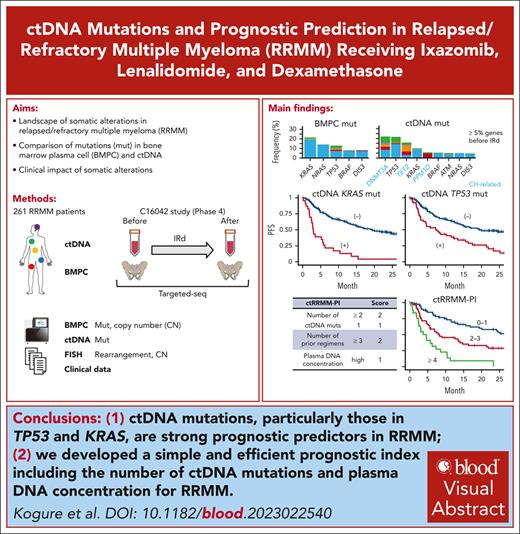
ctDNA mutations, particularly those in TP53 and KRAS, are stronger prognostic predictors than bone marrow plasma cell mutations in RRMM.
We develop a simple and efficient prognostic index including the number of ctDNA mutations and plasma DNA concentration for RRMM.
Visual Abstract
It remains elusive how driver mutations, including those detected in circulating tumor DNA (ctDNA), affect prognosis in relapsed/refractory multiple myeloma (RRMM). Here, we performed targeted-capture sequencing using bone marrow plasma cells (BMPCs) and ctDNA of 261 RRMM cases uniformly treated with ixazomib, lenalidomide, and dexamethasone in a multicenter, prospective, observational study. We detected 24 and 47 recurrently mutated genes in BMPC and ctDNA, respectively. In addition to clonal hematopoiesis–associated mutations, varying proportion of driver mutations, particularly TP53 mutations (59.2% of mutated cases), were present in only ctDNA, suggesting their subclonal origin. In univariable analyses, ctDNA mutations of KRAS, TP53, DIS3, BRAF, NRAS, and ATM were associated with worse progression-free survival (PFS). BMPC mutations of TP53 and KRAS were associated with inferior PFS, whereas KRAS mutations were prognostically relevant only when detected in both BMPC and ctDNA. A total number of ctDNA mutations in the 6 relevant genes was a strong prognostic predictor (2-year PFS rates: 57.3%, 22.7%, and 0% for 0, 1, and ≥2 mutations, respectively) and independent of clinical factors and plasma DNA concentration. Using the number of ctDNA mutations, plasma DNA concentration, and clinical factors, we developed a prognostic index, classifying patients into 3 categories with 2-year PFS rates of 57.9%, 28.6%, and 0%. Serial analysis of ctDNA mutations in 94 cases revealed that TP53 and KRAS mutations frequently emerge after therapy. Thus, we clarify the genetic characteristics and clonal architecture of ctDNA mutations and demonstrate their superiority over BMPC mutations for prognostic prediction in RRMM. This study is a part of the C16042 study, which is registered at www.clinicaltrials.gov as #NCT03433001.
Introduction
Multiple myeloma (MM) is the second most common hematological malignancy and remains largely incurable despite the development of novel therapeutics.1,2 MM is characterized by the expansion of clonal plasma cells that involve multiple sites of the bone marrow (BM) compartment. Cytogenetic analyses revealed that MM can be broadly divided into cases with recurrent chromosomal rearrangements involving the immunoglobulin heavy chain (IGH) locus, and those that are hyperdiploid (HRD) with trisomies of odd-numbered chromosomes.3 Recent next-generation sequencing (NGS) studies have delineated the entire landscape of genetic alterations in MM, reporting recurrent mutations in the RAS pathway (KRAS, NRAS, and BRAF), DNA repair pathway (TP53), and NF-κB pathway.4,5 Among these alterations, t(4;14), t(14;16), deletion of 17p13 [del(17p)], and gain/amplification of 1q21 [amp(1q)] are considered as high-risk genetic abnormalities by the International Myeloma Working Group.6 These structural variations and copy number alterations (CNAs) have been incorporated into current risk stratification systems, such as the Revised International Staging System and the Second Revision of the International Staging System.7,8 Although an increased number of driver mutations is associated with worse outcome, the predictive capacity of individual drivers is limited.4,5,9 Except for TP53 mutation associated with worse prognosis, there are no associations between patient outcome and mutation status in most of the frequently affected genes, including the RAS pathway genes.4,5,9-11
Nearly all patients with MM eventually relapse or become refractory to current treatments.1,2 Compared with newly diagnosed MM (NDMM), relapsed/refractory MM (RRMM) harbors more frequent mutations in the RAS pathway genes (KRAS, NRAS, and BRAF), TP53, and CRBN, a direct target of immunomodulatory drugs, such as lenalidomide and pomalidomide.12,13 However, the prognostic impacts of genetic alterations have not been well investigated in RRMM.
Liquid biopsy focusing on the analysis of circulating tumor cells and circulating tumor DNA (ctDNA) in the blood has received enormous attention because of its potential clinical implications for personalized medicine. Such implications include cancer detection, prognostic prediction, minimal residual disease monitoring, and treatment stratification.14 Prospective and large retrospective studies demonstrated that a high circulating tumor cell burden quantified by flow cytometry has a negative prognostic impact on patient survival in NDMM.15-17 NGS analysis showed high concordance in clonal somatic mutations between liquid and tumor biopsies, whereas there are mutations that exclusively exist in ctDNA, suggesting spatial heterogeneity of MM clones.18,19 However, the prognostic relevance of ctDNA mutations and its difference between BM plasma cells (BMPCs) and ctDNA have not been investigated well. Although sequential analysis of ctDNA mutations enables the identification of genetic alterations associated with disease progression and therapeutic resistance and the elucidation of the dynamics of clonal architecture, there have been only few such attempts in MM.19-21
A novel proteasome inhibitor, ixazomib, in combination with lenalidomide-dexamethasone (IRd), demonstrated an improvement of progression-free survival (PFS) in patients with RRMM, including cases with high-risk cytogenetic abnormalities,22,23 and accordingly current guidelines recommend IRd therapy as an option for RRMM.24,25 Here, we investigated the landscape of somatic alterations and their prognostic impacts by performing NGS analysis of BMPC and ctDNA samples in 261 RRMM cases uniformly treated with IRd therapy in a prospective cohort. We demonstrate a better predictive capacity of ctDNA mutations, particularly those involving KRAS, than BMPC mutations in RRMM, and developed a new prognostic scoring system incorporating ctDNA mutations, plasma DNA concentration, and clinical factors, which can better stratify patients with RRMM.
Methods
Patients and materials
A total of 295 patients with RRMM were enrolled in a multicenter, prospective, observational study of IRd therapy (C16042 study).26 Among them, 268 patients agreed to provide BMPC and/or ctDNA samples for sequencing analysis. IRd therapy was initiated at the physician's discretion, which was largely based on the diagnosis of RRMM according to the International Myeloma Working Group response criteria.27 BMPC and/or ctDNA were obtained at the start and end of IRd therapy. After quality control, data of 261 patients were analyzed. Clinical information was collected as per the C16042 study protocol. All patients provided written informed consent. This study was performed in accordance with the Declaration of Helsinki and was approved by the institutional ethics committees of National Cancer Center and other participating institutions.
Targeted-capture sequencing of BMPC and ctDNA
DNA from CD138+ cells in BM, and plasma ctDNA, were subjected to targeted-capture sequencing for 1085 genes at the Broad Institute (Cambridge, MA) and for 76 genes at PGDx (Baltimore, MD), respectively (supplemental Tables 1 and 2, available on the Blood website). Sequence alignment and mutation calling of BMPC DNA and ctDNA data were performed using the Genomon pipeline version 2.6.2, as previously described,28,29 and nonsilent mutations with allele frequency in tumor of ≥0.05 (for BMPC) and ≥0.01 (for ctDNA) were adopted. BMPC and ctDNA mutations were compared with respect to position and nucleotide change. Copy number (CN) and ploidy data were analyzed using CNACS.28 Driver gene list was curated from previous reports5,30 (supplemental Table 3). Detailed methods are provided in supplemental Materials.
FISH analysis
Fluorescence in situ hybridization (FISH) analysis was performed according to the protocol of the C16042 study.26 Briefly, t(4;14), t(11;14), t(14;16), amp(1q), and del(17p) were assessed using CD138+ cells in the BM collected before IRd. The cutoff was defined as 5% positive cells for del(17p) and 3% positive cells for the other 4 abnormalities, as used in the TOURMALINE-MM1 trial.22
Statistical analysis
All statistical analyses were performed using R version 4.1 (https://www.r-project.org/). P values <.05 were considered statistically significant.
PFS data were available for all 261 patients. Survival analyses for PFS were performed using R package survival version 4.1 and survminer version 0.4.9. Univariable analysis was performed using the Kaplan-Meier method with log-rank test. We subsequently performed multivariable analyses based on Cox proportional hazards (PH) model to adjust for clinical and cytogenetic covariates which were statistically significant (P < .05) in univariable analysis, and optimal combinations of covariates were selected with backward variable selection using Akaike’s information criterion with stepAIC function of R package MASS version 7.3-57. To validate the robustness of our model construction, we also used L1-regularized Cox PH model using R package glmnet version 4.1-4. Tenfold crossvalidation with a partial-likelihood deviance metric using the cv.glmnet function was performed to select a λ value, which minimized the metric. Variables identified as having nonzero coefficient values were selected, and estimates were recalculated using a nonregularized Cox PH model. To develop prognostic indices, we used the relative values of coefficients (log[hazard ratio]) from the Cox PH model (detailed in supplemental Materials). Moreover, to perform independent validation of our ctDNA-only model (ctRRMM-PI), we used a split-sample approach, dividing cases with ctDNA sequencing (ctDNA-seq) data based on the availability of BMPC sequencing (BMPC-seq) data. We used cases with BMPC-seq data (n = 161) for model construction, and those without BMPC-seq data (n = 98) for model validation.
Results
Somatic alterations detected by BMPC-seq and ctDNA-seq in RRMM
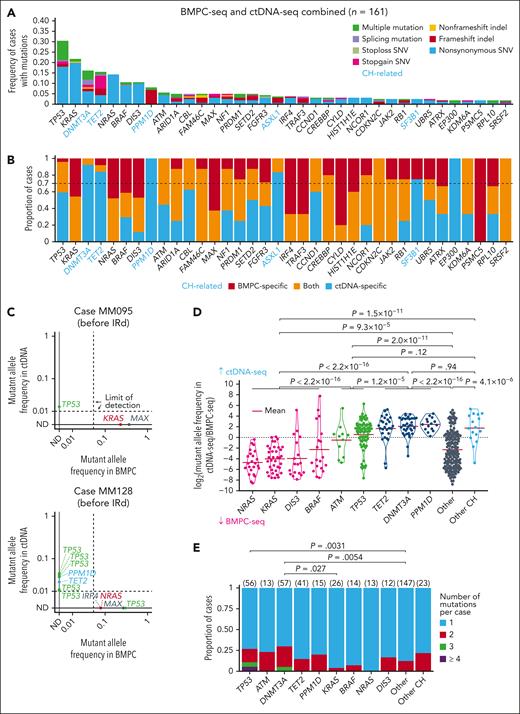
We investigated the entire overview of somatic alterations using BMPC and plasma ctDNA samples obtained from 261 patients with RRMM enrolled in the C16042 study. These patients had received a median of 2 prior treatment regimens, and most of them had prior exposure to bortezomib and/or lenalidomide (supplemental Table 4). We obtained 163 BM and 259 ctDNA samples at the initiation of IRd therapy (supplemental Figure 1A). We purified CD138+ tumor cells from the BM and performed targeted-capture sequencing (BMPC-seq), with a mean depth of 1229× (supplemental Figure 1B). BMPC-seq identified 1192 nonsilent mutations (0-25 mutations per case), including 1015 single-nucleotide variants (SNVs) and 177 insertion-deletions (indels) (supplemental Table 5). Recurrent mutations (observed in ≥3 cases) were detected in 24 driver genes (Figure 1A). Among them, KRAS was most frequent (21.5%), followed by NRAS (14.1%), TP53 (12.9%), BRAF (8.0%), and DIS3 (8.0%). At least 1 driver mutation was detected in 66.3% of cases.
Landscape of somatic alterations in BMPC and ctDNA in RRMM. (A) Frequency of driver mutations detected by BMPC-seq in RRMM cases (n = 163). Twenty-four genes detected in ≥3 cases are shown. (B) Frequency of driver mutations detected by ctDNA-seq in RRMM cases (n = 259). Forty-seven genes detected in ≥3 cases are shown. Reported CH-related genes are shown in blue.31,32
Landscape of somatic alterations in BMPC and ctDNA in RRMM. (A) Frequency of driver mutations detected by BMPC-seq in RRMM cases (n = 163). Twenty-four genes detected in ≥3 cases are shown. (B) Frequency of driver mutations detected by ctDNA-seq in RRMM cases (n = 259). Forty-seven genes detected in ≥3 cases are shown. Reported CH-related genes are shown in blue.31,32
CN analysis identified 901 arm-level CNAs, including 602 amplifications and 299 deletions, as well as 454 focal CNAs including 216 amplifications and 238 deletions (supplemental Figure 2A; supplemental Table 6). Among them, HRD was observed in 53 (32.5%) cases, and amp(1q) (CKS1B) and del(17p) (TP53) were detected in 74 (45.4%) and 27 (16.6%) cases, respectively (supplemental Figure 2A-B; supplemental Table 6). FISH analyses identified additional 31 (19.0%) and 12 (7.4%) cases with amp(1q) and del(17p), respectively, whereas, among those detected by BMPC-seq, only 1 and 3 additional cases with amp(1q) and del(17p), respectively, were not detected by FISH (supplemental Figure 2C).
We also performed targeted-capture sequencing of ctDNA (ctDNA-seq), with a mean depth of 2979× (supplemental Figure 1B). ctDNA-seq identified 580 nonsilent mutations (0-40 mutations per sample), including 464 SNVs and 116 indels (supplemental Table 7). Recurrent mutations (observed in ≥3 cases) were detected in 47 genes (Figure 1B). Among them, mutations related to clonal hematopoiesis (CH) were frequently observed, including those affecting DNMT3A (22.0%), TET2 (15.8%), PPM1D (5.8%), and ASXL1 (3.9%).31,32 Besides them, TP53 was most common (21.6%), followed by KRAS (10.0%), BRAF (5.4%), ATM (5.0%), NRAS (5.0%), and DIS3 (4.6%).
Plasma DNA concentration was variable, ranging from 3.2 to 1162.8 ng/mL, with a median of 9.5 ng/mL, and weakly but significantly correlated with BMPC fraction and the number of ctDNA mutations per case (supplemental Figure 3A-C). When cases were divided by a median value, only KRAS mutation was enriched in cases with high plasma DNA concentration (supplemental Figure 3D). These results suggest that plasma DNA concentration can reflect the tumor burden but is also affected by other mechanisms.
Comparison of mutations between BMPC-seq and ctDNA-seq in RRMM
The number of detected mutations correlated well between BMPC-seq and ctDNA-seq (supplemental Figure 4A). When both data were combined (n = 161), recurrent mutations (observed in ≥3 cases) were detected in 36 genes (Figure 2A; supplemental Figure 4B). Among them, TP53 was most frequent (30.4%), followed by KRAS (21.7%), DNMT3A (16.1%), TET2 (15.5%), NRAS (14.3%), BRAF (10.6%), DIS3 (10.6%), PPM1D (8.1%), and ATM (5.6%). At least 1 recurrent mutation was detected in 88.8% of cases (supplemental Figure 4C).
Comparison of driver and CH-related mutations between BMPC and ctDNA. (A) Frequency of mutations detected by combined BMPC-seq and ctDNA-seq analyses in RRMM cases (n = 161). Thirty-six genes detected in ≥3 cases are visualized. Reported CH-related genes are shown in blue. (B) Proportion of cases harboring mutations that are BMPC specific (red), ctDNA specific (blue), and detected by both (yellow). Cases with different BMPC-specific and ctDNA-specific mutations were counted as ctDNA-seq–specific. Thirty-six genes detected in ≥3 cases are visualized. (C) Diagonal plots showing mutant allele frequency in BMPC and ctDNA in representative cases. Mutations with a depth of ≥50× in both BMPC-seq and ctDNA-seq are shown; ND, not detected. (D) Comparison of mutant allele frequency between BMPC and ctDNA. An offset of 0.001 was added to each allele frequency before calculating log2 ratio. Genes with mutation frequency of ≥5% in panel A and/or ctDNA-specific frequency of ≥0.7 in panel B are shown. Other CH consists of ASXL1, SF3B1, and EP300. Two-sided Brunner-Munzel test. (E) Number of mutations for each gene in each case detected by ctDNA-seq (n = 261). Two-sided Fisher exact test.
Comparison of driver and CH-related mutations between BMPC and ctDNA. (A) Frequency of mutations detected by combined BMPC-seq and ctDNA-seq analyses in RRMM cases (n = 161). Thirty-six genes detected in ≥3 cases are visualized. Reported CH-related genes are shown in blue. (B) Proportion of cases harboring mutations that are BMPC specific (red), ctDNA specific (blue), and detected by both (yellow). Cases with different BMPC-specific and ctDNA-specific mutations were counted as ctDNA-seq–specific. Thirty-six genes detected in ≥3 cases are visualized. (C) Diagonal plots showing mutant allele frequency in BMPC and ctDNA in representative cases. Mutations with a depth of ≥50× in both BMPC-seq and ctDNA-seq are shown; ND, not detected. (D) Comparison of mutant allele frequency between BMPC and ctDNA. An offset of 0.001 was added to each allele frequency before calculating log2 ratio. Genes with mutation frequency of ≥5% in panel A and/or ctDNA-specific frequency of ≥0.7 in panel B are shown. Other CH consists of ASXL1, SF3B1, and EP300. Two-sided Brunner-Munzel test. (E) Number of mutations for each gene in each case detected by ctDNA-seq (n = 261). Two-sided Fisher exact test.
Most (>70%) mutations in DNMT3A, TET2, PPM1D, ASXL1, SF3B1, and EP300 were detected only by ctDNA-seq (Figure 2B), confirming the association of these mutations with CH existing in nonmalignant hematopoietic cells.31-35 Besides these mutations, varying proportion of the mutations were detected only by ctDNA-seq, with TP53 being one of the highest (59.2% of mutated cases). By contrast, in several genes, including KRAS and NRAS, all the mutations were detected by BMPC-seq. Amino acid change patterns in KRAS and NRAS were similar between BMPC-seq and ctDNA-seq (supplemental Tables 5 and 7). On average, 60.6% of the BMPC mutations were also detected by ctDNA-seq. For example, approximately half of BMPC mutations involving KRAS and NRAS were detected in ctDNA, whereas the other half were BMPC-specific mutations, which have been sporadically reported in the literature.18 Some cases with BMPC-specific KRAS or NRAS mutations harbored other driver mutations, including TP53 mutations, detected only by ctDNA-seq (Figure 2C).
Next, we compared allele frequencies of these mutations between BMPC and ctDNA (Figure 2C-D). Among the frequently altered genes, NRAS, KRAS, DIS3, and BRAF showed a higher allele frequency in BMPC than in ctDNA, which suggests the clonal expansion of these mutations in BMPC. By contrast, TP53 and ATM exhibited a higher allele frequency in ctDNA than in BMPC. As expected, allele frequency was higher in ctDNA than BM for CH-related genes. In addition, ctDNA-seq identified multiple (≥2) mutations more frequently in TP53 and DNMT3A than in other genes (Figure 2E). The proportion of pathogenic mutations was comparable in cases harboring a single and multiple TP53 mutations (supplemental Figure 5A-D). Given that the CH-related TP53 mutations are reported to range from 0.6% to 3% in patients with MM at autologous transplantation, and ∼1% in patients with solid cancer with a history of chemotherapy, these results likely reflect the multiple TP53-mutated subclones in MM.33-36
Prognostic impact of somatic alterations detected by BMPC-seq in RRMM
We assessed the effects of somatic alterations on patient prognosis in RRMM. We first investigated the effects of recurrent mutations detected by BMPC-seq that were present in ≥5% of 163 RRMM cases. In a univariable analysis, TP53 and KRAS mutations were significantly associated with shorter PFS (P = 1.0 × 10−5 and P = .0027, respectively; supplemental Figure 6A-B; supplemental Table 8). These findings suggest a potential prognostic value of KRAS mutation in RRMM, which is consistent with increased KRAS mutation frequency in RRMM but contrasts with no or minimal prognostic effect of KRAS mutation in NDMM.4,5,9,12
Regarding CNAs, HRD did not affect prognosis (supplemental Figure 7A). Among 19 amplifications and 6 deletions found in ≥10% of cases by BMPC-seq, only del(17p) was associated with worse prognosis (P = .0035; supplemental Figure 7B; supplemental Table 9). However, those detected only by FISH had no influence on survival (supplemental Figure 7C). In addition, although amp(1q) detected by BMPC-seq tended to be associated with worse prognosis, this association was lost for that detected only by FISH (supplemental Figure 7B-C). These results remained consistent when analyzed with a stricter FISH cutoff value of 20% (data not shown).26 These findings suggest that NGS analysis can more accurately detect CNAs associated with prognosis.
With respect to clinical factors, number of prior regimens (1, 2, or ≥3), treatment indication (clinical relapse and paraprotein relapse), and FISH results for t(11;14) rearrangements were significant predictors of PFS (supplemental Table 10). After adjusting for these factors using multivariable analysis, TP53 and KRAS mutations and del(17p) remained significant (supplemental Table 8), suggesting a critical role of these alterations in the progression and relapse of RRMM.
A stronger prognostic impact of mutations in ctDNA than in BMPC in RRMM
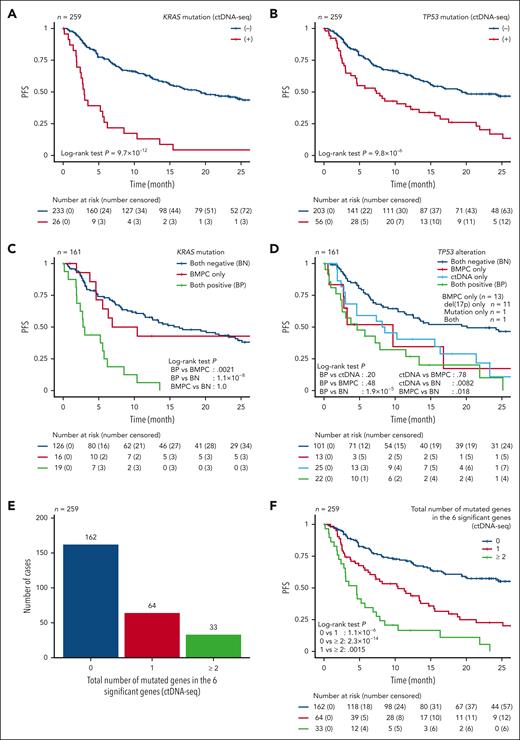
We subsequently evaluated the effect of plasma DNA concentration on prognosis and found that high plasma DNA concentration was associated with a shorter PFS (supplemental Figure 8A). We then assessed the effects of individual mutations detected by ctDNA-seq that were present in ≥3% of 259 RRMM cases. In a univariable analysis, 6 mutations, namely those involving TP53, KRAS, DIS3, BRAF, ATM, and NRAS, were significantly associated with worse prognosis (Figure 3A-B; supplemental Figure 8B-E; supplemental Table 11). Particularly, hazard ratio for KRAS mutation was substantially higher in ctDNA than in BMPC. In addition, combination of TP53 and KRAS mutation status more efficiently separated patient survival (supplemental Figure 8F). By contrast, CH-related gene mutations did not affect patient survival (supplemental Figure 8G).
Prognostic impact of mutations detected by BMPC and ctDNA analyses. (A-B) Kaplan-Meier survival curves of PFS of 259 RRMM cases with and without (A) KRAS and (B) TP53 mutation detected by ctDNA-seq. (C-D) Kaplan-Meier survival curves of PFS of 161 RRMM cases (having both BMPC-seq and ctDNA-seq data) without and with (C) KRAS mutation and (D) TP53 alteration present in only BMPC, only ctDNA, and both. (E) Total number of mutated genes in the 6 prognostically significant genes (KRAS, TP53, DIS3, BRAF, NRAS, and ATM) detected by ctDNA-seq in 259 RRMM cases. (F) Kaplan-Meier survival curves of PFS of 259 RRMM cases stratified by the total number of mutated genes in the 6 prognostically significant genes. Log-rank test.
Prognostic impact of mutations detected by BMPC and ctDNA analyses. (A-B) Kaplan-Meier survival curves of PFS of 259 RRMM cases with and without (A) KRAS and (B) TP53 mutation detected by ctDNA-seq. (C-D) Kaplan-Meier survival curves of PFS of 161 RRMM cases (having both BMPC-seq and ctDNA-seq data) without and with (C) KRAS mutation and (D) TP53 alteration present in only BMPC, only ctDNA, and both. (E) Total number of mutated genes in the 6 prognostically significant genes (KRAS, TP53, DIS3, BRAF, NRAS, and ATM) detected by ctDNA-seq in 259 RRMM cases. (F) Kaplan-Meier survival curves of PFS of 259 RRMM cases stratified by the total number of mutated genes in the 6 prognostically significant genes. Log-rank test.
Next, we separately compared the effects of mutations present in BMPC and ctDNA for the 6 significant genes in 161 RRMM cases with both BMPC-seq and ctDNA-seq data. Remarkably, compared with cases without KRAS mutations (both negative), cases having KRAS mutations in both BMPC and ctDNA (n = 19) demonstrated a substantially decreased survival (Figure 3C). In contrast, cases with BMPC-specific KRAS mutations showed a comparable survival with those without KRAS mutations. Similar trends were observed for DIS3 and BRAF (supplemental Figure 8H-K). These observations suggest that detection of ctDNA mutations has an influence on patient prognosis for these genes in RRMM. In contrast, compared with cases without TP53 alterations (both negative), cases harboring TP53 alterations in both BMPC and ctDNA (n = 22) showed an inferior survival (Figure 3D). Notably, TP53 alterations present in only either of BMPC (n = 13) or ctDNA (n = 25) were also associated with a significantly worse prognosis. TP53 alterations present in only BMPC were mostly (n = 12) del(17p), which was undetectable by ctDNA-seq, confirming that almost all TP53 mutations were found in ctDNA. These observations suggest that ctDNA-seq can detect many subclonal mutations of TP53, which are associated with clinical outcome in RRMM.
We also evaluated the effect of the total number of ctDNA mutations for these 6 prognostic genes on patient prognosis. Although no mutation was detected by ctDNA-seq in 162 (62.5%) cases, 1 and ≥2 mutations were found in 64 (24.7%) and 33 (12.7%) cases, respectively (Figure 3E). The number of mutations detected by ctDNA-seq significantly affected prognosis. Thus, the 2-year PFS rate was 57.3% for cases with 0 mutation, 22.7% for those with 1 mutation, and 0% for those with ≥2 mutations (P = 1.0 × 10−14; Figure 3F).
Multivariable risk stratification of patients with RRMM according to ctDNA mutations
Based on the superior predictive capacity of ctDNA mutations, we evaluated the relative effects of ctDNA mutations for the 6 genes using Cox PH model with a stepwise variable selection, incorporating clinical factors (treatment indication and number of prior therapies), IGH rearrangements (by FISH), 17p deletion (by BMPC-seq), and plasma DNA concentration as covariates. We found that KRAS, TP53, and ATM ctDNA mutations, t(11;14) rearrangement, increased number of prior therapies (≥3 regimens), and high plasma DNA concentration were independently associated with a shorter PFS, of which KRAS ctDNA mutation was the strongest predictor of clinical outcome of patients with RRMM (Table 1). We also assessed the prognostic impact of the total number of ctDNA mutations for the 6 genes in a Cox PH model incorporating the same factors. In this model, the number of ctDNA mutations, t(11;14) rearrangement, increased number of prior therapies (≥3 regimens), and high plasma DNA concentration were independently associated with worse prognosis, in which ≥2 ctDNA mutations was the strongest predictor of PFS (Table 2). Similar results were obtained by L1-regularized Cox PH models, confirming the results by different methods (supplemental Table 12).
Multivariable analysis incorporating individual ctDNA mutations, clinical factors, IGH rearrangements, del(17p), and plasma DNA concentration
| Variable . | Coefficient . | HR . | 95% lower CI . | 95% upper CI . | P value . |
|---|---|---|---|---|---|
| KRAS mutation (ctDNA-seq) | 1.1 | 3.1 | 1.7 | 5.7 | 1.8 × 10−4 |
| TP53 mutation (ctDNA-seq) | 0.7 | 2.0 | 1.3 | 3.1 | .0023 |
| ATM mutation (ctDNA-seq) | 1.3 | 3.8 | 1.6 | 9.1 | .0028 |
| t(11;14) (FISH) | 0.7 | 2.0 | 1.2 | 3.3 | .0065 |
| No. of prior regimens, n ≥ 3 | 1.1 | 2.9 | 1.7 | 5.1 | 1.5 × 10−4 |
| No. of prior regimens, n = 2 | 0.6 | 1.7 | 0.9 | 3.2 | .078 |
| Plasma DNA concentration: high | 0.6 | 1.8 | 1.2 | 2.7 | .0084 |
| Variable . | Coefficient . | HR . | 95% lower CI . | 95% upper CI . | P value . |
|---|---|---|---|---|---|
| KRAS mutation (ctDNA-seq) | 1.1 | 3.1 | 1.7 | 5.7 | 1.8 × 10−4 |
| TP53 mutation (ctDNA-seq) | 0.7 | 2.0 | 1.3 | 3.1 | .0023 |
| ATM mutation (ctDNA-seq) | 1.3 | 3.8 | 1.6 | 9.1 | .0028 |
| t(11;14) (FISH) | 0.7 | 2.0 | 1.2 | 3.3 | .0065 |
| No. of prior regimens, n ≥ 3 | 1.1 | 2.9 | 1.7 | 5.1 | 1.5 × 10−4 |
| No. of prior regimens, n = 2 | 0.6 | 1.7 | 0.9 | 3.2 | .078 |
| Plasma DNA concentration: high | 0.6 | 1.8 | 1.2 | 2.7 | .0084 |
HRs for PFS evaluated by Cox PH model incorporating the presence of KRAS, TP53, DIS3, BRAF, NRAS, and ATM ctDNA mutations and clinical factors (treatment indication and number of prior therapies), IGH rearrangements (by FISH), 17p deletion (by BMPC-seq), and plasma DNA concentration in 161 RRMM cases analyzed by both ctDNA-seq and BMPC-seq.
High and low plasma DNA concentration by a median split. Final Cox PH model after stepwise variable selection minimizing Akaike’s information criterion are shown.
CI, confidence interval; HR, hazard ratio.
Multivariable analysis incorporating the total number of ctDNA mutations in 6 significant genes, clinical factors, IGH rearrangements, del(17p), and plasma DNA concentration
| Variable . | Coefficient . | HR . | 95% lower CI . | 95% upper CI . | P value . | RRMM-PI score . |
|---|---|---|---|---|---|---|
| No. of ctDNA mutations, n ≥ 2 | 1.2 | 3.5 | 2.0 | 6.0 | 9.5 × 10−6 | 3 |
| No. of ctDNA mutations, n = 1 | 0.8 | 2.2 | 1.3 | 3.7 | .0018 | 2 |
| t(11;14) (FISH) | 0.7 | 2.0 | 1.2 | 3.3 | .0049 | 2 |
| No. of prior regimens, n ≥ 3 | 1.0 | 2.8 | 1.6 | 4.8 | 3.3 × 10−4 | 2 |
| No. of prior regimens, n = 2 | 0.4 | 1.5 | 0.8 | 2.8 | .17 | 0 |
| Plasma DNA concentration: high | 0.5 | 1.7 | 1.1 | 2.6 | .019 | 1 |
| Variable . | Coefficient . | HR . | 95% lower CI . | 95% upper CI . | P value . | RRMM-PI score . |
|---|---|---|---|---|---|---|
| No. of ctDNA mutations, n ≥ 2 | 1.2 | 3.5 | 2.0 | 6.0 | 9.5 × 10−6 | 3 |
| No. of ctDNA mutations, n = 1 | 0.8 | 2.2 | 1.3 | 3.7 | .0018 | 2 |
| t(11;14) (FISH) | 0.7 | 2.0 | 1.2 | 3.3 | .0049 | 2 |
| No. of prior regimens, n ≥ 3 | 1.0 | 2.8 | 1.6 | 4.8 | 3.3 × 10−4 | 2 |
| No. of prior regimens, n = 2 | 0.4 | 1.5 | 0.8 | 2.8 | .17 | 0 |
| Plasma DNA concentration: high | 0.5 | 1.7 | 1.1 | 2.6 | .019 | 1 |
HRs for PFS evaluated by Cox PH model incorporating the total number of ctDNA mutations in the 6 prognostically significant genes and clinical factors (treatment indication and number of prior therapies), IGH rearrangements (by FISH), 17p deletion (by BMPC-seq), and plasma DNA concentration in 161 RRMM cases analyzed by both ctDNA-seq and BMPC-seq.
High and low plasma DNA concentration by a median split. Final Cox PH model after stepwise variable selection minimizing Akaike’s information criterion are shown.
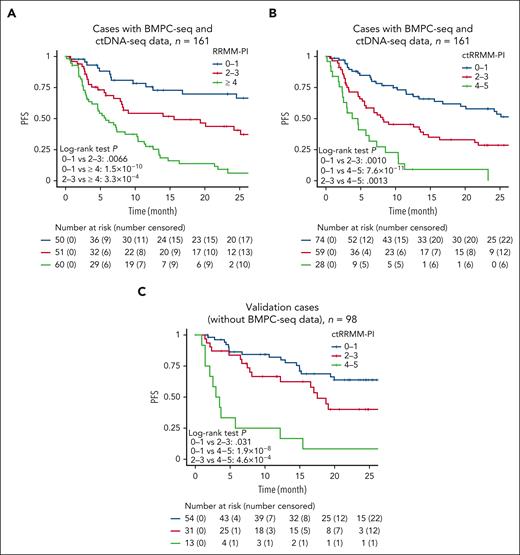
Based on the relative risks of these variables, we developed an RRMM prognostic index (RRMM-PI) by assigning 3 points to ≥2 ctDNA mutations; 2 points to 1 ctDNA mutation, ≥3 prior regimens, and t(11;14) rearrangement; and 1 point to high plasma DNA concentration (Table 2). According to the total score, patients with RRMM were classified into 3 risk categories showing significantly different outcomes (P = 3.3 × 10−9), with 2-year PFS rates of 69.8%, 43.8%, and 6.1% for scores of 0-1, 2-3, and ≥4, respectively (Figure 4A).
Multivariable risk classification of patients with RRMM according to ctDNA mutations. (A) Kaplan-Meier survival curves of 161 RRMM cases (having both BMPC-seq and ctDNA-seq data) stratified by the RRMM-PI score. This scoring system assigns 3 points to ≥2 ctDNA mutations; 2 points to 1 ctDNA mutation, ≥3 prior regimens, and t(11;14) rearrangement; and 1 point to high plasma DNA concentration. (B) Kaplan-Meier survival curves of 161 RRMM cases stratified by the ctRRMM-PI score. This scoring system assigns 2 points to ≥2 ctDNA mutations and ≥3 prior regimens; and 1 point to 1 ctDNA mutation and high plasma DNA concentration. (C) Kaplan-Meier survival curves of 98 validation cases (having BMPC-seq data but not ctDNA-seq data) stratified by the ctRRMM-PI score. Log-rank test.
Multivariable risk classification of patients with RRMM according to ctDNA mutations. (A) Kaplan-Meier survival curves of 161 RRMM cases (having both BMPC-seq and ctDNA-seq data) stratified by the RRMM-PI score. This scoring system assigns 3 points to ≥2 ctDNA mutations; 2 points to 1 ctDNA mutation, ≥3 prior regimens, and t(11;14) rearrangement; and 1 point to high plasma DNA concentration. (B) Kaplan-Meier survival curves of 161 RRMM cases stratified by the ctRRMM-PI score. This scoring system assigns 2 points to ≥2 ctDNA mutations and ≥3 prior regimens; and 1 point to 1 ctDNA mutation and high plasma DNA concentration. (C) Kaplan-Meier survival curves of 98 validation cases (having BMPC-seq data but not ctDNA-seq data) stratified by the ctRRMM-PI score. Log-rank test.
Simplified risk stratification model of patients with RRMM using ctDNA mutations
To use ctDNA mutations more efficiently and simplify the risk stratification model, we performed the similar multivariable analysis, but which did not incorporate somatic alterations in BMPC. We found that a higher number of ctDNA mutations (≥2 and 1 mutations), increased number of prior therapies (≥3 regimens), and high plasma DNA concentration were independently associated with a shorter PFS (Table 3). Based on the relative risks of these variables, we created a ctDNA-seq–only RRMM-PI (ctRRMM-PI) by assigning 2 points to ≥2 ctDNA mutations and ≥3 prior regimens; and 1 point to 1 ctDNA mutation and high plasma DNA concentration. The distribution of ctRRMM-PI scores was comparable between training and validation cases (supplemental Figure 9A). According to the total score, patients with RRMM were classified into 3 risk categories showing significantly different outcomes (P = 2.9 × 10−9), with 2-year PFS rates of 57.9%, 28.6%, and 0% for scores of 0 to 1, 2 to 3, and 4 to 5, respectively (Figure 4B). Subgroup analysis by the number of prior therapies confirmed the prognostic impact of the ctRRMM-PI (supplemental Figure 9B). The ctRRMM-PI was then applied to the validation cases for which only ctDNA-seq was performed (n = 98). This classification successfully separated the survival curves (P = 5.4 × 10−8) in the validation cases: 2-year PFS rates were 63.8%, 40.1%, and 8.3% for scores of 0 to 1, 2 to 3, and 4 to 5, respectively (Figure 4C). We also created and validated another ctDNA-seq–only prognostic index, which did not contain plasma DNA concentration (ctRRMM-PI2; supplemental Figure 9C-D; supplemental Table 13). Thus, the evaluation of the ctDNA mutations would be prognostically informative, independent of plasma DNA concentration and clinical factors, in RRMM.
Multivariable analysis incorporating number of ctDNA mutations out of 6 significant genes, clinical factors, and plasma DNA concentration
| Variable . | Coefficient . | HR . | 95% lower CI . | 95% upper CI . | P value . | ctRRMM-PI score . |
|---|---|---|---|---|---|---|
| No. of ctDNA mutations, n ≥ 2 | 1.2 | 3.5 | 2.0 | 6.0 | 1.0 × 10−5 | 2 |
| Number of ctDNA mutations, n = 1 | 0.8 | 2.2 | 1.3 | 3.6 | .0025 | 1 |
| No. of prior regimens, n ≥ 3 | 0.9 | 2.4 | 1.4 | 4.2 | .0015 | 2 |
| No. of prior regimens, n = 2 | 0.4 | 1.5 | 0.8 | 2.8 | .16 | 0 |
| Plasma DNA concentration: high | 0.5 | 1.7 | 1.1 | 2.6 | .020 | 1 |
| Variable . | Coefficient . | HR . | 95% lower CI . | 95% upper CI . | P value . | ctRRMM-PI score . |
|---|---|---|---|---|---|---|
| No. of ctDNA mutations, n ≥ 2 | 1.2 | 3.5 | 2.0 | 6.0 | 1.0 × 10−5 | 2 |
| Number of ctDNA mutations, n = 1 | 0.8 | 2.2 | 1.3 | 3.6 | .0025 | 1 |
| No. of prior regimens, n ≥ 3 | 0.9 | 2.4 | 1.4 | 4.2 | .0015 | 2 |
| No. of prior regimens, n = 2 | 0.4 | 1.5 | 0.8 | 2.8 | .16 | 0 |
| Plasma DNA concentration: high | 0.5 | 1.7 | 1.1 | 2.6 | .020 | 1 |
HRs for PFS evaluated by Cox PH model incorporating the total number of ctDNA mutations in the 6 prognostically significant genes, clinical factors (treatment indication and number of prior therapies), and plasma DNA concentration in 161 RRMM cases analyzed by both ctDNA-seq and BMPC-seq.
High and low plasma DNA concentration by a median split. Final Cox PH model after stepwise variable selection minimizing Akaike’s information criterion are shown.
Clonal architecture of BMPC and ctDNA mutations before and after IRd therapy
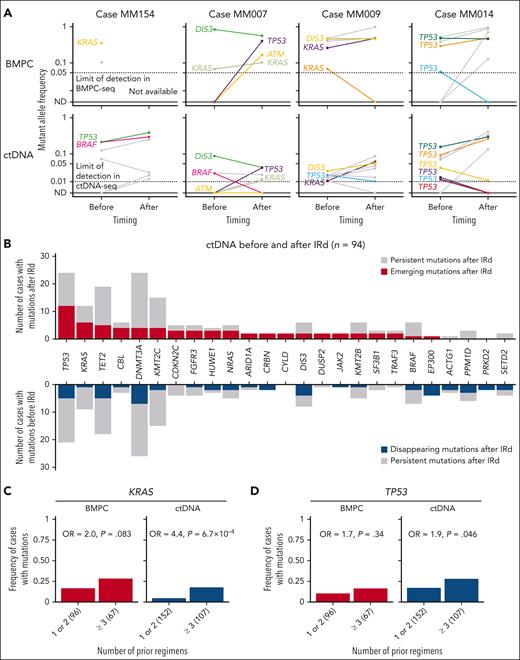
We also collected BMPC and plasma ctDNA samples from 25 and 94 RRMM cases, respectively, at discontinuation of IRd therapy in this cohort (supplemental Figure 10A). We performed targeted-capture sequencing of the BMPC and ctDNA samples, with a mean depth of 900× and 2948×, respectively (supplemental Figure 10B; supplemental Tables 14-16). Comparison of ctDNA and BMPC mutations before and after IRd therapy revealed the clonal dynamics in RRMM (Figure 5A). For example, in a representative case (MM154), TP53 and BRAF mutations persisted in ctDNA before and after treatment. In contrast, in another case (MM007), a TP53 mutation emerged in both ctDNA and BMPC after treatment. Interestingly, a KRAS mutation also appeared in ctDNA, although the allele frequency of this mutation in BMPC was comparable before and after treatment, suggesting that emergence of KRAS mutations in ctDNA is associated with progression or therapeutic resistance of RRMM. Serial assessment of ctDNA and BMPC mutations delineated a more complex clonal architecture. In a case (MM009), a KRAS mutation emerged in the same manner as in case MM007, whereas a TP53 mutation decreased in allele frequency in ctDNA after treatment. In a more complex case (MM014), among 5 TP53 mutations present before treatment, 2 mutations persisted after treatment, whereas 3 mutations decreased or even became undetectable. A similar clonal behavior was also observed in BMPC, although the number of TP53 mutations was limited.
Clonal architecture of BMPC and ctDNA mutations before and after IRd. (A) Changes in mutant allele frequencies in BMPC and ctDNA before and after IRd in 4 representative cases. The same mutations of KRAS, TP53, DIS3, BRAF, NRAS, and ATM are shown in the same color in BMPC and ctDNA within the same case. Open circles represent undetectable levels by each sequencing method; ND, not detected. (B) Number of emerging and disappearing mutations in ctDNA before and after IRd therapy in 94 cases. Emerging mutations are defined as those (1) which were present only after IRd, or (2) whose allele frequency after IRd was ≥4-times higher than before IRd. Disappearing mutations were defined as those (1) which were present only before IRd, or (2) whose allele frequency after IRd was ≥4-times lower than before IRd. Genes with emerging and/or disappearing mutations detected in ≥2 cases are shown. (C-D) Frequency of cases with (C) KRAS and (D) TP53 mutations according to the number of prior regimens, detected by BMPC-seq (n = 163) and ctDNA-seq (n = 259). The number of cases is shown in parenthesis. Odds ratio (OR) and P value are calculated using 2-sided Fisher exact test.
Clonal architecture of BMPC and ctDNA mutations before and after IRd. (A) Changes in mutant allele frequencies in BMPC and ctDNA before and after IRd in 4 representative cases. The same mutations of KRAS, TP53, DIS3, BRAF, NRAS, and ATM are shown in the same color in BMPC and ctDNA within the same case. Open circles represent undetectable levels by each sequencing method; ND, not detected. (B) Number of emerging and disappearing mutations in ctDNA before and after IRd therapy in 94 cases. Emerging mutations are defined as those (1) which were present only after IRd, or (2) whose allele frequency after IRd was ≥4-times higher than before IRd. Disappearing mutations were defined as those (1) which were present only before IRd, or (2) whose allele frequency after IRd was ≥4-times lower than before IRd. Genes with emerging and/or disappearing mutations detected in ≥2 cases are shown. (C-D) Frequency of cases with (C) KRAS and (D) TP53 mutations according to the number of prior regimens, detected by BMPC-seq (n = 163) and ctDNA-seq (n = 259). The number of cases is shown in parenthesis. Odds ratio (OR) and P value are calculated using 2-sided Fisher exact test.
In 94 cases with paired ctDNA samples, 43 cases had emerging mutations and 41 cases had disappearing mutations after treatment. The number of emerging mutations was the highest in TP53, followed by KRAS (Figure 5B). By contrast, CH-related genes, such as DNMT3A and TET2, as well as TP53 showed a larger number of disappearing mutations. In connection with this, when ctDNA and BMPC mutation frequency was evaluated according to the number of prior regimens, KRAS and TP53 mutations in ctDNA were significantly more common in more heavily treated cases (Figure 5C-D). These results are consistent with negative prognostic impacts and clonal evolution pattern of TP53 and KRAS mutations in RRMM.
Discussion
Through the combined analysis of BMPC and ctDNA mutations in a large-scale prospective cohort, we have delineated the differences of somatic alterations between BMPC and ctDNA in RRMM, demonstrating frequent ctDNA-specific mutations of TP53. In addition, we have identified ctDNA mutations, particularly those involving TP53 and KRAS, as more powerful prognostic factors in RRMM, compared with BMPC mutations. Clonal dynamics analysis supports such clinicogenetic aspects of TP53 and KRAS mutations. Based on these findings, we have proposed a new prognostic scoring system (ctRRMM-PI), which can successfully separate patients with RRMM into different risk categories using only clinical factors, plasma DNA concentration, and ctDNA mutations. In general, ctDNA analysis is gaining prominence because of its minimally invasive characteristics and suitability for multiple testing. Because ctDNA abundance can reflect entire tumor burden, as also suggested in this study, ctDNA detection and quantification is useful for minimal residual disease monitoring and prognostic prediction.14 In addition, owing to its high correlation with tumor tissues, ctDNA genotyping can be exploited for tumor genetic profiling.14 Our study points out a new prognostic value of detecting ctDNA mutations in a spectrum of driver genes, which is independent of plasma DNA concentration and outperforms that of BMPC mutations.
Frequent ctDNA-specific TP53 mutations would reflect substantial spatial heterogeneity in which multiple TP53-mutated subclones independently evolve in multiple lesions, as demonstrated by multiregion sequencing in MM,37 although there is a possibility that some of them are CH related. Consistent with a previous report showing the association between subclonal TP53 deletion and shorter survival,38 ctDNA-specific TP53 mutation confers a significant negative impact on prognosis in RRMM. These findings demonstrate the clinical utility of ctDNA analysis that can reduce sampling bias and capture prognostically relevant mutations that may be overlooked by BM sampling at a single site. TP53 alteration occurs not only as a mutation but also as a CN deletion. We found that BMPC-seq (NGS) can more accurately identify del(17p) associated with worse prognosis than conventional FISH. However, in this study, the capture probes for ctDNA-seq were not designed to estimate genome-wide CN. Because low-pass whole-genome sequencing was reported to faithfully detect CNAs from ctDNA in MM,39 a combined analysis of mutations and CNAs in ctDNA would have more clinical relevance in MM.
KRAS mutations were enriched in cases with high plasma DNA concentration. But unlike TP53 mutations, ctDNA-seq detected the same mutations in only half of patients harboring KRAS mutations in BMPC. Interestingly, only those harboring KRAS mutations in both BMPC and ctDNA show a worse outcome in RRMM, in contrast to those in NDMM.4 Although ctDNA is generally thought to be released reflecting tumor turnover, our understanding of the mechanisms underlying ctDNA release and clearance is still preliminary.14 Not only a multitude of release machineries including necrosis, apoptosis, and senescence, but also active secretion mechanisms, such as those in association with extracellular vesicles, have been described.40,41 Thus, KRAS mutation may be involved in such mechanisms, which can be related to aggressive cellular phenotype contributing to disease progression and therapeutic resistance. Consistent with the worst prognosis in KRAS-mutated RRMM cases in our cohort, reduced ixazomib sensitivity was demonstrated in KRAS-mutated lung and colon cancer xenograft models.42 In addition, activating RAS mutations were reported to promote proteasome inhibitor resistance by enhancing proteasome capacity and reducing endoplasmic reticulum stress in MM.43 Further understanding of mechanisms underlying KRAS mutation–mediated ixazomib resistance and development of therapeutic approaches targeting such mechanisms would be required. Besides ctDNA mutations, t(11;14) rearrangement is associated with worse prognosis in RRMM, consistent with recent reports in NDMM, which may be overcome by BCL2 inhibition.44
This study has several limitations. There is heterogeneity in patient background and clinical decision making because of the nature of the noninterventional study, although our cohort is more likely to reflect the real-world population of RRMM than those in randomized clinical trials. The validation of ctRRMM-PI based on a split-sample approach within our cohort may be subject to potential biases. Because we analyzed RRMM cases treated with IRd therapy, it is not evident that the prognostic impacts of ctDNA mutations can be applied to patients with NDMM or to patients with RRMM receiving other treatments, including anti-CD38 antibody. Although novel proteasome inhibitors and lenalidomide constitute key components of current treatment options for RRMM, further investigation of ctDNA mutations is warranted in MM. To implement the ctRRMM-PI in the clinical setting, the analytical methods, including plasma DNA quantification, ctDNA mutation analysis, and CNA detection, need to be optimized. In conclusion, we have clarified the genetic properties of ctDNA mutations and their superiority over BMPC mutations in prognostic prediction in RRMM. These findings have led to the development of a new prognostic scoring system incorporating ctDNA mutations, plasma DNA concentration, and clinical factors. Therefore, molecular profiling with ctDNA mutations can accurately identify low-risk patients who benefit from current treatment approaches, including IRd therapy, and high-risk patients suitable for clinical trials, thereby improving patient management strategy for RRMM.
Acknowledgments
Supercomputing resources were provided by the Human Genome Center, Institute of Medical Science, The University of Tokyo, Tokyo, Japan. Visual abstract illustrations were created using images from TogoTV (©2016 DBCLS TogoTV, CC-BY-4.0). The authors thank the patients and their families, as well as the physicians, nurses, study coordinators, and research staff, for their participation in this study.
This work was supported by Takeda Pharmaceutical Company Limited (K.K.).
Authorship
Contribution: Y.K., H.H., M.R., T.M., and K.K. conceptualized the study; Y.K., Y.I., T.S., T.Y., I.M., and K.K. were responsible for data curation; Y.K., Y.I., and K.K. were responsible for formal analysis; K.K. acquired funding; Y.K. and Y.I. were responsible for investigation; K.K. supervised the study, and was responsible for project administration; H.H., M.R., Y. Horigome, M.I., Y. Harazaki, T.K., M.A., T.I., S. Ito, H.I., J.K., H.S., K.S., H. Takamatsu, H. Tamura, T.H., K.A., S. Iida, and T.M. provided resources; Y.K., Y.I., and K.K. were responsible for data visualization; Y.K. and K.K. wrote the original manuscript draft; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: Y.K. received honoraria from Takeda, Daiichi Sankyo, Nippon Shinyaku, and Kyowa Kirin. H.H. received honoraria from Takeda, Janssen, Bristol Myers Squibb, Ono, Sanofi, AbbVie, and Novartis; served in consulting or advisory roles for Takeda, Janssen, and Bristol Myers Squibb; is part of the speakers’ bureau for Takeda, Janssen, Bristol Myers Squibb, Ono, Sanofi, AbbVie, and Novartis; and received research funding from Takeda, Bristol Myers Squibb, Kyowa Kirin, and Chugai. Y. Horigome received honoraria from Janssen, Takeda, Ono, Sanofi, Bristol Myers Squibb, Novartis, Nippon Kayaku, and Otsuka; and served in a consulting or advisory role for Janssen. T.K. is part of the speakers' bureau for Ono, Bristol Myers Squibb, Takeda, Fujimoto, Janssen, and Sanofi. M.A. received honoraria from Daiichi Sankyo, Janssen, Takeda, Sanofi, Bristol Myers Squibb, and Ono; and received research funding from Chugai (to the institution), GlaxoSmithKline (to the institution), Sanofi (to the institution), Kyowa Kirin (to the institution), Nippon Shinyaku (to the institution), Teijin (to the institution), and Ono (to the institution). T.I. is part of the speakers’ bureau for Ono, Sanofi, Bristol Myers Squibb, Janssen, and Takeda; and received research funding from Pfizer, Sanofi, Bristol Myers Squibb, Janssen, and Takeda. S. Ito received honoraria from Takeda and Bristol Myers Squibb; and received research funding from Bristol Myers Squibb (to the institution). H.I. received honoraria from Sanofi, SymBio, Janssen, Takeda, Chugai, Kyowa Kirin, Daiichi Sankyo, and LSI Medience; and received research funding from Kyowa Kirin (to the institution). J.K. served in consulting or advisory roles for Janssen, Bristol Myers Squibb, Asahi Kasei, and Pfizer; is part of the speakers’ bureau for Kyowa Kirin, Chugai, Japan Blood Products Organization, Daiichi Sankyo, Takeda, Ono, Sanofi, Eisai, Bristol Myers Squibb, Pfizer, Astellas, Novartis, AstraZeneca, SymBio, Meiji Seika, AbbVie, Nippon Shinyaku, and Otsuka; and received research funding from Kyowa Kirin, Chugai, Japan Blood Products Organization, Daiichi Sankyo, Mochida, Ono, Eisai, Taiho, Sumitomo, Asahi Kasei, Otsuka, Takeda, Shionogi, Nihon Pharmaceutical, Nippon Shinyaku, Bristol Myers Squibb, CSL Behring, Sysmex, AbbVie, and Teijin. H.S. served in consulting or advisory roles for Takeda, Janssen, Fujimoto, AbbVie, Eisai, Sanofi, AstraZeneca, and Chugai; is part of the speakers’ bureau for Takeda, Ono, Chugai, Janssen, Sanofi, AstraZeneca, Nippon Shinyaku, Meiji Seika Pharma, and Eisai; and received research funding from Bristol Myers Squibb, Ono, and AbbVie. K.S. received honoraria from Ono, Bristol Myers Squibb, Takeda, and Sanofi; and received research funding from Ono, Celgene, AbbVie, Takeda, Sanofi, Bristol Myers Squibb, GlaxoSmithKline, Chugai, Otsuka, and Janssen. H. Takamatsu received honoraria from Janssen, Ono, Sanofi, and Bristol Myers Squibb; served in a consulting or advisory role for SRL; and received research funding from Bristol Myers Squibb. H. Tamura received honoraria from Sanofi, Bristol Myers Squibb, Ono, Janssen, Chugai, and Takeda. T.H. is part of the speakers’ bureau for Takeda, Bristol Myers Squibb, Ono, Sanofi, and Fujimoto. K.A. is part of the speakers’ bureau for Takeda, MSD, Chugai, AstraZeneca, Bristol Myers Squibb, Taiho, Mochida, and Roche Diagnostics. T.S. served in a consulting or advisory role for Santen and Takeda; and is part of the speakers’ bureau for Novartis. T.Y. and I.M. are employed by Takeda. S. Iida received honoraria from Sanofi, Janssen, Bristol Myers Squibb, Takeda, and Ono; served in consulting or advisory roles for Takeda, Janssen, Pfizer, AbbVie, Bristol Myers Squibb, and Novartis; and received research funding from Takeda, Janssen, Bristol Myers Squibb, Daiichi Sankyo, Pfizer, Novartis, AbbVie, Sanofi, Ono, Otsuka, Caelum, Amgen Astellas, and Chugai. T.M. received honoraria from Chugai, Kyowa Kirin, Otsuka, AbbVie, Astellas, Novartis, and Takeda; served in consulting or advisory roles for Illumina, Sysmex, Otsuka, and AbbVie; and received research funding from Kyowa Kirin. K.K. has stock in Asahi Genomics; received honoraria from Ono, Eisai, Novartis, Chugai, AstraZeneca, Sumitomo, Kyowa Kirin, Janssen, Takeda, Otsuka, SymBio, Bristol Myers Squibb, Pfizer, Nippon Shinyaku, Daiichi Sankyo, Alexion, AbbVie, Meiji Seika Pharma, and Sanofi; served in a consulting or advisory role for Meiji Seika Pharma; and received research funding from Chordia Therapeutics, Asahi Kasei, Eisai, Otsuka, Kyowa Kirin, Shionogi, Daiichi Sankyo, Takeda, Sumitomo, Chugai, Teijin, Japan Blood Products Organization, Mochida, JCR Pharmaceuticals, Nippon Shinyaku, and Nippon Kayaku; and holds a patent to genetic alterations as a biomarker in T-cell lymphomas licensed and a patent to PD-L1 abnormalities as a predictive biomarker for immune checkpoint blockade therapy licensed. The remaining authors declare no competing financial interests.
Correspondence: Keisuke Kataoka, Division of Molecular Oncology, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo 104-0045, Japan; email: kkataoka-tky@umin.ac.jp.
References
Author notes
Presented in poster form at the EHA2023 Hybrid Congress of the European Hematology Association, Frankfurt, Germany, 9 June 2023; and presented as an oral presentation at the 85th Annual Meeting of the Japanese Society of Hematology (JSH2023), Tokyo, Japan, 13 October 2023.
The data sets, including the redacted study protocol, redacted statistical analysis plan, and individual participants’ data supporting the results reported in this article, will be made available within 3 months from initial request, to researchers who provide a methodologically sound proposal. The data will be provided after their deidentification, in compliance with applicable privacy laws, data protection and requirements for consent and anonymization.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal