Key Points
Single-cell multiomics delineates the maturation block in childhood ALL with DUX4-r cases exhibiting a high degree of lineage infidelity.
DUX4-r ALL displays high sensitivity to PI3K inhibitors and susceptibility to CD371 CAR T cell killing.
Visual Abstract
B-cell progenitor acute lymphoblastic leukemia (BCP-ALL) is the most common childhood malignancy and is driven by multiple genetic alterations that cause maturation arrest and accumulation of abnormal progenitor B cells. Current treatment protocols with chemotherapy have led to favorable outcomes but are associated with significant toxicity and risk of side effects, highlighting the necessity for highly effective, less toxic, targeted drugs, even in subtypes with a favorable outcome. Here, we used multimodal single-cell sequencing to delineate the transcriptional, epigenetic, and immunophenotypic characteristics of 23 childhood BCP-ALLs belonging to the BCR::ABL1+, ETV6::RUNX1+, high hyperdiploid, and recently discovered DUX4-rearranged (DUX4-r) subtypes. Projection of the ALL cells along the normal hematopoietic differentiation axis revealed a diversity in the maturation pattern between the different BCP-ALL subtypes. Although the BCR::ABL1+, ETV6::RUNX1+, and high hyperdiploidy cells mainly showed similarities to normal pro-B cells, DUX4-r ALL cells also displayed transcriptional signatures resembling mature B cells. Focusing on the DUX4-r subtype, we found that the blast population displayed not only multilineage priming toward nonhematopoietic cells, myeloid, and T-cell lineages, but also an activation of phosphatidylinositol 3-kinase (PI3K)/AKT signaling that sensitized the cells to PI3K inhibition in vivo. Given the multilineage priming of DUX4-r blasts with aberrant expression of myeloid marker CD371 (CLL-1), we generated chimeric antigen receptor T cells, which effectively eliminated DUX4-r ALL cells in vivo. These results provide a detailed characterization of BCP-ALL at the single-cell level and reveal therapeutic vulnerabilities in the DUX4-r subtype, with implications for the understanding of ALL biology and new therapeutic strategies.
Introduction
B-cell progenitor acute lymphoblastic leukemia (BCP-ALL) is initiated by genetic and epigenetic changes that cause uncontrolled expansion of immature B cells.1-3 Although the outcome is favorable in children,1,2 treatment with cytostatic drugs is associated with toxicity and long-term side effects.4,5 Recently, kinase inhibitors have dramatically improved the outcome for a limited set of patients,1,6 and immunotherapy using chimeric antigen receptor (CAR) T cells targeting CD19 has shown remarkable efficacy in treating relapsed/refractory BCP-ALL. However, some patients fail the latter because of loss of CD19.7,8 Hence, there is a need for novel and highly effective therapies with low toxicity.
BCP-ALL is classified into risk-stratifying subtypes based on recurring mutations and chromosomal aberrations.1,9,10 Here, we used a multimodal single-cell sequencing strategy to delineate the transcriptional, epigenetic, and immunophenotypic characteristics of some of the most common genetic subtypes in children, namely high hyperdiploid (HeH; 30%), ETV6::RUNX1+ (25%), DUX4-rearranged (DUX4-r; 4%), and BCR::ABL1+ (3%) BCP-ALL,9,11 with a focus on the recently described DUX4-r subtype.12-14 This subtype is defined by rearrangements that affect early embryonic transcription factor (TF) DUX4. Most commonly, a truncated DUX4 is inserted into the immunoglobulin heavy chain (IGH) gene locus, which leads to altered expression of DUX4 due to loss of suppression.11-14
The single-cell sequencing strategy allowed us to delineate the maturation block at a high resolution across different subtypes. In DUX4-r ALLs, we uncovered a high degree of lineage infidelity in leukemic blasts and sensitivity to phosphatidylinositol 3-kinase (PI3K) inhibitors in vivo. The lineage infidelity of DUX4-r blasts, for example, manifests as an aberrant expression of the myeloid marker CD371 (CLL-1). Based on this, we developed CD371 targeting CAR T cells that demonstrate efficient killing of DUX4-r leukemic cells.
Methods
Study samples
Twenty-three childhood BCP-ALLs were included: 4 each of BCR::ABL1+, ETV6::RUNX1+, and HeH ALLs, and 11 DUX4-r ALLs (Table 1; supplemental Table 1, available on the Blood website). Mononuclear cells from bone marrow (BM) samples from patients with BCP-ALL and 5 healthy donors were collected and cryopreserved at Skåne University and Copenhagen University Hospitals. ALLs were classified according to the recent International Consensus Classification (ICC) classification.9 Sorted CD34+, CD19+, and unsorted mononuclear cells from healthy donors were used (Table 1; supplemental Table 1). Informed consent was obtained according to the Declaration of Helsinki. The study was approved by the Ethics Committees of the Lund and Copenhagen Universities.
Summary of the study samples
| Characteristics . | Study samples . |
|---|---|
| No. of BCP-ALL cases | 23∗,† |
| Mean age, y | 8.5 |
| Female/male, n | 12/11 |
| BM blasts, mean % | 89 |
| Molecular subgroup/WHO classification, no. of patients | |
| BCR::ABL1/BCP-ALL with (9;22)(q34.1;q11.2);BCR::ABL1 | 4 |
| ETV6::RUNX1/t(12;21)(p13.2;q22.1);ETV6::RUNX1 | 4 |
| HeH/hyperdiploidy | 4 |
| DUX4-r/BCP-ALL with other defined genetic abnormalities | 11 |
| scRNA-seq, no. of BCP-ALL cases (no. of cells) | 23 (149 731) |
| BCR::ABL1 | 4 (25 706) |
| ETV6::RUNX1 | 4 (29 772) |
| HeH | 4 (21 346) |
| DUX4-r | 11 (72 907) |
| scATAC-seq, no. of BCP-ALL cases (no. of nuclei) | 19 (35 225) |
| BCR::ABL1 | 3 (7 181) |
| ETV6::RUNX1 | 4 (7 712) |
| HeH | 4 (3 623) |
| DUX4-r | 8 (15 657) |
| No. of NBM samples (donors) | 9 (5) |
| MNC | 3 |
| CD34+-enriched | 3 |
| CD19+-enriched | 3 |
| scRNA-seq, no. of NBM samples (no. of cells) | 9 (38 815) |
| MNC | 3 (16 717) |
| CD34+-enriched | 3 (11 131) |
| CD19+-enriched | 3 (10 967) |
| scATAC-seq, no. of NBM samples (no. of nuclei) | 4 (6 667) |
| MNC | 3 (4 623) |
| CD34+-enriched | 1 (3 096) |
| Characteristics . | Study samples . |
|---|---|
| No. of BCP-ALL cases | 23∗,† |
| Mean age, y | 8.5 |
| Female/male, n | 12/11 |
| BM blasts, mean % | 89 |
| Molecular subgroup/WHO classification, no. of patients | |
| BCR::ABL1/BCP-ALL with (9;22)(q34.1;q11.2);BCR::ABL1 | 4 |
| ETV6::RUNX1/t(12;21)(p13.2;q22.1);ETV6::RUNX1 | 4 |
| HeH/hyperdiploidy | 4 |
| DUX4-r/BCP-ALL with other defined genetic abnormalities | 11 |
| scRNA-seq, no. of BCP-ALL cases (no. of cells) | 23 (149 731) |
| BCR::ABL1 | 4 (25 706) |
| ETV6::RUNX1 | 4 (29 772) |
| HeH | 4 (21 346) |
| DUX4-r | 11 (72 907) |
| scATAC-seq, no. of BCP-ALL cases (no. of nuclei) | 19 (35 225) |
| BCR::ABL1 | 3 (7 181) |
| ETV6::RUNX1 | 4 (7 712) |
| HeH | 4 (3 623) |
| DUX4-r | 8 (15 657) |
| No. of NBM samples (donors) | 9 (5) |
| MNC | 3 |
| CD34+-enriched | 3 |
| CD19+-enriched | 3 |
| scRNA-seq, no. of NBM samples (no. of cells) | 9 (38 815) |
| MNC | 3 (16 717) |
| CD34+-enriched | 3 (11 131) |
| CD19+-enriched | 3 (10 967) |
| scATAC-seq, no. of NBM samples (no. of nuclei) | 4 (6 667) |
| MNC | 3 (4 623) |
| CD34+-enriched | 1 (3 096) |
MNC, mononuclear cells; no., number; NOS, not otherwise specified; scRNA-seq, single-cell RNA sequencing; WHO, World Health Organization.
Including 22 diagnostic samples and 1 relapse sample.
Including 1 mixed phenotype acute leukemia.
Multimodal single-cell sequencing
Cells were stained with TotalSeqC antibodies to detect fluorescence-activated cell sorting markers for diagnostics or minimal residual disease monitoring in the NOPHO ALL2008, EsPhALL, or ALLtogether protocols15,16 (supplemental Table 2). Sorted cells were used for single-cell antibody-derived tag sequencing (scADT-seq), single-cell B-cell receptor sequencing (scBCR-seq), and single-cell RNA-sequencing (scRNA-seq) library preparation according to the manufacturer’s protocol (10X Genomics). In addition, a separate single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) was performed with nuclei isolation and scATAC-seq library preparation according to the manufacturer’s instructions (10X Genomics). All libraries were sequenced on a NovaSeq 6000 System (Illumina).
Single-cell data analysis
Sequencing data were converted to the FASTQ format using bcl2fastq (Illumina). Cellranger multi (10X Genomics, v.5.1.0) and Cellranger atac count (10X Genomics, v.1.2.0) were used to generate read count matrices. Further analysis was performed using Seurat (v.4.0.0) and Signac (v1.5.0).17,18 SingleCellProjections.jl (https://github.com/rasmushenningsson/SingleCellProjections.jl) was used to create normal bone marrow (NBM) reference force–directed k-nearest neighbors (knn) graphs19 and to project the BCP-ALL data.
Cell culture and dose-response analysis of PI3K treatment
The BCP-ALL cell lines NALM6, 697, REH, SEM, TOM-1, and AT-1 were cultured as previously described.20 The cells were treated for 24 or 48 hours with 1 to 10 000 nM BYL-719 (alpelisib; MedChemExpress) and 1 to 10 000 nM BAY 80-6946 (copanlisib; MedChemExpress). DUX4-r ALL patient-derived xenografts (PDXs) were obtained by propagating diagnostic samples in NOD.Cg-Prkdcscid Il2r1gtm1Wjl/SzJ (NSG) mice, and the effect of alpelisib was tested in vivo.
Design of CD371 targeted CAR T cells
CAR constructs were generated as previously described.21 Primary human T cells were isolated from anonymous healthy donors. The CAR T-cell effects were tested on the DUX4-r cell line NALM6 and PDX samples ex vivo and in vivo.
Results
Multimodal single-cell sequencing reveals distinct genetic and epigenetic landscapes of BCP-ALL subtypes
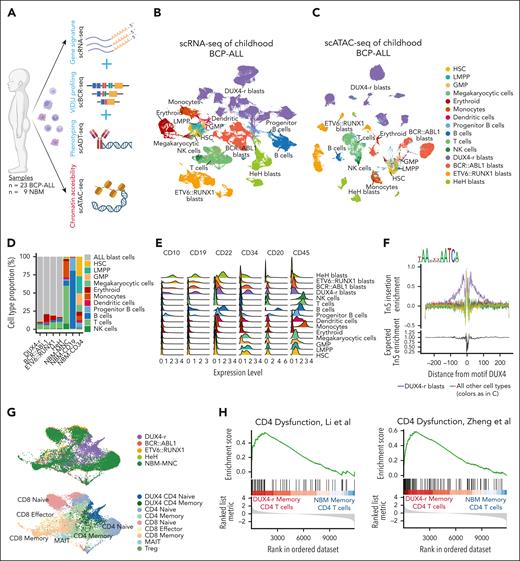
Using multimodal single-cell sequencing, we profiled 188 546 single cells from 23 BCP-ALLs and 9 NBM samples (Table 1; Figure 1A). All the cells were annotated using an external single-cell data set of NBM cells as reference22,23 (supplemental Figure 1A). The single-cell RNA sequencing data revealed an expanded aberrant cell population from each patient sample, visualized by uniform manifold approximation and projection (UMAP) as separate clusters sublocalized by genetic subtype (Figure 1B; supplemental Figure 1B-C). Epigenetic profiling by scATAC-seq of 41 892 nuclei revealed a cellular BCP-ALL landscape closely mirroring the transcriptomic landscape (Figure 1C; supplemental Figure 1D). Reference-based annotation of cells in the expanded aberrant populations classified most cells as “progenitor B cells” (supplemental Figure 1B). However, the proportion (40%-99% in each sample; Figure 1D; supplemental Figure 1E-F) and immunophenotypes of the expanded populations were concordant with the classifying these as BCP-ALL blast cells24,25 (Figure 1E; supplemental Figure 2). Interestingly, when studying the V(D)J rearrangements, we could also identify DUX4 gene segments present in the IGH region in 9 of 11 DUX4-r cases (supplemental Figure 3A-B). Motif footprinting confirmed that the DUX4 binding motif was exclusively active in these populations (Figure 1F). Consequently, all cells within the expanded populations were labeled “ALL blast cells.”
Multimodal single-cell sequencing reveals a severely altered leukemic BM. (A) Schematic illustration of multiomic profiling of BCP-ALL (n = 23) and NBM (n = 9) samples. Illustration created with BioRender.com. (B) UMAP visualization of scRNA-seq gene expression data representing 188 546 cells from the BCP-ALL and NBM samples. Each cell is colored according to the cell type. (C) UMAP visualization of scATAC-seq data representing 41 892 nuclei from the BCP-ALL and NBM samples. (D) Cell composition based on scRNA-seq data in DUX4-r, BCR::ABL1+, ETV6::RUNX1+, and HeH ALL samples, as well as mononuclear cells (MNC), CD34+-enriched cells, and CD19+-enriched cells from NBM samples. ALL blast cells, regardless of subtype, are marked in gray. (E) Protein expression by scADT-seq of CD10, CD19, CD22, CD34, CD20, and CD45 on BCP-ALL blast cells and nonleukemic cells from BCP-ALL and NBM samples, visualized using ridge plots. (F) TF footprinting of the DUX4-binding motif shows that DUX4 is exclusively active in DUX4-r blast cells (marked in purple). The annotation “all other cell types” denotes all other detected cell types using the color legend from C. (G) UMAP visualization of T cells extracted from the larger scRNA-seq data set colored by sample type (upper) and predicted cell type (lower) showing a specific cell cluster of an accumulation of CD4+ T cells exclusively from DUX4-r ALL. (H) Gene set enrichment scores using defined gene sets from dysfunctional CD4+ T cells in cancer by Li et al (left) and Zheng et al (right).31,32 The gene set enrichment scores are based on differentially expressed genes between memory CD4+ T cells in DUX4-r ALL and NBM-MNC. GMP, granulocyte-macrophage progenitors; LMPP, lympho-myeloid primed progenitor; NK, natural killer; scBCR-seq, single-cell BCR sequencing; scRNA-seq, single-cell RNA sequencing; UMAP, uniform manifold approximation and projection.
Multimodal single-cell sequencing reveals a severely altered leukemic BM. (A) Schematic illustration of multiomic profiling of BCP-ALL (n = 23) and NBM (n = 9) samples. Illustration created with BioRender.com. (B) UMAP visualization of scRNA-seq gene expression data representing 188 546 cells from the BCP-ALL and NBM samples. Each cell is colored according to the cell type. (C) UMAP visualization of scATAC-seq data representing 41 892 nuclei from the BCP-ALL and NBM samples. (D) Cell composition based on scRNA-seq data in DUX4-r, BCR::ABL1+, ETV6::RUNX1+, and HeH ALL samples, as well as mononuclear cells (MNC), CD34+-enriched cells, and CD19+-enriched cells from NBM samples. ALL blast cells, regardless of subtype, are marked in gray. (E) Protein expression by scADT-seq of CD10, CD19, CD22, CD34, CD20, and CD45 on BCP-ALL blast cells and nonleukemic cells from BCP-ALL and NBM samples, visualized using ridge plots. (F) TF footprinting of the DUX4-binding motif shows that DUX4 is exclusively active in DUX4-r blast cells (marked in purple). The annotation “all other cell types” denotes all other detected cell types using the color legend from C. (G) UMAP visualization of T cells extracted from the larger scRNA-seq data set colored by sample type (upper) and predicted cell type (lower) showing a specific cell cluster of an accumulation of CD4+ T cells exclusively from DUX4-r ALL. (H) Gene set enrichment scores using defined gene sets from dysfunctional CD4+ T cells in cancer by Li et al (left) and Zheng et al (right).31,32 The gene set enrichment scores are based on differentially expressed genes between memory CD4+ T cells in DUX4-r ALL and NBM-MNC. GMP, granulocyte-macrophage progenitors; LMPP, lympho-myeloid primed progenitor; NK, natural killer; scBCR-seq, single-cell BCR sequencing; scRNA-seq, single-cell RNA sequencing; UMAP, uniform manifold approximation and projection.
The most abundant nonmalignant cell type in leukemic BM was T cells (Figure 1D). In general, BCP-ALL T cells were transcriptionally similar to the NBM T cells (supplemental Figure 4A). However, a distinct cell cluster was observed containing CD4+ naïve and memory T cells almost exclusively from DUX4-r ALLs (representing 8/11 DUX4-r cases; Figure 1G; supplemental Figure 4B-D). The aberrant CD4+ T cells did not contain leukemia-specific immunoglobulin rearrangements in the 3 examined cases (supplemental Figure 4F-H) and did not exhibit DUX4 activity (supplemental Figure 4E), suggesting that they were altered by external factors rather than originating from DUX4-r cells. Differential gene expression (DGE) analysis of CD4+ T cells in DUX4-r ALL and NBM indicated the upregulation of genes involved in epigenetic enforcement of exhaustion, such as FOS, JUN, and NR4A- and TOX-family genes26-30 (supplemental Figure 4I; supplemental Tables 3 and 4). The presence of transcriptionally dysfunctional memory CD4+ T cells was also supported by gene set enrichment analysis (GSEA) using gene sets defining dysfunctional CD4+ T cells in cancer31,32 (Figure 1H).
In conclusion, single-cell profiling provided a detailed overview of the cellular landscape of BCP-ALL, allowed the identification of distinct clusters of patient-specific blast cells, and unexpectedly identified a CD4+ T-cell population in DUX4-r ALL that displayed transcriptional signs of dysfunction.
B-cell differentiation pattern in BCP-ALL differs between genetic subtypes
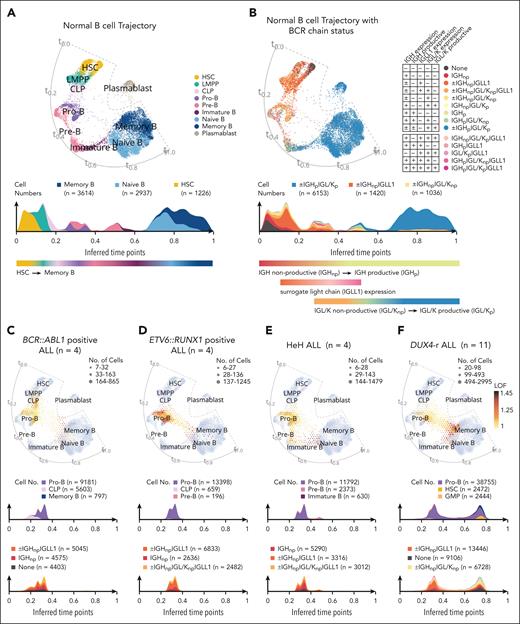
To delineate the differentiation block in BCP-ALL blast cells, the annotation for NBM samples was extended along the B-cell lineage (supplemental Figure 5A-E). We then constructed a force–directed knn graph19 that recapitulated the maturation trajectories present in normal B cells. Based on this, we created cell density plots along the inferred time points (t0.0-t1.0) of the trajectory (Figure 2A-B). The normal B-cell maturation trajectory was further delineated by assigning developmental stages to B-cell receptor (BCR) status. As expected, BCR rearrangements occurred gradually during B-cell differentiation and first appeared as nonproductive rearrangements accompanied by the surrogate light chain (IGLL1), followed by productive rearrangements and loss of IGLL1 expression (Figure 2B).33 Next, BCP-ALL blast cells were projected onto a normal B-cell maturation trajectory. The blast cells covered the entire B-cell maturation axis, from hematopoietic stem cells (HSC) to memory B cells, but with unique distribution patterns for different BCP-ALL samples (Figure 2C-F; supplemental Figures 6-11). Despite the observed heterogeneity, specific patterns were observed within each subtype: BCR::ABL1+ ALLs contained cells with an immature profile, with a distribution mainly covering common lymphoid progenitor (CLP) and pro-B cells (Figure 2C; supplemental Figure 6A-D). The ETV6::RUNX1+ cells mainly resembled pro-B cells, but with substantial gene expression differences between the NBM pro-B cells and ALL blasts, as indicated by high local outlier factor scores (Figure 2D; supplemental Figure 7A-D). HeH cells projected onto a region from pro-B cells to immature B cells, seemingly including a maturation path similar to that of normal cells (Figure 2E; supplemental Figure 8A-D). Studying BCR chain status, BCR::ABL1+, ETV6::RUNX1+, and 2 cases of HeH ALL preferentially displayed a BCR chain status consistent with the pro–B-cell stage (nonproductive IGH, often together with IGLL1 expression; Figure 2C-E; supplemental Figures 6-8). The remaining cases of HeH (HeH-2 and HeH-3) expressed BCR chains consistent with the early pre–B-cell stage (nonproductive IGH with nonproductive IGL/IGK and IGLL1 expression; supplemental Figure 8A-D). Analyzing the co-occurrence of expressed immunoglobulin rearrangements allowed us to infer the clonal structure of leukemia. Each case was typically dominated by a major clone with at least 1 rearranged IGH chain. However, BCR recombination was found to continue in leukemic cells, with small clones expressing additional rearranged IGH, IGL, and IGK chains (supplemental Figures 6-8). Interestingly, the inferred clonal trees indicated that the cell of origin for 2 of the 3 diagnostic BCR::ABL1+ ALLs lacked IGH rearrangements, in contrast to all other studied cases (supplemental Figure 6).
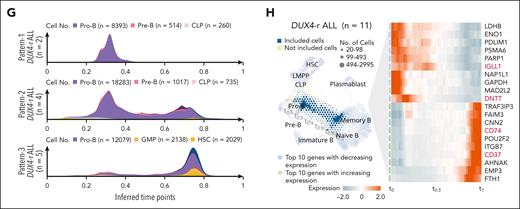
Projection of blast cells onto normal B-cell maturation trajectory shows variable maturation blocks between different subtypes of BCP-ALL. (A) A force–directed knn graph of cells involved in B-cell maturation from NBM samples, based on scRNA-seq data, colored by cell type with an accompanying cell density plot of numbers of cells along the trajectory. Inferred time points (t0.0-t1.0) are indicated along the trajectory. The 3 most common annotations are visualized at the top of the cell density plot. This force–directed knn graph was used as a reference for the projection of ALL blast cells in (C-F). (B) The force–directed knn graph of NBM B-cell trajectory colored by BCR chain status with an accompanying cell density plot of numbers of cells along the trajectory. The minus sign (−) indicates a feature that is not detected, the plus sign (+) indicates a feature that is detected, and a plus/minus sign (±) indicates that this category does not rely on the detection of this feature (ie, it can be either detected or not detected). The colors for the 3 most common annotations are visualized at the top of the color-coded cell density plots. (C-F) Single-cell projections of blast cells from all patients representing BCR::ABL1+ (n = 4), ETV6::RUNX1+ (n = 4), HeH (n = 4), and DUX4-r ALL (n = 11) onto the NBM reference. The density of the projected cells is visualized by hexagon size. Gene expression differences compared with the NBM reference, based on the local outlier factor, are highlighted by the color gradient. Cell density plots below the projection illustrate the number of cells along the trajectory and are color-coded by cell type (upper) and BCR chain status (lower). The color for the 3 most common annotations is visualized on the top of the color-coded cell density plots. (G) Three cell density plots illustrating the numbers of DUX4-r blast cells along the trajectory color-coded by cell types, presenting 3 cellular differentiation patterns observed within DUX4-r ALL denoted Pattern-1 (upper), Pattern-2 (middle), and Pattern-3 (lower). The color for the 3 most common annotations is visualized on the top of the color-coded cell density plots. (H) NBM reference knn graph projection of blast cells from all DUX4-r ALL samples, with time point path (t0.0-t1.0) indicated in gray, and an associated heat map showing the gradual gene expression changes along the path (with genes selected by variance filtering and linear regression). The genes marked in red represent lineage-defining genes for B cells. CLP, common lymphoid progenitor; IGH, heavy chain; IGLL1, surrogate light chain; IGL/K, light chain; p, productive; np, nonproductive.
Projection of blast cells onto normal B-cell maturation trajectory shows variable maturation blocks between different subtypes of BCP-ALL. (A) A force–directed knn graph of cells involved in B-cell maturation from NBM samples, based on scRNA-seq data, colored by cell type with an accompanying cell density plot of numbers of cells along the trajectory. Inferred time points (t0.0-t1.0) are indicated along the trajectory. The 3 most common annotations are visualized at the top of the cell density plot. This force–directed knn graph was used as a reference for the projection of ALL blast cells in (C-F). (B) The force–directed knn graph of NBM B-cell trajectory colored by BCR chain status with an accompanying cell density plot of numbers of cells along the trajectory. The minus sign (−) indicates a feature that is not detected, the plus sign (+) indicates a feature that is detected, and a plus/minus sign (±) indicates that this category does not rely on the detection of this feature (ie, it can be either detected or not detected). The colors for the 3 most common annotations are visualized at the top of the color-coded cell density plots. (C-F) Single-cell projections of blast cells from all patients representing BCR::ABL1+ (n = 4), ETV6::RUNX1+ (n = 4), HeH (n = 4), and DUX4-r ALL (n = 11) onto the NBM reference. The density of the projected cells is visualized by hexagon size. Gene expression differences compared with the NBM reference, based on the local outlier factor, are highlighted by the color gradient. Cell density plots below the projection illustrate the number of cells along the trajectory and are color-coded by cell type (upper) and BCR chain status (lower). The color for the 3 most common annotations is visualized on the top of the color-coded cell density plots. (G) Three cell density plots illustrating the numbers of DUX4-r blast cells along the trajectory color-coded by cell types, presenting 3 cellular differentiation patterns observed within DUX4-r ALL denoted Pattern-1 (upper), Pattern-2 (middle), and Pattern-3 (lower). The color for the 3 most common annotations is visualized on the top of the color-coded cell density plots. (H) NBM reference knn graph projection of blast cells from all DUX4-r ALL samples, with time point path (t0.0-t1.0) indicated in gray, and an associated heat map showing the gradual gene expression changes along the path (with genes selected by variance filtering and linear regression). The genes marked in red represent lineage-defining genes for B cells. CLP, common lymphoid progenitor; IGH, heavy chain; IGLL1, surrogate light chain; IGL/K, light chain; p, productive; np, nonproductive.
DUX4-r+ ALLs had a proportion of blast cells with an expression profile resembling pro-B cells, but in contrast to other subtypes, they also contained cells similar to both pro-B cells and mature B cells. Consequently, these cells overlaid a region between pro-B and mature B, sparsely occupied by NBM cells (Figure 2F; supplemental Figures 9-11). Moreover, DUX4-r ALL showed heterogeneous maturation profiles with 3 main patterns: an accumulation of cells at the pro–B-cell stage (Pattern-1; n = 2), a bimodal distribution with cells at the pro–B-cell stage, a second population of mature B cells (Pattern-2, n = 4), or a more differentiated stage corresponding to mature B cells (Pattern-3, n = 5; Figure 2G; supplemental Figures 9-11 and 12A-C). Cases with Pattern-1 displayed a BCR chain status consistent with the pro–B-cell stage, with a dominant clone expressing 1 rearranged IGH allele together with IGH::DUX4 (supplemental Figure 9A-B). Pattern-2 and -3 cases displayed highly diverse BCR chain expression profiles. Most cases (6/9) exhibited a dominant clone, with both heavy (IGH) and light chain (IGK/IGL) rearrangements at the root of the clonal tree (supplemental Figures 9-11). This indicated that the transforming event in these cases occurred in a more mature cell with a rearranged light chain.
Interestingly, DUX4-r blast cells resembling mature B cells displayed a high local outlier factor score, indicating gene expression differences between ALL blasts and NBM mature B cells. Visualizing the gradual change in gene expression of DUX4-r cells from pro-B cells (t0.0) to mature B cells (t1.0) revealed the downregulation of early B-lineage markers and the emergence of mature B-lineage markers (Figure 2H; supplemental Figure 12D; supplemental Tables 5 and 6). Overrepresentation analysis was performed on the expression of early and late time points using cell-type gene sets.34 At early time points, only B-cell genes were enriched. At later time points, genes associated with several different cell types were enriched (supplemental Figure 12E), indicating that DUX4-r cells can acquire the transcriptional features of nonlymphoid cell types, such as dendritic cells and monocytes.
Single-cell multiomic profiling of DUX4-r ALL identifies deregulated transcriptional programs and susceptibility to PI3K inhibition
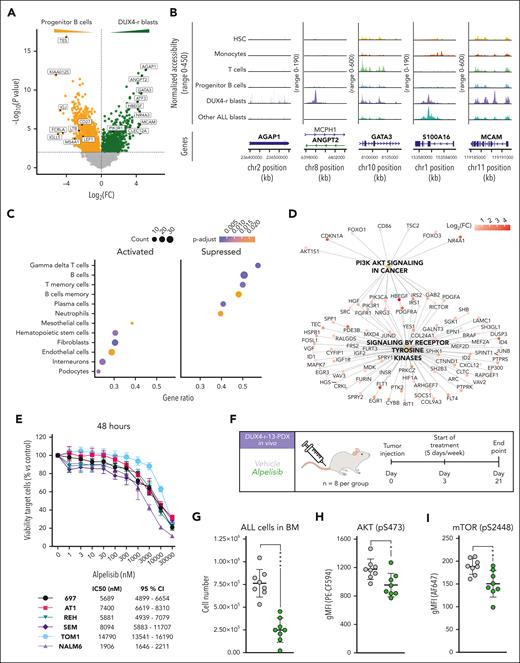
To further explore the altered pathways in DUX4-r blast cells, we performed a DGE analysis between DUX4-r blast cells and normal progenitor B cells (supplemental Table 7). This revealed the overexpression of 871 genes in DUX4-r blast cells, with the top hits including AGAP1, ANGPT2, GATA3, S100A16, MCAM, and NR4A3 (Figure 3A). These genomic regions also exhibited high levels of open chromatin (Figure 3B). GSEA of the differentially expressed genes using cell type marker genes34 highlighted the upregulation of marker genes for HSC, fibroblasts, and endothelial cells (Figure 3C). The most downregulated gene sets included B- and T-cell markers. Collectively, these data suggest that DUX4-r blast cells, which ectopically express the embryonal TF DUX4, activate pathways involved in nonlymphoid lineages and deregulate the programs involved in lymphoid development.
DUX4-r ALL cells are dependent on the PI3K/AKT pathway. (A) Volcano plot representing genes significantly overexpressed in DUX4-r blast cells (green) and normal progenitor B cells (yellow). Top genes based on fold changes are indicated with a label. P value cutoff is 0.01 (B) Chromatin accessibility of the gene regions of AGAP1, ANGPT2, GATA3, S100A16, and MCAM, the top 5 expressed genes based on fold change in DUX4-r blast cells. (C) Top 10 cell-type marker gene sets from GSEA of differentially expressed genes between DUX4-r blast cells and normal progenitor B cells. (D) Network plot visualizing GSEA of reactome pathways when comparing DUX4-r blast cells and progenitor B cells. (E) In vitro viability of BCP-ALL cell lines treated with a selective p110α (PIK3CA) inhibitor, alpelisib, with half-maximal inhibitory concentration (IC50) and 95% CI. (F) Experimental design for in vivo treatment of the DUX4-r-13 PDX sample in immunodeficient mice using alpelisib (35 mg/kg, 5 days on/2 days off for 3 weeks). (G) Number of human DUX4-r blast cells (ALL cells) in the BM after treatment. (H) Phosphorylation of AKT (pS473) in human DUX4-r blast cells in the mouse BM after treatment. (I) Phosphorylation of mammalian target of rapamycin (mTOR) (pS2448) in human DUX4-r blast cells in the mouse BM after treatment. CI, confidence interval.
DUX4-r ALL cells are dependent on the PI3K/AKT pathway. (A) Volcano plot representing genes significantly overexpressed in DUX4-r blast cells (green) and normal progenitor B cells (yellow). Top genes based on fold changes are indicated with a label. P value cutoff is 0.01 (B) Chromatin accessibility of the gene regions of AGAP1, ANGPT2, GATA3, S100A16, and MCAM, the top 5 expressed genes based on fold change in DUX4-r blast cells. (C) Top 10 cell-type marker gene sets from GSEA of differentially expressed genes between DUX4-r blast cells and normal progenitor B cells. (D) Network plot visualizing GSEA of reactome pathways when comparing DUX4-r blast cells and progenitor B cells. (E) In vitro viability of BCP-ALL cell lines treated with a selective p110α (PIK3CA) inhibitor, alpelisib, with half-maximal inhibitory concentration (IC50) and 95% CI. (F) Experimental design for in vivo treatment of the DUX4-r-13 PDX sample in immunodeficient mice using alpelisib (35 mg/kg, 5 days on/2 days off for 3 weeks). (G) Number of human DUX4-r blast cells (ALL cells) in the BM after treatment. (H) Phosphorylation of AKT (pS473) in human DUX4-r blast cells in the mouse BM after treatment. (I) Phosphorylation of mammalian target of rapamycin (mTOR) (pS2448) in human DUX4-r blast cells in the mouse BM after treatment. CI, confidence interval.
In addition, GSEA of the differentially expressed genes using cellular pathway genes35,36 highlighted the upregulation of genes involved in PI3K/AKT signaling in cancer, general receptor tyrosine kinase (RTK) signaling, and signaling of specific RTKs such as FGFR1, FGFR3, and NTRKs (Figure 3D; supplemental Figure 13A-D). In addition, DGE highlighted several RTKs among the overexpressed genes, with the highest expressing RTKs being FLT1, PDGFRA, and ROR2 (supplemental Figure 13B). The PI3K/AKT pathway can be activated by cell surface signaling through RTKs, as well as through BCR and pre-BCR.37-42 In DUX4-r ALL blast cells, the PI3K units encoded by PIK3CA (p110α) and PIK3R1 (p85α) were highly expressed (supplemental Figure 13E). However, DUX4-r ALLs did not exhibit the characteristic signature of ALLs with functional pre-BCR signaling41,42 (supplemental Figure 13F), indicating that the PI3K/AKT pathway is activated by another mechanism. Thus, the above-implicated RTKs are more likely activators of the PI3K/AKT pathway.37-39
To specifically target p110α, 6 BCP-ALL cell lines were cultured in increasing concentrations of the Food and Drug Administration-approved PI3K inhibitors copanlisib (inhibiting p110α/β/δ/γ) and alpelisib (inhibiting p110α selectively). The DUX4-r cell line NALM6 was significantly more sensitive to these inhibitors than the BCP-ALL cell lines with other genetic aberrations (Figure 3E; supplemental Figure 14A-K). This was also indicated by the increased number of cells in the G1/G0 phase, the level of drug-induced apoptosis, and the decrease in phosphorylation of downstream targets (supplemental Figure 14A-C and E-F). In vivo drug testing using alpelisib in a PDX model of primary DUX4-r cells resulted in a profound antileukemic effect (Figure 3F-I), demonstrating the functional relevance of the upregulated PI3K/AKT signaling in DUX4-r ALL.
DUX4-r ALLs exhibit transcriptional and epigenetic signatures of lineage infidelity
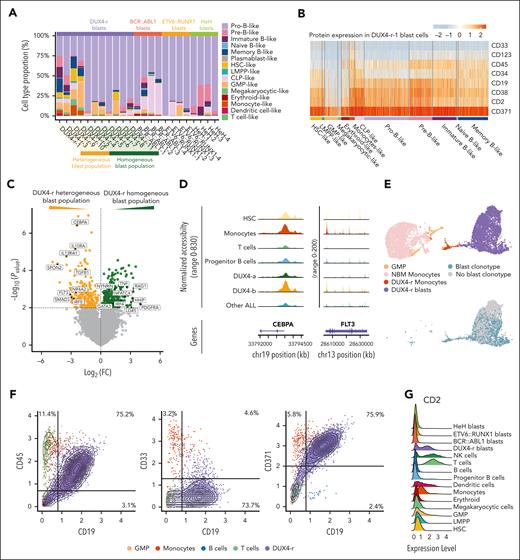
To study cellular characteristics specifically within the blast populations, all blast cells were reclassified based on the previously established NBM reference with extended B-cell subsets (supplemental Figure 5A-C). In most BCP-ALL cases, most blast cells were classified as pro-B cells. The DUX4-r cases instead formed 2 groups, 1 with a heterogeneous blast population and 1 with a homogeneous blast population. In the group with a heterogeneous population (4 cases), other cell type predictions than pro-B cells dominated, such as HSC-like, granulocyte-macrophage progenitors-like, and monocyte-like (Figure 4A; supplemental Figure 15A-G). These cases also exhibited higher proportions of blast cells classified as pre-B cells, immature B cells, and memory B cells, as highlighted by the projection onto the NBM B cell trajectory (supplemental Figures 9C, 10C, and 11B-C). Different cell type predictions within the blast population were also associated with distinct aberrant immunophenotypes, as determined using scADT-seq. For example, DUX4-r blast cells classified as granulocyte-macrophage progenitor-like were CD34–CD19–CD371+, whereas the corresponding memory B-like cells were CD34+CD19+CD371+ (Figure 4B), lending support to gene expression-based cell classifications at the protein level.
Lineage infidelity with an emphasis toward myeloid lineages detected in the DUX4-r blast population. (A) Cell type classification of the BCP-ALL blast cell population. DUX4-r ALL cases with only a minority of cells classified as pro-B cells (heterogeneous blast population) are grouped together on the left. (B) Cell surface protein expression in the predicted cell types, as determined by scADT-seq within the blast population of DUX4-r-1. (C) Volcano plot representing differentially expressed genes between the blast population of DUX4-r ALL samples with heterogeneous blast population (yellow) and homogeneous blast population (green). The P value cutoff is .01. (D) Chromatin accessibility of the gene regions of FLT3 and CEBPA in monocytes, progenitor B cells, DUX4-a, DUX4-b, and other ALL subtype blast cells (BCR::ABL1+, ETV6::RUNX1+, and HeH ALL). (E) (Upper figure) UMAP visualization of normal GMPs, normal monocytes, monocyte-like blast cells, and lymphoid blast cells from the samples NBM-MNCs and DUX4-r-1, respectively, colored by cell type. (Lower figure) UMAP visualization of GMPs, monocyte-like blast cells, and lymphoid blast cells from the sample DUX4-r-1 only, colored by the presence of the blast cell clonotypic V(D)J rearrangement. (F) Expression of the cell surface markers CD45, CD33, and CD371 vs CD19 on all single cells in the individual case of DUX4-1 ALL, as determined by scADT-seq. Cells are colored by cell type. (G) T-cell–associated cell surface marker CD2 expression on BCP-ALL blast cells and other cell types, as determined by scADT-seq.
Lineage infidelity with an emphasis toward myeloid lineages detected in the DUX4-r blast population. (A) Cell type classification of the BCP-ALL blast cell population. DUX4-r ALL cases with only a minority of cells classified as pro-B cells (heterogeneous blast population) are grouped together on the left. (B) Cell surface protein expression in the predicted cell types, as determined by scADT-seq within the blast population of DUX4-r-1. (C) Volcano plot representing differentially expressed genes between the blast population of DUX4-r ALL samples with heterogeneous blast population (yellow) and homogeneous blast population (green). The P value cutoff is .01. (D) Chromatin accessibility of the gene regions of FLT3 and CEBPA in monocytes, progenitor B cells, DUX4-a, DUX4-b, and other ALL subtype blast cells (BCR::ABL1+, ETV6::RUNX1+, and HeH ALL). (E) (Upper figure) UMAP visualization of normal GMPs, normal monocytes, monocyte-like blast cells, and lymphoid blast cells from the samples NBM-MNCs and DUX4-r-1, respectively, colored by cell type. (Lower figure) UMAP visualization of GMPs, monocyte-like blast cells, and lymphoid blast cells from the sample DUX4-r-1 only, colored by the presence of the blast cell clonotypic V(D)J rearrangement. (F) Expression of the cell surface markers CD45, CD33, and CD371 vs CD19 on all single cells in the individual case of DUX4-1 ALL, as determined by scADT-seq. Cells are colored by cell type. (G) T-cell–associated cell surface marker CD2 expression on BCP-ALL blast cells and other cell types, as determined by scADT-seq.
DGE analysis highlighted the expression of CEBPA, FLT3, and IL10RA in the heterogeneous blast population group and the expression of IRF4, NFATC4, and RAG1 in the remaining DUX4-r cases (Figure 4C; supplemental Table 8). Interestingly, a similar subdivision of DUX4-r cases was recently reported based on bulk RNA sequencing, where 2 classes (DUX4-a and DUX4-b) were described.3 Applying the published expression profiles for these subclasses in our cohort confirmed that the DUX4-r ALLs with heterogeneous blast populations matched the DUX4-b class (supplemental Figure 15H-I), indicating that DUX4-a/b subdivision is associated with heterogeneity in the cellular characteristics of the blasts. In concordance with the gene expression, the DUX4-b blast cells also displayed higher levels of chromatin accessibility in the gene regions of CEBPA and FLT3 than DUX4-a (Figure 4D). In fact, DUX4-b blast cells showed a similar level of chromatin accessibility to normal monocytes, suggesting myeloid potential. Interestingly, enforced expression of CEBPA in the DUX4-r+ cell line NALM6 was sufficient to prime the cells toward a monocytic immunophenotype, as revealed by upregulation of the monocytic markers CD371, CD33, and CD14 and downregulation of CD19 (supplemental Figure 15J). DUX4 was still expressed in cells with a monocytic immunophenotype, although it was lower than that in the lymphoid population (supplemental Figure 15K).
Most DUX4-r ALLs have previously been described to undergo a monocytic switch during treatment, in which the leukemic CD10+CD20–CD34+CD2+CD371+ cells switch to a monocytic CD19low/negCD33+CD14+ immunophenotype, which is also present in a small subset of patients already before treatment.43 We observed 1 case (DUX4-r-1) that already at diagnosis contained a cluster of cells classified as monocytes, notably expressing the blast clonotypic V(D)J rearrangement, as detected by single-cell BCR sequencing, confirming that these monocyte-like cells were derived from the blast cells (Figure 4E). DGE analysis of the monocytic and lymphoid blast populations showed upregulation of monocytic markers, including CD14, CD33, and LYZ, and downregulation of CD19, CD79A/B, and DNTT (supplemental Figure 15L; supplemental Table 9), as also observed by scADT-seq (Figure 4F). Interestingly, the expression level of DUX4 was lower in monocytic blast cells than in lymphoid blast cells (supplemental Figure 15M).
Although DUX4-r ALLs with heterogeneous blast populations showed features of monocytic lineage priming, other signs of lineage priming were observed across all DUX4-r cases. For example, DUX4-r blast cells displayed high expression and an open chromatin architecture of NR4A genes and GATA3 (Figure 3A-B), encoding TFs mediating differentiation and maintenance in T cells.29,44,45 Moreover, the T-cell marker CD2 was observed to be expressed on the cell surface of 9 of 11 DUX4-r cases (Figure 4G).
DUX4-r ALL cells can be effectively targeted by CAR T cells directed against the myeloid–associated marker CD371
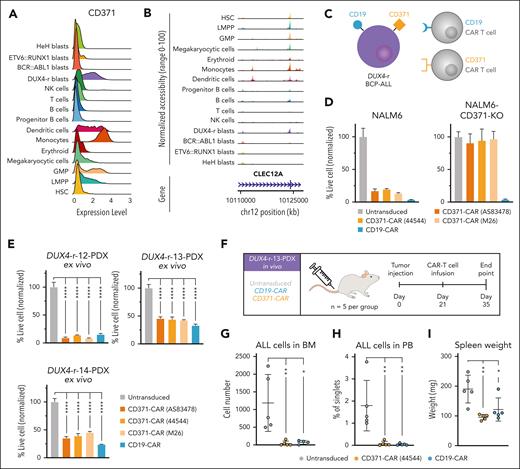
Due to the lineage infidelity in DUX4-r cases and their propensity to switch to a myeloid phenotype, targeting the entire blast population would require a marker expressed in both the B-cell and myeloid compartments. To identify such a marker, we performed DGE using a gene list encoding surface molecules.46 This revealed that CD371 (encoded by CLEC12A) is expressed in both lymphoid and monocytic blast cells (Figure 4F; supplemental Figure 16A). CD371 is a well-known marker of acute myeloid leukemia (AML) stem cells47,48 and has previously been reported to be expressed in DUX4-r cells.9,49 In our cohort, we observed high expression of CD371 in DUX4-r cases and, as expected, in healthy monocytes (Figure 5A). Consistent with this, a peak indicating open chromatin was observed in CLEC12A in DUX4-r blast cells and normal monocytes as well as in early progenitors (Figure 5B). To explore the possibility of targeting CD371 using CAR T cells in DUX4-r ALL, we generated 3 CD371-targeting second-generation 4-1BB-based CARs21 (Figure 5C; supplemental Figure 16B). Primary human T cells were engineered to express (1) 1 of the 3 CD371 CARs or a CD19 CAR as a positive control, or (2) both CD371 and CD19 CARs for dual targeting. Coincubation with the CD371+ cell line NALM6 elicited robust antitumor activity for both CD371 CAR T cells and CD19 CAR T cells, whereas only CD19 CAR T cells had cytotoxic activity against NALM6 with ablated CD371 expression (Figure 5D; supplemental Figure 16C-D), validating the specificity of our CD371 CARs. The dual CARs displayed antileukemic activity at a similar level as the monospecific CARs (supplemental Figure 16F). To further confirm these findings, we established PDX models of 3 DUX4-r ALLs and harvested the cells to test the efficacy of our CAR T cells on patient-derived ALL cells ex vivo and in vivo. All PDX cases demonstrated expression of CD371 and CD19 in 95% to 100% of blast cells (supplemental Figure 17A-C). We observed an effective reduction in PDX cells when coincubated ex vivo as well as in vivo with CD371 CAR T cells, at similar levels to that of the CD19 CAR T cells (Figure 5E-I; supplemental Figures 17 and 18).
DUX4-r ALL is susceptible to killing by CAR T cells directed against the myeloid–associated marker CD371. (A) Cell surface marker CD371 expression in BCP-ALL blast cells and residual nonleukemic hematopoiesis as determined by scADT-seq. (B) Open chromatin in the gene region of the CLEC12A gene (that encodes CD371) is visualized in BCP-ALL blast cells and other cell types. (C) Schematic illustration of targeting of DUX4-r ALL blast cells expressing CD371 and CD19 on the cell surface by CD19 and CD371 CAR T cells, respectively. (D) Coincubation with wild-type NALM6 displayed antitumor activity against CD371 and CD19 CAR T cells. Only CD19 CAR T cells retained the cytotoxic activity in NALM6-CD371-KO lacking CD371 expression. (E) Effective ex vivo targeting of DUX4-r ALL blast cells in 3 independently derived DUX4-r PDX samples. The proportion of live PDX cells after CAR T cell treatment. (F) Experimental design for in vivo targeting of the DUX4-r-13 PDX sample in immunodeficient mice using CD371-CAR (44544) T cells, CD19-CAR T cells, or untransduced T cells. (G) The number of human DUX4-r blast cells (ALL cells) in BM at the end point. (H) Number of human DUX4-r blast cells (ALL cells) in PB at the end point. (I) Spleen weight at the end point. PB, peripheral blood.
DUX4-r ALL is susceptible to killing by CAR T cells directed against the myeloid–associated marker CD371. (A) Cell surface marker CD371 expression in BCP-ALL blast cells and residual nonleukemic hematopoiesis as determined by scADT-seq. (B) Open chromatin in the gene region of the CLEC12A gene (that encodes CD371) is visualized in BCP-ALL blast cells and other cell types. (C) Schematic illustration of targeting of DUX4-r ALL blast cells expressing CD371 and CD19 on the cell surface by CD19 and CD371 CAR T cells, respectively. (D) Coincubation with wild-type NALM6 displayed antitumor activity against CD371 and CD19 CAR T cells. Only CD19 CAR T cells retained the cytotoxic activity in NALM6-CD371-KO lacking CD371 expression. (E) Effective ex vivo targeting of DUX4-r ALL blast cells in 3 independently derived DUX4-r PDX samples. The proportion of live PDX cells after CAR T cell treatment. (F) Experimental design for in vivo targeting of the DUX4-r-13 PDX sample in immunodeficient mice using CD371-CAR (44544) T cells, CD19-CAR T cells, or untransduced T cells. (G) The number of human DUX4-r blast cells (ALL cells) in BM at the end point. (H) Number of human DUX4-r blast cells (ALL cells) in PB at the end point. (I) Spleen weight at the end point. PB, peripheral blood.
Discussion
Herein, we used multimodal single-cell sequencing to dissect the molecular and cellular characteristics of 23 BCP-ALL cases, with a focus on the recently described DUX4-r ALL.12-14 Previous single-cell studies on BCP-ALL have mainly focused on KMT2A-rearranged and ETV6::RUNX1+ subtypes.50-55 A recent study investigated ALL-associated nonleukemic cells and identified an aberrant nonclassical monocytic cell population that supported the growth of the leukemic clone.56 Here, we identified a characteristic CD4+ T-cell subset within the leukemic niche in most DUX4-r ALL cases that displayed transcriptional signatures of dysfunction.26-28,31,32 The impact of CD4+ T-cell exhaustion in cancer31,32 and hematological malignancies57-60 is becoming increasingly recognized and could present a novel immune escape mechanism in DUX4-r ALL. Functional follow-up studies assessing CD4+ T-cell function in DUX4-r ALLs are needed to fully disclose the nature of this T-cell subset. Previous studies have inferred a maturation block in BCP-ALL by immunophenotyping using a restricted set of cell surface markers and reported that the blast cells were arrested at the B-precursor stage.24,25,61 Developmental states within BCP-ALL have recently been studied by projecting bulk RNA sequencing data of BCP-ALL onto single-cell-defined normal maturation trajectories.54,62 This highlights a similar point of arrest in the pro–B-cell compartment.54,62 Our single-cell projections identified diverse maturation patterns between the BCP-ALL subtypes. The BCR::ABL1+, ETV6::RUNX1+, and HeH subtypes harbored a partial maturation block, positioning most blast cells in the pro–B-cell stage. Interestingly, the maturation patterns seem to be related to the cell of origin, in which leukemias with a pro–B-cell phenotype were derived from a cell with IGH rearrangement. Leukemias containing blast cells with a more mature appearance (maturation pattern-2 and -3 in DUX4-r ALL), however, were likely derived from cells with both IGH- and IGK/L-rearrangements.
Notably, mature cells in DUX4-r ALL displayed downregulation of B-cell markers and upregulation of marker genes from other lineages. Comparison of DUX4-r blast cells with normal progenitor B cells revealed further signs of lineage infidelity, as DUX4-r blast cells exhibited transcriptional features of fibroblasts and endothelial cells. This analysis also revealed the expression of T-cell markers, including GATA3, described to be essential for repressing the B-cell potential in mice.63 An additional feature of multilineage priming was highlighted by the cell type prediction of the blast populations, where a greater heterogeneity correlated with the DUX4-a/DUX4-b subdivision.3DUX4-b cases expressed CEBPA and FLT3, previously described to be overexpressed in patients in which blast cells undergo a monocytic switch during treatment.43 Notably, we identified 1 DUX4-b case with monocytic cells expressing clonal IG-rearrangement, consistent with a monocytic switch present already at diagnosis. Overall, these features of lineage infidelity are consistent with DUX4, which has the capacity to prime multiple cellular lineages, as indicated by its normal expression during early embryogenesis.64,65 However, in BCP-ALL, DUX4 expression is, in most cases, regulated by the IGH locus and is active only in B cells,33 which potentially limits the plasticity of DUX4-r blast cells.
DUX4-r blast cells displayed a gene signature associated with enhanced PI3K/AKT signaling. The PI3K/AKT pathway directs multiple biological processes that can be aberrantly activated in cancer.38,40-42 PI3K/AKT signaling is normally involved in B-cell development and survival.40-42 However, because DUX4-r ALL neither showed signs of pre-BCR signaling nor expressed PIK3CD but showed enrichment of gene sets associated with RTK signaling, we hypothesized that PI3K/AKT signaling is activated by increased RTK signaling.37-39 This would be consistent with the detected upregulation of PIK3CA. Clinically, PI3K inhibitors are used as a treatment for follicular lymphoma and chronic lymphocytic leukemia.37 We demonstrated that DUX4-r cells are sensitive to PI3K/AKT inhibition, suggesting that targeting this pathway could offer a less toxic treatment alternative with the potential for de-escalating current chemotherapeutic regimens in DUX4-r BCP-ALL.
CAR T-cell–based therapies targeting CD19 have emerged as highly efficient treatment alternatives for relapsed BCP-ALL,7 although resistance may occur as a result of the loss of CD19 expression7,8 or lineage escape.66,67 Given that DUX4-r ALL has a propensity to switch to the monocytic lineage,43 thereby losing CD19 expression,43 CAR T-cell–based therapies against CD371 aberrantly expressed on DUX4-r cells may provide an attractive treatment alternative for this subtype. In fact, it has been reported that DUX4-r can relapse as an AML with a monocytic phenotype,43 although it should be noted that the leukemic potential of these switched monocytic cells remains to be investigated. Here, we showed that CAR T cells directed against CD371 on DUX4-r leukemic cells elicit highly effective cell killing in vivo. Targeting CD371, which is normally expressed on monocytes,68,69 could be associated with distinct clinical challenges compared with targeting CD19.70 However, CD371 is being explored as a CAR T target in childhood and adult AML, with clinical trials showing promising responses and low levels of severe toxicities.71-74
In conclusion, this multimodal single-cell characterization of BCP-ALL allowed for highly granular characterization of the differentiation block among the most common subtypes. Within DUX4-r ALL, we uncovered heterogeneous maturation patterns with a high degree of lineage infidelity as well as sensitivity to PI3K inhibitors and CD371 CAR T-cell killing. These results should aid further efforts to develop more specific and less toxic therapies for BCP-ALL.
Acknowledgments
The authors thank the Center for Translational Genomics at Lund University and Clinical Genomics Lund, SciLifeLab, for providing single-cell sequencing services.
This work was supported by the Swedish Childhood Cancer Fund, Sweden (grant PR2023-0097); Swedish Cancer Society, Sweden (23-2953 Pj 01); Swedish Research Council, Sweden (2023-02376); Governmental Funding of Clinical Research within the National Health Service (ALF-grant), Sweden; interregional Childhood Oncology Precision medicine Exploration (iCOPE), Sweden and Denmark; Knut and Alice Wallenberg Foundation (2022-0232), Sweden; Cancera Foundation, Sweden; Mats Paulsson Foundation, Sweden; and Interogo Foundation, Lichtenstein.
Authorship
Contribution: H.T., C.O.-P., H.L., and T.F. designed the study; H.T., R.H., N.P.-M., A.H.-W., and H.L. made the figures; H.T., N.P.-M., L.S., H.Å., P.P.-M., C.S., A.H.-W., and M.R. performed experiments; H.T., R.H., N.P.-M., L.S., A.H.-W., C.O.-P., H.L., and T.F. analyzed the data; A.C., H.M., S.M., K.P., C.J.P., K.S., C.O.-P., H.L., and T.F. collected the patient material and clinical data; H.T., C.O.-P., H.L., and T.F. wrote the manuscript; and all authors contributed valuable inputs.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hanna Thorsson, Division of Clinical Genetics, Department of Laboratory Medicine, Lund University, BMC C13, Klinikgatan 28, SE-221 84 Lund, Sweden; email: hanna.thorsson@med.lu.se; and Thoas Fioretos, Division of Clinical Genetics, Department of Laboratory Medicine, Lund University, BMC C13, Klinikgatan 28, SE-221 84 Lund, Sweden; email: thoas.fioretos@med.lu.se.
References
Author notes
The data sets generated during the current study fall under the General Data Protection Regulation (GDPR) regulations for sharing of personal data and will therefore, be deposited in Federated European Genome-phenome Archive (FEGA) Sweden once this repository is complete. Until then, the annotated data sets are publicly available, and raw sequencing reads are available on request from the corresponding authors (Hanna Thorsson and Thoas Fioretos) through the following DOIs: https://doi.org/10.17044/scilifelab.23592270 (multi∖modal single-cell RNA sequencing data set) and https://doi.org/10.17044/scilifelab.23592303 (single-cell assay for transposase-accessible chromatin using sequencing data set).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal