Key Points
A ML model, available as an interactive web application, was created to predict survival after transplant in MF.
This tool is a step toward personalized medicine, enabling the identification of 25% of patients with poor transplantation outcomes.
Visual Abstract
With the incorporation of effective therapies for myelofibrosis (MF), accurately predicting outcomes after allogeneic hematopoietic cell transplantation (allo-HCT) is crucial for determining the optimal timing for this procedure. Using data from 5183 patients with MF who underwent first allo-HCT between 2005 and 2020 at European Society for Blood and Marrow Transplantation centers, we examined different machine learning (ML) models to predict overall survival after transplant. The cohort was divided into a training set (75%) and a test set (25%) for model validation. A random survival forests (RSF) model was developed based on 10 variables: patient age, comorbidity index, performance status, blood blasts, hemoglobin, leukocytes, platelets, donor type, conditioning intensity, and graft-versus-host disease prophylaxis. Its performance was compared with a 4-level Cox regression–based score and other ML-based models derived from the same data set, and with the Center for International Blood and Marrow Transplant Research score. The RSF outperformed all comparators, achieving better concordance indices across both primary and postessential thrombocythemia/polycythemia vera MF subgroups. The robustness and generalizability of the RSF model was confirmed by Akaike information criterion and time-dependent receiver operating characteristic area under the curve metrics in both sets. Although all models were prognostic for nonrelapse mortality, the RSF provided better curve separation, effectively identifying a high-risk group comprising 25% of patients. In conclusion, ML enhances risk stratification in patients with MF undergoing allo-HCT, paving the way for personalized medicine. A web application (https://gemfin.click/ebmt) based on the RSF model offers a practical tool to identify patients at high risk for poor transplantation outcomes, supporting informed treatment decisions and advancing individualized care.
Introduction
Myelofibrosis (MF) is a chronic myeloproliferative neoplasm that appears de novo (primary MF [PMF]) or after a diagnosis of essential thrombocythemia or polycythemia vera (secondary MF [SMF]). Managing MF is complex because of its diverse clinical manifestations including an inherent risk of progression to acute myeloid leukemia (AML). Although the median overall survival (OS) is ∼6 years, it varies significantly among patients. Medical treatment focuses on symptom control and quality of life but is not curative and does not reduce the risk of AML.1,2
Allogeneic hematopoietic cell transplantation (allo-HCT) remains the only curative option for MF.3 However, its significant morbidity and mortality require a careful risk-benefit analysis to identify appropriate candidates. This has become particularly critical recently, as several effective therapies for MF have been incorporated into clinical practice,4-6 with others showing promising results in clinical trials.4 Existing prognostic models for OS after allo-HCT have been instrumental in guiding clinical decisions.7,8 However, they do not account for key factors such as patient comorbidities7 and emerging transplant strategies, such as haploidentical transplants or posttransplant cyclophosphamide use.9,10 Furthermore, there is significant room for improving their ability to accurately identify patients at high risk of posttransplant mortality, who may benefit more from alternative treatments or clinical trials.
Machine learning (ML) is a field of artificial intelligence in which prediction is based on modeling of outcomes considering the complex interactions among multiple variables, rather than relying on predefined human-made rules. These techniques have demonstrated their utility to provide accurate personalized survival predictions for patients with MF undergoing conventional drug treatment.11,12 In this study, we aim to assess whether ML techniques can similarly improve prognostication of OS in the setting of allo-HCT for MF. The ultimate goal is to enhance transplant decision-making by providing more precise and individualized survival predictions.
Methods
Data source
We retrieved data from adult patients with MF (PMF or SMF) who underwent first allo-HCT between 2005 and 2020 in European Society for Blood and Marrow Transplantation (EBMT) centers. Patients who received transplantation from umbilical cord blood and those with AML transformation were excluded. Myeloablative or reduced intensity conditioning regimens were defined by standard EBMT criteria.13 Matched unrelated transplants were matched at allele level for HLA-A, -B, -C, -DRB1, and -DQB1. Centers ensured informed consent in compliance with local regulations to report pseudonymized data to the EBMT. The study was approved by the Chronic Malignancies Working Party of EBMT and conducted in accordance with the Declaration of Helsinki. Informed consent for inclusion in the EBMT registry was obtained for all patients. The study database included 52 variables selected for their prognostic significance based on previous studies.7,14,15
Main study outcomes
The primary goal was to develop a prognostic model for OS using ML techniques and compare its performance with that of a Cox regression–based score developed in the same data set, and with the Center for International Blood and Marrow Transplant Research (CIBMTR) model.8 The models were also applied for predicting the secondary outcome of nonrelapse mortality (NRM). Progression-free survival (PFS) and cumulative incidence of relapse were estimated for descriptive purposes. Median follow-up was determined using the reverse Kaplan-Meier method. In patients who died after disease relapse, relapse was considered the primary cause of death.16
Statistical analysis
OS and PFS were estimated by the Kaplan-Meier method. NRM was defined as the time from the date of transplantation to the date of death (uncensored) or to the date of disease relapse (censored). The cumulative incidences of relapse/NRM (as competing risk for each other) were analyzed separately in a competing risks framework.17
Two independent statisticians applied distinct methodologies to evaluate the factors influencing OS. One used a conventional multivariate Cox proportional hazards regression model, whereas the other used a range of ML techniques. Both approaches were based on the same random distribution of the patient cohort into a training set (75% of the cohort, n = 3887) and a test set (25% of the cohort, n = 1296). Each statistician independently determined the optimal cutoff points for the newly derived risk scores to stratify patients into distinct prognostic groups. The resulting risk classifications were subsequently compared and contextualized to assess their clinical relevance for treatment decision-making.
Multivariate Cox regression model
Factors potentially associated with OS were entered into a Cox proportional hazards model. Selection of variables in the final model was based on expert clinical advice and data availability, to assess the independent effect of each covariate. Variables with a high degree of missingness (defined as >50%) were not considered for inclusion in the model, whereas a missing category was created for those variables with a low degree of missingness. Hazard ratios (HRs) were provided, and corresponding P values were calculated using the Wald test. A score of 1 or 2 was assigned to each significant variable for OS based on the HRs obtained from multivariable analysis. The cutoff values were arbitrarily defined as follows: HR of <1.25 = 0 points, HR of 1.25 to 1.50 = 1 point, HR of >1.50 = 2 points. A prognostic scoring system was subsequently developed considering the sum of risk points to discriminate 4 patient risk groups with significant differences in OS. All P values were 2-sided and P < .05 was considered significant.
RSF model
Random survival forests (RSF) were created with 1000 trees. For cross-validation, sampling was performed without replacement, which, by default, takes 0.632 times the sample size. Missing variables were imputed in the training and test cohorts separately using a missing data algorithm developed by Ishwaran et al.22 Predictions were cross-validated in the training set (using the out-of-bag method) and then validated in the test cohort. This was done to rule out overfitting of performance metrics in the training set related to either variable selection or the imputation process. Dimensionality reduction was performed using variable-importance estimation analysis. Redundant and dependent variables were discarded applying variable-importance estimations and clinical knowledge, achieving a minimally dimensioned yet effective model.
Hyperparameter tuning was explored to optimize the performance of the RSF model. Specifically, a grid search was used to explore the impact of key hyperparameters, including “mtry” (the number of variables randomly selected at each split) and “node size” (the minimum number of samples required in terminal nodes). Partial dependence plots were used to evaluate the marginal effect of individual covariates on survival estimates, while accounting for the average influence of all other variables. These plots help visualize nonlinear relationships and potential threshold effects, providing insight into the contribution of specific predictors to survival probability over time. This approach enhances the model’s interpretability by isolating the independent effect of each covariate on the predicted survival outcomes.
Comparison of different ML techniques for survival analysis
In addition to the baseline RSF model, we evaluated the performance of 3 complementary methods using the 10 variables included in the final model: oblique RSF (ORSF), gradient-boosted survival trees using XGBoost, and a deep neural network–based survival model (DeepSurv).23-25
A detailed explanation of the different ML techniques is outlined in the supplemental Data, available on the Blood website. All 3 approaches, ORSF, XGBoost-based survival modeling, and DeepSurv, were evaluated using cross-validation for performance estimation. Early stopping was applied when computationally practical to limit overfitting. The performance of each model was assessed using standard survival metrics (eg, C-index) to facilitate a rigorous comparative analysis.
Comparison of the discriminative capacity between the ML and the Cox-derived models for survival prediction
The discriminative capacity of the ML and Cox OS models was compared in the training and test sets using the Harrell C-index.26 Time-dependent receiver operating characteristic (ROC) area under the curves (AUCs) for OS were calculated using the timeROC package.27 Quantitative scores (continuous risk estimates) and categorical risk groups were analyzed separately for each model. For risk groups, categorical labels were converted into numeric values for compatibility. Time-dependent ROC-AUCs assessed model performance at specific time points, allowing a robust evaluation of prognostic accuracy over time of the RSF and Cox models in terms of their quantitative and group-based predictions.
Because the C-index is not an optimal metric for competing risk models, we also included the Akaike information criterion (AIC) scores for both NRM and OS.28
Results
Patient and transplant characteristics
A total of 5183 patients from 288 centers fulfilling the selection criteria were included. Baseline characteristics of all patients, along with the training (n = 3887) and validation (n = 1296) cohorts, are presented in Table 1. Median follow-up was 58.2 months (95% confidence interval [CI], 55.6-59.8) in the training set and 60.0 months (95% CI, 55.7-63.2) in the test set. Median OS was 79.4 months (95% CI, 69.2-89.6) in the training set and 73.7 months (95% CI, 54.7-92.7) in the test set. No significant differences in characteristics were observed between the cohorts, apart from a higher platelet count at allo-HCT and less antithymocyte globulin use in the test cohort.
Main characteristics of a series of 5183 patients with MF undergoing allo-HCT and of the training and test cohorts
| Characteristic . | Group . | Missing (%) . | Total cohort . | Training set . | Test set . | P value . |
|---|---|---|---|---|---|---|
| N (%) . | n (%) . | n (%) . | ||||
| No. of patients | 5183 (100) | 3887 (100) | 1296 (100) | |||
| Age at allo-HCT, y∗ | 58.3 (52-63.5) | 58.2 (51.8-63.4) | 58.6 (52.7-63.8) | .072 | ||
| Age at allo-HCT | <60 y | 3003 (57.9) | 2274 (58.5) | 729 (56.2) | .164 | |
| Patient sex | Male | 3242 (62.6) | 2412 (62.1) | 830 (64.0) | .21 | |
| Year of MF diagnosis | <2000 | 615 (11.9) | 455 (11.7) | 160 (12.3) | .21 | |
| 2000-2010 | 1765 (34.1) | 1310 (33.7) | 455 (35.1) | |||
| 2010-2015 | 1422 (27.4) | 1057 (27.2) | 365 (28.2) | |||
| ≥2015 | 1381 (26.6) | 1065 (27.4) | 316 (24.4) | |||
| MF type | PMF | 3743 (72.2) | 2807 (72.2) | 936 (72.2) | 1.00 | |
| SMF | 1440 (27.8) | 1080 (27.8) | 360 (27.8) | |||
| JAK2 inhibitor treatment before allo-HCT | Yes | 1503 (29) | 1039 (28.2) | 764 (27.9) | 275 (29.2) | .459 |
| Genotype | JAK2+ | 2422 (46.7) | 2156 (78.1) | 1625 (77.7) | 531 (79.3) | .43 |
| MPL+ | 94 (3.4) | 68 (3.3) | 26 (3.9) | |||
| CALR+ | 377 (13.7) | 290 (13.9) | 87 (13.0) | |||
| Triple negative | 134 (4.9) | 108 (5.2) | 26 (3.9) | |||
| Constitutional symptoms at allo-HCT | Yes | 3089 (59.6) | 911 (43.5) | 693 (44.3) | 218 (41.3) | .255 |
| Hemoglobin at allo-HCT, g/dL∗ | 2575 (49.7) | 9.2 (8.1-10.5) | 9.2 (8.1-10.5) | 9.2 (8.2-10.6) | .43 | |
| Leukocyte count at allo-HCT, ×109/L∗ | 2600 (50.2) | 6.9 (3.5-14.4) | 6.8 (3.5-14.3) | 7 (3.6-14.4) | .336 | |
| Blood blasts at allo-HCT, %∗ | 3095 (59.7) | 1 (0-3) | 1 (0-3) | 1 (0-3) | .637 | |
| Platelets at allo-HCT, ×109/L∗ | 2640 (50.9) | 117 (53-242.5) | 114.5 (53-237) | 125 (51-274) | .039 | |
| Splenectomy before allo-HCT | Yes | 2536 (48.9) | 324 (12.2) | 231 (11.7) | 93 (14.0) | .129 |
| Spleen size below costal margin, cm, by physical examination∗ | 3857 (74.4) | 5 (0-10) | 5 (1-11) | 5 (0-10) | .184 | |
| Spleen span by ultrasound or CT scan, max diameter, cm∗ | 4283 (82.6) | 20 (16-23) | 20 (16-23) | 20 (16.2-23) | .72 | |
| HCT-CI risk group | Low risk (0 points) | 1360 (26.2) | 2041 (53.4) | 1538 (53.7) | 503 (52.4) | .745 |
| Intermediate risk (1-2 points) | 910 (23.8) | 674 (23.5) | 236 (24.6) | |||
| High risk (≥3 points) | 872 (22.8) | 651 (22.7) | 221 (23.0) | |||
| KPS score at allo-HCT | 90-100 | 489 (9.4) | 3111 (66.3) | 2316 (65.8) | 795 (67.7) | .508 |
| 80 | 1232 (26.2) | 937 (26.6) | 295 (25.1) | |||
| <80 | 351 (7.5) | 266 (7.6) | 85 (7.2) | |||
| DIPSS risk group at allo-HCT | Low risk | 2715 (52.4) | 60 (2.4) | 40 (2.2) | 20 (3.2) | .286 |
| Intermediate-1 | 919 (37.2) | 695 (37.7) | 224 (36.0) | |||
| Intermediate-2 | 954 (38.7) | 702 (38.0) | 252 (40.4) | |||
| High risk | 535 (21.7) | 408 (22.1) | 127 (20.4) | |||
| CIBMTR risk score at allo-HCT | Low | 2640 (50.9) | 1020 (40.1) | 763 (39.6) | 257 (41.6) | .641 |
| Intermediate | 1313 (51.6) | 1004 (52.2) | 309 (50.0) | |||
| High | 210 (8.3) | 158 (8.2) | 52 (8.4) | |||
| Donor type | Identical sibling | 1534 (29.6) | 1147 (29.5) | 387 (29.9) | .919 | |
| MRD (other than sibling) | 45 (0.9) | 34 (0.9) | 11 (0.8) | |||
| MMRD | 339 (6.5) | 249 (6.4) | 90 (6.9) | |||
| MUD | 2175 (42.0) | 1646 (42.3) | 529 (40.8) | |||
| MMUD | 673 (13.0) | 504 (13.0) | 169 (13.0) | |||
| Unrelated, number of mismatches unknown | 417 (8.0) | 307 (7.9) | 110 (8.5) | |||
| Recipient-donor match | Recipient male with donor male | 76 (1.5) | 2297 (45.0) | 1714 (44.7) | 583 (45.9) | .614 |
| Recipient male with donor female | 895 (17.5) | 666 (17.4) | 229 (18.0) | |||
| Recipient female with donor male | 1147 (22.5) | 878 (22.9) | 269 (21.2) | |||
| Recipient female with donor female | 768 (15.0) | 578 (15.1) | 190 (14.9) | |||
| Donor age, y∗ | 1132 (21.8) | 36.6 (27.2-49.8) | 36.7 (27.2-49.6) | 36.1 (27.1-50.1) | .902 | |
| CMV serology in patient/donor | −/− | 233 (4.5) | 1437 (29.0) | 1056 (28.4) | 381 (30.8) | .174 |
| −/+ | 464 (9.4) | 348 (9.4) | 116 (9.4) | |||
| +/− | 1002 (20.2) | 776 (20.9) | 226 (18.3) | |||
| +/+ | 2047 (41.4) | 1533 (41.3) | 514 (41.6) | |||
| Stem cell source | Bone marrow | 409 (7.9) | 305 (7.8) | 104 (8.0) | .884 | |
| Peripheral blood | 4774 (92.1) | 3582 (92.2) | 1192 (92.0) | |||
| Busulfan- or melphalan-based conditioning regimen | Busulfan based | 89 (1.7) | 3522 (69.1) | 2642 (69.2) | 880 (69.0) | .861 |
| Melphalan based | 718 (14.1) | 533 (14.0) | 185 (14.5) | |||
| Other regimen | 854 (16.8) | 644 (16.9) | 210 (16.5) | |||
| Conditioning drugs | BuCy w/wo others (MAC) | 146 (2.8) | 166 (3.3) | 137 (3.6) | 29 (2.3) | .324 |
| FluBu w/wo others (MAC) | 1153 (22.9) | 858 (22.7) | 295 (23.4) | |||
| FluTreo w/wo others (MAC) | 160 (3.2) | 121 (3.2) | 39 (3.1) | |||
| TBI w/wo Cy w/wo others (MAC) | 135 (2.7) | 111 (2.9) | 24 (1.9) | |||
| Others (MAC) | 155 (3.1) | 116 (3.1) | 39 (3.1) | |||
| FluBu w/wo others (RIC) | 2135 (42.4) | 1588 (42.1) | 547 (43.3) | |||
| FluMel w/wo others (RIC) | 579 (11.5) | 428 (11.3) | 151 (12.0) | |||
| FluTreo w/wo others (RIC) | 143 (2.8) | 105 (2.8) | 38 (3.0) | |||
| TBI w/wo Cy w/wo Flu w/wo others (RIC) | 275 (5.5) | 206 (5.5) | 69 (5.5) | |||
| Others (RIC) | 136 (2.7) | 105 (2.8) | 31 (2.5) | |||
| Conditioning regimen intensity | MAC | 75 (1.4) | 1802 (35.3) | 1366 (35.7) | 436 (34.1) | .32 |
| RIC | 3306 (64.7) | 2463 (64.3) | 843 (65.9) | |||
| TBI | Yes | 37 (0.7) | 420 (8.2) | 326 (8.4) | 94 (7.3) | .219 |
| T-cell depletion | No | 148 (2.9) | 1356 (26.9) | 1010 (26.8) | 346 (27.4) | .451 |
| Yes in vivo, no ex vivo | 3585 (71.2) | 2696 (71.4) | 889 (70.5) | |||
| Yes ex vivo, no in vivo | 17 (0.3) | 10 (0.3) | 7 (0.6) | |||
| Yes in vivo + ex vivo | 77 (1.5) | 58 (1.5) | 19 (1.5) | |||
| ATG | Yes | 120 (2.3) | 3387 (66.9) | 2568 (67.7) | 819 (64.5) | .038 |
| Alemtuzumab | Yes | 167 (3.2) | 306 (6.1) | 218 (5.8) | 88 (7.0) | .146 |
| GVHD prophylaxis group | Post-Cy | 112 (2.2) | 415 (8.2) | 311 (8.2) | 104 (8.2) | .064 |
| ATG + CNI + MMF | 1398 (27.6) | 1064 (28.0) | 334 (26.3) | |||
| ATG + CNI + MTX | 1352 (26.7) | 1021 (26.9) | 331 (26.0) | |||
| ATG + CNI | 354 (7.0) | 276 (7.3) | 78 (6.1) | |||
| ATG w/wo other(s) | 195 (3.8) | 150 (3.9) | 45 (3.5) | |||
| Post-Cy + ATG | 88 (1.7) | 57 (1.5) | 31 (2.4) | |||
| CNI only | 184 (3.6) | 129 (3.4) | 55 (4.3) | |||
| CNI + MMF w/wo other | 434 (8.6) | 311 (8.2) | 123 (9.7) | |||
| CNI + MTX w/wo other | 522 (10.3) | 393 (10.3) | 129 (10.1) | |||
| Other | 129 (2.5) | 88 (2.3) | 41 (3.2) |
| Characteristic . | Group . | Missing (%) . | Total cohort . | Training set . | Test set . | P value . |
|---|---|---|---|---|---|---|
| N (%) . | n (%) . | n (%) . | ||||
| No. of patients | 5183 (100) | 3887 (100) | 1296 (100) | |||
| Age at allo-HCT, y∗ | 58.3 (52-63.5) | 58.2 (51.8-63.4) | 58.6 (52.7-63.8) | .072 | ||
| Age at allo-HCT | <60 y | 3003 (57.9) | 2274 (58.5) | 729 (56.2) | .164 | |
| Patient sex | Male | 3242 (62.6) | 2412 (62.1) | 830 (64.0) | .21 | |
| Year of MF diagnosis | <2000 | 615 (11.9) | 455 (11.7) | 160 (12.3) | .21 | |
| 2000-2010 | 1765 (34.1) | 1310 (33.7) | 455 (35.1) | |||
| 2010-2015 | 1422 (27.4) | 1057 (27.2) | 365 (28.2) | |||
| ≥2015 | 1381 (26.6) | 1065 (27.4) | 316 (24.4) | |||
| MF type | PMF | 3743 (72.2) | 2807 (72.2) | 936 (72.2) | 1.00 | |
| SMF | 1440 (27.8) | 1080 (27.8) | 360 (27.8) | |||
| JAK2 inhibitor treatment before allo-HCT | Yes | 1503 (29) | 1039 (28.2) | 764 (27.9) | 275 (29.2) | .459 |
| Genotype | JAK2+ | 2422 (46.7) | 2156 (78.1) | 1625 (77.7) | 531 (79.3) | .43 |
| MPL+ | 94 (3.4) | 68 (3.3) | 26 (3.9) | |||
| CALR+ | 377 (13.7) | 290 (13.9) | 87 (13.0) | |||
| Triple negative | 134 (4.9) | 108 (5.2) | 26 (3.9) | |||
| Constitutional symptoms at allo-HCT | Yes | 3089 (59.6) | 911 (43.5) | 693 (44.3) | 218 (41.3) | .255 |
| Hemoglobin at allo-HCT, g/dL∗ | 2575 (49.7) | 9.2 (8.1-10.5) | 9.2 (8.1-10.5) | 9.2 (8.2-10.6) | .43 | |
| Leukocyte count at allo-HCT, ×109/L∗ | 2600 (50.2) | 6.9 (3.5-14.4) | 6.8 (3.5-14.3) | 7 (3.6-14.4) | .336 | |
| Blood blasts at allo-HCT, %∗ | 3095 (59.7) | 1 (0-3) | 1 (0-3) | 1 (0-3) | .637 | |
| Platelets at allo-HCT, ×109/L∗ | 2640 (50.9) | 117 (53-242.5) | 114.5 (53-237) | 125 (51-274) | .039 | |
| Splenectomy before allo-HCT | Yes | 2536 (48.9) | 324 (12.2) | 231 (11.7) | 93 (14.0) | .129 |
| Spleen size below costal margin, cm, by physical examination∗ | 3857 (74.4) | 5 (0-10) | 5 (1-11) | 5 (0-10) | .184 | |
| Spleen span by ultrasound or CT scan, max diameter, cm∗ | 4283 (82.6) | 20 (16-23) | 20 (16-23) | 20 (16.2-23) | .72 | |
| HCT-CI risk group | Low risk (0 points) | 1360 (26.2) | 2041 (53.4) | 1538 (53.7) | 503 (52.4) | .745 |
| Intermediate risk (1-2 points) | 910 (23.8) | 674 (23.5) | 236 (24.6) | |||
| High risk (≥3 points) | 872 (22.8) | 651 (22.7) | 221 (23.0) | |||
| KPS score at allo-HCT | 90-100 | 489 (9.4) | 3111 (66.3) | 2316 (65.8) | 795 (67.7) | .508 |
| 80 | 1232 (26.2) | 937 (26.6) | 295 (25.1) | |||
| <80 | 351 (7.5) | 266 (7.6) | 85 (7.2) | |||
| DIPSS risk group at allo-HCT | Low risk | 2715 (52.4) | 60 (2.4) | 40 (2.2) | 20 (3.2) | .286 |
| Intermediate-1 | 919 (37.2) | 695 (37.7) | 224 (36.0) | |||
| Intermediate-2 | 954 (38.7) | 702 (38.0) | 252 (40.4) | |||
| High risk | 535 (21.7) | 408 (22.1) | 127 (20.4) | |||
| CIBMTR risk score at allo-HCT | Low | 2640 (50.9) | 1020 (40.1) | 763 (39.6) | 257 (41.6) | .641 |
| Intermediate | 1313 (51.6) | 1004 (52.2) | 309 (50.0) | |||
| High | 210 (8.3) | 158 (8.2) | 52 (8.4) | |||
| Donor type | Identical sibling | 1534 (29.6) | 1147 (29.5) | 387 (29.9) | .919 | |
| MRD (other than sibling) | 45 (0.9) | 34 (0.9) | 11 (0.8) | |||
| MMRD | 339 (6.5) | 249 (6.4) | 90 (6.9) | |||
| MUD | 2175 (42.0) | 1646 (42.3) | 529 (40.8) | |||
| MMUD | 673 (13.0) | 504 (13.0) | 169 (13.0) | |||
| Unrelated, number of mismatches unknown | 417 (8.0) | 307 (7.9) | 110 (8.5) | |||
| Recipient-donor match | Recipient male with donor male | 76 (1.5) | 2297 (45.0) | 1714 (44.7) | 583 (45.9) | .614 |
| Recipient male with donor female | 895 (17.5) | 666 (17.4) | 229 (18.0) | |||
| Recipient female with donor male | 1147 (22.5) | 878 (22.9) | 269 (21.2) | |||
| Recipient female with donor female | 768 (15.0) | 578 (15.1) | 190 (14.9) | |||
| Donor age, y∗ | 1132 (21.8) | 36.6 (27.2-49.8) | 36.7 (27.2-49.6) | 36.1 (27.1-50.1) | .902 | |
| CMV serology in patient/donor | −/− | 233 (4.5) | 1437 (29.0) | 1056 (28.4) | 381 (30.8) | .174 |
| −/+ | 464 (9.4) | 348 (9.4) | 116 (9.4) | |||
| +/− | 1002 (20.2) | 776 (20.9) | 226 (18.3) | |||
| +/+ | 2047 (41.4) | 1533 (41.3) | 514 (41.6) | |||
| Stem cell source | Bone marrow | 409 (7.9) | 305 (7.8) | 104 (8.0) | .884 | |
| Peripheral blood | 4774 (92.1) | 3582 (92.2) | 1192 (92.0) | |||
| Busulfan- or melphalan-based conditioning regimen | Busulfan based | 89 (1.7) | 3522 (69.1) | 2642 (69.2) | 880 (69.0) | .861 |
| Melphalan based | 718 (14.1) | 533 (14.0) | 185 (14.5) | |||
| Other regimen | 854 (16.8) | 644 (16.9) | 210 (16.5) | |||
| Conditioning drugs | BuCy w/wo others (MAC) | 146 (2.8) | 166 (3.3) | 137 (3.6) | 29 (2.3) | .324 |
| FluBu w/wo others (MAC) | 1153 (22.9) | 858 (22.7) | 295 (23.4) | |||
| FluTreo w/wo others (MAC) | 160 (3.2) | 121 (3.2) | 39 (3.1) | |||
| TBI w/wo Cy w/wo others (MAC) | 135 (2.7) | 111 (2.9) | 24 (1.9) | |||
| Others (MAC) | 155 (3.1) | 116 (3.1) | 39 (3.1) | |||
| FluBu w/wo others (RIC) | 2135 (42.4) | 1588 (42.1) | 547 (43.3) | |||
| FluMel w/wo others (RIC) | 579 (11.5) | 428 (11.3) | 151 (12.0) | |||
| FluTreo w/wo others (RIC) | 143 (2.8) | 105 (2.8) | 38 (3.0) | |||
| TBI w/wo Cy w/wo Flu w/wo others (RIC) | 275 (5.5) | 206 (5.5) | 69 (5.5) | |||
| Others (RIC) | 136 (2.7) | 105 (2.8) | 31 (2.5) | |||
| Conditioning regimen intensity | MAC | 75 (1.4) | 1802 (35.3) | 1366 (35.7) | 436 (34.1) | .32 |
| RIC | 3306 (64.7) | 2463 (64.3) | 843 (65.9) | |||
| TBI | Yes | 37 (0.7) | 420 (8.2) | 326 (8.4) | 94 (7.3) | .219 |
| T-cell depletion | No | 148 (2.9) | 1356 (26.9) | 1010 (26.8) | 346 (27.4) | .451 |
| Yes in vivo, no ex vivo | 3585 (71.2) | 2696 (71.4) | 889 (70.5) | |||
| Yes ex vivo, no in vivo | 17 (0.3) | 10 (0.3) | 7 (0.6) | |||
| Yes in vivo + ex vivo | 77 (1.5) | 58 (1.5) | 19 (1.5) | |||
| ATG | Yes | 120 (2.3) | 3387 (66.9) | 2568 (67.7) | 819 (64.5) | .038 |
| Alemtuzumab | Yes | 167 (3.2) | 306 (6.1) | 218 (5.8) | 88 (7.0) | .146 |
| GVHD prophylaxis group | Post-Cy | 112 (2.2) | 415 (8.2) | 311 (8.2) | 104 (8.2) | .064 |
| ATG + CNI + MMF | 1398 (27.6) | 1064 (28.0) | 334 (26.3) | |||
| ATG + CNI + MTX | 1352 (26.7) | 1021 (26.9) | 331 (26.0) | |||
| ATG + CNI | 354 (7.0) | 276 (7.3) | 78 (6.1) | |||
| ATG w/wo other(s) | 195 (3.8) | 150 (3.9) | 45 (3.5) | |||
| Post-Cy + ATG | 88 (1.7) | 57 (1.5) | 31 (2.4) | |||
| CNI only | 184 (3.6) | 129 (3.4) | 55 (4.3) | |||
| CNI + MMF w/wo other | 434 (8.6) | 311 (8.2) | 123 (9.7) | |||
| CNI + MTX w/wo other | 522 (10.3) | 393 (10.3) | 129 (10.1) | |||
| Other | 129 (2.5) | 88 (2.3) | 41 (3.2) |
ATG, antithymocyte globulin; Bu, busulfan; CNI, calcineurin inhibitor; CT, computed tomography; Cy, cyclophosphamide; DIPSS, Dynamic International Prognostic Scoring System; Flu, fludarabine; MAC, myeloablative conditioning; Mel, Melphalan; MMF, mycophenolate mofetil; MMRD, mismatched related donor; max, maximum; MMUD, mismatched unrelated donor; MRD, matched related donor; MTX, methotrexate; MUD, matched unrelated donor; RIC, reduced intensity conditioning; TBI, total body irradiation; Treo, treosulfan; w/wo, with/without.
Values are median (interquartile range).
Transplantation outcomes
The estimated OS rate at 1, 5, and 10 years was 70% (95% CI, 69-71), 53% (95% CI, 51-54), and 43% (95% CI, 41-45), respectively.
The probability of PFS after 1, 5, and 10 years was 62% (95% CI, 60-63), 44% (95% CI, 43-46), and 35% (95% CI, 33-37), respectively. The estimated NRM rate at 1, 5, and 10 years was 23% (95% CI, 22-24), 32% (95% CI, 31-33), and 36% (95% CI, 35-38), respectively. Cumulative incidence of relapse at 1, 5, and 10 years was 15% (95% CI, 14-16), 24% (95% CI, 23-25), and 29% (95% CI, 27-31), respectively.
The graphical representation of the main study outcomes in the training and test cohorts can be seen in supplemental Figure 1.
Risk model for OS using Cox regression analysis
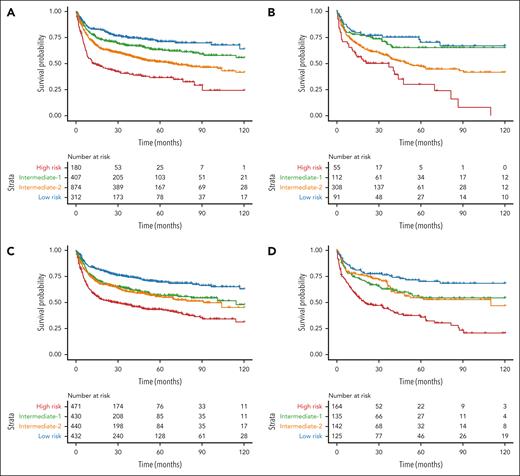
Factors associated with OS in the univariable analysis are shown in supplemental Table 1. In the multivariable analysis, 7 independent factors significantly predicted reduced OS: older patient age, HLA-mismatched donor type, lower Karnofsky performance status (KPS), higher HCT-specific comorbidity index (HCT-CI), JAK2/triple-negative genotype, graft from a female donor to a male patient, and graft from a donor who is cytomegalovirus (CMV) positive to a recipient who is CMV negative (Table 2). Based on the HRs, a score of 2 was assigned to patient age of ≥60 years and the JAK2/triple-negative genotype; and a score of 1 to patient age of 50 to 59 years, haploidentical or mismatched unrelated donors, KPS of <90, and HCT-CI of ≥3. Because the HRs for sex match and CMV serostatus were only modestly increased, no score was assigned to these factors. The total score ranged from 0 to 7 points, with 4 risk categories: low risk (0-1 points), intermediate-1 risk (2-3 points), intermediate-2 risk (4-5 points), and high risk (6-7 points). The corresponding 5-year OS of each category in the training and test set were 82% (95% CI, 74-90) and 65% (95% CI, 41-89) for low risk (6% of the cohort); 62% (95% CI, 58-66) and 65% (IC 95%, 57-73) for intermediate-1 (36% of the cohort); 52% (95% CI, 48-56) and 47% (95% CI, 40-54) for intermediate-2 (48% of the cohort); and 39% (95% CI, 30-47) and 30% (95% CI, 13-47) for high risk (10% of the cohort), respectively (Figure 1A-B).
Cox regression analysis of factors associated with OS in the training cohort
| . | HR (95% CI) . | (Overall) P . |
|---|---|---|
| Conditioning intensity | ||
| MAC | 1.00 | |
| RIC | 1.08 (0.97-1.20) | .17 |
| Donor type | ||
| MRD/MUD | 1.00 | |
| HD/MMUD | 1.34 (1.19-1.51) | <.0001 |
| Age at allo-HCT, y | ||
| <49 | 1.00 | |
| 50-59 | 1.36 (1.17-1.58) | <.0001 |
| ≥60 | 1.67 (1.44-1.94) | <.0001 |
| Sex | ||
| Male | 1.00 | |
| Female | 0.89 (0.80-0.99) | .04 |
| KPS at allo-HCT | ||
| 90-100 | 1.00 | |
| <90 | 1.33 (1.19-1.48) | <.0001 |
| HCT-CI | ||
| 0-2 | 1.00 | |
| ≥3 | 1.36 (1.19-1.55) | <.0001 |
| CMV donor/patient | ||
| +/− | 1.16 (1.03-1.32) | .01 |
| Other | 1.00 | |
| Sex match donor/patient | ||
| Female to male | 1.15 (1.00-1.31) | .04 |
| Other | 1.00 | |
| Genotype | ||
| CALR+/MPL+ | 1.00 | |
| JAK2+/triple negative | 1.56 (1.26-1.92) | <.0001 |
| . | HR (95% CI) . | (Overall) P . |
|---|---|---|
| Conditioning intensity | ||
| MAC | 1.00 | |
| RIC | 1.08 (0.97-1.20) | .17 |
| Donor type | ||
| MRD/MUD | 1.00 | |
| HD/MMUD | 1.34 (1.19-1.51) | <.0001 |
| Age at allo-HCT, y | ||
| <49 | 1.00 | |
| 50-59 | 1.36 (1.17-1.58) | <.0001 |
| ≥60 | 1.67 (1.44-1.94) | <.0001 |
| Sex | ||
| Male | 1.00 | |
| Female | 0.89 (0.80-0.99) | .04 |
| KPS at allo-HCT | ||
| 90-100 | 1.00 | |
| <90 | 1.33 (1.19-1.48) | <.0001 |
| HCT-CI | ||
| 0-2 | 1.00 | |
| ≥3 | 1.36 (1.19-1.55) | <.0001 |
| CMV donor/patient | ||
| +/− | 1.16 (1.03-1.32) | .01 |
| Other | 1.00 | |
| Sex match donor/patient | ||
| Female to male | 1.15 (1.00-1.31) | .04 |
| Other | 1.00 | |
| Genotype | ||
| CALR+/MPL+ | 1.00 | |
| JAK2+/triple negative | 1.56 (1.26-1.92) | <.0001 |
Overall P values were obtained using the Wald test. A total of 3559 patients and 1607 events were included in the model.
HD, haploidentical donor; MAC, myeloablative conditioning; MMUD, mismatched unrelated donor; MRD, matched related donor; MUD, matched unrelated donor; RIC, reduced intensity conditioning.
Kaplan-Meier curves illustrating OS after transplant based on risk groups defined by the prognostic models. (A-B) Kaplan-Meier plots showing OS according to the Cox regression statistical model in the training (A) and test (B) sets. (C-D) Kaplan-Meier plots displaying OS according to the ML model in the training (C) and test (D) sets. Patients were split according to the predicted quartile of risk. Each branch represents a quartile of patients with either low (blue), intermediate-low (green), intermediate-high (orange), and high risk (red).
Kaplan-Meier curves illustrating OS after transplant based on risk groups defined by the prognostic models. (A-B) Kaplan-Meier plots showing OS according to the Cox regression statistical model in the training (A) and test (B) sets. (C-D) Kaplan-Meier plots displaying OS according to the ML model in the training (C) and test (D) sets. Patients were split according to the predicted quartile of risk. Each branch represents a quartile of patients with either low (blue), intermediate-low (green), intermediate-high (orange), and high risk (red).
Risk model for OS using RSF
A RSF model was created to predict OS using the 52 initial variables of the data set. This model achieved a C-index of 0.603 in the training set and 0.632 in the test set. The variable-importance metrics for the model in the training set are shown in supplemental Figure 2. After dimensionality reduction, the model was refined to a smaller set of key prognostic variables: patient age, HCT-CI, KPS, blood blasts percentage, hemoglobin level, leukocyte and platelet counts, donor type, conditioning intensity, and graft-versus-host disease (GVHD) prophylaxis. This model achieved a C-index of 0.599 in the training set and 0.623 in the test set. Despite having performed hyperparameter tuning, no improvement in C-index was achieved (supplemental Table 2). To further elucidate these relationships, we have also included partial dependence plots in supplemental Figure 3.
Comparison of the RSF with other ML techniques
As shown in supplemental Table 3, RSF achieved higher concordance indices for OS and NRM predictions in both training and test sets compared with 3 alternative methods (ORSF, DeepSurv, and XGBoost). The consistent and superior performance of RSF across both data partitions justified its selection as the primary approach for downstream analyses.
Comparison of the ML model with the Cox regression–derived model
This analysis was performed on the subset of patients who had complete information on the variables included in the Cox-derived score to minimize biases (training set: n = 1773; test set: n = 566). In mortality prediction, the ML model demonstrated modestly better performance in the training set, achieving a C-index of 0.603 compared with the Cox-derived score of 0.594. The test set results confirmed the better discriminative capacity of the ML model, with a score of 0.612, surpassing the Cox-derived score C-index of 0.587 (Table 3). These findings were corroborated by the AIC scores, with the ML model showing lower values than the Cox-derived score in the test set, indicating a better overall model fit (Table 3).
Comparison of the performance of the ML model, the Cox-derived score, and the CIBMTR model
| . | ML model . | ML model . | Cox-derived score . | Cox-derived score . |
|---|---|---|---|---|
| 4 groups . | 4 groups . | |||
| OS risk score for mortality prediction (Harrell C-index) . | ||||
| Training set (n = 1773)∗ | 0.603 | 0.596 | 0.594 | 0.589 |
| Test set (n = 566)∗ | 0.612 | 0.608 | 0.587 | 0.580 |
| ML model | ML model | CIBMTR model | ||
| 4 groups | 3 groups | |||
| Training set (n = 1925)† | 0.608 | 0.599 | 0.557 | |
| Test set (n = 618)† | 0.654 | 0.650 | 0.581 | |
| OS risk score for mortality prediction (AIC) | ||||
| ML model | ML model | Cox-derived score | Cox-derived score | |
| 4 groups | 4 groups | |||
| Training set (n = 1773)∗ | 9 944 | 9 954 | 9945 | 9 954 |
| Test set (n = 566)∗ | 2 757 | 2 759 | 2765 | 2 767 |
| ML model | ML model | CIBMTR model | ||
| 4 groups | 3 groups | |||
| Training set (n = 1925)† | 12 573 | 12 588 | 12 647 | |
| Test set (n = 618)† | 3 318 | 3 322 | 3 374 | |
| OS risk score for NRM prediction (AIC) | ||||
| ML model | ML model | Cox-derived score | Cox-derived score | |
| 4 groups | 4 groups | |||
| Training set (n = 1763)∗ | 7 210 | 7 210 | 7208 | 7 214 |
| Test set (n = 566)∗ | 2 006 | 2 002 | 2008 | 2 006 |
| ML model | ML model | CIBMTR model | ||
| 4 groups | 3 groups | |||
| Training set (n = 1925)† | 8 738 | 8 738 | 8 762 | |
| Test set (n = 618)† | 2 154 | 2 160 | 2 182 | |
| . | ML model . | ML model . | Cox-derived score . | Cox-derived score . |
|---|---|---|---|---|
| 4 groups . | 4 groups . | |||
| OS risk score for mortality prediction (Harrell C-index) . | ||||
| Training set (n = 1773)∗ | 0.603 | 0.596 | 0.594 | 0.589 |
| Test set (n = 566)∗ | 0.612 | 0.608 | 0.587 | 0.580 |
| ML model | ML model | CIBMTR model | ||
| 4 groups | 3 groups | |||
| Training set (n = 1925)† | 0.608 | 0.599 | 0.557 | |
| Test set (n = 618)† | 0.654 | 0.650 | 0.581 | |
| OS risk score for mortality prediction (AIC) | ||||
| ML model | ML model | Cox-derived score | Cox-derived score | |
| 4 groups | 4 groups | |||
| Training set (n = 1773)∗ | 9 944 | 9 954 | 9945 | 9 954 |
| Test set (n = 566)∗ | 2 757 | 2 759 | 2765 | 2 767 |
| ML model | ML model | CIBMTR model | ||
| 4 groups | 3 groups | |||
| Training set (n = 1925)† | 12 573 | 12 588 | 12 647 | |
| Test set (n = 618)† | 3 318 | 3 322 | 3 374 | |
| OS risk score for NRM prediction (AIC) | ||||
| ML model | ML model | Cox-derived score | Cox-derived score | |
| 4 groups | 4 groups | |||
| Training set (n = 1763)∗ | 7 210 | 7 210 | 7208 | 7 214 |
| Test set (n = 566)∗ | 2 006 | 2 002 | 2008 | 2 006 |
| ML model | ML model | CIBMTR model | ||
| 4 groups | 3 groups | |||
| Training set (n = 1925)† | 8 738 | 8 738 | 8 762 | |
| Test set (n = 618)† | 2 154 | 2 160 | 2 182 | |
Analyses performed on the subset of patients with complete data on the variables included in the Cox-derived score and the CIBMTR model to minimize biases.
Higher C-index values reflect better model performance in ranking predictions whereas lower AIC values indicate a better fit to the data.
Cox-derived score.
CIBMTR model.
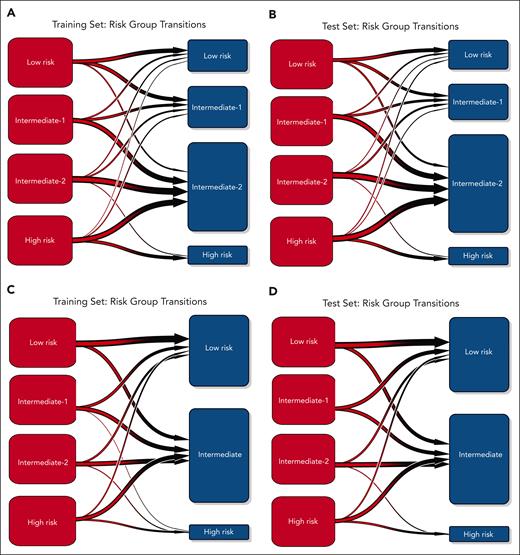
In refining our analysis, we segmented the ML score into 4 equal groups within the training set and applied the same classification thresholds to the test set (Figure 1C-D). The ML model maintained similar C-indices after segmentation, indicating that the model's prognostic accuracy is resilient to simplification (Table 3). A substantial reassignment of patients from the intermediate-2 risk group of the Cox score to other risk groups by the ML model was noted (Figure 2A-B). The time-dependent ROC AUCs comparing both models are presented in supplemental Figure 4.
Transition plots illustrating the flow of patients between the ML model and Cox-derived scoring systems. (A-B) Flow of patients between the ML model (red) and the Cox-derived score (blue) in the training (A) and test (B) cohorts. (C-D) Flow of patients between the ML model (red) and the CIBMTR model (blue) in the training (C) and test (D) cohorts.
Transition plots illustrating the flow of patients between the ML model and Cox-derived scoring systems. (A-B) Flow of patients between the ML model (red) and the Cox-derived score (blue) in the training (A) and test (B) cohorts. (C-D) Flow of patients between the ML model (red) and the CIBMTR model (blue) in the training (C) and test (D) cohorts.
Comparison of the ML model with the CIBMTR model
We compared the performance of the ML model with the CIBMTR scoring system8 in a comparable subset of patients with complete annotations for the CIBMTR score. By integrating patient age, hemoglobin level at transplant, and donor type, this model defined 3 risk categories in the original series, with a 3-year posttransplant OS of 69%, 51%, and 34% for low, intermediate, and high-risk groups, respectively.
The ML model achieved better performance, with a C-index of 0.608 vs 0.557 in the training set (n = 1925) and 0.654 vs 0.581 in the test set (n = 618; Table 3; supplemental Figure 5). Additionally, the lower AIC scores observed with the ML approach further validated these findings (Table 3). The time-dependent ROC AUCs comparing both models are presented in supplemental Figure 6. The difference between the CIBMTR score and ML method was mostly driven by prognostic refinement within the CIBMTR intermediate risk group, for which the ML algorithm reclassified most patients into different risk categories (Figure 2C-D).
Notably, the ML model exhibited a consistently better discriminative performance than the Cox-derived models for patients with PMF and patients with SMF, with the advantage being more pronounced in the test set (supplemental Table 4).
Application of the ML model to predict NRM
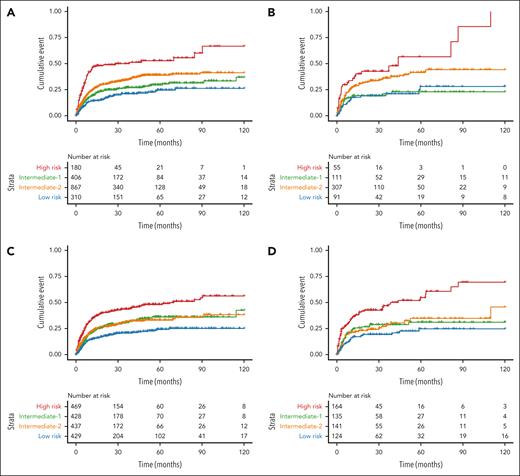
In predicting NRM, the ML model achieved comparable AIC scores to those of the Cox-based model in the training set but substantially lower AIC scores in the test set, indicating better overall performance (Table 3). Furthermore, when compared with the CIBMTR score, the ML model demonstrated an even more pronounced improvement in overall model fit (Table 3; Figure 3).
Cumulative incidence of NRM after transplant based on risk groups defined by the prognostic models. (A-B) Cumulative incidence of NRM according to the Cox-derived score in the training (A) and test (B) sets, and according to the ML model in the training (C) and test (D) sets. Patients were divided into 4 quartile groups according to their risk.
Cumulative incidence of NRM after transplant based on risk groups defined by the prognostic models. (A-B) Cumulative incidence of NRM according to the Cox-derived score in the training (A) and test (B) sets, and according to the ML model in the training (C) and test (D) sets. Patients were divided into 4 quartile groups according to their risk.
Ability of the ML model to identify patients at high risk of posttransplant mortality
The clinical utility of the ML model was evident in its ability to stratify patients into risk groups. Notably, it assigned 25% of patients to the high-risk group, significantly more than the 10.1% in the Cox-derived score and 8.2% in the CIBMTR model (Figure 2). Moreover, the ML model not only identified a larger proportion of high-risk patients but also showed consistent and generalizable results across training and test sets (Figure 1). In the training set, the 12- and 24-month OS rates for the ML high-risk group were 58.9% and 51.5%, respectively, closely aligning with the Cox-derived scores of 58.3% and 52.7%, respectively. In the test set, the ML high-risk group had OS rates of 61.0% at 12 months and 48.1% at 24 months, closely matching the 61.8% and 50.1% of the Cox model high-risk group.
The ML model also identified a larger high-risk population for NRM compared with the Cox-derived score (Figure 3). In the training set, the ML high-risk group had 12- and 24-month NRM rates of 34.9% and 40.7%, respectively, lower than the 46.3% and 48.8% observed in the Cox model high-risk group. However, in the test set, the ML high-risk group showed 12- and 24-month NRM rates of 36.4% and 42.6%, respectively, nearly matching the Cox score rates of 36.0% and 42.8%.
The comparison of patient distribution between the Cox-derived method and the CIBMTR model is elicited in supplemental Figure 7.
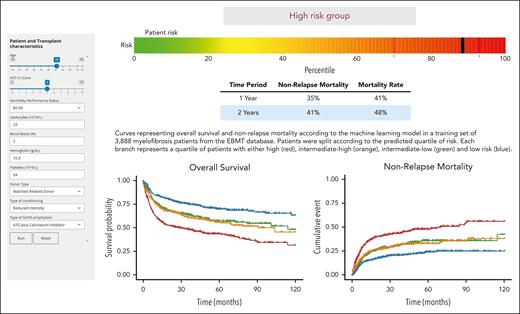
To predict OS after allo-HCT in MF, we developed a prognostic tool based on the RSF model, accessible as an interactive web application (https://gemfin.click/ebmt). Figure 4 illustrates the web-based calculator, showing the risk score for a hypothetical transplant candidate.
Illustration of the web-based calculator for the ML model. Risk score for a hypothetical MF patient candidate for transplantation. ATG, antithymocyte globulin.
Illustration of the web-based calculator for the ML model. Risk score for a hypothetical MF patient candidate for transplantation. ATG, antithymocyte globulin.
Impact of modifiable key factors of the transplantation procedure on OS
We compared OS after allo-HCT in patients who received the “optimal” donor type, conditioning intensity, or GVHD prophylaxis with those who did not. Optimal strategies were defined as those that maximize the survival probability according to the model’s predictions for a given patient, conditional on the other individual and disease characteristics. To ensure reliability, these predictions were evaluated exclusively on the test set.
For the optimal donor type, the univariable Cox proportional hazards model indicated that receiving transplantation from a donor type predicted as optimal by the ML model was associated with a HR of 0.76 (95% CI, 0.64-0.89; P = .001) compared with nonoptimal donor type, suggesting a statistically significant survival benefit. However, after adjusting for potential confounding factors in the multivariable analysis using inverse probability weighting (IPW),29 the HR was 0.96 (95% CI, 0.56-1.65; P = .89), indicating no significant survival advantage for ML-predicted optimal donor type.
The univariable Cox model revealed that patients receiving the conditioning regimen intensity predicted as optimal by the ML model had a HR of 0.89 (95% CI, 0.76-1.06; P = .19) compared with those receiving nonoptimal intensities. However, the IPW-adjusted analysis showed a HR of 1.025 (95% CI, 0.07-14.80; P = .99), indicating the lack of predictive value of the ML model to select the optimal conditioning regimen intensity.
Regarding GVHD prophylaxis, neither the unadjusted Cox model nor the IPW-adjusted analysis showed any significant difference in survival between patients receiving the ML-predicted optimal GVHD prophylaxis and those who did not. The unadjusted Cox model yielded a HR of 0.95 (95% CI, 0.79-1.15; P = .62), whereas the IPW-adjusted analysis resulted in a HR of 0.95 (95% CI, 0.43-2.09; P = .90). These results indicate no discernible impact of the ML-predicted optimal GVHD prophylaxis on patient survival.
Discussion
In this study, we have developed a ML model to enhance risk stratification for patients with MF undergoing allo-HCT, using a large database of 5183 patients with MF from the EBMT registry. Notably, this model is particularly comprehensive, because it considers the broad spectrum of current transplant practices, including diverse conditioning regimens, GVHD prophylaxis approaches, and donor types, such as haploidentical transplants. After dimensionality reduction, the model was simplified to a set of 10 key variables maintaining a notable discriminative capacity for both OS and NRM in both training and test sets.
Comparative analyses demonstrated a better performance of the ML model over a risk score developed within the same cohort using Cox regression methods. It also showed better discriminative capacity than the CIBMTR score,8 along with improved generalizability and an enhanced ability to identify a larger group of patients at high risk for posttransplant mortality. Notably, the improved performance of the ML model was evident in both patients with PMF and those with SMF. The ML model’s discriminative capacity remained higher after dividing patients into equally sized risk groups based on individual risk predictions, making the method comparable with traditional risk grouping strategies used in clinical practice. However, the intermediate risk categories identified by the ML model had similar OS and should be consolidated into a single, broader intermediate category, because the model lacks sufficient discrimination within this range. Although the C-indices of the ML model may be deemed moderate in terms of discriminative capacity, our data support its integration into clinical prognostics, offering a more refined and nuanced approach to managing the complexities of patient risk assessment before allo-HCT.
The clinical relevance of our ML model is evident in its ability to stratify allo-HCT candidates into well-defined risk categories. Notably, it identifies 25% of the cohort as high-risk with poor outcome after allo-HCT (∼35% NRM rate and 40% overall mortality at 1 year). Moreover, the web-based calculator permits the identification of a subset of very high-risk patients with a predicted 1-year OS of <50%, allowing for more tailored therapeutic interventions where needed. Accurate risk stratification is essential for optimizing allo-HCT outcomes, enabling physicians to select candidates who are most likely to benefit from transplantation, thereby improving treatment efficacy and patient survival.3 The ML model’s robust capability to classify these patients enhances its utility in clinical decision-making, ultimately fostering more personalized and effective patient management strategies in the complex landscape of allo-HCT.30
Although we have proved the feasibility of modeling risk using certain prognostic variables in the context of allo-HCT, the observational nature of the data caution against using these variables to guide the optimal transplantation procedure. The complexity of allo-HCT, characterized by numerous interacting factors that were not all included in our models, limits the practical utility of predictive models for determining the most effective treatment approaches.31 This underscores the need for continued research, randomized clinical studies, as well as sophisticated modeling techniques that can account for the dynamic and multifactorial nature of such medical interventions before they can be reliably implemented in clinical decision-making processes.
Our study has several limitations that warrant consideration. Firstly, allo-HCT is a multifaceted procedure influenced by numerous interrelated and independent variables, including differences in patient and disease characteristics and center-specific protocols. This complexity can significantly constrain the power of prognostic models, especially for detecting early posttransplant mortality, which may be influenced by acute and unforeseeable clinical events. We recognize that further investigation into potential, less obvious correlations among pretransplant risk factors could provide additional insights. However, our primary focus was on constructing a clinically actionable prognostic tool rather than conducting an in-depth mechanistic analysis of risk determinants. Additionally, the EBMT data set had a substantial rate of missing data for some variables. The ML method addressed this through data imputation, whereas our Cox model used the missing indicator method for variables with missing values, excluding those with a high degree of missingness (eg, hematologic parameters and spleen size) from score development. Although cross-validation was used to reduce overfitting in the ML model, it was not applied to the risk score developed using Cox regression. The lack of molecular annotation regarding additional somatic mutations, which has been shown to provide prognostic information after allo-HCT in some studies7,32 but not others,33,34 prevented a comparison with the Myelofibrosis Transplant Scoring System.7 Furthermore, data on the grade of bone marrow fibrosis and the variant allele frequency of driver mutations at transplant were not available. Future research could benefit from enhancing data completeness to potentially refine the model’s prognostic accuracy.
In conclusion, this investigation compared the effectiveness of 2 different Cox regression–derived models with a ML-driven approach for stratifying risk in patients with MF undergoing allo-HCT. The results demonstrate that the ML-driven model outperformed traditional statistical approaches by providing enhanced generalizability and identifying a broader subset of patients at high risk for adverse outcomes. ML methods facilitate the modeling of complex interactions and nonlinear associations more effectively than traditional statistical methods. However, our findings also underscore the challenges in predicting early posttransplant mortality based on conventional baseline characteristics, which remain difficult to anticipate with current prognostic tools. To improve clinical decision-making, we have developed a novel prognostic tool using ML techniques that can identify 25% of patients at high risk for mortality after transplantation. The web-based calculator (https://gemfin.click/ebmt) represents a significant advance toward personalized medicine for patients with MF, enabling better strategic planning and potentially improving outcomes. As we move forward, refining this tool through the integration of more comprehensive data and ongoing validation will be crucial to fully realize its clinical potential.
Acknowledgments
The authors are grateful to all the centers and patients contributing to the European Society for Blood and Marrow Transplantation database.
Authorship
Contribution: J.C.H.-B. and A.M.-O. conceived the idea and developed the project proposal; A.M.-O. performed the machine learning analysis, created figures and tables, and cowrote the first draft of the manuscript with J.C.H.-B.; L.G. and J.C.H.-B. developed the Cox regression–based prognostic model for survival; L.K. and J.T. managed the study data; J.R. contributed to the tables and cowrote part of the first draft; C.P.M. and D.C. designed the online calculator; and all other coauthors contributed data to the study, critically revised the paper, and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the centers from the Chronic Malignancies Working Party of the EBMT that participated in this study appears in the supplemental Appendix.
Correspondence: Juan Carlos Hernández-Boluda, Hematology Department, Hospital Clínico Universitario, Avd Blasco Ibáñez 17, 46010 Valencia, Spain; email: hernandez_jca@gva.es; and Donal P. McLornan, Department of Haematology and Stem Cell Transplantation, University College London Hospitals NHS Trust, 3rd Floor W Wing, 250 Euston Rd, London NW1 2PG, United Kingdom; email: donal.mclornan@nhs.net.
References
Author notes
J.C.H.-B. and A.M.-O. contributed equally to this study.
The data that support the findings of this study are available on request from the corresponding authors, Juan Carlos Hernández-Boluda (hernandez_jca@gva.es) and Donal P. McLornan (donal.mclornan@nhs.net). The data are not publicly available because of privacy or ethical restrictions.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal