Key Points
Alternative splicing that is controlled by the cotranscriptional splicing complex may be therapeutically tractable in high-risk B-ALL.
Models of high-risk B-ALL are susceptible to pharmacologic and genetic inhibition of RBM39.
Visual Abstract
There are only a few options for patients with relapsed/refractory B-cell acute lymphoblastic leukemia (B-ALL), thus, this is a major area of unmet medical need. In this study, we reveal that the inclusion of a poison exon in RBM39, which could be induced by both CDK9 or CDK9 independent cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinases, CDC-like kinases (CMGC) kinase inhibition, is recognized by the nonsense-mediated messenger RNA decay pathway for degradation. Targeting this poison exon in RBM39 with CMGC inhibitors led to protein downregulation and the inhibition of ALL growth, particularly in relapsed/refractory B-ALL. Mechanistically, disruption of cotranscriptional splicing by the inhibition of CMGC kinases, including DYRK1A, or inhibition of CDK9, which phosphorylate the C-terminal domain of RNA polymerase II (Pol II), led to alteration in the SF3B1 and Pol II association. Disruption of SF3B1 and the transcriptional elongation complex altered Pol II pausing, which promoted the inclusion of a poison exon in RBM39. Moreover, RBM39 ablation suppressed the growth of human B-ALL, and targeting RBM39 with sulfonamides, which degrade RBM39 protein, showed strong antitumor activity in preclinical models. Our data reveal that relapsed/refractory B-ALL is susceptible to pharmacologic and genetic inhibition of RBM39 and provide 2 potential strategies to target this axis.
Introduction
Hematopoietic malignancies frequently harbor heterozygous mutations in splicing factors, such as SF3B1, which lead to alterations in the splicing patterns that promote oncogenesis.1-3 Heterozygous mutations in splicing factors can alter their function, and the remaining wildtype allele is crucial for tumor cell survival. Consequently, this increases the sensitivity of tumor cells to splicing inhibitors.4-6 For example, acute myeloid leukemia (AML) cells with a splicing factor mutation are more sensitive to indisulam, a small molecule that causes selective degradation of RBM39,7 a splicing factor that promotes cassette exon inclusion.8 Although splicing factor mutations are rarely observed in acute lymphoblastic leukemia (ALL), several studies have revealed that these malignant cells show prominent changes in messenger RNA (mRNA) splicing when compared with normal lymphoid cells, suggesting that aberrant splicing might also be a potential target in ALL.9,10 Although frontline therapy for B-cell acute lymphoblastic leukemia (B-ALL) leads to high remission rates, treatment of relapsed/refractory disease remains a major unmet need with long-term survival only approaching 50%.11 Poor outcomes are enriched in certain molecular subtypes of B-ALL, including Ph-like, KMT2A-rearranged, MEF2D fusions, and Down syndrome–associated ALL (DS-ALL).12-14
Posttranslational modification of the splicing machinery and RNA polymerase II (Pol II), including the phosphorylation of its intrinsically disordered C-terminal domain (CTD), influences the association between splicing and transcriptional complexes.15,16 Recent studies have implicated phosphorylation of Ser5 within the CTD of Pol II as a regulatory step that promotes its association with the spliceosome and therefore with the process of cotranscriptional splicing.17 However, the landscape of Pol II CTD phosphorylation events by serine/threonine kinases and the downstream effects on alternative splicing have yet to be fully defined.
The cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinases, CDC-like kinases (CMGC) kinase family, which has been implicated in multiple cellular processes including cell cycling, transcription, and splicing, is a promising therapeutic target in several tumor types.18 In recent studies, we demonstrated that the chromosome 21 kinase DYRK1A, which belongs to the CMGC family, is a therapeutic target in DS-ALL and in high-risk subtypes, including Ph-like ALL, through regulation of its substrates FOXO1 and STAT3.19 DYRK1A is also reported to phosphorylate splicing factors, such as SF3B1,20,21 leading us to investigate the extent to which targeting DYRK1A affects alternative splicing in B-ALL. In this study, we report that the small molecule inhibitors EHT1610 and GNF2133, both of which inhibit DYRK1A and related CMGC kinases, lead to substantial changes in alternative splicing, especially of splicing factors genes. We focused on alternative splicing changes in RBM39 and showed that CMGC kinase inhibition leads to the inclusion of a poison exon that is recognized by the nonsense-mediated mRNA decay (NMD) pathway for degradation. Interestingly, inclusion of this poison exon in RBM39 via EHT1610 was antagonized by a compensatory response mediated by CDK9, consistent with previous reports that demonstrated that the NMD pathway can generate mRNA fragments that bind to the complementary nucleotides in the original and/or related genes associated with the activation of transcription.22,23 Thus, the combination of CDK9 inhibition and EHT1610 leads to RBM39 protein downregulation and subsequent cell death. Furthermore, both CDK9 inhibition and EHT1610 disrupt a Pol II and SF3B1 complex during cotranscriptional splicing, which leads to the inclusion of a poison exon in RBM39. Finally, we provide preclinical rationale for targeting RBM39 in B-ALL with CMGC inhibitors and sulfonamides.
Methods
Human subjects and vertebrate animals
Animal research was approved by the St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee. Male and female mice were included. Studies with patient-derived xenograft (PDX) samples obtained from the Public Resource of Patient-derived and Expanded Leukemias repository and with T-cell acute lymphoblastic leukemia (T-ALL) and DS-ALL samples were approved by the institutional review board of St Jude.
Statistical analysis
Statistical comparisons were made using the Student unpaired, 2-sided t test. P values <.05 were considered statistically significant. Western blot and polymerase chain reaction (PCR) bands were quantified using ImageJ. Log-rank (Mantel-Cox) tests were used to compare mouse survival curves. Drug synergy was assessed using Synergy Finder.24
Additional methods are provided in an online supplemental Data (available on the Blood website).
Results
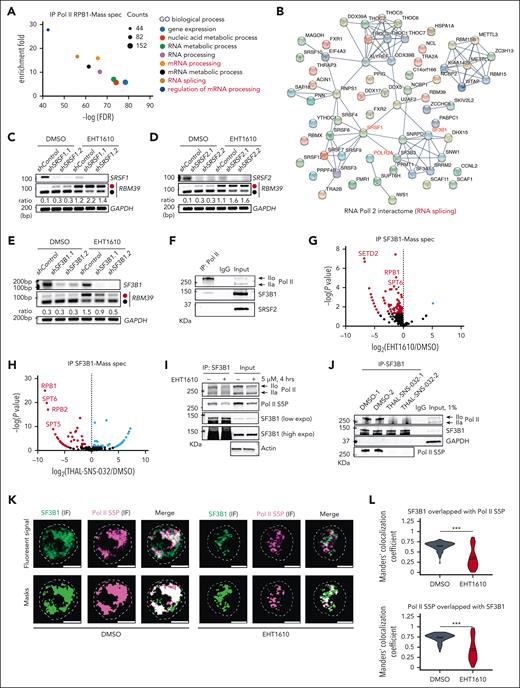
Inhibition of CMGC kinases alters alternative splicing and leads to inclusion of a poison exon in RBM39
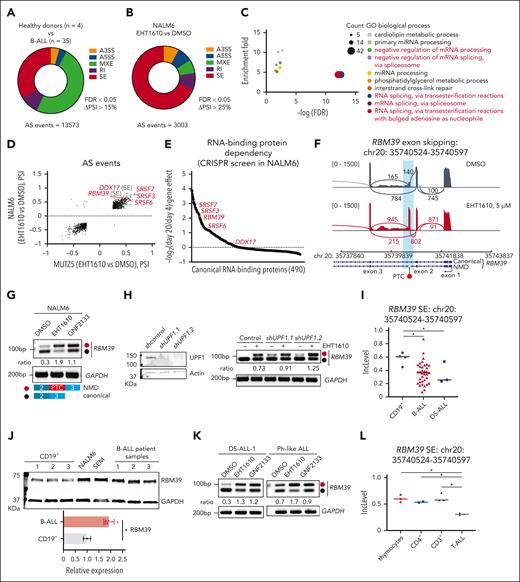
To test the hypothesis that DYRK1A inhibition affects alternative splicing and that alternative splicing is a therapeutic target in lymphoid malignancies, we performed an alternative splicing analysis in which normal human CD19+ B cells were compared with samples from patients with B-ALL.25-29 We detected 13 573 differential splicing events with a Δpercentage splice-in (PSI) of >15% that were largely composed of mutually exclusive exon and exon skipping events (Figure 1A; supplemental Table 1). We then treated 2 B-ALL cell lines, namely NALM6 and MUTZ-5, with 2 DYRK1A inhibitors, EHT161030 and GNF2133,31 to analyze alternative splicing. Using a stringent ΔPSI cutoff of >25%, we identified 3003 and 1799 alternative splicing events with EHT1610 treatment of NALM6 or MUTZ-5 cells, respectively (Figure 1B; supplemental Figure 1A). Likewise, we detected 6100 or 4513 alternative splicing events following treatment with GNF2133 in NALM6 or MUTZ-5 cells, respectively (supplemental Figure 1B). A striking overlap in alternative splicing events between EHT1610 and GNF2133 treatment in NALM6 or MUTZ-5 cells was observed (supplemental Figure 1C-D). Gene ontology analysis of the overlapping alternative splicing events between NALM6 and MUTZ-5 cells after EHT1610 treatment revealed enrichment in genes involved in RNA-related biologic processes, such as RNA splicing (Figure 1C). The top differential alternative spliced events included SRSF7, SRSF6, SRSF3, DDX17 (exon skipping), and RBM39 (exon skipping) (Figure 1D). To dissect the essentiality of downstream alternative splicing events using DYRK1A inhibitors, we performed a CRISPR/Cas9 screen that targeted 490 classical RNA binding proteins in NALM6 cells. Of note, SRSF3, SRSF7, and RBM39 showed a >fourfold depletion on day 20 when compared with day 4 of culture (Figure 1E; supplemental Table 2). Interestingly, one of the most significant splicing changes caused inclusion of a 73-bp poison exon (chr20: 35740524-35740597) that contains a premature termination codon (PTC) and led to the formation of an RBM39 transcript predicted to be degraded by the NMD pathway (referred to as the RBM39 NMD isoform) (Figure 1F). This RBM39 NMD isoform was induced with both EHT1610 and GNF2133 treatments in B-ALL cell lines (Figure 1G; supplemental Figure 1E-H). Knocking down UPF1, a core NMD factor that decays mRNA with a PTC,32 stabilized the RBM39 NMD isoform (Figure 1H) and demonstrated that this transcript is recognized and degraded by the NMD pathway in line with a previous report.33 Given the essentiality of RBM39 in ALL, the induction of the RBM39 NMD isoform by DYRK1A inhibitors, and previous studies that showed that RBM39 is a potential therapeutic target in AML,8 we focused on this alternative splicing event.
Identification of an alternatively spliced RBM39 transcript in ALL. (A) Pie chart showing differential splicing events in B-ALL (n = 35) vs normal human CD19+ B cells (n = 4). (B) Pie chart showing differential splicing events in NALM6 cells treated with DMSO or with EHT1610 (5 μM) for 4 hours. (C) GO analysis of overlapping differentially spliced genes between (B) and supplemental Figure 1A. (D) Scatter plot comparison of the differential splicing events analyzed in (B) and supplemental Figure 1A. Red gene names depict the top differentially spliced events. Transcripts presenting ΔPSI > 25% are shown. (E) Fold-change (day 20/day 4) in sgRNA abundance in pooled RBP-focused negative selection screen in NALM6 cells. Red dots indicate RBPs that are the top differentially spliced genes shown in (D). Each dot represents the average of all sgRNAs that targeted an RBP. (F) Sashimi plots depicting splicing and exon-exon junctions for an alternative splicing event in the RBM39 transcript in NALM6 cells treated with EHT1610 for 4 hours. The gene is shown along the horizontal axis. Thicker sections represent exons that code for protein sequence. Numbers over the lines that are connecting exons represent the number of reads mapped to that exon-exon junction. (G) NALM6 cells were treated with EHT1610 (5 μM) or GNF2133 (5 μM) for 4 hours. PCR reactions were performed for the detection of an RBM39 splicing event (upper). Schematic plot showing the detection of a PTC insertion between exon 2 and exon 3 of RBM39. Red and black dots represent the NMD and canonical isoforms, respectively (lower). (H) Western blot for UPF1 upon silencing of UPF1 (shUPF1.1 and shUPF1.2) in NALM6 cells (left). PCR reactions were performed for the detection of RBM39 splicing events in indicated cells with EHT1610 (1 μM) for 4 hours (right). (I) Inclusion level in normal human CD19+ B cells (n = 4) and B-ALL (n = 35) for the identical alternative spliced event in RBM39 transcript shown in (F). ∗P < .05. (J) Western blot analysis of RBM39 in CD19+ B cells from 3 different donors, ALL cell lines, and B-ALL PDX samples (upper). Quantification of the RBM39 protein normalized to GAPDH is shown (lower). (K) B-ALL PDX samples were treated with EHT1610 (5 μM) and GNF2133 (5 μM) for 4 hours. PCR reactions were performed for the detection of RBM39 splicing events. (L) Inclusion level in thymocytes (n = 3), CD4+ (n = 2), CD3+ (n = 3), and T-ALL (n = 3) for the identical alternative spliced event in the RBM39 transcript is shown in (F). ∗P < .05. Data were acquired from GSE139622.9A3SS, alternative 3′ splice sites; A5SS, alternative 5′ splice sites; AS, alternative splicing; FDR, false discovery rate; PSI, percent spliced in; PTC, premature stop codon; RI, intron retention.
Identification of an alternatively spliced RBM39 transcript in ALL. (A) Pie chart showing differential splicing events in B-ALL (n = 35) vs normal human CD19+ B cells (n = 4). (B) Pie chart showing differential splicing events in NALM6 cells treated with DMSO or with EHT1610 (5 μM) for 4 hours. (C) GO analysis of overlapping differentially spliced genes between (B) and supplemental Figure 1A. (D) Scatter plot comparison of the differential splicing events analyzed in (B) and supplemental Figure 1A. Red gene names depict the top differentially spliced events. Transcripts presenting ΔPSI > 25% are shown. (E) Fold-change (day 20/day 4) in sgRNA abundance in pooled RBP-focused negative selection screen in NALM6 cells. Red dots indicate RBPs that are the top differentially spliced genes shown in (D). Each dot represents the average of all sgRNAs that targeted an RBP. (F) Sashimi plots depicting splicing and exon-exon junctions for an alternative splicing event in the RBM39 transcript in NALM6 cells treated with EHT1610 for 4 hours. The gene is shown along the horizontal axis. Thicker sections represent exons that code for protein sequence. Numbers over the lines that are connecting exons represent the number of reads mapped to that exon-exon junction. (G) NALM6 cells were treated with EHT1610 (5 μM) or GNF2133 (5 μM) for 4 hours. PCR reactions were performed for the detection of an RBM39 splicing event (upper). Schematic plot showing the detection of a PTC insertion between exon 2 and exon 3 of RBM39. Red and black dots represent the NMD and canonical isoforms, respectively (lower). (H) Western blot for UPF1 upon silencing of UPF1 (shUPF1.1 and shUPF1.2) in NALM6 cells (left). PCR reactions were performed for the detection of RBM39 splicing events in indicated cells with EHT1610 (1 μM) for 4 hours (right). (I) Inclusion level in normal human CD19+ B cells (n = 4) and B-ALL (n = 35) for the identical alternative spliced event in RBM39 transcript shown in (F). ∗P < .05. (J) Western blot analysis of RBM39 in CD19+ B cells from 3 different donors, ALL cell lines, and B-ALL PDX samples (upper). Quantification of the RBM39 protein normalized to GAPDH is shown (lower). (K) B-ALL PDX samples were treated with EHT1610 (5 μM) and GNF2133 (5 μM) for 4 hours. PCR reactions were performed for the detection of RBM39 splicing events. (L) Inclusion level in thymocytes (n = 3), CD4+ (n = 2), CD3+ (n = 3), and T-ALL (n = 3) for the identical alternative spliced event in the RBM39 transcript is shown in (F). ∗P < .05. Data were acquired from GSE139622.9A3SS, alternative 3′ splice sites; A5SS, alternative 5′ splice sites; AS, alternative splicing; FDR, false discovery rate; PSI, percent spliced in; PTC, premature stop codon; RI, intron retention.
We subsequently investigated the extent to which this poison exon of RBM39 is present in samples of patients with primary ALL. Of note, the identical alternative splicing event in RBM39 that leads to increased inclusion of the poison exon in cell lines was more prominent in normal human CD19+ B cells than in samples of patients with B-ALL, including those of patients with Down syndrome who are trisomic for DYRK1A (Figure 1I). Importantly, there was no difference in the levels of RBM39 mRNA between normal human CD19+ B cells and samples of patients with B-ALL, whereas an increased level of RBM39 protein was observed in B-ALL cells when compared with normal human CD19+ B cells, suggesting that RBM39 expression is regulated by the alternative splicing–triggered NMD pathway (Figure 1J; supplemental Figure 1I). Moreover, treatment of 5 B-ALL PDX samples with EHT1610 and GNF2133 increased the production of the RBM39 NMD isoform (Figure 1K; supplemental Figure 1J). DYRK1A inhibition similarly increased the levels of the RBM39 NMD isoform in T-ALL cells (supplemental Figure 1K). In contrast, EHT1610 had little effect on the induction of the RBM39 NMD isoform in normal CD19+ B cells or CD3+ T cells, suggesting that dysregulation of this RBM39 alternative splicing event in malignant lymphocytes could be exploited therapeutically (supplemental Figure 1L-N). Finally, higher inclusion of the RBM39 poison exon was observed in normal T cells when compared with samples of patients with T-ALL (Figure 1L).
Next, to assess the extent to which induction of the RBM39 NMD isoform is DYRK1A dependent, we employed the FKBP12-based degron system34 using a CRISPR/Cas9 knock-in of an FKBP12F36V tag on the C-terminus of DYRK1A to degrade the endogenous FKBP12F36V tagged DYRK1A (supplemental Figure 2A). However, we observed a very small increase in the RBM39 NMD isoform upon DYRK1A degradation, indicating that inhibition of DYRK1A alone is not sufficient to induce the alternative splicing event (supplemental Figure 2B). Moreover, we investigated several potential EHT1610 targets (supplemental Tables 3 and 4) by genetic abrogation of glycogen synthase kinase 3A/B (GSK3A/B) and using 2 GSK3-α/β inhibitors, namely laduviglusib and SB 216763,35 a pan-CLK inhibitor (T025),36 a CK2 inhibitor (silmitasertib),37 and a CDK8 inhibitor (CCT251545).38 Unfortunately, no induction of the RBM39 NMD isoform was observed in NALM6 cells with these manipulations (supplemental Figure 2C-I). Together, we identified an alternative splicing event in RBM39 that is repressed in B- and T-cell ALL when compared with normal lymphocytes and is induced by CMGC inhibitors.
Alternative splicing of RBM39 is associated with inhibition of phosphorylation of RNA Pol II during cotranscriptional splicing
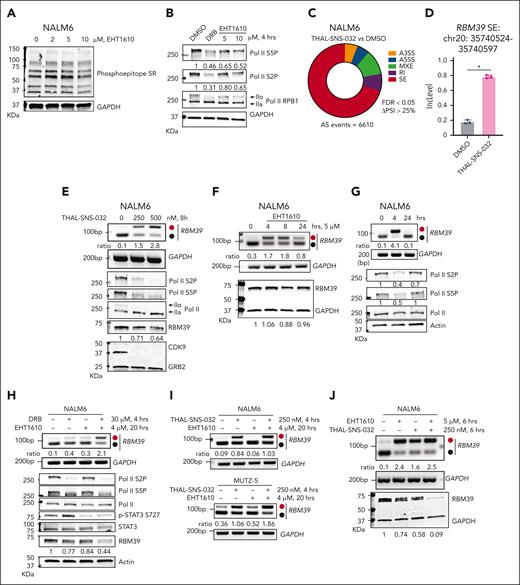
We next investigated the downstream substrates that were affected by CMGC kinases inhibition and that mediates the alternative splicing of RBM39. EHT1610, in contrast to the pan Cdc2-like kinase (CLK)/DYRK inhibitor SM09419,39 failed to alter the phosphorylation of serine/arginine (SR)-rich splicing factors in NALM6 cells (Figure 2A; supplemental Figure 3A). Furthermore, inhibition of CDK11, which mediates SF3B1 phosphorylation–dependent alterative splicing40 using the inhibitor OTS964, also failed to induce this RBM39 NMD isoform. These results suggest that phosphorylation of splicing factors alone is unlikely to be the key regulatory mechanism for the alternative splicing effect of EHT1610 (supplemental Figure 3B).
Previous studies have shown that DYRK1A, DYRK1B, homeodomain-interating protein kinase (HIPK), casein kinase, and the GSK3 family kinases control Pol II phosphorylation at Ser2 and Ser5 and its associated activity.41-45 Because RNA splicing occurs in parallel with transcription,46,47 we hypothesized that inhibition of CMGC kinases impedes Pol II phosphorylation and its association with the splicing complex. Indeed, treatment of B-ALL cell lines and PDX samples with EHT1610 for 4 hours decreased phosphorylation of both the Ser2 and Ser5 residues of RNA Pol II (Figure 2B; supplemental Figure 3C-F). Because CDK9 is a master controller of Pol II CTD phosphorylation,48 we employed the positive transcription elongation factor b (p-TEFB) inhibitor 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) and a CDK9 proteolysis targeting chimera (PROTAC), THAL-SNS-032,49 in our assays. Treatment of NALM6 cells with THAL-SNS-032 for 8 hours produced 6610 alternative splicing changes, including an increase in the RBM39 NMD isoform, along with a decrease in Pol II phosphorylation (Figure 2C-E). Similar results were observed with DRB treatment (Figure 2B; supplemental Figure 3G). Notably, CDK9 was not targeted by EHT1610 as evidenced by kinase cellular selectivity profiling of EHT1610 in 293 T cells and kinase profiling (468 kinases) with EHT1610 in vitro (supplemental Tables 3 and 4).
Pol II phosphorylation mediates cotranscriptional splicing of an RBM39 poison exon. (A) Western blot to detect changes in phosphorylated SR-rich proteins after treatment of NALM6 cells with increasing concentrations of EHT1610 for 8 hours. (B) Western blot analysis of extracts from NALM6 cells treated with DMSO, DRB (30 μM), and EHT1610 for 4 hours. IIo and IIa indicate hyperphosphorylated and hypophosphorylated CTD52, respectively. (C) Pie chart showing differential splicing events in NALM6 cells treated with DMSO when compared with those treated with THAL-SNS-032 (500 nM) for 8 hours. (D) The inclusion level in NALM6 cells treated with DMSO (n = 3) and THAL-SNS-032 (n = 3) for the identical alternative spliced event in RBM39 transcript shown in Figure 1F. ∗P < .05. (E) The NALM6 cells were treated with increasing doses of THAL-SNS-032 for 8 hours. PCR reactions were performed for the detection of RBM39 splicing events (upper), and western blot analysis of extracts were shown (lower). (F) NALM6 cells were treated with EHT1610 in a time-dependent manner. PCR reactions were performed for the detection of the RBM39 splicing event (upper), and western blot analysis of extracts were shown (lower). (G) NALM6 cells were treated with EHT1610 (4 μM) for 4 and 24 hours. PCR reactions were performed for detection of the RBM39 splicing event (upper), and western blot analysis of extracts are shown (lower). (H) NALM6 cells were treated with EHT1610 for 16 hours, followed by additional treatment of DRB for 4 hours. PCR reactions were performed for the detection of RBM39 splicing event (upper), and western blot analysis of extracts were shown (lower). (I) NALM6 cells (upper) or MUTZ-5 cells (lower) were treated with EHT1610 for 16 hours, followed by additional treatment with THAL-SNS-032 for 4 hours. PCR reactions were performed to detect the RBM39 splicing event. (J) NALM6 cells were treated with EHT1610, THAL-SNS-032 or a combination for 6 hours. PCR reactions were performed for the detection of an RBM39 splicing event (upper) and representative western blot analysis of 3 biological replicates of extracts are shown (lower).
Pol II phosphorylation mediates cotranscriptional splicing of an RBM39 poison exon. (A) Western blot to detect changes in phosphorylated SR-rich proteins after treatment of NALM6 cells with increasing concentrations of EHT1610 for 8 hours. (B) Western blot analysis of extracts from NALM6 cells treated with DMSO, DRB (30 μM), and EHT1610 for 4 hours. IIo and IIa indicate hyperphosphorylated and hypophosphorylated CTD52, respectively. (C) Pie chart showing differential splicing events in NALM6 cells treated with DMSO when compared with those treated with THAL-SNS-032 (500 nM) for 8 hours. (D) The inclusion level in NALM6 cells treated with DMSO (n = 3) and THAL-SNS-032 (n = 3) for the identical alternative spliced event in RBM39 transcript shown in Figure 1F. ∗P < .05. (E) The NALM6 cells were treated with increasing doses of THAL-SNS-032 for 8 hours. PCR reactions were performed for the detection of RBM39 splicing events (upper), and western blot analysis of extracts were shown (lower). (F) NALM6 cells were treated with EHT1610 in a time-dependent manner. PCR reactions were performed for the detection of the RBM39 splicing event (upper), and western blot analysis of extracts were shown (lower). (G) NALM6 cells were treated with EHT1610 (4 μM) for 4 and 24 hours. PCR reactions were performed for detection of the RBM39 splicing event (upper), and western blot analysis of extracts are shown (lower). (H) NALM6 cells were treated with EHT1610 for 16 hours, followed by additional treatment of DRB for 4 hours. PCR reactions were performed for the detection of RBM39 splicing event (upper), and western blot analysis of extracts were shown (lower). (I) NALM6 cells (upper) or MUTZ-5 cells (lower) were treated with EHT1610 for 16 hours, followed by additional treatment with THAL-SNS-032 for 4 hours. PCR reactions were performed to detect the RBM39 splicing event. (J) NALM6 cells were treated with EHT1610, THAL-SNS-032 or a combination for 6 hours. PCR reactions were performed for the detection of an RBM39 splicing event (upper) and representative western blot analysis of 3 biological replicates of extracts are shown (lower).
We next assessed the downstream protein changes following treatment with either EHT1610 or THAL-SNS-032. Although we saw that THAL-SNS-032 treatment decreased the RBM39 protein level by a modest degree, EHT1610 reduced RBM39 protein levels at 10 μM but not at 5 μM (Figure 2E-F; supplemental Figure 3H). A dose-dependent inhibition of ALL growth with EHT1610 was also observed (supplemental Figure 3I). Furthermore, the increase in the NMD isoform was reversed, along with the reduction in RNA Pol II phosphorylation, at 24 hours when compared with 4 hours, suggesting that a compensatory mechanism restores Pol II phosphorylation and normalizes splicing after EHT1610 treatment (Figure 2G). Previous publications indicated that PTC-bearing mRNA stimulated the NMD pathway through genetic modulation, followed by a compensatory mechanism that triggered transcriptional activation of its origin and/or a related gene to counteract the genetic phenotype.22,23 Consistent with this, treatment of NALM6 cells with EHT1610 for 16 hours, followed by an additional 4 hours with DRB, increased the RBM39 NMD isoform when compared with 20 hours of EHT1610 treatment alone, and this was accompanied by a dramatic decrease in the RBM39 protein levels (Figure 2H). Furthermore, we corroborated these findings with THAL-SNS-032, suggesting that inhibition of multiple CDK9-associated transcription elongation complexes prevents the compensatory response of the NMD pathway that is induced by EHT1610 (Figure 2I). Similarly, THAL-SNS-032 and EHT1610 combination treatment dramatically decreased the RBM39 protein level when compared with treatment with each of 2 inhibitors individually (Figure 2J; supplemental Figure 3J). Unlike EHT1610, the CDK9 PROTAC led to a similar increase in the RBM39 NMD isoform after 8 and 20 hours treatment (supplemental Figure 3K), indicating that the CMGC kinases were unable to restore Pol II phosphorylation under these conditions.
Inhibition of Pol II phosphorylation alters Pol II distribution and pausing
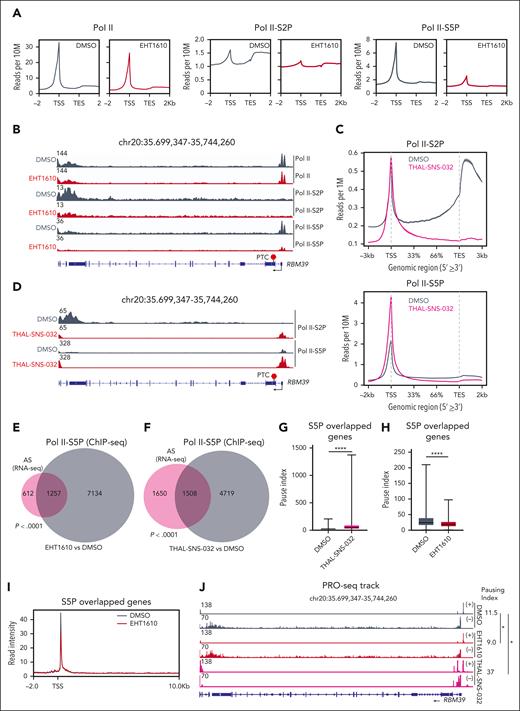
To gain additional insight into the regulation of RBM39 alternative splicing induced by Pol II phosphorylation, we performed chromatin immunoprecipitation sequencing (ChIP-seq) for total Pol II and the p-Ser2 and p-Ser5 forms upon EHT1610 or THAL-SNS-032 treatment in NALM6 cells. The averages of the genomic p-Ser2 and p-Ser5 Pol II occupancy were decreased following EHT1610 treatment, whereas the total Pol II occupancy remained largely unaffected (Figure 3A). Moreover, we witnessed a reduction in p-Ser5 Pol II binding at the transcription start site (TSS) of RBM39 and a reduction in p-Ser2 Pol II at the transcription end site (TES) of RBM39 (Figure 3B). In contrast, THAL-SNS-032 treatment led to an increase in genomic p-Ser5 Pol II occupancy at the TSS of genes and a decrease in genomic p-Ser2 occupancy at the TES of genes, including RBM39 (Figure 3C-D), which is consistent with a previous publication that suggested the importance of CDK9 in the regulation of transcriptional pausing and elongation.48 A previous report demonstrated that p-Ser5 Pol II is associated with the catalytic spliceosome machinery during cotranscriptional splicing.17 To further explore this relationship, we investigated the concordance of the p-Ser5 changes and alternative splicing events upon EHT1610 or THAL-SNS-032 treatment. p-Ser5 Pol II ChIP-seq following EHT1610 treatment identified 8391 genes that showed decreased genomic occupancy at their TSS or TES sites. Similarly, p-Ser5 Pol II ChIP-seq following THAL-SNS-032 treatment identified 6227 genes that showed differential genomic occupancy at their TSS or TES sites. A significant overlap between differential p-Ser5 Pol II binding sites and alternatively spliced genes with either EHT1610 or THAL-SNS-032 treatment was observed (Figure 3E-F).
Pol II distribution and pausing are altered with disruption of cotranscriptional splicing. (A) ChIP-seq signal of Pol II (left), p-Ser2 Pol II (middle), and p-Ser5 Pol II (right) from NALM6 cells treated with DMSO and EHT1610 (5 μM). The average profiles of DMSO and EHT1610 that covered ±2 kb of the gene were plotted. (B) Snapshots of RBM39 loci (IGV browser) with ChIP-seq signals for Pol II, p-Ser2 Pol II, and p-Ser5 Pol II upon EHT1610 treatment in NALM6 cells. (C) ChIP-seq signal of p-Ser2 Pol II (left) and p-Ser5 Pol II (right) from NALM6 cells treated with DMSO and THAL-SNS-032 (500 nM). The average profiles of DMSO and THAL-SNS-032 were plotted and covered ±3kb of the gene. (D) Snapshots of RBM39 loci (IGV browser) with ChIP-seq signals for p-Ser2 Pol II and p-Ser5 Pol II upon THAL-SNS-032 treatment in NALM6 cells. (E) Venn diagram showing overlapped alterative spliced genes (analyzed from Figure 1B) and decreased p-Ser5 Pol II ChIP-seq signal genes upon EHT1610 treatment. (F) Venn diagram showing overlapped alternatively spliced genes (analyzed from Figure 2C) and differential p-Ser5 Pol II ChIP-seq signal genes upon THAL-SNS-032 treatment. (G-J) NALM6 cells were treated with DMSO and EHT1610 (5 μM) for 4 hours and with THAL-SNS-032 (500 nM) for 8 hours followed by PRO-seq. (G) Comparison of the pausing indexes of the overlapping genes in (F). (H) Comparison of pausing indexes of those overlapped genes in (E). (I) Metagene plot showing PRO-seq signal in NAML6 cells treated with DMSO and EHT1610 for those overlapped genes in (E). (J) Snapshot of PRO-seq track on the RBM39 locus in NALM6 cells with DMSO, EHT1610, and THAL-SNS-032 treatment. (+) indicates transcription from left to right; (−) indicates transcription from right to left. The pausing indexes for the RBM39 locus are indicated to the right (n = 2; ∗P < .05).
Pol II distribution and pausing are altered with disruption of cotranscriptional splicing. (A) ChIP-seq signal of Pol II (left), p-Ser2 Pol II (middle), and p-Ser5 Pol II (right) from NALM6 cells treated with DMSO and EHT1610 (5 μM). The average profiles of DMSO and EHT1610 that covered ±2 kb of the gene were plotted. (B) Snapshots of RBM39 loci (IGV browser) with ChIP-seq signals for Pol II, p-Ser2 Pol II, and p-Ser5 Pol II upon EHT1610 treatment in NALM6 cells. (C) ChIP-seq signal of p-Ser2 Pol II (left) and p-Ser5 Pol II (right) from NALM6 cells treated with DMSO and THAL-SNS-032 (500 nM). The average profiles of DMSO and THAL-SNS-032 were plotted and covered ±3kb of the gene. (D) Snapshots of RBM39 loci (IGV browser) with ChIP-seq signals for p-Ser2 Pol II and p-Ser5 Pol II upon THAL-SNS-032 treatment in NALM6 cells. (E) Venn diagram showing overlapped alterative spliced genes (analyzed from Figure 1B) and decreased p-Ser5 Pol II ChIP-seq signal genes upon EHT1610 treatment. (F) Venn diagram showing overlapped alternatively spliced genes (analyzed from Figure 2C) and differential p-Ser5 Pol II ChIP-seq signal genes upon THAL-SNS-032 treatment. (G-J) NALM6 cells were treated with DMSO and EHT1610 (5 μM) for 4 hours and with THAL-SNS-032 (500 nM) for 8 hours followed by PRO-seq. (G) Comparison of the pausing indexes of the overlapping genes in (F). (H) Comparison of pausing indexes of those overlapped genes in (E). (I) Metagene plot showing PRO-seq signal in NAML6 cells treated with DMSO and EHT1610 for those overlapped genes in (E). (J) Snapshot of PRO-seq track on the RBM39 locus in NALM6 cells with DMSO, EHT1610, and THAL-SNS-032 treatment. (+) indicates transcription from left to right; (−) indicates transcription from right to left. The pausing indexes for the RBM39 locus are indicated to the right (n = 2; ∗P < .05).
Next, we performed precision run-on sequencing (PRO-seq) in NALM6 cells treated with dimethylsulfoxide (DMSO), EHT1610, or THAL-SNS-032 to assess nascent mRNA expression and Pol II pausing. The differential gene expression analysis for nascent mRNA showed 3362 downregulated genes and 2977 upregulated genes following THAL-SNS-032 treatment when compared with DMSO treatment, highlighting the essential role of CDK9 in transcription regulation (supplemental Figure 4A). By contrast, EHT1610 treatment had a modest effect on nascent mRNA expression and was characterized by 114 upregulated and 109 downregulated genes (supplemental Figure 4B). Of note, EHT1610 treatment showed no effect on nascent RBM39, whereas THAL-SNS-032 treatment increased nascent RBM39 expression, suggesting that the decrease in RBM39 protein is not primarily caused by a transcriptional defect (Figure 2J; supplemental Figure 4C). Furthermore, the CDK9 PROTAC led to inhibition of release of promoter-proximal Pol II, which is in line with a previous publication50 (supplemental Figure 4D), especially on those genes that had both differential p-Ser5 occupancy and alternative splicing (Figure 3G). In contrast, treatment with EHT1610 decreased promoter Pol II density and reduced the pausing duration on the genes that undergo both alternatively splicing and differential p-Ser5 occupancy (Figure 3H-I), including RBM39 (Figure 3J). Taken together, we found that disruption of Pol II cotranscriptional splicing by inhibition of Pol II phosphorylation led to alterations in promoter-proximal Pol II pausing on alternatively spliced genes.
Inhibition of phosphorylation of Pol II alters the Pol II/SF3B1 association
To identify potential splicing factors that regulate the alternative splicing of RBM39 in concert with Pol II during cotranscriptional splicing, we performed immunoprecipitation of RNA Pol II, followed by mass spectrometry (supplemental Table 5). Gene ontology enrichment analysis revealed that proteins involved in RNA-related biologic processes, including mRNA processing, the regulation of mRNA processing, and RNA splicing, were enriched in the Pol II interactome (Figure 4A). Of note, 66 RNA splicing-related interactors were identified, including U2AF2, SRSF1, and SF3B1 (Figure 4B). Because the SRSF family members bind exonic splicing enhancers to stimulate splicing and U2 small nuclear ribonucleoprotein particle (snRNP) is involved in the activation of the spliceosome for the first transesterification splicing reaction,51 we silenced SF3B1, SRSF1, and SRSF2 with and without EHT1610 treatment in B-ALL cells. Knockdown of SRSF1 and SRSF2 led to increased levels of the NMD transcript, even in the absence of EHT1610, indicating that they normally function to exclude this poison exon (Figure 4C-D). In contrast, knocking down SF3B1 rescued the increased RBM39 NMD isoform induced by EHT1610 (Figure 4E), suggesting that SF3B1 is critical for regulating the cotranscriptional splicing of RBM39.
SF3B1 interacts with phosphorylated Pol II that regulates RBM39 poison exon inclusion. (A) GO analysis of the identified proteins from Pol II immunoprecipitation (IP)-mass spectrometry. Pathways with by -log (FDR) and enrichment >5 are shown. (B) Protein-protein interaction network of 66 RNA splicing proteins identified in the Pol II IP-mass spectrometry. (C-E) PCR analysis for RBM39 alternative splicing upon SRSF1 (with shSRSF1.1 and shSRSF1.2) (C), SRSF2 (with shSRSF2.1 and shSRSF2.2) (D), and of SF3B1 (with shSF3B1.1 and shSF3B1.2) (E) silencing in NALM6 cells. PCR reactions were performed for detection of RBM39 splicing event in indicated cells with EHT1610 (2 μM) treatment for 4 hours. (F) Western blot analysis following IP for Pol II in NALM6 cells. (G-H) The NALM6 cells were treated with DMSO, EHT1610 (5 μM), and THAL-SNS-032 (500 nM) for 4 hours, followed by SF3B1 IP-mass spectrometry. Scatter plot showing the differential proteins in comparison with DMSO and EHT1610 (G) or DMSO and THAL-SNS-032 (H). (I-J) Western blot analysis following IP for SF3B1 in NALM6 cells treated with EHT1610 (I) or THAL-SNS-032 (500 nM) (J) for 4 hours. (K-L) Immunofluorescence analysis of p-Ser5 Pol II and SF3B1 in NALM6 cells treated with DMSO or EHT1610 (5 μM) (K). Scale bars depict 5 microns. Quantification of Manders' colocalization coefficient values between SF3B1 and p-Ser5 Pol II Immunofluorescence signal (L) (See methods). ∗∗∗P < .001.
SF3B1 interacts with phosphorylated Pol II that regulates RBM39 poison exon inclusion. (A) GO analysis of the identified proteins from Pol II immunoprecipitation (IP)-mass spectrometry. Pathways with by -log (FDR) and enrichment >5 are shown. (B) Protein-protein interaction network of 66 RNA splicing proteins identified in the Pol II IP-mass spectrometry. (C-E) PCR analysis for RBM39 alternative splicing upon SRSF1 (with shSRSF1.1 and shSRSF1.2) (C), SRSF2 (with shSRSF2.1 and shSRSF2.2) (D), and of SF3B1 (with shSF3B1.1 and shSF3B1.2) (E) silencing in NALM6 cells. PCR reactions were performed for detection of RBM39 splicing event in indicated cells with EHT1610 (2 μM) treatment for 4 hours. (F) Western blot analysis following IP for Pol II in NALM6 cells. (G-H) The NALM6 cells were treated with DMSO, EHT1610 (5 μM), and THAL-SNS-032 (500 nM) for 4 hours, followed by SF3B1 IP-mass spectrometry. Scatter plot showing the differential proteins in comparison with DMSO and EHT1610 (G) or DMSO and THAL-SNS-032 (H). (I-J) Western blot analysis following IP for SF3B1 in NALM6 cells treated with EHT1610 (I) or THAL-SNS-032 (500 nM) (J) for 4 hours. (K-L) Immunofluorescence analysis of p-Ser5 Pol II and SF3B1 in NALM6 cells treated with DMSO or EHT1610 (5 μM) (K). Scale bars depict 5 microns. Quantification of Manders' colocalization coefficient values between SF3B1 and p-Ser5 Pol II Immunofluorescence signal (L) (See methods). ∗∗∗P < .001.
We then confirmed the interaction between Pol II and SF3B1 (Figure 4F) and performed a reciprocal immunoprecipitation of SF3B1, followed by mass spectrometry in NALM6 cells treated with DMSO, EHT1610, and THAL-SNS-032. Disruption of SF3B1 binding with Pol II and its associated transcriptional elongation complex components, like SPT6,52 was observed in response to both EHT1610 and THAL-SNS-032 treatment when compared with DMSO (Figure 4G-H; supplemental Table 6). These observations were further confirmed by immunoprecipitation of SF3B1, which showed a decreased interaction between SF3B1 and hyperphosphorylated Pol II upon either EHT1610 or THAL-SNS-032 treatment, suggesting that inhibition of Pol II phosphorylation alters the interactome (Figure 4I-J). We then performed immunofluorescence of SF3B1 and p-Ser5 Pol II in NALM6 cells to assess their intracellular association. We observed a striking decrease in the association upon EHT1610 treatment, as confirmed by analysis of the Manders’ colocalization coefficient values between SF3B1 and p-Ser5 Pol II (Figure 4K-L). Taken together, we found that reducing Pol II phosphorylation alters the Pol II/SF3B1 association, which contributes to the aberrant cotranscriptional splicing, including of RBM39.
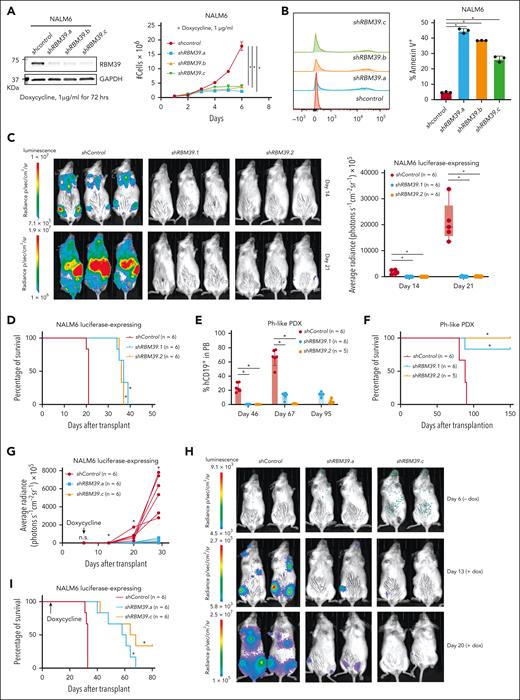
RBM39 is a therapeutic target in B-ALL
We then assessed the extent to which RBM39 is a therapeutic target in B-ALL. We knocked down RBM39 in NALM6 cells using 2 independent short hairpin RNAs, namely shRBM39.1 and shRBM39.2, and found that its depletion led to prominent inhibition of NALM6 cell growth as a consequence of increased apoptosis and cell cycle arrest (supplemental Figure 5A-C). Knockdown of RBM39 in SUPB15 cells, which are a Ph-ALL cell line, showed a similar in vitro dependency (supplemental Figure 5D-F). Furthermore, we also employed a doxycycline-inducible system in NALM6 cells and observed a similar cell proliferation defect (Figure 5A-B; supplemental Figure 6A-B). We subsequently transplanted shRBM39-expressing NALM6 cells into immunocompromised mice and monitored tumor growth. RBM39 silencing led to a marked inhibition of tumor growth and enhanced survival (Figure 5C-D). We also transplanted Ph-like ALL PDX cells that harbored short hairpin RNAs that targeted RBM39 and found that recipient mice had a significantly decreased tumor burden and a significant survival advantage (Figure 5E-F). To determine whether RBM39 is also required for the maintenance of B-ALL, we employed doxycycline-inducible shRBM39 expression (shRBM39.a and shRBM39.c) after disease establishment (supplemental Figure 6C). RBM39 ablation after B-ALL establishment inhibited tumor growth and significantly extended mouse survival (Figure 5G-I).
RBM39 is required for progression and maintenance of ALL. (A) Western blot for RBM39 (left) and the growth curves (right) from the inducible control hairpin RNA, shRBM39.a, shRBM39.b, and shRBM39.c expressing NALM6 cells with doxycycline treatment. (B) Annexin V staining of NALM6 cells that expressed either an inducible control short hairpin RNA or shRBM39 after 72 hours. A representative example (left) and quantification of Annexin V+ cells (n = 3, right; ∗P < .05) are shown. (C) Representative bioluminescence pictures of immunocompromised animals (left) that were transplanted with luciferase-expressing NALM6 cells that were previously transduced with a lentiviral vector expressing either a control hairpin RNA, shRBM39.1, or shRBM39.2, and selected using puromycin for a period of 3 days. Quantification of tumor growth by total flux in vivo (right). ∗P < .05. (D) Survival analysis of the immunocompromised mice that received a transplant of control hairpin RNA-, shRBM39.1-, or shRBM39.2-expressing NALM6 cells. The P value (∗P < .05) was calculated using a log-rank (Mantel-Cox) test. (E) Immunocompromised animals were transplanted with Ph-like ALL PDX cells that were previously transduced with a lentiviral vector expressing either a control hairpin RNA, shRBM39.1, or shRBM39.2, and selected using puromycin for a period of 3 days. Human CD19+ cells in peripheral blood (PB) were monitored to assess disease burden. ∗P < .05. (F) Survival analysis of immunocompromised mice transplanted with the control hairpin, shRBM39.1-, or shRBM39.2-expressing Ph-like ALL PDX cells. ∗P < .05. (G-H) The total flux from each day as indicated in immunocompromised animals that were transplanted with luciferase-expressing NALM6 cells that were previously transduced with a lentiviral vector expressing either a control hairpin RNA, shRBM39.a, or shRBM39.c (G). ∗P < .05. The relative bioluminescence intensity is shown for 3 representative mice per group on day 6 (before doxycycline administration), day 13, and day 20 (after doxycycline administration) (H). (I) Survival analysis of mice transplanted with control hairpin RNA, shRBM39.a, or shRBM39.c-expressing NALM6 cells. (∗P < .05).
RBM39 is required for progression and maintenance of ALL. (A) Western blot for RBM39 (left) and the growth curves (right) from the inducible control hairpin RNA, shRBM39.a, shRBM39.b, and shRBM39.c expressing NALM6 cells with doxycycline treatment. (B) Annexin V staining of NALM6 cells that expressed either an inducible control short hairpin RNA or shRBM39 after 72 hours. A representative example (left) and quantification of Annexin V+ cells (n = 3, right; ∗P < .05) are shown. (C) Representative bioluminescence pictures of immunocompromised animals (left) that were transplanted with luciferase-expressing NALM6 cells that were previously transduced with a lentiviral vector expressing either a control hairpin RNA, shRBM39.1, or shRBM39.2, and selected using puromycin for a period of 3 days. Quantification of tumor growth by total flux in vivo (right). ∗P < .05. (D) Survival analysis of the immunocompromised mice that received a transplant of control hairpin RNA-, shRBM39.1-, or shRBM39.2-expressing NALM6 cells. The P value (∗P < .05) was calculated using a log-rank (Mantel-Cox) test. (E) Immunocompromised animals were transplanted with Ph-like ALL PDX cells that were previously transduced with a lentiviral vector expressing either a control hairpin RNA, shRBM39.1, or shRBM39.2, and selected using puromycin for a period of 3 days. Human CD19+ cells in peripheral blood (PB) were monitored to assess disease burden. ∗P < .05. (F) Survival analysis of immunocompromised mice transplanted with the control hairpin, shRBM39.1-, or shRBM39.2-expressing Ph-like ALL PDX cells. ∗P < .05. (G-H) The total flux from each day as indicated in immunocompromised animals that were transplanted with luciferase-expressing NALM6 cells that were previously transduced with a lentiviral vector expressing either a control hairpin RNA, shRBM39.a, or shRBM39.c (G). ∗P < .05. The relative bioluminescence intensity is shown for 3 representative mice per group on day 6 (before doxycycline administration), day 13, and day 20 (after doxycycline administration) (H). (I) Survival analysis of mice transplanted with control hairpin RNA, shRBM39.a, or shRBM39.c-expressing NALM6 cells. (∗P < .05).
Next, given that indisulam (E7070) and E7820 degrade RBM39 protein via ubiquitination through the DCAF15 associated E3 complex,8 we investigated whether these compounds would suppress B-ALL development by targeting RBM39. We found that both E7070 and E7820 caused a dose-dependent degradation of RBM39 protein in the ALL cell lines (supplemental Figure 7A). Furthermore, treatment of NAML6 and Kasumi-7 with these RBM39 degraders increased apoptosis and arrested the cell cycle in the S phase (supplemental Figure 7B-E).
To determine the sensitivity of various ALL subtypes to these drugs, we evaluated the correlation between RBM39 and DCAF15 mRNA expression in various leukemias and found that the DUX4 fusion, KMT2A-rearranged, and the MEF2D fusion subtypes displayed the strongest Pearson correlation (Figure 6A). We then analyzed public data for the 50% inhibitory concentration (IC50) of E7070 across 758 cancer cell lines53 and observed that B-ALL, T-ALL, and AML cell lines are more sensitive to E7070 than other cancer cell types (Figure 6B). In accordance with the correlation between RBM39 and DCAF15 mRNA expression based on the Pearson correlation analysis, SEM (KMT2A-rearranged) and NALM6 (DUX4 fusion) cells showed the greatest sensitivity to E7070 (Figure 6B). To confirm the data obtained from public databases, we determined the IC50 for E7070 and E7820 in several ALL cell lines and PDX samples in vitro. The Kasumi-7 and Kasumi-9 cell lines, which contain an MEF2D fusion, and the NALM6 and SEM cells were highly sensitive to both RBM39 degraders (Figure 6C; supplemental Figure 7F). With respect to the PDX specimens, those that contained an MEF2D rearrangement were sensitive to low micromolar doses (Figure 6D).
High-risk subtypes of ALL are sensitive to RBM39 degradation. (A) Pearson correlation analysis between RBM39 mRNA and DCAF15 mRNA across the indicated subtypes of B-ALL. Patients with B-ALL mRNA expression data were obtained from the Pediatric Cancer Genome Project data portal (PeCan, St Jude, Memphis). (B) Indisulam area under the curve (AUC) of B-ALL, T-ALL and AML in comparison with nonleukemia cell lines. Data were obtained from the CTD2 network53 (left). The indisulam AUC for the indicated B-ALL cell lines (right). (C) IC50 of E7820 (n = 3; the mean value ± standard deviation [SD] is shown) across different B-ALL cell lines as indicated. MEF2D fusion subtypes include Kasumi-7 and -9. (D) The IC50 of E7820 (n = 3; the mean value ± SD is shown) across different B-ALL PDX samples as indicated. (E) Western blot analysis of RBM39 in Kasumi-7 and SUPB15 treated with increasing concentration of E7820 for 4 h. One representative blot is shown on left and quantification of 3 biological replicates is shown on right. (F) Bar graph showing different types of splicing events for patients cells with MEF2D-BCL9 ALL treated with DMSO or E7820 (1 μM) and patient cells with MEF2D-HNRNPUL1 ALL treated with DMSO or E7820 (1 μM). (G-I) MEF2D-HNRNPUL1 ALL PDX cells were transplanted into NSG mice and randomized to E7820 (50 mg/kg) or vehicle, which were orally administrated to mice beginning on day 14 for 6 weeks. The disease burden was monitored by human CD19+ cells in peripheral blood (G). ∗P < .05. (H) Spleen weight of mice when the humane end points were reached or on day 75 (end of the study, left; ∗P < .05). Representative images of spleens (right). (I) Survival analysis of the mice administrated with E7820 (50 mg/kg) or vehicle. ∗P < .05.
High-risk subtypes of ALL are sensitive to RBM39 degradation. (A) Pearson correlation analysis between RBM39 mRNA and DCAF15 mRNA across the indicated subtypes of B-ALL. Patients with B-ALL mRNA expression data were obtained from the Pediatric Cancer Genome Project data portal (PeCan, St Jude, Memphis). (B) Indisulam area under the curve (AUC) of B-ALL, T-ALL and AML in comparison with nonleukemia cell lines. Data were obtained from the CTD2 network53 (left). The indisulam AUC for the indicated B-ALL cell lines (right). (C) IC50 of E7820 (n = 3; the mean value ± standard deviation [SD] is shown) across different B-ALL cell lines as indicated. MEF2D fusion subtypes include Kasumi-7 and -9. (D) The IC50 of E7820 (n = 3; the mean value ± SD is shown) across different B-ALL PDX samples as indicated. (E) Western blot analysis of RBM39 in Kasumi-7 and SUPB15 treated with increasing concentration of E7820 for 4 h. One representative blot is shown on left and quantification of 3 biological replicates is shown on right. (F) Bar graph showing different types of splicing events for patients cells with MEF2D-BCL9 ALL treated with DMSO or E7820 (1 μM) and patient cells with MEF2D-HNRNPUL1 ALL treated with DMSO or E7820 (1 μM). (G-I) MEF2D-HNRNPUL1 ALL PDX cells were transplanted into NSG mice and randomized to E7820 (50 mg/kg) or vehicle, which were orally administrated to mice beginning on day 14 for 6 weeks. The disease burden was monitored by human CD19+ cells in peripheral blood (G). ∗P < .05. (H) Spleen weight of mice when the humane end points were reached or on day 75 (end of the study, left; ∗P < .05). Representative images of spleens (right). (I) Survival analysis of the mice administrated with E7820 (50 mg/kg) or vehicle. ∗P < .05.
Consistent with the positive correlation between RBM39 and DCAF15 mRNA expression in the MEF2D fusion subtype of ALL, MEF2D fusion ALL cells showed greater degradation of RBM39 protein than Ph-ALL or Ph-like ALL cells (Figure 6E; supplemental Figure 8A). This might be because of the unique expression profile observed in MEF2D fusion ALL.12 Furthermore, it is known that the MEF2D-HNRNPUL1 fusion leads to dysregulation of HNRNPUL1, which plays an important role in mRNA splicing.12,54 We then compared the alternative splicing profile of MEF2D-BCL9 fusion PDX cells with MEF2D-HNRNPUL1 fusion PDX cells following E7820 treatment. The increased alternative splicing events included mutually exclusive exons and exon skipping, conforming to the alternative splicing events of mutually exclusive exons and exon skipping upon knockdown of RBM3955 and suggesting that MEF2D-HNRNPUL1 fusion potentiates the effect of RBM39 loss on alternative splicing, which is associated with a lower IC50 for E7820 in these patient cells (Figure 6D,F).
We also assayed the ability of E7820 to suppress disease progression in a PDX model derived from a sample of a patient with MEF2D-HNRNPUL1 fusion. Strikingly, treatment with 25 mg/kg or 50 mg/kg E7820 relieved the disease burden both in peripheral blood and spleen, ultimately leading to an impressive extension of survival (Figure 6G-I; supplemental Figure 8B-C). The combination treatment of EHT1610 and E7820 in NALM6 cells showed synergistic effects on the inhibition of cell growth and deregulated RBM39 protein, suggesting crosstalk between CMGC kinases and RBM39 networks in ALL (supplemental Figure 8D-E).
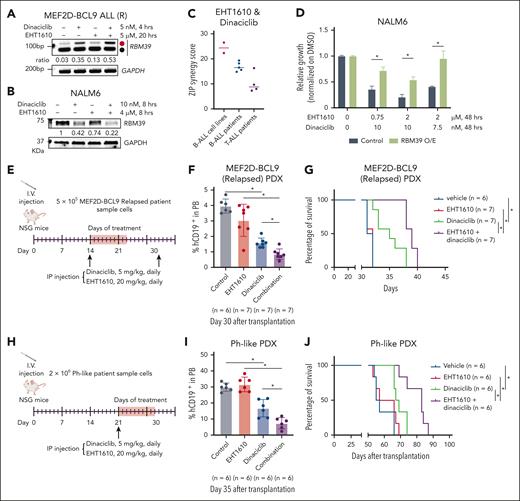
Targeting cotranscriptional splicing in ALL with CMGC inhibitors
Given that dinaciclib has been reported to have in vivo efficacy in chronic lymphoblastic leukemia,56 we focused on dinaciclib as a CDK9 inhibitor for the in vivo studies. Because dinaciclib targets CDK9 and other CDKs,57 we conducted a CRISPR screen with a kinase-focused library in NALM6 cells. CDK9 ranked seventh of 455 kinases based on the Robust Rank Aggregation score,58 whereas other top-ranked kinases were not targeted by dinaciclib (supplemental Figure 9A; supplemental Table 7). This suggests that CDK9 plays a key role in dinaciclib-induced cellular effects in B-ALL. Moreover, we confirmed induction of the RBM39 NMD isoform with dinaciclib alone and in combination with EHT1610 in both ALL cell lines and patient samples (Figure 7A; supplemental Figure 9B-C). Consistent with the strong effect of the combination on the production of the NMD isoform, we observed a concomitant striking decrease in RBM39 protein (Figure 7B; supplemental Figure 9D). Furthermore, dinaciclib and EHT1610 showed strong synergistic effects in B-ALL cell lines and samples from patients with B-ALL or T-ALL in vitro (Figure 7C; supplemental Figures 10 and 11). Similar results were observed with the combination of THAL-SNS-032 and EHT1610 treatments (supplemental Figure 9E). To verify the importance of downregulation of the RBM39 protein in the growth suppressive effect of EHT1610 and dinaciclib treatment, ectopic expression of an RBM39 complimentary DNA lacking the poison exon, and therefore not prone to NMD, partially rescued the growth inhibition of NALM6 cells upon EHT1610 and dinaciclib treatment (Figure 7D; supplemental Figure 9F). Meanwhile, the combination of EHT1610 and dinaciclib showed a minimal effect on the viability of healthy human CD34+ cells (supplemental Figure 9G). We then tested the efficacy of dinaciclib and EHT1610 in vivo with a high-risk ALL PDX model. Although single treatment with dinaciclib significantly reduced the disease burden in peripheral blood and extended survival, we found that the combination of dinaciclib and EHT1610 led to an even greater reduction in disease burden in the peripheral blood and extension of survival (Figure 7E-J).
Targeting the poison exon of RBM39 with CMGC kinase inhibitors in ALL. (A) Relapsed MEF2D-BCL9 PDX cells were treated with EHT1610 (5 μM) for 16 hours, followed by additional treatment with dinaciclib (5 nM) for 4 hours. PCR reactions were performed for detection of the RBM39 splicing event. (B) Representative western blot analysis of 3 biologic replicates for RBM39 in NALM6 cells treated with dinaciclib, EHT1610, or the combination for 8 hours. (C) ZIP synergy score for EHT1610 and dinaciclib for 3 days in B-ALL cell lines, B-ALL PDX samples, and T-ALL PDX samples. (D) Cell viability assay for control or RBM39 O/E NALM6 cells treated with dinaciclib and EHT1610 for 72 hours (n = 2; ∗P < .05). (E-J) Schematic showing details of dinaciclib and EHT1610 treatment in relapsed MEF2D-BCL9 ALL (E) or Ph-like (H) ALL PDX models. Disease burden was monitored by assessing the human CD19+ cells in the peripheral blood (F, I). Survival analysis of relapsed MEF2D-BCL9 ALL (G) or Ph-like (J) ALL PDX. ∗P < .05. O/E, overexpression; ZIP, Zero interaction potency.
Targeting the poison exon of RBM39 with CMGC kinase inhibitors in ALL. (A) Relapsed MEF2D-BCL9 PDX cells were treated with EHT1610 (5 μM) for 16 hours, followed by additional treatment with dinaciclib (5 nM) for 4 hours. PCR reactions were performed for detection of the RBM39 splicing event. (B) Representative western blot analysis of 3 biologic replicates for RBM39 in NALM6 cells treated with dinaciclib, EHT1610, or the combination for 8 hours. (C) ZIP synergy score for EHT1610 and dinaciclib for 3 days in B-ALL cell lines, B-ALL PDX samples, and T-ALL PDX samples. (D) Cell viability assay for control or RBM39 O/E NALM6 cells treated with dinaciclib and EHT1610 for 72 hours (n = 2; ∗P < .05). (E-J) Schematic showing details of dinaciclib and EHT1610 treatment in relapsed MEF2D-BCL9 ALL (E) or Ph-like (H) ALL PDX models. Disease burden was monitored by assessing the human CD19+ cells in the peripheral blood (F, I). Survival analysis of relapsed MEF2D-BCL9 ALL (G) or Ph-like (J) ALL PDX. ∗P < .05. O/E, overexpression; ZIP, Zero interaction potency.
Discussion
We have demonstrated that the inhibition of the CMGC family kinases affects cotranscriptional splicing and disrupts the association between Pol II and the spliceosome machinery, which represents a therapeutic target in ALL. Specifically, EHT1610, which targets DYRKs, GSK3s, casein kinases, and HIPKs but not CDK9, decreased RNA Pol II CTD phosphorylation. This is consistent with previous reports that demonstrated the potential of these kinases to regulate Pol II CTD phosphorylation.41-45 Consequently, the inhibition of Pol II CTD phosphorylation led to alterations in the SF3B1 and Pol II association and the release of Pol II pausing to promote upstream RBM39 poison exon inclusion. The modulation of Pol II CTD phosphorylation during cotranscriptional splicing could also be achieved by the inhibition of CDK9. Notably, although EHT1610 and CDK9 inhibition both induced inclusion of the RBM39 poison exon through mediation of cotranscriptional splicing, their mechanisms are distinct. This is evidenced by the opposite effects that the THAL-SNS-032 and EHT1610 treatments had on Pol II pausing.
A recent publication demonstrated that DYRK/CLK inhibition impeded the phosphorylation of SR-rich splicing factors, which contributed to the alternative splicing dysregulation in AML.39 However, in our studies, the inhibition of CMGC kinases including DYRK1A or CDK9 impaired the interaction between the phosphorylated Pol II CTD and the SF3B1-associated splicing complex, which contributes to the regulation of cotranscriptional splicing. Our results align with a recent study that underscores the critical role of U2 snRNP in efficient RNA Pol II pause release.59 DYRK1A has been shown to phosphorylate both splicing factors, including SF3B1, SRSF1, and SRSF2, and Ser2 and Ser5 in the Pol II CTD.20,41,60-62 Future studies to dissect the connection between RNA Pol II and splicing factor phosphorylation in the control of mRNA splicing are warranted.
Several studies have highlighted RBM39 as a promising target in cancer, antitumor immunity, and graft-versus-host disease.55,63-66 In this study, we extend RBM39 targeting to high-risk ALL. In addition to targeting RBM39 with the degraders E7070 and E7820, we have uncovered a novel mechanism for targeting RBM39 via the modulation of cotranscriptional splicing and provided strong preclinical data that showed favorable in vivo efficacy. Targeting RBM39 by the combination of EHT1610 and dinaciclib may be effective in patients with Ph-like ALL or relapsed MEF2D-ALL. Although there was no meaningful clinical response when E7820 was administered as a single agent in a phase 2 study of patients with relapsed and refractory myeloid malignancies who had splicing factor mutations, the drug was well tolerated, thereby prompting future combination studies.67 In fact, a phase 2 clinical trial is currently underway to test the combination of the RBM39 degrader E7820 with venetoclax in splicing-mutant myeloid malignancies.68 Therefore, clinical trials that investigate E7820 either as a single agent or in combination with conventional ALL therapies for relapsed or refractory B-ALL is something to consider.
Acknowledgments
The authors thank Sridhar Rao for helpful discussions; Eric Wang, Jun Yang, and Iannis Aifantis for providing the CRISPR/Cas9 library; Biosplice for providing SM09419; and Novartis Biomedical Research for providing GNF2133. The visual abstract was created with BioRender.com.
J.D.C. was supported by the National Institutes of Health (NIH), National Cancer Institute (NCI) grant (R35CA253096), the Samuel Waxman Cancer Research Foundation, and the St. Jude/American Lebanese Syrian Associated Charities. R.S.B. was supported by the Hematopoiesis Training Program at the University of Pennsylvania (grant T32DK07780). A.Y. was partly supported by funding from Japan Society for the Promotion of Science KAKENHI (grant-in-aid no. 21H04828) and the Japan Agency for Medical Research and Development (grant no. JP22jm0210085). The Center for Advanced Genome Engineering, the Hartwell Center, the protein production shared resource, and the flow cytometry and cell sorting shared resource at St. Jude are supported in part by NIH/NCI (grant P30 CA021765). E.A.O. is a recipient of an American Society of Hematology Scholar Award and a Gabriel’s Angel Foundation Medical Research Award. The Ntziachristos laboratory was supported by the Research Foundation Flanders (grants G0F4721N and G0A8B24N), start-up funds from the Department of Biomolecular Medicine, Ghent University, a Flanders interuniversity consortium grant (BOF.IBO.2023.0006.02), and a Cancer Research Institute Ghent partnership grant. S.M. was supported by a fellowship from the Cancer Council Western Australia and by the Child Cancer Research Foundation.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Authorship
Contribution: Q.J., E.H., H.K.S., A.B.S., R.S.B., J.K., S.N., T.P., A.Y., and S.M.P.-M. performed the experiments, analyzed the data, and contributed to writing of the manuscript; J.A.M., R.M., and D.W.B. provided bioinformatic analyses and contributed to writing of the manuscript; A.C. and J.Q.W. helped with the in vivo studies; R.K., O.A.-W., S.M., P.N., E.A.O., and J.D.C. conceived the experiments, analyzed the data, and contributed to writing of the manuscript.
Conflict-of-interest disclosure: A.Y. reports receiving research grants from Eisai Inc and Chugai Pharmaceutical Co, Ltd. J.D.C. reports receiving research support from Syndax, consulting fees from Cellarity, and serving as a scientific advisor for Alethiomics. O.A.-W. has served as a consultant for Foundation Medicine Inc, Merck, Prelude Therapeutics, and Janssen; is on the Scientific Advisory Board of Envisagenics Inc, AIChemy, Harmonic Discovery Inc, and Pfizer Boulder; has received prior research funding from H3 Biomedicine, Nurix Therapeutics, Minovia Therapeutics, and Loxo Oncology unrelated to the current manuscript; and is a founder of Codify Therapeutics for which he also serves as a consultant and receives research support. R.S.B. reports previously receiving consulting fees from Alva10. The remaining authors declare no competing financial interests.
Correspondence: John D. Crispino, Department of Hematology, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, MS341, Memphis, TN 38105; email: john.crispino@stjude.org.
References
Author notes
The RNA sequencing data reported in this article have been deposited in the Gene Expression Omnibus/Sequence Read Archive database (accession number GSE252550), and the Proteomic Data in the Proeomics Identification (PRIDE) database (accession number PXD048846).
Original data are available on request from the corresponding author, John D. Crispino (john.crispino@stjude.org). This study used data generated by the St. Jude Children’s Research Hospital Genomes for Kids Study, the Pediatric Cancer Genome Project, and the Pan-Acute Lymphoblastic Leukemia Data Set. Additional materials and methods are described in the supplemental Data. A list of reagents, including antibodies, primers, short hairpin RNAs, drugs, and assay kits, is described in supplemental Table 9.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![High-risk subtypes of ALL are sensitive to RBM39 degradation. (A) Pearson correlation analysis between RBM39 mRNA and DCAF15 mRNA across the indicated subtypes of B-ALL. Patients with B-ALL mRNA expression data were obtained from the Pediatric Cancer Genome Project data portal (PeCan, St Jude, Memphis). (B) Indisulam area under the curve (AUC) of B-ALL, T-ALL and AML in comparison with nonleukemia cell lines. Data were obtained from the CTD2 network53 (left). The indisulam AUC for the indicated B-ALL cell lines (right). (C) IC50 of E7820 (n = 3; the mean value ± standard deviation [SD] is shown) across different B-ALL cell lines as indicated. MEF2D fusion subtypes include Kasumi-7 and -9. (D) The IC50 of E7820 (n = 3; the mean value ± SD is shown) across different B-ALL PDX samples as indicated. (E) Western blot analysis of RBM39 in Kasumi-7 and SUPB15 treated with increasing concentration of E7820 for 4 h. One representative blot is shown on left and quantification of 3 biological replicates is shown on right. (F) Bar graph showing different types of splicing events for patients cells with MEF2D-BCL9 ALL treated with DMSO or E7820 (1 μM) and patient cells with MEF2D-HNRNPUL1 ALL treated with DMSO or E7820 (1 μM). (G-I) MEF2D-HNRNPUL1 ALL PDX cells were transplanted into NSG mice and randomized to E7820 (50 mg/kg) or vehicle, which were orally administrated to mice beginning on day 14 for 6 weeks. The disease burden was monitored by human CD19+ cells in peripheral blood (G). ∗P < .05. (H) Spleen weight of mice when the humane end points were reached or on day 75 (end of the study, left; ∗P < .05). Representative images of spleens (right). (I) Survival analysis of the mice administrated with E7820 (50 mg/kg) or vehicle. ∗P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/23/10.1182_blood.2024024281/2/m_blood_bld-2024-024281-gr6.jpeg?Expires=1771166864&Signature=xtV7uWszW6W44efiw5hKKcHK0gHEnEYnVx1-Fuq0YYzaUfqAEA0pa6FNQY4H5qYLaQ9wvY6oGTZITpEVpsvbuWeKzB9W3QmGbgSjO3DlXRJgWKt9FxDdZxAcahbwBHnxykyz-1A6fXrs6~iy80oBYJjaqkRIfIYB0OyzUYVtbcxZeXUCcPa7~0G1VX~AOLg5r5tJdcTt7lmblwWlu5-uIzANOESRwxP1~Y8jCX7TJAidTcOV-XpdiDuPycqnuk1rA7BZDdAvJFoilRIpwXqXrCwNxH0JHCJh3TVER206K7fadbvyQnUK5pJ7~~6qzQWpK1hJPqs--nyC7uc0zL98Lw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal