Abstract
The Lutheran (Lu) blood group antigens and the B-cell adhesion molecule (B-CAM) epithelial cancer antigen are carried by recently cloned integral glycoproteins that belong to the Ig superfamily. We have previously shown that the Lu and B-CAM antigens are encoded by the same gene, LU, and that alternative splicing of the primary transcript most likely accounts for the presence of both antigens on two isoforms that differ by the length of their cytoplasmic tails. In the present report, we isolated the human LU gene by cloning a 20-kb HindIII fragment from Lu(a − b+) genomic DNA. The LU gene is organized into 15 exons distributed over 12.5 kb. Alternative splicing of intron 13 generates the 2.5- and 4.0-kb transcript spliceoforms encoding the long tail and the short tail Lu polypeptides, respectively. Sequencing of the major mRNA species (2.5 kb) amplified from human bone marrow, kidney, placenta, and skeletal muscle did not suggest the presence of tissue-specific Lu glycoprotein isoforms. The same transcription initiation point, located 22 bp upstream from the initiation codon, was characterized in several tissues. In agreement with the wide tissue distribution of the Lu messengers, the GC-rich proximal 5′ flanking region of the LU gene does not contain TATA or CAAT boxes, but includes several potential binding sites for the ubiquitous Sp1 transcription factor. In addition, the distal 5′ region, encompassing nucleotides −673 to −764, contains clustered binding sequences for the GATA, CACCC, and Ets transcription factors. Analysis of the coding sequences amplified from genomic DNA of Lu(a + b−) or Lu(a − b+) donors showed a single nucleotide change in exon 3 (A229G) that correlates with an Aci I restriction site polymorphism and results in a His77Arg amino-acid substitution. Polymerase chain reaction/restriction fragment length polymorphism analysis indicated that the A229G mutation is associated with the Lua/Lub blood group polymorphism. When expressed in Chinese hamster ovary (CHO) cells, Lu cDNAs carrying the A229 or the G229 produced cell surface proteins that reacted with anti-Lua or anti-Lub antibodies, respectively, showing that these nucleotides specify the Lua and Lub alleles of the Lutheran blood group locus. CHO cells expressing recombinant short-tail or long-tail Lu glycoproteins reacted as well with anti-Lu as with anti–B-CAM antibodies, providing the definitive proof that the Lu blood group and B-CAM antigens are carried by the same molecules.
THE LUTHERAN blood group antigens were originally represented by two allelic antigens, Lua and Lub,1,2 with a frequency of 5% and 95%, respectively,3 carried by two membrane glycoproteins (gps) of 85 kD (major species) and 78 kD (minor species).4,5 The 85-kD Lu gp has been recently cloned from human placenta and was shown to represent a new member of the Ig superfamily, with extracellular parts consisting of two variable (V) region and three constant region (C2 set) domains.6
Comparison with nucleotide databases indicated that the 85-kD Lu gp is virtually identical to the B-cell adhesion molecule (B-CAM) epithelial cancer antigen cloned from the colon cancer HT29 cell line.7 The gp encoded by the HT29 clone differed from that encoded by the placental cDNA only by the lack of the last 40 amino acids of the cytoplasmic tail, which carries a consensus binding site for SH3 motifs.6
Independent studies indicated that the Lu and B-CAM antigens are expressed in a broad range of human cells and tissues.6,7 Furtheremore, the B-CAM antigen identified by monoclonal antibodies (MoAbs) raised against human tumor cells was shown to be overexpressed in ovarian carcinomas in vivo and upregulated after malignant transformation in some cell types,8-10 whereas immunostaining of human tissues with anti-Lu MoAbs indicated that the Lu antigens are under developmental control in liver.6 However, the function of these gps has not been yet identified, but homology with MUC18 (31%), a human metastatic melanoma cell surface protein that probably contributes to metastasis,7,11 as well as studies with neuroblastoma cell lines that can grow in suspension culture or as substrate-adherent monolayers,9 suggested a possible role both in cell-cell and cell-substrate binding events.8
We previously showed that the gps isolated from placenta and the HT29 cell line represent isoforms of the same protein encoded by two mRNA spliceoforms of a unique gene, LU, on chromosome 19q13.2-13.3. The presence or the absence of a 977-bp sequence, alternatively spliced from the 3′ end of the LU gene, resulted in a stop codon at the first triplet after the divergence site between the two mRNA species, or to an open reading frame leading to a 40-amino acid longer polypeptide, respectively.
The 2.5-kb transcript encoding the long-tail Lu gp is highly expressed when compared with the 4.0-kb transcript encoding the short-tail isoform in various tissues and cells except in the colon carcinoma cell line HT29. This result suggested that differential regulation of the alternative splicing of the Lu primary transcript could be associated with malignant transformation.12
In the present study, the complete characterization of the LU gene, including its promoter region and the molecular basis of the Lua/Lub polymorphism, are reported. In addition, we provide the definitive proof that the Lu and B-CAM antigens are carried by the same gps.
MATERIALS AND METHODS
Genomic library.A restricted genomic library enriched with the 20-kb HindIII genomic restriction fragment was constructed from 300 μg of leukocyte DNA from one Lu(a − b+) donor as described13 except that the DNA was digested with HindIII restriction enzyme and ligated with HindIII arms of λ DASHII (Stratagene, La Jolla, CA).
Intron size determination and sequencing of intron/exon junctions.Intron sizes were characterized by polymerase chain reaction (PCR) using exon-specific oligonucleotide primers (Table 1). PCRs were performed on DNA isolated from positive clones using the Expand Long template PCR System (Boehringer Mannheim, Mannheim, Germany) with the Taq/Pwo mix to avoid errors caused by the Taq polymerase and to amplify potential large fragments in the following conditions: 10 cycles at 94°C (10 seconds), 60°C (30 seconds), and 68°C (3 minutes); 20 cycles at 94°C (10 seconds), 60°C (30 seconds), and 68°C (3 minutes plus 20 seconds extension at each cycle). The PCR products were then analyzed by hybridization with a Lu cDNA probe or internal oligonucleotide probes. DNA fragments up to 3 kb were subcloned into pUC 18 vector and sequenced with Taq polymerase kit (Pharmacia, Uppsala, Sweden) using an automatic sequencer (Alf-Express; Pharmacia); large PCR fragments were directly sequenced.
Sequence and Position of the Exon-Specific Primers Used to Determine the Structure of the LU Gene
| Primers . | Position* (bp) . | Introns Encompassed . |
|---|---|---|
| Lu49: 5′ ATATATACGGATCCGTGAACATGGAGCCCCCGGACGCACCGG 3′ | (−)20-22 | 1 to 5 |
| Lu28: 5′ TGCTGGTGAGGGAGAGCAGGCC 3′ | 661-640 | |
| Lu13: 5′ AGGTGCCCGTAGAGATGAACCC 3′ | 578-599 | 5, 6 |
| Lu22: 5′ TGGACAGTGTCACCCTCGCG 3′ | 865-846 | |
| Lu33: 5′ AGTTCTGGGTGGGCAGCCC 3′ | 803-821 | 7 |
| Lu57: 5′ ATAGGTCCCGCTCTGGCCCCGGGTCACTCC 3′ | 1,005-976 | |
| Lu16: 5′ AGGGTGACACTGTCCAGCTGC 3′ | 848-868 | 7 to 14 |
| Lu48: 5′ AGGGACAGCCTCTAGGAGGTTCTT 3′ | 1,914-1,891 | |
| Lu17: 5′ TGAACTGCTCCGTGCACGGCC 3′ | 1,145-1,165 | 9 to 11 |
| Lu11: 5′ AGTTTGGGGTCTGGATGGCC 3′ | 1,448-1,429 | |
| Lu12: 5′ AAGGCAGATGGCAGCTGGA 3′ | 1,372-1,390 | 11, 12 |
| Lu42: 5′ AGGAGGCCCACGCTGACG 3′ | 1,682-1,665 | |
| Lu23: 5′ TGGCCGTCAGCGTGGGCCT 3′ | 1,655-1,673 | 13, 14 |
| Lu48: 5′ AGGGACAGCCTCTAGGTTCTT 3′ | 1,914-1,891 | |
| Lu36: 5′ AAGAACCTCCTAGAGGCTGTCCC 3′ | 1,891-1,913 | |
| Lu60: 5′ ATTTATTCCAGACTCCAGTGTCCACAGATGATGGGGTGGG 3′ | 2,365-2,326 |
| Primers . | Position* (bp) . | Introns Encompassed . |
|---|---|---|
| Lu49: 5′ ATATATACGGATCCGTGAACATGGAGCCCCCGGACGCACCGG 3′ | (−)20-22 | 1 to 5 |
| Lu28: 5′ TGCTGGTGAGGGAGAGCAGGCC 3′ | 661-640 | |
| Lu13: 5′ AGGTGCCCGTAGAGATGAACCC 3′ | 578-599 | 5, 6 |
| Lu22: 5′ TGGACAGTGTCACCCTCGCG 3′ | 865-846 | |
| Lu33: 5′ AGTTCTGGGTGGGCAGCCC 3′ | 803-821 | 7 |
| Lu57: 5′ ATAGGTCCCGCTCTGGCCCCGGGTCACTCC 3′ | 1,005-976 | |
| Lu16: 5′ AGGGTGACACTGTCCAGCTGC 3′ | 848-868 | 7 to 14 |
| Lu48: 5′ AGGGACAGCCTCTAGGAGGTTCTT 3′ | 1,914-1,891 | |
| Lu17: 5′ TGAACTGCTCCGTGCACGGCC 3′ | 1,145-1,165 | 9 to 11 |
| Lu11: 5′ AGTTTGGGGTCTGGATGGCC 3′ | 1,448-1,429 | |
| Lu12: 5′ AAGGCAGATGGCAGCTGGA 3′ | 1,372-1,390 | 11, 12 |
| Lu42: 5′ AGGAGGCCCACGCTGACG 3′ | 1,682-1,665 | |
| Lu23: 5′ TGGCCGTCAGCGTGGGCCT 3′ | 1,655-1,673 | 13, 14 |
| Lu48: 5′ AGGGACAGCCTCTAGGTTCTT 3′ | 1,914-1,891 | |
| Lu36: 5′ AAGAACCTCCTAGAGGCTGTCCC 3′ | 1,891-1,913 | |
| Lu60: 5′ ATTTATTCCAGACTCCAGTGTCCACAGATGATGGGGTGGG 3′ | 2,365-2,326 |
Numbers indicate positions on the Lu cDNA sequence. +1 taken as the first nucleotide of the initiation codon.
5′ mapping of mRNA transcripts.Primer extensions were performed as described14 with 0.5 μg of poly(A+) RNA (Clontech, Palo Alto, CA) from bone marrow, fetal liver, placenta, skeletal muscle and brain as templates, and 32[P] labeled oligonucleotide Lu25 (nt 126-150).
5′ RACE-PCR was performed with marathon cDNAs (Clontech) from the same tissues used in primer extension experiments. PCR reactions were performed between the specific primer Lu25 (nt 126-150) and the anchor oligonucleotide as reverse primer. The PCR products were analyzed by agarose gel electrophoresis and hybridization with an internal oligonucleotide probe. The hybridizing band was sequenced after subcloning in PCR2 vector using the T-A cloning procedure (In Vitrogen Corp, San Diego, CA).
Amplification of Lua exons and restriction fragment length polymorphism (RFLP) analysis.Overlapping PCR amplifications were performed on genomic DNA of an Lu (a + b−) individual with exon-specific oligonucleotide primers and sequenced as described previously for the Lub gene. For the Lua/Lub polymorphism, PCR was performed with 200 ng of leukocyte DNA between primers Lu91 (nt 205-227) and Lu92 (nt 290-273) under the following conditions: 30 cycles of 10 seconds at 94°C, 20 seconds at 60°C, and 30 seconds at 72°C with 2.5 U of Taq polymerase (Perkin Elmer, Norwalk, CT). PCR products were purified on Microcon (Amicon, Danvers, MA), and half were digested for 2 hours with 10 U of Aci I. Restriction fragments were directly analyzed in 15% acrylamide minigels.
Expression of Lua and Lub cDNAs.Lub cDNA was isolated by amplification with kidney cDNAs (Clontech) as template and primers Lu49 and Lu60 surrounding the coding sequence of the Lu cDNA (Table 1). PCR products of the expected size were subcloned into pcDNA3 expression vector (In Vitrogen Corp). The Lua and B-CAM cDNAs were constructed by in vitro mutagenesis (Quick-change site-directed mutagenesis kit; Stratagene) from pcDNA3 Lub double-strand recombinant DNA according to the supplier. Primers used were Lu96 (sens primer, nt 217-243; 5′GGAGCTCGCCCCCACCTAGCCTCGGCT3′) and Lu97 (antisens primer, nt 243-217) for Lua cDNA and Lu98 (sens primer, nt 1751-1778; 5′GAAGGGGGCTCCGTAGCCAGGGGAGCCA3′) and Lu99 (antisens primer, nt 1778-1751) for B-CAM cDNA. Briefly, complementary primers (22 nmol/L) and 50 ng of pcDNA3 Lub cDNA template were used in PCR reactions under the following conditions: 12 cycles of denaturation for 30 seconds at 95°C and primer annealing and extension at 68°C for 15 minutes.
Transient expression of Lu in Chinese hamster ovary (CHO) cells was performed by transfecting the cells with 50 μg of pcDNA3-Lua, pcDNA3-Lub, or pcDNA3-B-CAM vectors using the lipofectine procedure (GIBCO-BRL, Gaithersburg, MD). After 48 hours, transfected CHO cells were detached from culture dishes by incubation in phosphate-buffered saline (PBS) buffer supplemented with 0.2 g/L ethylenediamine tetraacetate (EDTA) and expression of Lu antigens was tested by staining 5 × 105 cells with Lua human antisera (Biotest, Dreieich, Germany), LM342 monoclonal anti-Lub (Scotland Blood Transfusion Service, Glasgow, Scotland), and G253 monoclonal anti–B-CAM.15 After washing, the cells were incubated with fluorescein-conjugated antimouse and antihuman IgG (H + L) (Immunotech, Marseille, France). Propidium iodide-negative cells (living cells) were selected and analyzed on a FACScan flow cytometer (Becton Dickinson, San Jose, CA), as described.16
RESULTS
Cloning and structure of the LU gene.Previous genomic Southern blot analysis performed with a cDNA probe common to the Lu and B-CAM cDNAs detected a HindIII RFLP associated with the Lua/Lub blood group polymorphism and indicated that the single copy LU gene was entirely carried by 20-kb or 23-kb HindIII fragments in Lu(a − b+) or Lu(a + b−) individuals, respectively.12 From these results, a partial genomic library in λ DASHII was prepared from Lu(a − b+) leukocyte DNA preparation enriched with the 20-kb HindIII genomic restriction fragments (see Materials and Methods). Among 3 × 105 plaques, 2 positive clones ( λ Lub1.2 and λ Lub3.1) were further analyzed to characterize the exon/intron organization of the LU gene. Restriction maps were established after digestion with BamHI, EcoRI, and Xho I restriction enzymes followed by hybridization of the restriction fragments with the total Lu cDNA probe and oligonucleotides specific of the 5′ and 3′ ends of the cDNA probe (Fig 1A). Intron lengths and locations were determined by comparing the sizes and partial sequences of eight overlapping PCR fragments, covering the entire transcribed sequence, and obtained with λ Lu or cDNA templates between primer sets deduced from the Lu cDNA sequence (Table 1). These analyses indicated that the LU gene is composed of 15 exons ranging in size from 71 bp to 498 bp and distributed over 12.5 kb (Table 2). All intron/exon junction sequences fitted with the gt-ag rule, and the extended sequences around the splice donor and acceptor sites were consistent with the consensus sequences established for mammalian genes (Table 2).17 These results indicated that the presence or absence of a 977-bp sequence, previously shown to generate the 4.0- and 2.5-kb Lu transcripts, respectively,12 corresponded to alternative splicing of intron 13 of the LU gene (Fig 1B).
Restriction map, organization and transcripts of the LU gene. (A) Restriction map and organization of the LU gene. The organization of the LU gene was established from analysis of the λ Lub1.2 and λ Lub3.1 genomic clones. Exons 1 to 15 are numbered and represented by open boxes. The hatched box indicates the alternative spliced intron and the interrupted black boxes represent the two l DASH arms. (B) Schematic representation of the alternative splicing of intron 13. Alternative splicing of intron 13 results in the synthesis of two transcripts of 2.5 kb and 4.0 kb encoding the long-tail (85 kD) and the short-tail (78 kD) Lu polypeptides, respectively. The exons are represented by open boxes. The hatched box indicates the alternative spliced intron.
Restriction map, organization and transcripts of the LU gene. (A) Restriction map and organization of the LU gene. The organization of the LU gene was established from analysis of the λ Lub1.2 and λ Lub3.1 genomic clones. Exons 1 to 15 are numbered and represented by open boxes. The hatched box indicates the alternative spliced intron and the interrupted black boxes represent the two l DASH arms. (B) Schematic representation of the alternative splicing of intron 13. Alternative splicing of intron 13 results in the synthesis of two transcripts of 2.5 kb and 4.0 kb encoding the long-tail (85 kD) and the short-tail (78 kD) Lu polypeptides, respectively. The exons are represented by open boxes. The hatched box indicates the alternative spliced intron.
Exon and Intron Organization of the LU Gene
| Exon . | Nucleotide Number* . | Amino Acid . | Exon Size (bp) . | Splice Acceptor Site Splice Donor Site . | Intron Size (bp) . | ||
|---|---|---|---|---|---|---|---|
| 1 | −23-82 | 1-27 | 105 | CCCAG/gtatggctcgggctg | >2,000 | ||
| 2 | 83-204 | 28-68 | 122 | gttgctctcttgcag/ATGCC | .... | TCCTT/gtgagtgcttggggc | ∼500 |
| 3 | 205-433 | 69-144 | 229 | gccccgcgcccacag/ACCGA | .... | GTTTG/gtaagtgtcctcggg | 85 |
| 4 | 434-504 | 145-168 | 71 | acgtctttttcacag/CAAAG | .... | AGGAG/gtacctctcgggtgg | ∼650 |
| 5 | 505-601 | 169-200 | 97 | ccgtgtctgcctcag/ATCGC | .... | CCCAG/gtgagcagcgcagga | 91 |
| 6 | 602-784 | 201-261 | 183 | cccgatctctcccag/AGGGC | .... | GCACT/gttgagtcttctggc | ∼500 |
| 7 | 785-921 | 262-307 | 137 | ctgcccttcccttag/ATCCC | .... | TTCAG/gtgacccacccaagg | 315 |
| 8 | 922-1,078 | 308-359 | 157 | tcccccgtctcccag/GATGA | .... | GGCCT/gtgagagccctgggt | ∼3,500 |
| 9 | 1,079-1,194 | 360-398 | 116 | tgtcccccactgcag/ATCTG | .... | CCAAG/gtgagagggagagga | 103 |
| 10 | 1,195-1,336 | 399-445 | 142 | ctctgcccttcccag/GACTC | .... | CCAAG/gttcagggggcaggg | 173 |
| 11 | 1,337-1,473 | 446-491 | 137 | ccccaccacctacag/GCTCG | .... | GCAGC/gtaagggaccttcct | 153 |
| 12 | 1,474-1,618 | 492-539 | 145 | tcggagcctccatag/CCCGC | .... | CGCCG/gtgagtgactgaggt | 91 |
| 13 | 1,619-1,763 | 540-588 | 145 | atcctgtccctgcag/TGAGC | .... | GCTCC/gtgagtggcctgcta | 977 |
| 14 | 1,764-1,881 | 589-627 | 118 | gcctcctccccccag/GCCGC | .... | ACGAG/gtgggtgagggcctg | 93 |
| 15 | 1,882-2,380 | 628 | 498 | ccatccgctccccag/TGCTG | .... |
| Exon . | Nucleotide Number* . | Amino Acid . | Exon Size (bp) . | Splice Acceptor Site Splice Donor Site . | Intron Size (bp) . | ||
|---|---|---|---|---|---|---|---|
| 1 | −23-82 | 1-27 | 105 | CCCAG/gtatggctcgggctg | >2,000 | ||
| 2 | 83-204 | 28-68 | 122 | gttgctctcttgcag/ATGCC | .... | TCCTT/gtgagtgcttggggc | ∼500 |
| 3 | 205-433 | 69-144 | 229 | gccccgcgcccacag/ACCGA | .... | GTTTG/gtaagtgtcctcggg | 85 |
| 4 | 434-504 | 145-168 | 71 | acgtctttttcacag/CAAAG | .... | AGGAG/gtacctctcgggtgg | ∼650 |
| 5 | 505-601 | 169-200 | 97 | ccgtgtctgcctcag/ATCGC | .... | CCCAG/gtgagcagcgcagga | 91 |
| 6 | 602-784 | 201-261 | 183 | cccgatctctcccag/AGGGC | .... | GCACT/gttgagtcttctggc | ∼500 |
| 7 | 785-921 | 262-307 | 137 | ctgcccttcccttag/ATCCC | .... | TTCAG/gtgacccacccaagg | 315 |
| 8 | 922-1,078 | 308-359 | 157 | tcccccgtctcccag/GATGA | .... | GGCCT/gtgagagccctgggt | ∼3,500 |
| 9 | 1,079-1,194 | 360-398 | 116 | tgtcccccactgcag/ATCTG | .... | CCAAG/gtgagagggagagga | 103 |
| 10 | 1,195-1,336 | 399-445 | 142 | ctctgcccttcccag/GACTC | .... | CCAAG/gttcagggggcaggg | 173 |
| 11 | 1,337-1,473 | 446-491 | 137 | ccccaccacctacag/GCTCG | .... | GCAGC/gtaagggaccttcct | 153 |
| 12 | 1,474-1,618 | 492-539 | 145 | tcggagcctccatag/CCCGC | .... | CGCCG/gtgagtgactgaggt | 91 |
| 13 | 1,619-1,763 | 540-588 | 145 | atcctgtccctgcag/TGAGC | .... | GCTCC/gtgagtggcctgcta | 977 |
| 14 | 1,764-1,881 | 589-627 | 118 | gcctcctccccccag/GCCGC | .... | ACGAG/gtgggtgagggcctg | 93 |
| 15 | 1,882-2,380 | 628 | 498 | ccatccgctccccag/TGCTG | .... |
Intron and exon sequences are shown in lowercase and uppercase, respectively. The 5′ donor gt and the 3′ acceptor ag are underlined.
+1 taken as the first nucleotide of the initiation codon in the cDNA sequence.
The major Lu mRNAs species (2.5 kb) amplified from human bone marrow, kidney, placenta, and skelatal muscle exhibited identical sequences and, thus, did not show tissue-specific spliceoforms (not shown).
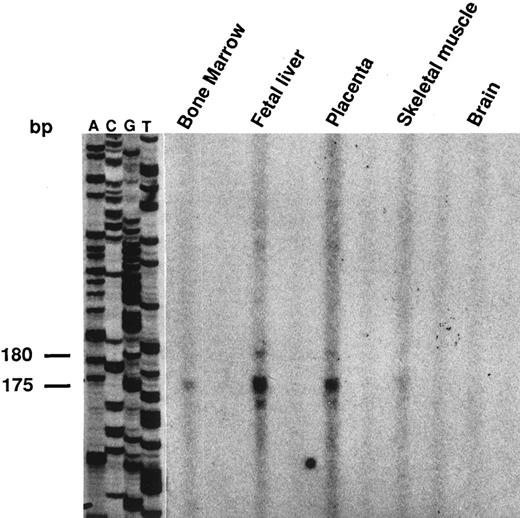
Structure of the LU 5′ flanking sequence.To determine the transcriptional initiation site(s) of the LU gene in different tissues, primer extension experiments were performed using the Lu25 oligonucleotide primer (nt 126-150) located 3′ of the initiator AUG and mRNA templates from human bone marrow, fetal liver, placenta, skeletal muscle, and brain. The same major product of 175/180 nt was identified in all tissues except in brain (Fig 2) in agreement with the Northern blot analysis, which indicated a very low expression of the Lu messengers in this tissue.12 To confirm these results, the 5′ ends of the Lu transcripts were amplified by the 5′ RACE-PCR procedure. Using the anchor and Lu25 primers, PCR products of about 180 bp were detected in all tissues after hybridization with a 5′ internal oligonucleotide probe, subcloned, and sequenced. Both experiments located the cap site for Lu mRNAs at position-22 upstream from the translation initiation codon. These results were consistent with preliminary analysis of the 5′ end of the placental Lu mRNA previously reported.6
Determination of the transcription initiation site by primer extension analysis in erythroid and nonerythroid cells. Primer extension was performed as described in Materials and Methods. A same primer-extended product of 175/180 bp was detected with all the poly(A+) RNAs, with a stronger intensity in fetal liver and placenta than in the other tissues. These products were clearly detected in bone marrow and skeletal muscle with an overexposure of the autoradiogram.
Determination of the transcription initiation site by primer extension analysis in erythroid and nonerythroid cells. Primer extension was performed as described in Materials and Methods. A same primer-extended product of 175/180 bp was detected with all the poly(A+) RNAs, with a stronger intensity in fetal liver and placenta than in the other tissues. These products were clearly detected in bone marrow and skeletal muscle with an overexposure of the autoradiogram.
A total of 1.1 kb of the 5′ flanking sequence of the LU gene, present in the λLu recombinant phages, were sequenced to identify putative cis-acting regulatory elements by comparison with a compilation of the binding sequences for vertebrate-encoded nuclear proteins18 (Fig 3). The proximal 5′ region is GC-rich (80% GC in the 200 bp upstream from the cap site) and contains three consensus Sp1 binding sites at positions −47/−55, −61/−69, and −66/−74 5′ of the cap site. Sequence deviating by one substitution from a consensus EGR-1 motif and one perfect AP2 binding sequence were identified at positions −104/−112 and −314/−321, respectively. No typical TATA and CAAT boxes were found. In addition, the distal 5′ flanking region (upstream nt −200) contains one binding sequence for the transcription factors of the GATA family (position −733/−742) surrounded by two CACCC sequences (positions −673/−681 and −758/−764) and one Ets binding site (position −749/−755).
Nucleotide sequence of the LU 5′ flanking region showing the putative regulatory cis-acting elements. +1 refers to the transcription initiation site. The first 40 nucleotides of exon 1 are in upper case and the initiation codon ATG is underlined. The boxes correspond to putative cis-acting regulatory elements (Sp1, EGR-1, AP2, GATA, CACCC, and Ets). Mismatches with consensus motifs are indicated by asterisks.
Nucleotide sequence of the LU 5′ flanking region showing the putative regulatory cis-acting elements. +1 refers to the transcription initiation site. The first 40 nucleotides of exon 1 are in upper case and the initiation codon ATG is underlined. The boxes correspond to putative cis-acting regulatory elements (Sp1, EGR-1, AP2, GATA, CACCC, and Ets). Mismatches with consensus motifs are indicated by asterisks.
Sequence analysis of the Lua allele.Using information on the exon/intron organization of the Lub allele described above, PCR fragments carrying all exons and splice junctions of the Lua allele were amplified from the genomic DNA of a homozygous Lu(a + b−) donor. When compared with the overall sequence of the Lub allele, the Lua allele exhibited only one nucleotide subtitution at position +229 (+1 taken as the first nucleotide of the initiation codon in the cDNA sequence) corresponding to a G→A base exchange resulting in the amino acid substitution Arg77His (Fig 4A). Polymorphism at this position carried by exon 3 of the LU gene was confirmed after amplification and sequencing of this exon in two other unrelated homozygous Lu(a + b−) and two unrelated homozygous Lu(a − b+) genomes.
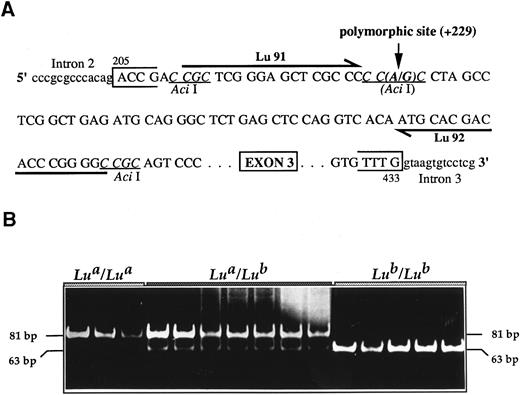
Lua/Lub typing. (A) Strategy of the PCR-RFLP. Primers Lu91 and Lu92 were designed to amplify an 81-bp LU gene fragment that encompasses the single base substitution (A229G) identified between the Lua and the Lub alleles and that correlates with an allele-specific Aci I restriction site. (B) Typical results of the PCR-RFLP assay. DNA from donors with the indicated Lu phenotypes was used in the PCR-RFLP assay. The sizes of the digested fragments are indicated on both sides.
Lua/Lub typing. (A) Strategy of the PCR-RFLP. Primers Lu91 and Lu92 were designed to amplify an 81-bp LU gene fragment that encompasses the single base substitution (A229G) identified between the Lua and the Lub alleles and that correlates with an allele-specific Aci I restriction site. (B) Typical results of the PCR-RFLP assay. DNA from donors with the indicated Lu phenotypes was used in the PCR-RFLP assay. The sizes of the digested fragments are indicated on both sides.
The G→A nucleotide substitution was correlated with an Aci I restriction site polymorphism (CG CC→CA CC), and this was used to develop a PCR-RFLP assay for the DNA typing of the Lu phenotypes. Primers Lu91 and Lu92 were designed to amplify an 81-bp fragment encompassing the polymorphic nucleotide at position +229 (Fig 4A). PCR-RFLP analyses were performed with genomic DNAs of 20 unrelated donors of each Lu phenotype and typical results are shown in Fig 4B. After Aci I digestion, the 81-bp PCR product was cleaved into two fragments of 63 and 18 bp (the 18-bp fragment migrated out of the gel and was not visualized) in all Lu(a − b+) samples, whereas only the uncleaved 81-bp fragment was observed with DNAs from the Lu(a + b−) donors. All 81-, 63-, and 18-bp fragments were detected in the heterozygous Lu(a + b+) samples, although the 63-bp fragment (Lub allele) was under-represented as compared with the 81-bp fragment (Lua allele). This might represent a preferential amplification of one allele in heterozygous samples as previously observed in blood group ABO genotyping experiments.19
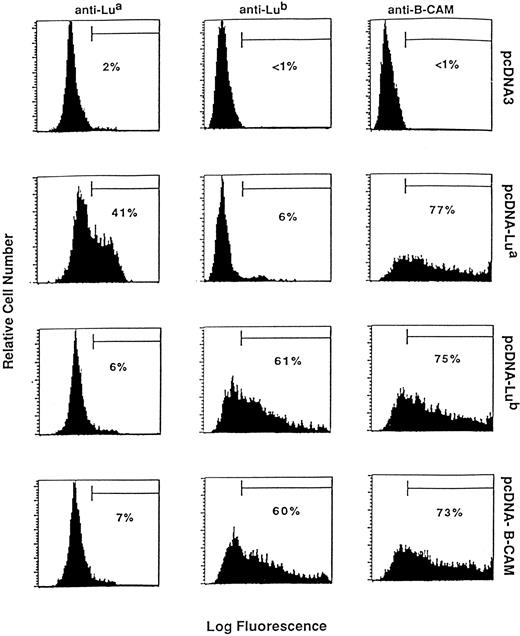
Transient expression of the Lu and B-CAM antigens in CHO cells.Transient expression of the Lu and B-CAM antigens in CHO cells were performed by introducing the Lua, Lub, and B-CAM cDNA constructs (see Materials and Methods) in the pcDNA3 expression vector. Flow cytometry analysis with the anti–B-CAM MoAb G253 revealed positive staining on cells transfected with all three pcDNA-Lua, pcDNA-Lub, or pcDNA–B-CAM constructs, whereas no staining could be detected on control cells transfected by the vector alone (Fig 5). In contrast, when anti-Lua or anti-Lub reagents were used, positive signals were obtained only with cells transfected with pcDNA-Lua or pcDNA-Lub, respectively. In addition, cells transfected with pcDNA–B-CAM carrying the Lub-specific G229 nucleotide were reactive with the anti-Lub but not with the anti-Lua reagents.
Expression of the Lu and B-CAM antigens in CHO cells. CHO cells were transfected with plasmid containing Lua (pcDNA-Lua), Lub (pcDNA-Lub), or B-CAM (pcDNA-B-CAM) cDNAs or with the pcDNA3 vector alone as a negative control. Expression of the Lua, Lub, and B-CAM antigens was determined by flow cytometry analysis using anti-Lua human antisera, LM342 monoclonal anti-Lub, and G253 monoclonal anti–B-CAM. The relative numbers of positive cells in the selected windows are indicated as percentages.
Expression of the Lu and B-CAM antigens in CHO cells. CHO cells were transfected with plasmid containing Lua (pcDNA-Lua), Lub (pcDNA-Lub), or B-CAM (pcDNA-B-CAM) cDNAs or with the pcDNA3 vector alone as a negative control. Expression of the Lua, Lub, and B-CAM antigens was determined by flow cytometry analysis using anti-Lua human antisera, LM342 monoclonal anti-Lub, and G253 monoclonal anti–B-CAM. The relative numbers of positive cells in the selected windows are indicated as percentages.
Before these experiments, the specificity of the anti-Lu and anti–B-CAM antibodies were controled on Lu-positive (homozygous or heterozygous for Lua and Lub) and Lunull (Lua-b-) human red blood cells. FACScan analysis indicated that the anti-Lu reagents gave positive staining when tested with red blood cells of the relevant Lu-positive phenotypes, whereas the anti–B-CAM G253 MoAb strongly reacted with red blood cells of all Lu-positive phenotypes. As a control, Lunull red blood cells were unreactive with all antibodies.
DISCUSSION
The LU gene is short (12.5 kb) but exhibits a rather complex organization with 15 exons ranging in size from 71 to 498 bp and separated by type 0 or type 1 introns. In contrast with most members of the Ig superfamily, the LU gene does not fit with the one domain: one exon rule.20 The signal peptide is encoded by exon 1. According to the membrane organization of the Lu gps deduced from hydropathy analysis,6 7 the first and second Ig-like variable domains are encoded by exons 2 to 3 and 4 to 6, respectively; the first, second, and third Ig-like constant domains are encoded by exons 7 to 8, 9 to 10, and 11 to 12, respectively; exon 13 encodes the transmembrane and the cytoplasmic domains common to the short- and long-tail Lu gps; exon 14 encodes either the 40 C-ter amino acid residues specific to the long-tail Lu isoform or the first 120 nt of the 3′ UT region of the 4.0-kb transcript. Finally, exon 15 encodes the 3′ UT domain common to the 2.5- and 4.0-kb Lu transcripts.
Our results indicate that the 2.5- and 4.0-kb Lu mRNA spliceoforms result from alternative splicing of intron 13. Both messengers are widely expressed but the level of the 4.0-kb transcript is very low as compared with that of the 2.5-kb transcript except in the colon carcinoma HT29 cell line.12 Because the 4.0-kb transcript directs the synthesis of a short-tail Lu gp isoform lacking a consensus binding site for an SH3 motif, we speculated that alternative splicing of the region coding for the cytoplasmic domain might be associated with malignant transformation and with the nonpolarized expression of B-CAM antigens in epithelial cancers8 and, thus, may affect some potential function(s) of the Lu polypeptides. Accordingly, it is assumed that a fine regulation of alternative splicing responding to some precise signals should occur. However, examination of exonic and intronic splice junctions did not show specific sequences, such as those that have been recently shown to be associated with the regulated alternative splicing of the 4.1 primary transcript during erythroid differenciation21 22 (F. Baklouti, personal communication, September 1996).
Sequence analysis of the major Lu mRNAs species (2.5 kb) amplified from human bone marrow, kidney, placenta, and skelatal muscle showed identical primary structures (not shown) and, therefore, suggested that alternative splicing events are not involved in the synthesis of tissue-specific Lu spliceoforms.
Primer extension and 5′–RACE-PCR analysis indicated that transcription of the LU gene is initiated at a position located −22 upstream from the translation initiation site in all tissues investigated, suggesting that the expression of the Lu gps in a wide, albeit restricted, variety of tissues and cells might be regulated by different elements within the same 5′ flanking region rather than by alternative promoters.
Sequence analysis of the proximal 5′ flanking region (positions −1 to −200) showed an organization typical of promoters of ubiquitous genes, which should account for the tissue distribution of the Lu messengers observed in Northern blot analysis.6,7,12 Indeed, the region is GC-rich and exhibits three clustered perfect consensus binding sites for the ubiquitous Sp1 protein, whereas no TATA nor CAAT boxes were identified. Clustered GC boxes in the absence of a TATA box are usually associated with a heterogeneity of transcriptional initiation, because the TATA box is generally believed to direct RNA polymerase II to a defined start position.23 Identification of a precise transcription start site in the LU gene in absence of a TATA box might be explained by the presence of the sequence CTCA GTCT around the initiation nucleotide that matches rather well with the core of the “initiator” control element that directs accurate transcription initiation in some TATA-less genes.24 The sequence between nucleotides −104 to −112 deviates by one substitution to the binding site for EGR-1, a ubiquitous protein encoded by an immediate early response gene and that has a broad role in signal transduction pathways.25,26 Whether this partial EGR-1 motif is involved in transcription regulation of the LU gene will justify further functional studies. Inspection of the distal 5′ flanking region (upstream nt −200) showed a binding sequence for the GATA transcription factor family located in the proximity of a CACCC sequence known to bind the CACCC or Sp1 transcription factors.27 Because GATA/Sp1 or GATA/CACCC association represents the central core of erythroid regulatory regions,28 29 further experiments should be performed to determine whether the GATA site within the LU 5′ region binds the erythroid member of the GATA family, hGATA-1 and, if so, whether the GATA/CACCC motif is involved in the expression of LU in erythroid cells.
Transfection of in vitro mutated forms of the Lu cDNAs in CHO cells showed the following. (1) the Lu blood group phenotypes given by the Lua and Lub alleles are produced by the His77Arg amino acid substitution resulting from the nucleotide polymorphism A229G. Thus, the PCR/RFLP assay based on this nucleotide polymorphism should be useful for DNA typing of the main Lu phenotypes. (2) The long-tail and short-tail Lu gps, previously characterized by anti-Lu and anti–B-CAM reagents, respectively,6,7 are reactive with both types of antibodies. This result provides the definitive proof that the Lu and B-CAM antigens are carried by the same molecules. This finding raises the question of whether potential functions attributed to the B-CAM antigens in cell-cell and cell-substrate adhesions8 are shared by the short-tail and the long-tail gps or whether each isoform might be differently involved in these processes. Based on previously published results indicating that the anti–B-CAM MoAb G253 did not react with red blood cells,8 we suggested that the epitope recognized by this antibody might be differently glycosylated in erythroid versus nonerythroid cells. However, in the course of the present study, we found that the MoAb G253 reacted strongly with human red blood cells of all Lu-positive phenotypes, whereas it was unreactive with Lunull erythrocytes, which lack all Lu antigens, expression. Thus, it is assumed that the same gps (85 and 78 kD) carry the Lu and B-CAM epitopes on red blood cells. However, a different glycosylation pattern might account for the presence of a Lu gp of higher apparent molecular weight (90 to 95 kD) in HT29 cells, which reacts with MoAb G25310 as well as with anti-Lu antibodies.12
In summary, we have clarified the structure of the LU gene in individuals that express the Lua and/or Lub blood group antigens and showed that the Lu and B-CAM antigens are carried by the same gp present under two isoforms that may be referred to as long-tail and short-tail Lu gps. These data, as well as the elucidation of the primary structure of the 5′ flanking region of the LU gene, should help to elucidate the molecular basis of the Lunull phenotype and to analyze the events that regulate the alternative splicing of the Lu messengers that lead to the overexpression of the short-tail Lu gp in some cancer cell lines.
ACKNOWLEDGMENT
We are grateful to Dr V. Van Huffel (GIP-INTS, Paris, France) for the generous gift of Lu-typed genomic DNAs and to Dr H. Fraser (Scottish National Blood Transfusion Service, Glascow, Scotland) and Dr W.J. Rettig (Ludwig Institute for Cancer Research, Biberach) for providing MoAbs LM342 and G253 MoAbs, respectively.
Address reprint requests to Caroline Le Van Kim, PhD, Unité INSERM U76, GIP-Institut National de la Transfusion Sanguine, 6 rue Alexandre Cabanel, 75015 Paris, France.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal