed by SSCP of p53 exons 4 to 9 showed the presence of p53 gene mutations that accounted for p53 protein accumulation. The other 19 cases, despite the presence of p53-reactive cells (10% to 80% of the neoplastic population), retained p53 in the wild-type status, as shown by SSCP analysis and as confirmed by the sequencing of the entire coding region in some cases (cases VR, F33H, N32, N609, N522, N615) (Table 2).
Only one case of 51 showed the presence of a p21 somatic gene mutation (case N613, C to T transition at codon 20, Arg to Ser); however, this case did not overexpress the p53 or p21 protein. The presence of p21 gene allelic variants, which has been suggested to correlate with differential biochemical properties of the p21 protein in Burkitt's lymphomas,17 was detected in some cases. In particular, a C to T transition at codon 22 was observed in case F2. Cases F10, VR, and N522, showed a C to A transversion at codon 31 and case L914 showed a A to T transversion at codon 91. In addition, a C to G allelic variant was observed in many cases at the 17th nucleotide of intron 2 (allelic frequencies: pC = 0.69, qG = 0.31). However, no preferential distribution of these alleles was evidenced with respect to p53 gene status or p21 accumulation (Fig 2).
Comparative analysis.To verify whether the accumulation of wt-p53 was associated with the transcriptional induction of the p21 gene, immunohistochemical and molecular data were compared (Table 2). p21 accumulation was mostly detected in p53 overexpressing cases (16 of 19 cases, 85%). Molecular analysis showed that in over 80% of the cases (13 of 16), the accumulation of p21 was associated with the overexpression of wt-p53, indicating a significative correlation between accumulation of wt-p53 and overexpression of p21 (χ2 test, P < .05). The association was particularly evident when considering the cases overexpressing wt-p53 in a large fraction of neoplastic cells: all but one of the cases with a number of p53-reactive nuclei greater than 20% showed the accumulation of p21 protein, as well. The strict association between wt-p53 accumulation and p21 induction was also confirmed by serial section analysis, which indicated that the p21-positive population was a fraction of the p53-positive one. This analysis also showed that p53-positive elements, and consequently the p21-positive ones, were almost confined to the actively proliferating cell population, as showed by the overlapping reactivity for anti-p53 antibody and MIB1 antibody.
DISCUSSION
The detection of a large amount of an apparently normal p53 protein in NHL cells prompted us to verify the biological significance of such overexpression. We, therefore, investigated the functionality of the wt-p53 protein in NHL by analyzing p21 induction as an indicator of the p53 ability to act as a transcriptional regulator.
p21 expression was detected in about 27% (19 of 70) of the NHLs analyzed. p21 accumulation prevailed in p53-overexpressing cases and significantly correlated with the accumulation of the wild-type p53 protein. Even if recent observations indicate the existence of p53-independent mechanisms of p21 induction18,19 (these mechanisms seem to be true for cases N315 and N519, which carry a homozygous mutation at p53 gene, but still express p21), the fact that p21 accumulates almost exclusively in the NHLs overexpressing wt-p53, with a pattern of immunoreactivity overlapping or included in that of p53, suggests that the wt-p53 overexpressed in most of these cases is actually a functional transcriptional regulator responsible for the induction of p21. This is consistent with our previous observations about a correlation between wt-p53 overexpression and accumulation of the MDM2 gene, which is also a transcriptional target of p53.9 Taken together, these results support the notion that a large fraction of NHLs overexpress a wild-type form of p53, which conserves its ability to function as a transcriptional regulator and to potentially evoke G1 arrest.
The fact that overexpression of wt-p53 and accumulation of the p21 CDK-inhibitor occur mainly in intermediate-grade and high-grade NHLs, which show proliferative indices significantly higher than low-grade NHLs, together with the observation of actively proliferative elements in the p21-positive population, suggests that the presumptive p21-dependent G1 arrest is somehow overridden in these tumors.
The analysis of the p21 gene status allowed us to exclude the presence of structural abnormalities that may account for the noninduction of G1 arrest, indicating that p21 is a potentially functional CDK inhibitor in these tumors. One possible explanation of this phenomenon is that p21 induction levels in NHLs are insufficient to inhibit CDK activity. Recent data indicate the existence of a dose-dependent effect with regard to the ability of p21 to inhibit cyclin-CDK complexes: at least two molecules of p21 are required to completely abolish the growth-promoting role of the CDK complexes.20 It is, therefore, possible that, even if induced by the overexpressed wt-p53, the quantity of p21 produced in NHL cells is under the level required to promote G1 arrest. Alternatively, the induction of p21 in NHL could vanish because of the presence of alterations in some p21 targets. Alterations in a CDK-inhibitor partner have been recently described in some cases of familial and sporadic melanoma. In these cases, the p16 CDK-inhibitor is normally induced, but its interaction with the cyclin-CDK4 complex is prevented by a structural modification of the kinase, as a consequence of CDK4 gene mutation.21,22 It is, therefore, possible that an equivalent phenomenon, involving one of the p21 partners, may occur in NHL. Finally, it cannot be excluded that the p53-dependent G1 arrest pathway is normally conserved and evoked in NHL cells, but the presence of other abnormalities may bypass the effect of this induction. In this regard, the existence of a heat-labile inhibitor of p21 which, if overexpressed, is able to abrogate the p53-induced cell cycle arrest in mouse fibroblast has recently been described.23 Further studies, specifically focused on these topics, will shed light on the molecular bases of wt-p53 overexpression in NHLs and evaluate the role of p53-pathway abnormalities in the development and progression of NHL.
A.G., C.D., and S.P. contributed equally to this study.
Supported in part by a grant from the Italian Association for Cancer Research, Milan.
Address reprint requests to Mauro Boiocchi, PhD, Division of Experimental Oncology 1, Centro di Riferimento Oncologico, via Pedemontana Occidentale 12, 33081 Aviano (PN) Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal