Key Points
CD86 is a promising immunotherapeutic target in cHL, and the CD86–CTLA-4 axis orchestrates an immunosuppressive cHL tumor microenvironment.
CD86-targeted immunotherapies induce a distinct reprogramming of cHL-associated T cells and are highly effective treatment options for cHL.
Visual Abstract
The success of targeted therapies for hematological malignancies has heralded their potential as both salvage treatment and early treatment lines, reducing the need for high-dose, intensive, and often toxic chemotherapeutic regimens. For young patients with classic Hodgkin lymphoma (cHL), immunotherapies provide the possibility to lessen long-term, treatment-related toxicities. However, suitable therapeutic targets are lacking. By integrating single-cell dissection of the tumor landscape and an in-depth, single-cell–based off-tumor antigen prediction, we identify CD86 as a promising therapeutic target in cHL. CD86 is highly expressed on Hodgkin and Reed-Sternberg cancer cells and cHL-specific tumor-associated macrophages. We reveal CD86–CTLA-4 as a key suppressive pathway in cHL, driving T-cell exhaustion. Cellular therapies targeting CD86 had extraordinary efficacy in vitro and in vivo and were safe in immunocompetent mouse models without compromising bacterial host defense in sepsis models. Our results prove the potential value of anti-CD86 immunotherapies for treating cHL.
Introduction
In hematological malignancies, targeted immunotherapies including chimeric antigen receptor (CAR) T cells were found to have unparalleled clinical efficacy1,2 and are now considered standard of care as second-line treatment in early relapsed non-Hodgkin lymphoma (NHL).3,4
As cHL most often occurs at a younger age,5 long-term, therapy-related toxicity is almost as important as therapeutic efficacy. Although the multimodal first-line treatment regimens in cHL are highly effective (complete response rates >90%6), up to 94% of long-term cHL survivors have treatment-related comorbidities, including treatment-related secondary neoplasm, cardiopulmonary toxicities, and infertility.7-9
As recently found in the HD-21 clinical trial,6 the development of novel immunotherapies and their application in early treatment lines possess the potential to reduce these toxicities, yet, alternatives to CD30-targeted immunotherapies have rarely been reported.10,11 Although immune checkpoint blockade is highly active12 and considered as a standard of care in relapsed/refractory cHL (R/R cHL), long-term remissions are scarce.13 Anti-CD30 CAR T-cell therapy was found to have promising results in a phase 1/2 clinical trial,14 but early reports also revealed high rates of toxicities and overall limited antitumor activity.15
To address this unmet medical need, we developed a cHL-tailored, multimodal screening algorithm to identify unrecognized and functionally relevant target antigens for cHL. We identified the central role of the CD86–CTLA-4 axis in orchestrating the immunosuppressive cHL tumor microenvironment (TME), with CD86 having high expression on HRS cells (HRSCs) and cHL-associated tumor-associated macrophages (cHL TAMs). CD86 blockade reverts the exhausted phenotype of cHL-associated T cells, and preclinical testing of anti-CD86 CAR T cells were found to have extraordinary treatment efficacy, without exhibiting relevant safety signals in immunocompetent mouse models.
Methods
Ethics approval and consent
Fixed, paraffin-embedded cHL specimens were obtained after written informed consent in accordance with the regulation of the Declaration of Helsinki. The use of the cHL samples for immunohistochemistry and multiplexed immunofluorescence was approved by the institutional review board of the Julius Maximilian University, Würzburg, Germany.
Software and statistical analysis
Flow cytometric data were analyzed using FlowJo version (v.)10.3 to v.10.9.0 software. Established ImageJ was used for the analysis of chip cytometric images using established protocols.16 Radiance calculation of images of bioluminescence imaging (BLI) was performed using Living Image 4.4 (PerkinElmer, Shelton, CT).
LASX (v.3.8) and deconvolution were performed with the fully integrated Leica lightening software plugin using standard settings. Image processing was done in Imaris (v.10.0). Live cell imaging analyses were performed with Incucyte S3 Software 2022B, Rev2. Immunohistochemical stainings were analyzed with QuPath (v.0.4.4). All statistical analyses were performed using GraphPad Prism software v.9.3.1 to v.9.4.1 (San Diego, CA).
A detailed description of all other methods can be found in the supplemental Material, available on the Blood website.
Results
Multimodal screening approach identifies CD86 as an immunotherapeutic target in cHL
To identify target antigens for cHL, we integrated publicly available microarray gene expression analysis of laser-dissected HRSCs and subsequent flow cytometric validation with single-cell analysis of the cHL TME. In addition, we carried out single-cell–based off-tumor antigen prediction using publicly available data sets (Figure 1A).
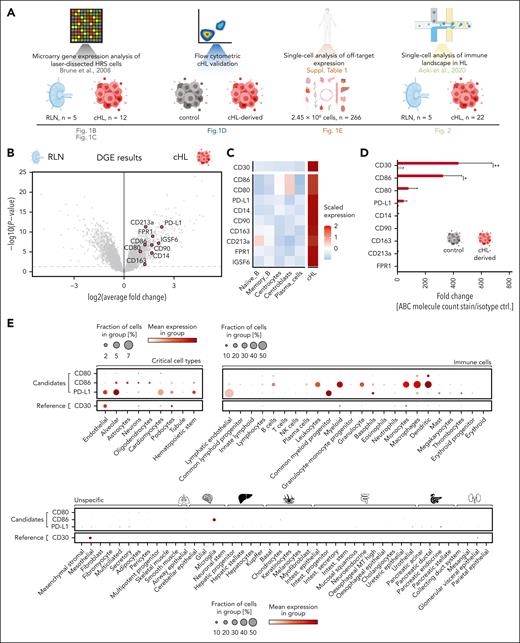
Identification of CD80, CD86, and PD-L1 as potential therapeutic targets in cHL. (A) Schematic overview of components of the used multimodal target screening approach in cHL. (B) Volcano plot illustrating the log2 average fold change and P values of surface antigens absent on T cells and differentially overexpressed on HRSCs compared with GCB. Microarray data (GSE12453) were obtained from the gene expression omnibus17 (cHL, n = 12; RLN, n = 5). Labeled genes in red passed all filters (surface expression, absent on T cells), whereas non-highlighted gray dots indicate overexpressed genes that did not pass the filter thresholds. (C) Heat map visualizing the expression of the identified target antigens on microdissected control cells (left) or HRSCs (right). (D) Comparison of absolute densities of indicated antigens measured using flow cytometry on a panel of cHL cell lines (L-428, L-540, KM-H2) with that of the control cell line Nalm-6. Plotted is the pooled fold change ± standard error of the mean (SEM) of absolute molecule count in comparison to an isotype control stain of 3 different cHL cell lines. Statistical significance was calculated using 2-way analysis of variance (ANOVA) with Sidak multiple comparison correction. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05. (E) Single-cell cross-organ off-target transcriptomic atlas screening for CD80, CD86, PD-L1, and CD30. The transcriptomic atlas consists of a total of 2.5 million sequenced cells comprising 11 different organs. A detailed summary of all used data sets is provided in the supplementary Material.
Identification of CD80, CD86, and PD-L1 as potential therapeutic targets in cHL. (A) Schematic overview of components of the used multimodal target screening approach in cHL. (B) Volcano plot illustrating the log2 average fold change and P values of surface antigens absent on T cells and differentially overexpressed on HRSCs compared with GCB. Microarray data (GSE12453) were obtained from the gene expression omnibus17 (cHL, n = 12; RLN, n = 5). Labeled genes in red passed all filters (surface expression, absent on T cells), whereas non-highlighted gray dots indicate overexpressed genes that did not pass the filter thresholds. (C) Heat map visualizing the expression of the identified target antigens on microdissected control cells (left) or HRSCs (right). (D) Comparison of absolute densities of indicated antigens measured using flow cytometry on a panel of cHL cell lines (L-428, L-540, KM-H2) with that of the control cell line Nalm-6. Plotted is the pooled fold change ± standard error of the mean (SEM) of absolute molecule count in comparison to an isotype control stain of 3 different cHL cell lines. Statistical significance was calculated using 2-way analysis of variance (ANOVA) with Sidak multiple comparison correction. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05. (E) Single-cell cross-organ off-target transcriptomic atlas screening for CD80, CD86, PD-L1, and CD30. The transcriptomic atlas consists of a total of 2.5 million sequenced cells comprising 11 different organs. A detailed summary of all used data sets is provided in the supplementary Material.
We first obtained available transcriptomic profiles of laser-dissected HRSCs and carried out differential gene expression (DGE) analysis.17 We identified 4293 genes that were overexpressed on HRSCs compared with germinal center B cells. To determine target accessibility to immunotherapies, we next used a library consisting of 4924 genes that encode cell surface proteins established by integrating different available surfaceome libraries.18-21 We manually removed genes encoding proteins that are primarily secreted and proteins with known intracellular functions from the candidate list. Finally, to ensure transferability to T-cell–based therapies, we excluded genes high on T cells, as measured by single-cell RNA-sequencing (scRNA-seq). Using this stringent approach, we identified 8 genes encoding potential targets highly expressed in HRSCs, which are as follows: CD14, CD80, CD86, THY1 (CD90), CD163, IL13RA1 (CD213a), CD274 (PD-L1), and IGSF6 (Figure 1B-C). CD30 was excluded due to high expression on cHL-infiltrating T cells, but it was used as a reference control for all further analyses.
To confirm these results, we quantified expression of the target antigens on 6 different cell lines using published microarray profiles. Three NHL cell lines were used as controls. Of the above-described candidates, CD86 was most highly expressed on cHL cell lines with overall comparable expression to CD30 (supplemental Figure 1A). Comparison to NHL cell lines revealed a higher expression of CD86, CD80, and PD-L1 on cHL cell lines, whereas CD90 expression was similar (supplemental Figure 1B). We next performed flow cytometric analysis for all candidate antigens, except for IGFS6, due to the lack of a suitable antibody, on cHL cell lines (L-428, L-540, KM-H2) and on control non-cHL B-cell line (Nalm-6). Only CD30 and CD86 were validated on all 3 cHL cell lines, PD-L1 and CD80 were only moderately expressed on 1 and 2 cell lines, respectively, whereas other antigens were undetectable by flow cytometry (Figure 1D).
To obtain off-tumor expression patterns of CD80, CD86, PD-L1, and CD30, we carried out in-depth, off-tumor antigen prediction using a total of 24 different scRNA-seq data sets of 11 healthy human tissues.16,22-44 This atlas consisted of nearly 2.5 million sequenced single cells from 266 individuals (supplemental Figure 1C). PD-L1 was most strongly expressed on cell clusters of tissues, reported as targets for on-target, off-tumor toxicities of CAR T cells (endothelial cells, alveolar cells, cardiomyocytes, podocytes, hematopoietic stem cells).45,46 In contrast, CD80, CD86, and CD30 were minimally expressed in these clusters. CD80 and CD86 expression were detected on lymphoid and myeloid immune cells, with CD86 having higher baseline expression than CD80. CD30 was minimally expressed on immune cells but was expressed on mesothelial and glomerular visceral epithelial cells (Figure 1E).
The CD80–CD86–CTLA-4–CD28 axis is active in cHL and shifted toward CTLA-4–mediated immunosuppression
Our analyses centered on HRSCs. Yet, HRSCs represent only 1% to 5% of the entire cHL TME. The remaining tumor mass consists of infiltrating immune cells.47 CD30, PD-L1, CD80, and CD86 are all central mediators of the adaptive immune response. The CD80-CD86 axis can provide either immunostimulatory or immunosuppressive stimuli, depending on the expression patterns of its ligands CTLA-4 and CD28, whereas PD-1–PD-L1/PD-L2 axis is primarily known for its immunosuppressive function. CTLA-4 binds to CD80 and CD86 with higher affinity than CD28, and high expression of CTLA-4 can tip the equilibrium toward immunosuppression.17,48
To evaluate the advantages and disadvantages of targeting these critical immunomodulatory molecules in cHL, we characterized functions of these axes at the tumor site by analyzing a scRNA-seq data set of 22 cHL specimens and 5 reactive lymph node (RLN) controls49 (Figure 2A).
scRNA-seq reveals dysregulated immunomodulatory pathways in cHL. (A) Uniform manifold approximation and projection (UMAP) visualization of the obtained cell clusters in RLN (left) or cHL (right). The data set was generated by Aoki et al and downloaded from the European genome-phenome archive (cHL, n = 22; RLN, n = 5; 127 686 sequenced cells). (B) Heat map visualization of the scaled averaged gene expression of CD30 vs CD86, CD80, and PD-L1 measured by scRNA-seq. (C) UMAP plots visualizing the expression of CD86 (left) or CD30 (right) on the respective cell clusters. (D) Fold changes based on the differential abundance of the respective cell cluster between RLN (left) and cHL (right). (E-F) Heat map illustrating significantly upregulated genes of hallmark immune inhibitory pathways (E) and activating pathways (F) in cHL (red) or RLN (blue) as the result of DGE analysis within each cell subset. Markers were adapted from Chen and Flies50 with permission.
scRNA-seq reveals dysregulated immunomodulatory pathways in cHL. (A) Uniform manifold approximation and projection (UMAP) visualization of the obtained cell clusters in RLN (left) or cHL (right). The data set was generated by Aoki et al and downloaded from the European genome-phenome archive (cHL, n = 22; RLN, n = 5; 127 686 sequenced cells). (B) Heat map visualization of the scaled averaged gene expression of CD30 vs CD86, CD80, and PD-L1 measured by scRNA-seq. (C) UMAP plots visualizing the expression of CD86 (left) or CD30 (right) on the respective cell clusters. (D) Fold changes based on the differential abundance of the respective cell cluster between RLN (left) and cHL (right). (E-F) Heat map illustrating significantly upregulated genes of hallmark immune inhibitory pathways (E) and activating pathways (F) in cHL (red) or RLN (blue) as the result of DGE analysis within each cell subset. Markers were adapted from Chen and Flies50 with permission.
We pooled and integrated sequencing data of all samples to offset batch effects during sample preparation. Uniform manifold approximation and projection (UMAP) visualization illustrate the successful integration of the obtained data (supplemental Figure 2A). We reannotated the data set based on upregulated functional signatures from DGE analysis and identified 25 different cell clusters (supplemental Figure 2B-C), spanning the most relevant innate and adaptive immune cell subtypes and a distinct “proliferative” cell cluster with high mitotic activity and positive for proliferative markers (Ki67; Figure 2A; supplemental Figure 2B-D). As previously reported by Aoki et al,49 in this data set, we failed to detect HRSC, most likely due to loss of these cells during preprocessing procedures and their overall rarity at the tumor site.
We next analyzed expression of CD30, CD80, CD86, and PD-L1 on infiltrating immune cells and found that all 4 were expressed on cHL TAMs. CD80 and CD86 were also expressed on B-cell subtypes (germinal center B cells; plasma cells; memory B cells, B_m), whereas PD-L1 was expressed on different regulatory T-cell (Treg) populations. CD30 was strongly expressed on Treg_3 and Treg_4 clusters and on the proliferative cell cluster (Figure 2B-C). To better understand characteristics of cHL-infiltrating immune cells, we next interrogated the difference in fractions of immune cell populations between RLN and cHL samples. Macrophages (log2 FC = 3.46), Treg_3 (log2 FC = 2.04), and proliferative cells (log2 FC = 1.62) were highly enriched in cHL compared with RLN controls (Figure 2D; supplemental Figure 2E-F). Importantly, these nearly cHL-exclusive immune cell subtypes highly expressed our genes of interest (Figure 2B).
Using DGE analysis, we compared differences in gene expression of known inhibitory (Figure 2E) and activating (Figure 2F) signaling pathways50 between cHL and RLN specimens. We observed upregulation of several hallmark inhibitory and activating axes in cHL compared with RLN controls (Figure 2E-F). As previously described, the human leukocyte antigen class II, DR alpha (HLA-DRA)–LAG3 (lymphocyte-activation gene 3) axis was highly active in cHL, with higher expression of LAG3 on CD4+ and CD8+ T-cell subsets in cHL compared with RLN.49 At the single-cell level, the CD80–CD86–CTLA-4 axis was the second most prominent axis, with high expression of CTLA-4 in Tregs and CD4 memory cells (CD4m_1, 2) and overexpression of CD86 in cHL TAMs (Figure 2E).
Collectively, in line with previous results,51 our single-cell analyses revealed overexpression of several hallmark immunosuppressive pathways in the TME of cHL compared with RLN.
CD86 is highly expressed in primary diagnosis and relapsed cHL
To confirm expression of the identified target antigens in a larger validation cohort and in R/R cHL, we obtained NanoString profiling data of 95 patients with primary diagnosis of cHL, who were treated within the NIVAHL phase 2 trial.52 In NIVAHL, patients with early stage unfavorable cHL were either randomized to receive 4 cycles of concomitant nivolumab-AVD (nivolumab, doxorubicin, vinblastine, dacarbazine) treatment or a sequential therapeutic regimen consisting of 4 cycles of nivolumab monotherapy, followed by 2 cycles of nivolumab-AVD and 2 cycles of AVD. We also obtained matched-pair samples of early treatment biopsies during first-line therapy (Figure 3A; n = 10). Matched-pair biopsies were split into samples revealing complete HRSC clearance (n = 4, left) or HRSC maintenance (n = 6, right). As with our previous data, CD30 was most highly expressed at primary diagnosis, followed by CD86, CD80, and PD-L1 (Figure 3B). In matched-pair biopsies, CD30 expression was lower in secondary tissue, independent of treatment response, whereas no change was observed for CD86, CD80, or PD-L1 (Figure 3C).
CD86 is highly expressed in primary diagnosis and R/R cHL. (A) Summary of treatment schedules and cohort distribution of the NIVHAL trial.52 (B) Heat map illustrating expression of reference control gene CD30 or candidate antigens CD86, CD80, and PD-L1 in the NIVHAL cohort (n = 95 primary diagnosis samples). Each line represents 1 patient. (C) Change of target antigen expression between primary (gray) and secondary (light green, red) biopsies after NIVHAL trial first-line therapy. Treatment response was evaluated by a board-certified pathologist and classified into HRSC clearance (light green, n = 4 primary patient samples) or HRSC maintenance (red, n = 6 primary patient samples). Statistical significance was calculated using 2-way ANOVA with Sidak multiple comparison correction. (D-E) H&E (left) or immunohistochemical staining of CD86 (middle) in comparison to CD30 (right) in primary cHL tissue. (D,F,G) Primary diagnosis cHL samples. n = 7 different patients. (E,H-I) R/R cHL samples. n = 10 different patients. (F,H) Representative images of CD86+ HRSCs. Arrowheads indicate HRSCs. (G,I) Count of CD86+ or CD30+ HRSCs in primary diagnosis cHL samples (G) or R/R cHL (I) quantified by machine learning classifier. Statistical significance was calculated using unpaired t test. (J-K) Expression of CD86 on HRSCs measured by multiplex immunofluorescence microscopy on chip-loaded primary diagnosis cHL samples (J) or R/R cHL (K). Chips were sequentially stained with antibodies against CD86 (yellow), CD30 (blue), and CD20 (red) and with a DNA Hoechst stain (blue). Between each staining step, images were acquired with a fluorescence microscope followed by a 30-second photobleaching procedure. For all panels: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05.
CD86 is highly expressed in primary diagnosis and R/R cHL. (A) Summary of treatment schedules and cohort distribution of the NIVHAL trial.52 (B) Heat map illustrating expression of reference control gene CD30 or candidate antigens CD86, CD80, and PD-L1 in the NIVHAL cohort (n = 95 primary diagnosis samples). Each line represents 1 patient. (C) Change of target antigen expression between primary (gray) and secondary (light green, red) biopsies after NIVHAL trial first-line therapy. Treatment response was evaluated by a board-certified pathologist and classified into HRSC clearance (light green, n = 4 primary patient samples) or HRSC maintenance (red, n = 6 primary patient samples). Statistical significance was calculated using 2-way ANOVA with Sidak multiple comparison correction. (D-E) H&E (left) or immunohistochemical staining of CD86 (middle) in comparison to CD30 (right) in primary cHL tissue. (D,F,G) Primary diagnosis cHL samples. n = 7 different patients. (E,H-I) R/R cHL samples. n = 10 different patients. (F,H) Representative images of CD86+ HRSCs. Arrowheads indicate HRSCs. (G,I) Count of CD86+ or CD30+ HRSCs in primary diagnosis cHL samples (G) or R/R cHL (I) quantified by machine learning classifier. Statistical significance was calculated using unpaired t test. (J-K) Expression of CD86 on HRSCs measured by multiplex immunofluorescence microscopy on chip-loaded primary diagnosis cHL samples (J) or R/R cHL (K). Chips were sequentially stained with antibodies against CD86 (yellow), CD30 (blue), and CD20 (red) and with a DNA Hoechst stain (blue). Between each staining step, images were acquired with a fluorescence microscope followed by a 30-second photobleaching procedure. For all panels: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05.
Our multimodal analyses revealed CD86 as the most relevant target antigen with overall comparable expression to CD30 (overview of all cohorts in supplemental Figure 3A). Thus, we decided to primarily investigate CD86 as a novel therapeutic target for cHL.
To gain deeper insights into expression profiles of CD86 in cHL, we carried out immunohistochemistry staining of either primary diagnosis cHL samples (Figure 3D,F-G) or R/R cHL (Figure 3E,H-I; supplemental Figure 3B). CD86 was highly expressed on HRSCs of both primary diagnosis and R/R cHL with an overall count of CD86+ tumor cells comparable to CD30 (Figure 3G,I). To assess coexpression patterns of CD86 and CD30, we conducted multiplexed immunofluorescence of formalin-fixed, paraffin-embedded primary cHL tissues using ChipCytometry (Figure 3J-K; supplemental Figure 3C-D). Chip-loaded cHL specimens from either primary diagnosis (Figure 3J) or R/R cHL (Figure 3K) were sequentially stained with anti-CD86, anti-CD30, and anti-CD20 antibodies and the Hoechst stain, for DNA, and then visualized by immunofluorescence microscopy. Staining intensities revealed high expression of both CD86 and CD30 in primary and R/R cHL, with general expression patterns of CD86 comparable to CD30. Importantly, CD86-expressing HRSCs were present in all stained samples, in both nodular sclerosis and mixed cellularity subtype (supplemental Figure 3C-D).
In summary, we reveal broad and conserved expression of CD86 on HRSCs both at primary diagnosis and in R/R cHL patients.
CD86-targeted immunotherapies reverse cHL-associated immunosuppression
Our scRNA-seq analyses highlighted a pivotal role of the CD86–CTLA-4 axis in cHL TME and suggested a shift toward CTLA-4–mediated immunosuppression. NanoString profiling of biopsies from patients treated within the NIVAHL trial confirmed these results (Figure 4A). Analyses of the NIVAHL matched-pair biopsies revealed a downregulation of CTLA-4 after PD-1 checkpoint inhibition, yet with still relevant expression levels in secondary biopsies (Figure 4B).
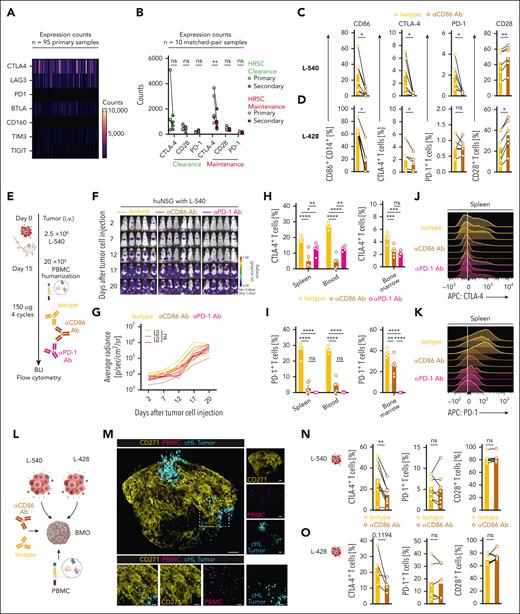
CD86 blockade reduces expression of inhibitory cell surface markers PD-1 and CTLA-4 on cHL-associated T cells. (A) Heat map illustrating expression of inhibitory signaling molecules of the NIVHAL cohort (n = 95 primary diagnosis samples). Each line represents 1 patient. (B) Change of inhibitory signaling molecules between primary (gray) and secondary (red) biopsies. Treatment response was evaluated by a board-certified pathologist and classified into HRSC clearance (light green, n = 4) or HRSC maintenance (red, n = 6). Statistical significance was calculated using 2-way ANOVA with Sidak multiple comparison correction. (C-D) Expression of indicated antigens on either CD14+ monocytes (left, CD86) or CD3+ T cells (middle left, CTLA-4; middle right, PD-1; right, CD28) measured by flow cytometry. cHL cell lines (L-540, C; L-428, D) and PBMCs were cocultured with either αCD86 antibody (mustard) or isotype control antibody (yellow). Data are mean ± SEM from 6 independent donors. Statistical significance was calculated using Wilcoxon signed rank test. (E) Summary of the treatment schedule used for humanized L-540 xenograft in vivo experiments. (F-G) BLI images (F) and fLuc-BLI–based quantification of tumor burden (G) of L-540 tumor-bearing mice injected with human PBMCs and treated with αCD86 antibody (mustard), isotype control antibody (yellow), or αPD-1 antibody (magenta). (H-I) Flow cytometric quantification of CTLA-4 (H) or PD-1 expression (I) in different organs on CD3+ T cells in different organs. There were 5 to 6 mice per group. Statistical significance was calculated using 2-way ANOVA with Tukey multiple comparison correction. (J-K) Representative histograms illustrating expression of CTLA-4 (J) or PD-1 (K) on CD3+ T cells in the spleen. (L) Overview of experimental scheme used to induce CD86 blockade in complex BMO. (M) Representative confocal images of cHL-PBMC-BMO cocultures. Yellow: mesenchymal tissue (CD271); violet: PBMC; turquoise: cHL tumor cells. Magnified images of the rectangle area are depicted at the bottom. (N-O) Expression of indicated antigens on CD3+ T cells (left, CTLA-4; middle PD-1; right CD28) in coculture with L-540 (N) or L-428 (O) cHL tumor cell lines measured by flow cytometry. cHL cell lines and BMO were cocultured either with αCD86 antibody or isotype control antibody. Data are mean ± SEM from n = 4-8 independent donors and BMO. Scale bar in panel M 100 µm (top), 30 µm (bottom). Statistical significance was calculated using Wilcoxon signed rank test. For all panels: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05.
CD86 blockade reduces expression of inhibitory cell surface markers PD-1 and CTLA-4 on cHL-associated T cells. (A) Heat map illustrating expression of inhibitory signaling molecules of the NIVHAL cohort (n = 95 primary diagnosis samples). Each line represents 1 patient. (B) Change of inhibitory signaling molecules between primary (gray) and secondary (red) biopsies. Treatment response was evaluated by a board-certified pathologist and classified into HRSC clearance (light green, n = 4) or HRSC maintenance (red, n = 6). Statistical significance was calculated using 2-way ANOVA with Sidak multiple comparison correction. (C-D) Expression of indicated antigens on either CD14+ monocytes (left, CD86) or CD3+ T cells (middle left, CTLA-4; middle right, PD-1; right, CD28) measured by flow cytometry. cHL cell lines (L-540, C; L-428, D) and PBMCs were cocultured with either αCD86 antibody (mustard) or isotype control antibody (yellow). Data are mean ± SEM from 6 independent donors. Statistical significance was calculated using Wilcoxon signed rank test. (E) Summary of the treatment schedule used for humanized L-540 xenograft in vivo experiments. (F-G) BLI images (F) and fLuc-BLI–based quantification of tumor burden (G) of L-540 tumor-bearing mice injected with human PBMCs and treated with αCD86 antibody (mustard), isotype control antibody (yellow), or αPD-1 antibody (magenta). (H-I) Flow cytometric quantification of CTLA-4 (H) or PD-1 expression (I) in different organs on CD3+ T cells in different organs. There were 5 to 6 mice per group. Statistical significance was calculated using 2-way ANOVA with Tukey multiple comparison correction. (J-K) Representative histograms illustrating expression of CTLA-4 (J) or PD-1 (K) on CD3+ T cells in the spleen. (L) Overview of experimental scheme used to induce CD86 blockade in complex BMO. (M) Representative confocal images of cHL-PBMC-BMO cocultures. Yellow: mesenchymal tissue (CD271); violet: PBMC; turquoise: cHL tumor cells. Magnified images of the rectangle area are depicted at the bottom. (N-O) Expression of indicated antigens on CD3+ T cells (left, CTLA-4; middle PD-1; right CD28) in coculture with L-540 (N) or L-428 (O) cHL tumor cell lines measured by flow cytometry. cHL cell lines and BMO were cocultured either with αCD86 antibody or isotype control antibody. Data are mean ± SEM from n = 4-8 independent donors and BMO. Scale bar in panel M 100 µm (top), 30 µm (bottom). Statistical significance was calculated using Wilcoxon signed rank test. For all panels: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05.
Although the expression analyses reveal a high abundance of CD86 and CTLA-4 expression in cHL, the functional consequences of modulating the CD86–CTLA-4 axis remain unknown.
Consequently, we first investigated whether CD86 blockade influences the proliferative capacity or the phenotype of cHL cell lines. Although CD86 was highly expressed on all tested cell lines (supplemental Figure 4A-B), the addition of a CD86-blocking antibody (αCD86) did not affect cHL proliferation as measured by live cell imaging (supplemental Figure 4C-D). Similarly, clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (CRISPR-Cas9) knockout of CD86 did not affect the proliferation of KM-H2 cells (supplemental Figure 4E-F). CD86 blockade also did not change the expression of several HRSC-associated surface molecules, except for CD86 (supplemental Figure 4G-H).
Next, we designed in vitro experiments that would inform on the impact of CD86-targeted immunotherapies on the cHL TME. We cocultured cHL cell lines L-540, L-428, KM-H2, or Nalm-6 control cells with peripheral blood mononuclear cells (PBMCs) from healthy donors for 5 days in the presence of either an αCD86 or an isotype antibody. In coculture with cHL cell lines, but not with Nalm-6 cells, CD14+ monocytes highly expressed CD86. Treatment with αCD86 antibody reduced surface CD86 expression (Figure 4C-D; supplemental Figure 5C-D, left). Interestingly, CD86 blockade diminished expression of CTLA-4 and PD-1 on T cells (Figure 4C-D; supplemental Figure 5C-D, middle). Conversely, surface CD28 expression increased on T cells in cocultures with L-540 and L-428 cHL cells, but not with KM-H2 or Nalm-6 cells (Figure 4C-D; supplemental Figure 5C-D, right).
To corroborate these finding, we developed a PBMC-humanized mouse model of cHL, using tumor-bearing–immunodeficient NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ or tm1/Rj (NSG/NXG) mice. Humanized mouse models were chosen due to the lack of relevant cHL models in immunocompetent mice. We first injected 2 × 106 L-540 cells transduced with a firefly luciferase (L-540-fLuc+) intravenously into NSG mice. Tumor growth was measured with BLI. After 15 days, we injected 2 × 107 PBMCs of healthy human donors into L-540 tumor-bearing mice and regularly treated with αCD86, isotype, or an anti–PD-1 antibody (nivolumab, αPD-1; Figure 4E). Antibody treatment did not inhibit tumor progression in any of the mice (Figure 4F-G). Seven days after PBMC injection, we sacrificed the mice and analyzed the phenotype of human immune cells with flow cytometry. Antibody treatment did not reduce tumor burden in the bone marrow, spleen, or blood of mice (supplemental Figure 5E).
Intriguingly, αCD86 antibody led to a striking reduction of CTLA-4 expression on CD3+ T cells in the spleen, blood, and bone marrow of the mice. αPD-1 treatment also resulted in a downregulation of CTLA-4 expression, however to a lower extent than αCD86 (Figure 4H, J). As expected, αPD-1 treatment induced a near-complete absence of measurable surface PD-1 expression. T cells of αCD86-treated mice revealed similar low PD-1 levels in the spleen and blood as αPD-1–treated mice. In the bone marrow, this reduction was less pronounced (Figure 4I,K). CD28 was lower in T cells in the spleen and blood, but not in the bone marrow of αCD86 antibody-treated mice (supplemental Figure 5F).
L-540 tumor cells were originally established from the bone marrow of a 20-year-old patient with advanced cHL, and it is known that the characteristic cHL microenvironment is also present in the case of primary bone marrow involvement.53 To provide proof of principle in a clinically relevant, 3-dimensional, fully human model of the cHL-bone marrow niche, we used a recently published induced pluripotent stem cell-based bone marrow organoid (BMO) model.54 The BMO was cocultured with L-540 or L-428 cells and PBMC (addition of PBMC is necessary, as BMO lacks mature T cells due to missing thymic development and priming steps) and treated with αCD86 or an isotype control (Figure 4L). BMO cocultures were then visualized with confocal microscopy or dissociated and analyzed by flow cytometry. Both cHL tumor cells and PBMC engrafted the BMO niche, visualized by CD271 expression on mesenchymal stromal cells (Figure 4M). Again, αCD86 antibody led to a striking reduction of CTLA-4 expression in L-540–PBMC–BMO cocultures (Figure 4N-O, left). In L-428 cocultures, similar trends were observed. Surface PD-1 expression in both models did not change (Figure 4N-O, middle). No changes in CD28 expression were also observed (Figure 4N-O, right).
Altogether, in several advanced disease models, we reveal a distinct reprogramming of cHL-associated T cells by CD86 blockade.
Anti-CD86 CAR T cells engage and lyse cHL cell lines in vitro
Given the high expression of CD86 on HRSCs, cHL TAMs, and the distinct TME reprogramming after CD86 blockade, we hypothesized that CD86-directed CAR T cells would have triple-effector functions by simultaneously targeting key, pathognomonic components of the cHL TME.
We designed a second-generation anti-CD86 CAR harboring CD28 and CD3 zeta stimulatory domains (CD86-28z), which was equivalent to that of the anti-CD30 CAR T cells (CD30-28z) currently used in clinical trials.55 Anti-CD19 CAR T cells (CD19-28z) were used as a negative control for all experiments (Figure 5A). All CAR constructs could be efficiently transduced into primary human T cells (Figure 5B) with average transduction efficiencies of approximately 60% (Figure 5C). Flow cytometric phenotyping of CAR T cells after viral transduction did not reveal relevant differences in the cellular composition of T cell subsets of the generated CAR T cell products or untransduced control cells (supplemental Figure 5G-H).
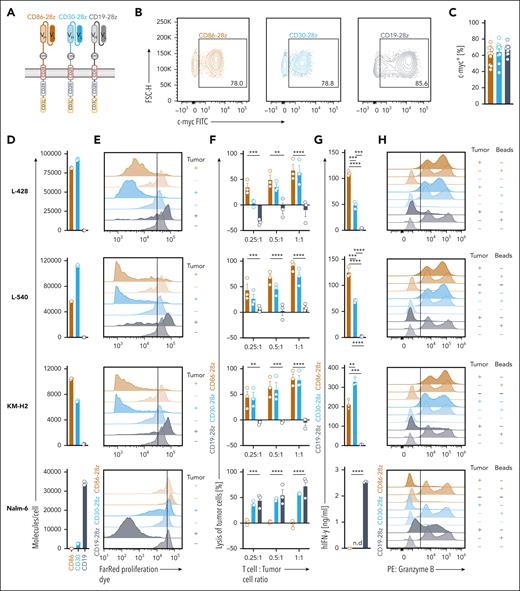
CD86-28z CAR T cells exhibit high potency toward cHL cell lines in vitro. (A) Summary of the composition of anti-CD86 (CD86-28z), anti-CD30 (CD30-28z), and anti-CD19 CAR (CD19-28z) constructs. (B-C) Representative flow cytometric images (B) and quantification (C; individual results and mean ± SEM of 10 different donors) of transduction efficiencies. Transduction efficiency was determined by staining for the extracellular c-Myc tag. (D) Absolute quantification of the molecule count per cell measured with quantitative flow cytometry. Molecule counts/cell of the indicated cell lines were calculated for CD86, CD30, and CD19, respectively. Molecule counts/cell of the isotype control were subtracted from total molecule counts. Depicted are 3 biological replicates. Data are representative of 2 independent experiments. (E-H) CD86-28z (mocha), CD30-28z (light blue), or CD19-28z (gray) CAR T cells were cocultured with the indicated cell lines (from top to bottom: L-428, L-540, KM-H2, Nalm-6, all transduced with fLuc-GFP). (E) Before cocultures, CAR T cells were stained with a Far Red proliferation dye, and antigen-specific proliferation was determined by trace dilution. Cocultures were analyzed by flow cytometry after 7 days. Top row of each color: CAR cocultured with tumor cells. Bottom row of each color: CAR only. Illustrated are representative histograms of in total 3 different donors. (F) Bioluminescence measurement of CAR-mediated lysis of tumor cells. Cell numbers were plated according to the indicated T-cell:tumor cell ratio. Tumor cell killing was determined after 72 hours. Specific lysis was calculated by normalizing to tumor cell-only controls. (G) IFN-γ release into coculture supernatant measured by ELISA. (F-G) Data are mean ± SEM from 3 independent donors. Statistical significance was calculated using 2-way ANOVA with Sidak multiple comparison correction. (H) Representative histograms depicting granzyme B-positive cells after 48 hours of coculture. Granzyme B was measured by intracellular staining after 12 hours of incubation with GolgiStop and GolgiPlug. Illustrated are representative histograms of in total 3 different donors. For all panels: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05.
CD86-28z CAR T cells exhibit high potency toward cHL cell lines in vitro. (A) Summary of the composition of anti-CD86 (CD86-28z), anti-CD30 (CD30-28z), and anti-CD19 CAR (CD19-28z) constructs. (B-C) Representative flow cytometric images (B) and quantification (C; individual results and mean ± SEM of 10 different donors) of transduction efficiencies. Transduction efficiency was determined by staining for the extracellular c-Myc tag. (D) Absolute quantification of the molecule count per cell measured with quantitative flow cytometry. Molecule counts/cell of the indicated cell lines were calculated for CD86, CD30, and CD19, respectively. Molecule counts/cell of the isotype control were subtracted from total molecule counts. Depicted are 3 biological replicates. Data are representative of 2 independent experiments. (E-H) CD86-28z (mocha), CD30-28z (light blue), or CD19-28z (gray) CAR T cells were cocultured with the indicated cell lines (from top to bottom: L-428, L-540, KM-H2, Nalm-6, all transduced with fLuc-GFP). (E) Before cocultures, CAR T cells were stained with a Far Red proliferation dye, and antigen-specific proliferation was determined by trace dilution. Cocultures were analyzed by flow cytometry after 7 days. Top row of each color: CAR cocultured with tumor cells. Bottom row of each color: CAR only. Illustrated are representative histograms of in total 3 different donors. (F) Bioluminescence measurement of CAR-mediated lysis of tumor cells. Cell numbers were plated according to the indicated T-cell:tumor cell ratio. Tumor cell killing was determined after 72 hours. Specific lysis was calculated by normalizing to tumor cell-only controls. (G) IFN-γ release into coculture supernatant measured by ELISA. (F-G) Data are mean ± SEM from 3 independent donors. Statistical significance was calculated using 2-way ANOVA with Sidak multiple comparison correction. (H) Representative histograms depicting granzyme B-positive cells after 48 hours of coculture. Granzyme B was measured by intracellular staining after 12 hours of incubation with GolgiStop and GolgiPlug. Illustrated are representative histograms of in total 3 different donors. For all panels: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05.
Quantitate measurements of CAR antigen densities illustrated highest CD86 expression on L-428 cells, followed by L-540 and KM-H2 cells. Nalm-6 cells did not express CD86 and expressed low levels of CD30. The density of CD19 was quantified as a reference control (Figure 5D).
To elucidate in vitro efficacy, we cocultured CD86-28z, CD30-28z, or CD19-28z CAR T cells with cHL cell lines or Nalm-6 cells (Figure 5E-H). CD86-28z CAR T cells showed a strong, antigen-dependent proliferation in the presence of cHL cells, similar to CD30-28z CAR T cells (Figure 5E). Comparable to CD30-28z, CD86-28z CAR T cells efficiently lysed cHL cells also at low effector-to-target cell ratios (Figure 5F). An alternative measurement using flow cytometry confirmed high lysis (supplemental Figure 5I) and proliferative capacity (supplemental Figure 5J) of CD86-28z CAR T cells. In cocultures with cHL cell lines, CD86-28z CAR T cells released high amounts of interferon gamma (IFN-γ; Figure 5G) and were as positive for granzyme B as cells stimulated with CD3-CD28 activation beads (Figure 5H).
cHL TAMs express CD86 and can be depleted by CD86-28z CAR T cells in vitro
Besides being strongly expressed on HRSCs, CD86 was expressed on cHL TAMs. In cHL, high infiltration of TAM has been associated with an adverse prognosis and lower overall survival of treated patients.56,57 Consequently, we characterized the phenotypic properties and functions of cHL TAMs. Reclustering using scRNA-seq data delineated 4 clusters of cHL TAMs that could be subdivided into M2-like (high expression of CSF1R, CD163, CD204, CD206; clusters 0, 1) or M1-like (high expression of CD80; clusters 2, 3) macrophages (supplemental Figure 6A-C). M2-like macrophages were more abundant than M1-like macrophages. CD86 was expressed in both M1-like and M2-like macrophages, with strongest expression in clusters 0 and 2. Microarray profiling of cHL cell lines and primary cHL samples confirmed high expression of M1- and M2-like stimulating cytokines (supplemental Figure 6D-E).
To characterize possible effects of CD86-28z CAR T cells toward cHL TAMs, we generated macrophages from CD14+ monocytes and polarized them toward different macrophage trajectories. Macrophages were either polarized using IFN-γ and LPS (M1 like) and IL-4 and IL-13 (M2 like) or stimulated solely with the supernatant of cHL cell lines without exogenous cytokines (MH like). In vitro-polarized, MH-like macrophages mimicked cHL TAMs of clusters 0 and 1, revealing high expression of CD163 and CD206 (supplemental Figure 6F). CD86 was expressed on all 3 subtypes (supplemental Figure 6G). Next, in vitro-polarized macrophages were cocultured with CD86-28z, CD30-28z, or CD19-28z CAR T cells. Only CD86-28z CAR T cells proliferated and selectively depleted in vitro-polarized macrophages (supplemental Figure 6H-I) and released high amounts of IFN-γ (supplemental Figure 6J), correlating with CD86 expression.
CD86-28z CAR T cells lyse cHL cell lines in vivo and retain efficacy in models of CD30-negative disease
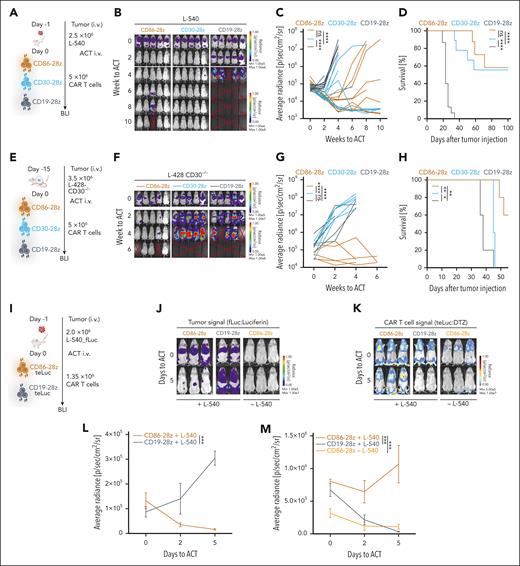
We then sought out to evaluate in vivo therapeutic efficacy of CD86-CD28z CAR T cells. We again injected L-540-fLuc+ cHL cells intravenously into NSG mice, which resulted in rapid tumor development in all mice. We treated tumor-bearing mice with a single injection of CD86-28z, CD30-28z, or CD19-28z CAR T cells (Figure 6A). CD86-28z and CD30-28z CAR T cells efficiently eradicated L-540 cHL burden (complete responses in 6 of 10 mice), whereas cHL rapidly progressed in mice injected with CD19-28z CAR T cells (Figure 6B-D). Similar results were obtained in a second cHL xenograft model using L-428 tumor cells (supplemental Figure 7D-G).
CD86-28z CAR T cells elicit a strong antitumor response toward cHL cell lines in vivo. (A) Summary of the treatment schedule used for L-540 xenograft in vivo experiments. (B-D) BLI images (B), fLuc-BLI–based quantification of tumor burden (C), and Kaplan-Meier estimation of overall survival (D) of L-540 tumor-bearing mice treated with CD86-28z, CD30-28z, or CD19-28z CAR T cells, respectively. (B) Representative fLuc-BLI images of 5 mice per group of 1 of 2 independent experiments. (C-D) Pooled data from 2 independent experiments of total 10 mice per group are depicted. (E) Summary of the in vivo treatment schedule used for L-428–CD30−/− cells xenografted into NSG mice. (F-H) BLI images (F), fLuc-BLI–based quantification of tumor burden (G), or Kaplan-Meier estimation of overall survival (H) of L-428–CD30−/− tumor-bearing mice, treated with CD86-28z, CD30-28z, or CD19-28z CAR T cells, respectively. There were 5 mice per group. (I) Treatment scheme used to determine antigen-specific proliferation of CD86-28z CAR in cHL xenograft models in vivo. (J-M) Representative BLI images of tumor cell proliferation (fLuc-BLI images; J) or T-cell proliferation (teLuc-BLI images; K). Quantification of fLuc (L) or teLuc signal (M), respectively. There were 2 to 3 mice per group. Error bars indicate SEM. For all panels, statistical significance was calculated using 2-way ANOVA with Sidak multiple comparison correction. For Kaplan-Meier curves, statistical significance was calculated with a log-rank test. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05.
CD86-28z CAR T cells elicit a strong antitumor response toward cHL cell lines in vivo. (A) Summary of the treatment schedule used for L-540 xenograft in vivo experiments. (B-D) BLI images (B), fLuc-BLI–based quantification of tumor burden (C), and Kaplan-Meier estimation of overall survival (D) of L-540 tumor-bearing mice treated with CD86-28z, CD30-28z, or CD19-28z CAR T cells, respectively. (B) Representative fLuc-BLI images of 5 mice per group of 1 of 2 independent experiments. (C-D) Pooled data from 2 independent experiments of total 10 mice per group are depicted. (E) Summary of the in vivo treatment schedule used for L-428–CD30−/− cells xenografted into NSG mice. (F-H) BLI images (F), fLuc-BLI–based quantification of tumor burden (G), or Kaplan-Meier estimation of overall survival (H) of L-428–CD30−/− tumor-bearing mice, treated with CD86-28z, CD30-28z, or CD19-28z CAR T cells, respectively. There were 5 mice per group. (I) Treatment scheme used to determine antigen-specific proliferation of CD86-28z CAR in cHL xenograft models in vivo. (J-M) Representative BLI images of tumor cell proliferation (fLuc-BLI images; J) or T-cell proliferation (teLuc-BLI images; K). Quantification of fLuc (L) or teLuc signal (M), respectively. There were 2 to 3 mice per group. Error bars indicate SEM. For all panels, statistical significance was calculated using 2-way ANOVA with Sidak multiple comparison correction. For Kaplan-Meier curves, statistical significance was calculated with a log-rank test. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05.
To investigate whether CD86-28z CAR T cells were effective against CD30-negative relapse, we used CRISPR-Cas9 to knockout CD30 in L-428 cells (L-428-CD30−/−), which was confirmed with flow cytometry (supplemental Figure 7A). In vitro cocultures of CD86-28z, CD30-28z, or CD19-28z CAR T cells with L-428-CD30−/− cells revealed lysis of tumor cells only by CD86-28z CAR T cells (supplemental Figure 7B). When CD30-28z CAR T cells were cocultured with L-428 wild type (WT) and L-428-CD30−/− cells, high production of granzyme B was only observed in cocultures with L-428 WT cells (supplemental Figure 7C). Only CD86-28z CAR T cells controlled tumor growth in L-428-CD30−/− tumor-bearing mice (Figure 6E-H).
We next challenged the efficacy of CD86-28z CAR T cells by injecting only 106 CAR T cells into immunodeficient mice with L-540 tumors (supplemental Figure 7H). CD86-28z CAR T cells retained their ability to control tumor progression, leading to prolonged survival in comparison to CD19-28z CAR T cell-treated mice (supplemental Figure 7H-K). No differences in treatment response were observed between CD86-28z and CD30-28z CAR T cells (supplemental Figure 7H-K).
To reveal antigen-dependent CAR T cell proliferation in vivo, we used the teLuc-system, which is optimized for deep-tissue imaging and is excited by a different substrate than the one used for fLuc, thus permitting simultaneous detection of 2 cell populations in the same mouse.58 The CD86-28z or CD19-28z CAR construct together with the teLuc-F2A-mCherry construct was efficiently introduced into primary human T cells with double-transduction efficiencies of 50% to 60% (supplemental Figure 7L). Decrease of tumor burden mediated by CD86-28z-teLuc, but not CD19-28z-teLuc CAR T cells, was paralleled by strong, antigen-dependent expansion of CD86-28z-teLuc CAR T cells (Figure 6J-M). Neither CD86-28z-teLuc CAR T cells injected into nontumor-bearing mice nor CD19-28z-teLuc CAR T cells injected into L-540 tumor-bearing mice expanded (Figure 6K,M).
Collectively, our results reveal remarkable efficacy of CD86-28z CAR T cells in cHL tumor models.
CD86-targeted immunotherapy is safe and does not affect bacterial host defense in vivo
Given the expression of CD86 on antigen-presenting cells such as B cells, monocytes, and dendritic cells, anti-CD86–targeted immunotherapies bear the prospect of increasing the risk for opportunistic infections and on-target–off-tumor toxicities.
Consequently, we investigated the impact of CD86-directed immunotherapies in immunocompetent, syngeneic mouse models. We generated second-generation, murine, anti-CD86 CAR T cells (mCD86-28z; supplemental Figure 8A). Anti-EpCAM CAR (mEpCAM-28z), green fluorescent protein (GFP)-transduced (GFP), or untransduced T cells were used as controls. mCD86-28z CAR T cells could be efficiently transduced into primary murine T cells at comparable efficiencies to mEpCAM CAR T cells (supplemental Figure 8B). mCD86-28z CAR T cells specifically recognized Fc-immobilized recombinant murine CD86 protein, determined by upregulation of activation and degranulation markers CD69 and CD107a (supplemental Figure 8C). Only mCD86-28z CAR T cells efficiently lysed the J774A.1 murine reticulum cell sarcoma cell line, which was highly positive for CD86 and secreted high amounts of IFN-γ (supplemental Figure 8D-G). We then used mCD86-28z CAR T cells to test their safety in syngeneic mouse models (supplemental Figure 8H). To this end, C57Bl/6 WT mice were irradiated with 3.5 Gy (total body irradiation) and injected with either mCD86-28z CAR- or GFP-transduced T cells. No weight loss was observed in any of the treated mice (supplemental Figure 8I). Flow cytometric analysis of the blood at day 8 revealed higher rates of engraftment and proliferation of mCD86-28z CAR T cells compared with GFP controls (supplemental Figure 8J). Yet, after analyzing the organs after 29 days, we did not observe measurable reduction of circulating B or myeloid cells and no mCD86-28z CAR T cells were detected in the spleen or the lymph node (supplemental Figure 8K-L). Overall, this suggests that these T cells did not persist in the mice.
Consequently, we designed a more intensive model, which included myeloablative total body irradiation with 5.5 Gy and injection of supraphysiological T cell doses (107 active CAR T cells per mouse; Figure 7A). Again, mice did not exhibit signs of weight loss (Figure 7B). mCD86-28z CAR T cells expanded rapidly in the blood (Figure 7C). Percentages of circulating CD86+CD19+ B cells were reduced in mCD86-28z–treated mice (Figure 7D, left). We did not observe differences in the percentages of circulating CD86+CD11b+ monocytes between the different treatment groups (Figure 7D, right). Expansion of mCD86-28z CAR T cells (measured by GFP expression) inversely correlated with percentages of circulating CD86+CD19+ B cells (Figure 7E). Furthermore, 28 days after T-cell injection, mice were sacrificed and organs were analyzed with flow cytometry. mCD86-28z CAR T cells were detected in large amounts in the spleen, lung, and liver of treated mice (Figure 7F). In organs, there was a trend toward lower percentages of CD19+ B cells, whereas the overall amount of CD11b+ myeloid cells did not differ between the treatment groups (Figure 7G). Interestingly, we observed higher percentages of CD86+ myeloid cells in the lung, most likely due to activation of the myeloid cell compartment resulting in upregulation of CD86 (Figure 7H).
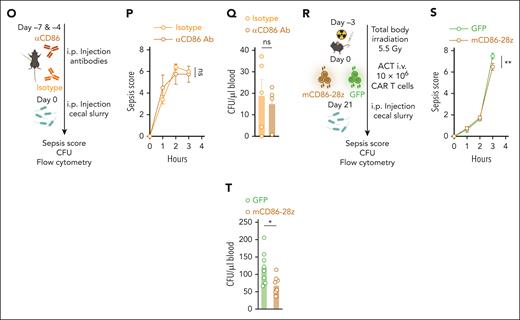
mCD86-28z CAR T cells are safe and do not interfere with bacterial host defense in polymicrobial sepsis models. (A) Summary of the treatment schedule used for toxicity assessment in C57Bl/6 mice. (B) Weight curves of mice treated with CD86-28z (mocha) or GFP control T cells (forest). There were 5 to 8 mice per group. As control mCD86-28z CAR T cells were injected in non-lymphodepleted mice (gray, n = 3). (C) Percentage of transferred T cells in the blood of C57Bl/6 mice at indicated time points after adoptive T-cell transfer (ACT) measured by flow cytometry. (D) Percentage of CD86+ B cells (left) or CD86+ monocytes (right) in the blood of C57Bl/6 mice at indicated time points after ACT. (E) Simple linear regression of CD3+CD8+ GFP+ T cells (x-axis) and CD86+ B cells (y-axis) in the blood of the mice over the different time points after ACT (days 6, 13, 23). r = Pearson correlation coefficient. (F-H) Mice were sacrificed 28 days after ACT, and organs were analyzed by flow cytometry. (F) Percentage of transferred T cells in the different organs of C57Bl/6 mice. (G) Immune cell composition in organs. (H) Percentage of CD86+ CD11b+ cells in different organs. (A-H) Data are mean ± SEM of 3 to 8 mice per group. Statistical significance was calculated using 2-way ANOVA with Tukey multiple comparison correction. (I,L) Summary of the treatment schedule used to assess formation of antigen-specific T cells in C57Bl/6 mice. (J,M) Percentage of OVA-specific T cells in the blood or spleen of antibody (J; n = 4-5 mice per group) or CAR T cell-treated mice (M; n = 7-8 mice per group). SINFEKL pentamer staining was used to measure antigen-specific T cells by flow cytometry. (K,N) IFN-γ–positive T cells measured by intracellular flow cytometry after restimulating harvested splenocytes with a SINFEKL peptide. (K,N) Mice were injected with aCD86 antibody or isotype control antibody on the indicated days (K) or treated with depicted amounts of mCD86-28z CAR T cells or GFP control T cells (N). (O,R) Summary of the treatment schedule used to assess bacterial host defense in C57Bl/6 mice. (O) 4 to 5 mice per group. (R) 8 mice per group. (P,S) Sepsis severity score after injection of cecal slurry. (Q,T) Bacterial colony counts in the blood (colony formation units per microliter) of mice IP injected with cecal slurry. For all panels, data are mean ± SEM of the indicated n number. Statistical significance was calculated using 2-way ANOVA with Sidak or Tukey multiple comparison correction or unpaired t test. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05. i.p., intraperitoneal.
mCD86-28z CAR T cells are safe and do not interfere with bacterial host defense in polymicrobial sepsis models. (A) Summary of the treatment schedule used for toxicity assessment in C57Bl/6 mice. (B) Weight curves of mice treated with CD86-28z (mocha) or GFP control T cells (forest). There were 5 to 8 mice per group. As control mCD86-28z CAR T cells were injected in non-lymphodepleted mice (gray, n = 3). (C) Percentage of transferred T cells in the blood of C57Bl/6 mice at indicated time points after adoptive T-cell transfer (ACT) measured by flow cytometry. (D) Percentage of CD86+ B cells (left) or CD86+ monocytes (right) in the blood of C57Bl/6 mice at indicated time points after ACT. (E) Simple linear regression of CD3+CD8+ GFP+ T cells (x-axis) and CD86+ B cells (y-axis) in the blood of the mice over the different time points after ACT (days 6, 13, 23). r = Pearson correlation coefficient. (F-H) Mice were sacrificed 28 days after ACT, and organs were analyzed by flow cytometry. (F) Percentage of transferred T cells in the different organs of C57Bl/6 mice. (G) Immune cell composition in organs. (H) Percentage of CD86+ CD11b+ cells in different organs. (A-H) Data are mean ± SEM of 3 to 8 mice per group. Statistical significance was calculated using 2-way ANOVA with Tukey multiple comparison correction. (I,L) Summary of the treatment schedule used to assess formation of antigen-specific T cells in C57Bl/6 mice. (J,M) Percentage of OVA-specific T cells in the blood or spleen of antibody (J; n = 4-5 mice per group) or CAR T cell-treated mice (M; n = 7-8 mice per group). SINFEKL pentamer staining was used to measure antigen-specific T cells by flow cytometry. (K,N) IFN-γ–positive T cells measured by intracellular flow cytometry after restimulating harvested splenocytes with a SINFEKL peptide. (K,N) Mice were injected with aCD86 antibody or isotype control antibody on the indicated days (K) or treated with depicted amounts of mCD86-28z CAR T cells or GFP control T cells (N). (O,R) Summary of the treatment schedule used to assess bacterial host defense in C57Bl/6 mice. (O) 4 to 5 mice per group. (R) 8 mice per group. (P,S) Sepsis severity score after injection of cecal slurry. (Q,T) Bacterial colony counts in the blood (colony formation units per microliter) of mice IP injected with cecal slurry. For all panels, data are mean ± SEM of the indicated n number. Statistical significance was calculated using 2-way ANOVA with Sidak or Tukey multiple comparison correction or unpaired t test. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ns, P > .05. i.p., intraperitoneal.
We next sought out to dissect the impact of anti-CD86–targeted immunotherapies on the formation of antigen-specific T cells. We injected C57Bl/6 mice with an anti-murine CD86-blocking antibody (αCD86 antibody, 2 injections, day [d]7 and d4). On d0 and d7, we subcutaneously inoculated mice with a complexed ovalbumin (OVA) vaccine. On d14, we measured the amount of OVA-specific T cells by SINFEKL-pentamer staining. We observed modest reduction of antigen-specific T cells in the blood, but not in the spleen of αCD86 antibody-treated mice (Figure 7J). Restimulation of splenocytes with a SINFEKL peptide did not reveal differences in the amount of IFN-γ–positive T cells (Figure 7K). When we injected mCD86-28z CAR or GFP control T cells into lymphodepleted mice, which were subcutaneously vaccinated with the OVA vaccine on d24 and d31 (Figure 7L), we did not observe differences in the percentages of OVA-specific T cells in the blood (Figure 7M) or the amount of IFN-γ–positive T cells on restimulation of splenocytes (Figure 7N). Adequate expansion of mCD86-28z CAR T cells and depletion of CD86+ B cells were found as reported previously (supplemental Figure 8M-N).
Next, we tested the impact of either αCD86 antibody or mCD86-28z CAR T cell therapy on bacterial host defense. We made use of a clinically relevant polymicrobial peritonitis model, which rapidly induces sepsis in mice.59 We first treated C57Bl/6 mice with an αCD86 or isotype antibody on d7 and d4. On d0, mice were intraperitoneally injected with cecal slurry, derived from healthy donor mice (Figure 7O). A sepsis severity score, which includes hallmark characteristics of sepsis in mice, did not differ between the treatment groups (Figure 7P). Similarly, bacterial colony formation units in the blood of mice were not different (Figure 7Q). We then repeated the experiment using mCD86-28z CAR or GFP control T cells. After injection into lymphodepleted mice, mCD86-28z CAR T cells again rapidly expanded in the blood of the mice (supplemental Figure 8O). After 21 days from T-cell injection, mice were intraperitoneally injected with cecal slurry (Figure 7R). Mice treated with mCD86-28z CAR T cells had a lower sepsis severity score (Figure 7S) and lower colony formation unit counts in the blood (Figure 7T). We again observed rapid expansion of CAR T cells and target antigen-specific depletion (CD86+CD19+ B cells) in mCD86-28z–treated mice (supplemental Figure 8Q).
Overall, CD86-targeted immunotherapies seem to have acceptable safety profiles in mice.
Discussion
We used multidimensional target screening to identify CD86 as the single most promising target antigen in cHL and illustrate its expression in different stages of cHL. Yet, the exact underlying molecular pathways leading to high CD86 expression on HRSCs remain elusive. Although genetic alterations (eg, copy number gain of 9p24.1) resulting in high PD-L1–PD-L2 expression on HRSCs are found in up to 55% of patients with cHL,60 no genetic alterations in the CD86-coding region have been reported.60 Our data reveal that CD86 blockade or genetic knockout does not influence the proliferative capacity of HRSCs. It can be hypothesized that known activation of proinflammatory signaling cascades resulting in JAK/STAT and NF-κB signaling induces CD86 expression on HRSCs. Furthermore, EBV positivity is found in up to 30% of patients with cHL.61 The EBV-encoded genes LMP2a and LMP1 induce a proliferative cell state by either directly inducing B-cell receptor pathways through structural similarity (LMP2a) or mimicking CD40 downstream signaling (LMP1). These pathways are known to increase CD86 expression. Other environmental factors such as high secretion of IL-4 with subsequent auto- or paracrine engagement of IL-4R may further contribute to upregulation of CD86 expression, both in the cHL TME and on HRSCs. CD86+-HRSCs and CD86+-nontumor cells of the cHL TME then in turn seem to interact with CTLA-4–expressing T cells, to create immunologically privileged niches in which HRSCs are preferentially located.51
We believe that our results lay the basis for other treatment modalities such as antibody-drug conjugates or T-cell–engaging antibodies.62 It should be noted that although we did not observe slowing of tumor progression in humanized mouse models with anti-CD86 antibodies, this does not exclude the possibility of treatment efficacy in humans. A major drawback of these models is the lack of critical immune cell interactions in lymphoid organs, preventing deep functional insights into human immunological processes.63 This is highlighted by the fact that highly effective treatments for cHL in humans such as PD-1 blockade do not induce relevant treatment responses in these models.64
Exploring different treatment modalities could also aid in overcoming possible limitations of CD86-directed CAR T cells. CD86 is expressed on antigen-presenting cells, including microglia, and is upregulated in inflammatory conditions.65 This might cause on-target–off-tumor toxicities, immune-effector cell-associated neurotoxicity (due to microglia expression) or predispose to opportunistic viral or bacterial infections. These theoretical considerations could, however, not be confirmed in various syngeneic mouse models.
These risks should also be interpreted in the clinical context, in which CD86-targeted immunotherapies could be used. Although most patients with cHL can be cured with novel, less toxic multimodal treatment regimens,6 in R/R cHL, treatment alternatives include high-dose chemotherapy and autologous/allogeneic stem cell transplantation. Both short-term and long-term toxicities of these therapies (therapy-related secondary malignancies, graft-versus-host disease) are expected to be more detrimental than those of CD86-directed immunotherapies. Our results confirm high expression of CD86 in R/R cHL, underpinning the value of CD86-targeted immunotherapy as an option for salvage therapies.
Collectively, our results reveal that CD86-targeted immunotherapies act on several hallmark characteristics of cHL and could serve as a first-in-class dual cytolytic- and checkpoint-inhibitory agents in cHL.
Acknowledgments
Cytometry data were obtained in the Core Facility Flow Cytometry of the University Hospital, Ludwig Maximilian University (LMU) Munich, using a BD LSRFortessa II or Beckman Coulter CytoFLEX. The authors acknowledge Life Science editors for their editing services. Figure illustrations were created with BioRender.com under a paid subscription.
This study was supported by the Förderprogramm für Forschung und Lehre of the Medical Faculty of the LMU Munich (grants 1138 [A.G.] and 1168 [S.S.]), the Bavarian Cancer Research Center (BZKF) (A.G. and TANGO to S.K.), the Deutsche Forschungsgemeinschaft (DFG, grant GO 3823/1-1 to A.G.; grants KO5055-2-1 and KO5055/3-1 to S.K., Walter Benjamin-Program to D.S.), the DKTK School of Oncology (A.G. and S.S.), the international doctoral program “i-Target: immunotargeting of cancer” (funded by the Elite Network of Bavaria; S.K. and S.E.), the Melanoma Research Alliance (grant 409510 to S.K.), Marie Skłodowska-Curie Training Network for Optimizing Adoptive T Cell Therapy of Cancer (funded by the Horizon 2020 program of the European Union; grant 955575 to S.K.) and Marie Skłodowska-Curie Doctoral Network Tracking and controlling therapeutic immune cells in cancer (TRAFIC) (grant agreement 101168810), Else Kröner-Fresenius-Stiftung (2023_EKEA.19 to A.G., 2024 Excellence Stipend to P.J.B., and IOLIN to A.G., S.S., and S.K.), German Cancer Aid (Deutsche Krebshilfe, grant 70115514 to A.G., AvantCAR.de to S.K., and Mildred-Scheel-Doktorandenprogramm to P.J.M.), the Wilhelm Sander-Stiftung (S.K.), Ernst Jung Stiftung (S.K.), Institutional Strategy LMUexcellent of LMU Munich (within the framework of the German Excellence Initiative; to S.E. and S.K.), the Go-Bio-Initiative (C.M. and S.K.), the m4-Award of the Bavarian Ministry for Economic Affairs (S.E. and S.K.), Bundesministerium für Bildung und Forschung (S.E. and S.K., Eurostars to A.O.), European Research Council (Starting grant 756017, CoG 101124203, and PoC grant 101100460 to S.K.), DFG (grants KO5055-2-1 and 510821390 to S.K.), the SFB-TRR 338/1 2021-452881907 (S.K.), SFB 1123/Z1 (R.T.A.M.), Fritz Bender Foundation (S.K.), Deutsche José Carreras Leukämie-Stiftung (S.K.), Hector Foundation (S.K.), Monika-Kutzner Foundation for Cancer Research (S.K.), Bavarian Research Foundation (BAYCELLATOR to S.K.), the Bruno and Helene Jöster Foundation (360° CAR to S.K.), and DFG (German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy: ID 390857198; E.B.). M. Thomas is funded by the Volkswagen Foundation (project OntoTime). I.P. was supported by grants from the Bavarian Cancer Research Center (BZKF), the Stiftungen zu Gunsten der Medizinischen Fakultät (Cluster 1), and the Novartis Foundation (InCa prize). C.K. and S.F.-W. have received funding from the Care for Rare Foundation, the Reinhard Frank-Stiftung (S.F.-W.), the Hector Foundation, the DFG (grant TRR332/B01 to C.K.) and the German Federal Ministry of Education and Research (BMBF) as part of the DZKJ (grant 01GL2406A to C.K.). C.M. has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement 866411) and support from the Hightech Agenda Bayern. R.G., G.V.H., J.J., V.I., T.A.J., T.L., A.Ö., and S.M. were supported by a grant from the Förderprogramm für Forschung und Lehre of the Medical Faculty of the LMU Munich. S.S. was supported by the Else Kröner-Fresenius Clinician Scientist Program Cancer Immunotherapy, the Munich Clinician Scientist Program, and the SFB-TRR 338 (startup funding). In vivo imaging device was funded by the DFG (German Research Foundation)–INST 409/231-1, and the Leica SP8 3X confocal microscope was funded by LMUexcellent of LMU Munich and by the DFG INST 409/150-1 FUGG (R.T.A.M.).
Authorship
Contribution: A.G., R.G., G.V.H., S.N., A.Ö., R.K., J.G., I.P., H.S., V.I., D.B., L.d.F., M.M., T.A.J., E.C., C.A.T., M.P.T., P.M., M. Thelen, T.L., T.S., F.M., S.S., and S.M. performed or assisted with the in vitro and in vivo experiments; V.K., M. Thomas, D.S., R.B., C.M., and E.B. performed computational analyses; P.J.M. and J.J. performed multiplex immunohistochemistry; S.F.-W. and S.D.F. performed confocal imaging and provided bone marrow organoid; A.G., R.G., G.V.H., S.F., S.D.F., V.K., M. Thomas, D.S., M. Thelen, S.R., and R.B. analyzed the data; J.H., M. Thelen, S.R., W.K., P.F.G., L.N., E.B., R.T.A.M., C.K., F.B.-M., A.R., M.v.B.-B., and P.J.B. provided critical reagents or critical infrastructure; I.J., A.R., C.M., E.B., M.v.B.-B., S.E., and S.K. supported the project; A.G., R.G., G.V.H., S.N., and S.K. designed the experiments; A.G., C.M., E.B., M.v.B.-B., S.E., and S.K. supervised the project and acquired the funding; A.G., R.G., G.V.H., and S.K. conceptualized the data and prepared the figures; A.G., R.G., G.V.H., S.N., and S.K. wrote the manuscript; and all authors critically read and approved the final manuscript.
Conflict-of-interest disclosure: A.G., M. Thomas, R.G., S.E., C.M., and S.K. submitted patent applications related to this work filed by Ludwig Maximilians Universität München, the University Hospital of LMU Munich, or the Helmholtz Centre Munich. S.K. has received honoraria from Cymab, Plectonic, TCR2 Inc, Novartis, Bristol Myers Squibb, Miltenyi, and GlaxoSmithKline. S.K. and S.E. are inventors of several patents in the field of immunooncology. S.K. and S.E. received license fees from TCR2 Inc and Carina Biotech. A.G. received research support from Tabby Therapeutics and Plectonic for work unrelated to the manuscript. P.J.B. reports advisory roles for Hexal, Merck Sharp & Dohme, Need Inc, Stemline, and Takeda; holds stock options in Need Inc; has received honoraria from AstraZeneca, BeiGene, Bristol Myers Squibb/Celgene, Lilly, Merck Sharp & Dohme, Need Inc, Stemline, and Takeda; and has received research funding from BeiGene (Institution), Bristol Myers Squibb (Institution), Merck Sharp & Dohme (Institution), and Takeda (Institution); all unrelated to the manuscript. S.K. and S.E. received research support from TCR2 Inc, Plectonic GmBH, CatalYm GmBH, and Arcus Bioscience for work unrelated to the manuscript. S.F.-W. and C.K. are inventors on a patent application covering bone marrow organoids (WO2023/194370A1). The remaining authors declare no competing financial interests.
Correspondence: Sebastian Kobold, Division of Clinical Pharmacology, LMU University Hospital, LMU Munich, Lindwurmstr 2a, 80337 Munich, Germany; email: sebastian.kobold@med.uni-muenchen.de; and Adrian Gottschlich, Department of Medicine III, LMU University Hospital, LMU Munich, Marchioninistr 15, 81377, Munich, Germany, or Division of Clinical Pharmacology, LMU University Hospital, LMU Munich, Lindwurmstr 2a, 80337 Munich, Germany; email: adrian.gottschlich@med.uni-muenchen.de.
References
Author notes
A.G., R.G., G.V.H., and S.N. contributed equally to this work.
Microarray data set of laser-dissected HRS cells is publicly accessible under the Gene Expression Omnibus database (accession number GSE12453). Single-cell data sets of cHL specimens are deposited in the European Genome-phenome Archive (EGAS00001004085).
The transcriptomic single-cell atlas was generated using published data sets and can be accessed through the respective studies 27 to 50; scripts for replicating the figures from the scRNA-seq analysis will be made available in the following GitHub repositories on publication: https://gitfront.io/r/vlkavaka/298HUSeN1dik/hodgkin-2023/ and https://gitfront.io/r/MThomas/hXvhiqhnRjjy/HL-off-tumor-antigen-expression/. Supplemental Figure 1C summarizes all used publicly available scRNA-seq studies. Accession codes of microarray data and cHL-scRNA-Seq data are provided in the detailed method descriptions. Python and R scripts for replicating the figures from the scRNA-seq data can be found in the GitHub repository at https://github.com/beltranLab/hodgkin_2023 and https://github.com/MoritzTh/HL_off-tumor-antigen-expression/. Count matrices of processed scRNA-seq data will be made available upon reasonable request. All raw data generated in this study will be uploaded to the LMU open data repository (https://data.ub.uni-muenchen.de) and can be accessed by using the article’s digital object identifier (DOI) in accordance with the principles of open science of the European Research Council (ERC). All reagents and biological material will be made available upon reasonable request to the authors given the agreement by the providing institution.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal