The histone demethylase PHF8 is a master regulator of cell-intrinsic immune response in AML.
Pharmacologically induced PHF8 phosphorylation activates RNA sensors, triggering IFN-I response and offering potential for AML immunotherapy.
Visual Abstract
Epigenetic modulation of the cell-intrinsic immune response holds promise as a therapeutic approach for leukemia. However, current strategies designed for transcriptional activation of endogenous transposons and subsequent interferon type-I (IFN-I) response, show limited clinical efficacy. Histone lysine methylation is an epigenetic signature in IFN-I response associated with suppression of IFN-I and IFN-stimulated genes, suggesting histone demethylation as key mechanism of reactivation. In this study, we unveil the histone demethylase PHF8 as a direct initiator and regulator of cell-intrinsic immune response in acute myeloid leukemia (AML). Site-specific phosphorylation of PHF8 orchestrates epigenetic changes that upregulate cytosolic RNA sensors, particularly the TRIM25-RIG-I-IFIT5 axis, thereby triggering the cellular IFN-I response-differentiation-apoptosis network. This signaling cascade largely counteracts differentiation block and growth of human AML cells across various disease subtypes in vitro and in vivo. Through proteome analysis of over 200 primary AML bone marrow samples, we identify a distinct PHF8/IFN-I signature in half of the patient population, without significant associations with known clinically or genetically defined AML subgroups. This profile was absent in healthy CD34+ hematopoietic progenitor cells, suggesting therapeutic applicability in a large fraction of patients with AML. Pharmacological support of PHF8 phosphorylation significantly impairs the growth in samples from patients with primary AML. These findings provide novel opportunities for harnessing the cell-intrinsic immune response in the development of immunotherapeutic strategies against AML.
Introduction
Avoiding immune destruction is a core hallmark of cancer cells.1 Although modern therapies have revolutionized immune-oncology treatments by targeting T-cell inhibitory or activating pathways, the benefits are limited to a minority of patients.2 Emerging evidence from solid tumors and hematological malignancies suggests that cancer cell–intrinsic genetic and epigenetic alterations affect tumor immune landscapes.3-6 Therefore, exploiting the reversibility of those epigenetic marks that control cell-intrinsic immune response presents an attractive platform for the development of more tailored cancer immunotherapies. Epigenetic control of the genome operates through a complex network of regulatory layers and interconnections, which actively facilitate modifications in both DNA and histone tails. Histone lysine methylation is an epigenetic signature in interferon type-I (IFN-I) response associated with suppression of IFN-I and IFN-stimulated genes (ISG).7,8 Thus, histone demethylation could be a key mechanism to activate cell-intrinsic immune response. The Jumonji C (JmjC) family of histone lysine demethylases (KDMs) are a novel class of epigenetic enzymes9 that act in the center of cellular differentiation and proliferation,10,11 contributing to both normal hematopoiesis12,13 and leukemogenesis. The JmjC family member PHF8 (KDM7B) specifically targets those histone lysine methylation marks that have been associated with the repression of IFN-I response.7,14,15 We hypothesized that controlling histone demethylation of these specific marks could potentiate antiviral response unleashing the IFN-I response network against leukemia.
This study uncovers a trigger of cell-intrinsic immune response: phosphorylated PHF8 directly initiates and regulates cell-intrinsic immune response through histone demethylation. PHF8 activates key initiators of innate immune response including IFIT5, leading to NF-κB–driven IFN-I response, differentiation, and apoptosis network against acute myeloid leukemia (AML). Proteome analyses of samples of >200 patients with AML who were uniformly treated identifies a distinct PHF8/IFN type-I signature in half the population that is absent in healthy CD34+ hematopoietic progenitor cells. Thus, our findings provide a novel strategy harnessing cell-intrinsic immune response for cancer immunotherapy.
Methods
A detailed description of all methods is presented in the supplemental Methods, available on the Blood website.
Human/patients samples
Pharmacological studies with primary samples from patients with AML were approved by the ethics committee of the medical faculty of the University of Heidelberg (S-169/2017) before the study began.
Bone marrow biopsies from patients with AML for proteomics analyses were obtained and collected before and after treatment. All patients gave informed consent according to the Declaration of Helsinki to participate in the collection of samples. Use of bone marrow aspirates of patients with AML and CD34+ hematopoietic progenitor cells from healthy volunteers was approved by the Ethics Committee of University Hospital Frankfurt (approval number: SHN-12-2016, amend 01 2021 and 02 2022) and the Study Alliance Leukemia (approval number: EK98032010).16
Cell characterization assays
Retroviral/lentiviral transduction and transformation assays were performed on primary murine hematopoietic stem and progenitor cells (HSPCs) and human AML cell lines (from DSMZ) as previously described.11 Briefly, after transduction, cells were plated in methylcellulose medium supplemented with cytokines and the suitable antibiotics for selection. Colonies were counted and replated every 5 to 7 days.
Apoptosis analyses were performed with the PE Annexin V Apoptosis Detection Kit (Becton Dickinson), according to the manufacturer’s instructions. Flow cytometry was performed on Attune NxT flow cytometer.
Xenograft transplantation
All in vivo studies were performed as described before11 and approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen under the file number 84-02.04.2015.A231. Briefly, NOD/SCID/IL2rγnull (NSG) mice were injected via tail vein injection with 1 × 105 THP1 transduced cells, after 5 days of antibiotic selection. Before transplantation, mice were conditioned with sublethal radiation (2.5 Gy).
Drugs
The following drugs were used: okadaic acid (OKA; Merck; catalog no. 495609-25UG) and all-trans retinoic acid (ATRA; Merck; catalog no. R2625-50mg).
RNA-sequencing and analyses
Deep sequencing of the RNA libraries (30 × 106 reads per sample) was carried out on the Illumina NextSeq 500 at the Core Facility Genomics at the Medical Faculty of the University of Muenster.
The alignment was conducted with STAR 2.7.0c against the human reference genome hg38 with default parameters. Read counts per gene were counted with the R package bam signals 3.8. DESeq2 was used to identify differentially expressed genes per condition.
ChIP-seq and analyses
Cells were processed as described before.11 Briefly, AML cells were formaldehyde crosslinked, sodium dodecyl sulfate (SDS) lysed, and sonicated. The size of the DNA fragments was checked on the Agilent 2100 Bioanalyzer before the library preparation. For each chromatin immunoprecipitation (ChIP) library, the NEBNext Ultra II DNA Library Prep Kit and the NEBNext Multiplex Oligos for Illumina were used. Deep sequencing was carried out on the Illumina NextSeq 500.
Raw ChIP-seq reads were aligned to the human reference genome hg38 with BWA 0.7.17 and subsequently sorted with SAMtools 1.9. Peaks were identified with MACS2 and default parameters.
Deep proteome peptide fractionation
Peptides prepared from THP1 cells with PHF8 AA and PHF8 DD were separated into 8 fractions each using the high-pH reversed-phase “spider fractionator” as described elsewhere.17
Data-dependent acquisition proteomics and data analysis
A nanoflow HPLC (EASY-nLC1000, Thermo Fisher Scientific) coupled online to an Orbitrap Exploris480 mass spectrometer via a nano electrospray ion source was used for sample analysis. Mass spectra was acquired in a data dependent mode.
MS raw files were processed using Maxquant version 1.5.5.2 supported by Andromeda search engine. The data were searched for proteins and peptides using a target-decoy approach with a reverse database against Uniprot Human (version 2018) FASTA file, with a false discovery rate of <1% at the levels of protein and peptide.
Phosphoproteome and phosphopeptide enrichment
DIA phosphoproteomics and data analysis
For data-independent data acquisition (DIA), the Thermo Xcalibur Qual Browser (4.0.27.19) software was used for the Exploris 480 instrument.
DIA raw files were analyzed using Spectronaut (Biognosys; catalog no. Sw-3001, version 14) using the default settings for targeted DIA analysis. Direct DIA search using pulser against the human Uniprot reference proteome database (version August 2018, containing 21.039 entries) was used.
Proteome and phosphoproteome imputation
We performed data imputation using the in-house open-source Perseus environment (version 1.5.2.11) and R (version 4.0.2).
Pharmacological studies with primary samples from patients with AML were approved by the ethics committee of the medical faculty of the University of Heidelberg (S-169/2017) before the study began. Bone marrow biopsy samples from patients with AML for proteomics analyses were obtained and collected before and after treatment. All patients gave informed consent according to the Declaration of Helsinki to participate in the collection of samples. Use of bone marrow aspirates of patients with AML and CD34+ hematopoietic progenitor cells from healthy volunteers was approved by the Ethics Committee of University Hospital Frankfurt (approval number SHN-12-2016, amend 01 2021 and 02 2022) and the Study Alliance Leukemia (approval number EK98032010). All animal studies were approved by the LANUV NRW under the file number 84-02.04.2015.A231.
Results
Epigenetic activity of phosphorylated PHF8 counteracts differentiation block via induction of cell-intrinsic immune response in AML
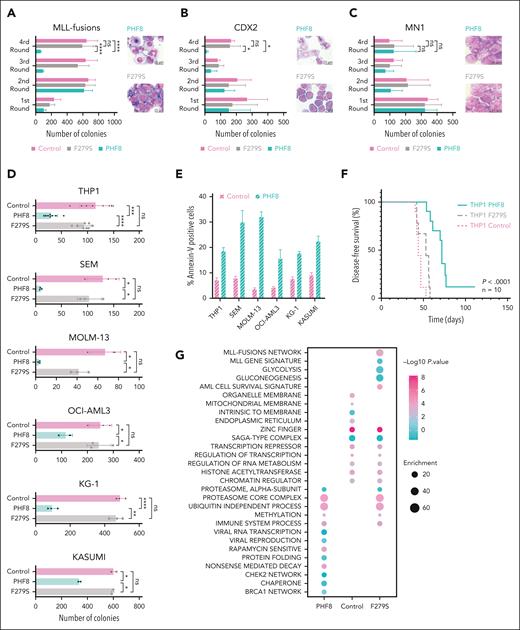
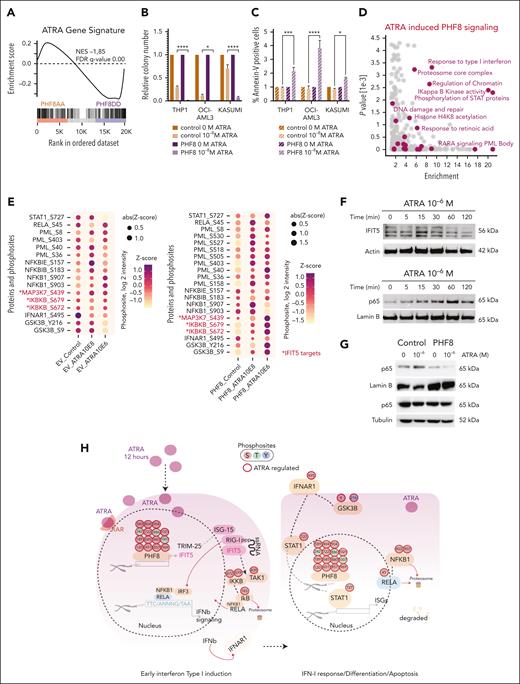
To investigate the functional significance of the PHF8 histone demethylase activity in leukemogenesis, we cotransduced primary murine HSPCs (mHSPCs) with typical AML-associated oncogenes or oncofusions and either empty vector (Control), PHF8 wild type (wt), or its catalytically inactive JmjC domain point mutant (F279S) that has been discovered in patients with X-linked mental retardation (supplemental Figure 1A).20,21 Only expression of PHF8 with intact histone demethylase activity compromised leukemic transformation and increased cellular differentiation of primary mHSPCs expressing different AML-associated oncofusions or oncogenes (Figure 1A-B; supplemental Figure 1B-C). No effect was observed in cells transduced with the MN1 oncogene known to prevent immune response pathway activation22 or with E2A-HLF, a chimeric oncoprotein in B-cell lymphoblastic leukemia (Figure 1C; supplemental Figure 1D-E). Importantly, MLL-fusions, RUNX1-RUNX1T1, CDX2, and CBX7 cells cotransduced with wt PHF8 only formed few small third colonies with diffuse colony morphology (supplemental Figure 1F). Additionally, we demonstrated that wt PHF8 significantly reduced clonogenic growth and increased apoptosis of human AML cells (Figure 1D-E). NSG mice that received transplantation with human THP1 AML cells ectopically expressing wt PHF8 showed significantly prolonged disease-free survival compared with mice that received transplantation with THP1 cells transduced with F279S or controls (Figure 1F). Transcriptomic and proteomic analyses of human THP1 cells under the same 3 different conditions confirmed that AML cells overexpressing wt PHF8 showed a significantly distinct gene signature (supplemental Figure 1G-H). Systematic pathway analyses of differentially expressed proteins among the 3 groups revealed that AML cells expressing the F279S mutant were enriched in oncogenic signatures, whereas AML cells with wt PHF8 displayed enrichment of signaling associated with viral transcription, viral reproduction, immunogenic cell death pathways, and tumor suppression (Figure 1G). These data collectively show that PHF8 histone demethylation activity strongly inhibits leukemogenesis and clonogenic growth in a broad subset of different AML subtypes through the induction of differentiation and apoptosis and is potentially associated with antiviral immune response. However, loss of PHF8 did not affect leukemic transformation, colony morphology, or proliferation of mHSPCs cotransduced with AML-associated oncogenes (Figure 1H; supplemental Figure 1I-K) or clonogenic growth of human AML cells (Figure 1I). Thus, changes in expression levels alone do not determine PHF8 function in leukemia, suggesting that posttranslational modifications of the protein are required. In fact, we found significantly increased PHF8 phosphorylation levels upon its overexpression in THP1 cells (supplemental Figure 1L). PHF8 phosphorylation levels affect its functions,23,24 including cellular responses to retinoic acid.11 Therefore, we conducted a deep phospho-proteome analysis of THP1 AML cells under ATRA treatment. We identified specific phospho-residues with significant increase in phosphorylation levels of endogenous PHF8 under increasing concentrations of ATRA (Figure 1J). Most of these residues exhibited further increased phosphorylation upon higher ATRA concentrations. These results indicate that low levels of ATRA are sufficient to trigger site-specific phosphorylation of PHF8.
Epigenetic activity of phosphorylated PHF8 counteracts differentiation block via induction of cell-intrinsic immune response in AML. (A-C) Bar charts represent colony numbers in a serial replating assay and typical morphology of cells from fourth round of replating; primary mHSPCs were cotransduced with the indicated AML oncogenes or oncofusions as well as with wt PHF8 (PHF8), PHF8-F279S (F279S), or empty vector (control). Error bars indicate standard deviation (SD) of 3 independent experiments. Significance was tested using the Tukey multiple comparison test (2-way analysis of variance [ANOVA], ∗P < .04; ∗∗∗∗P < .000, nonsignificant [ns]). (D) Bar charts depict colony numbers of human AML cells transduced with PHF8 vs F279S vs Control. Error bars indicate SD of at least 4 independent experiments. Significance was tested using the Tukey multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001; ∗∗∗P < .0004; ∗∗P < .006; ∗P < .04). (E) Bar charts represent early apoptosis as percentage of Annexin-V–positive cells from flow cytometry analysis. Error bars indicate SD of 4 independent experiments. Sidak multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001 each cell group). (F) Kaplan-Meier curves for disease-free survival (DFS) of NSG mice (Charles River Laboratories) that received transplantation with human THP1 cells transduced with indicated constructs (n = 10 mice per group). Six-to-8-week-old NSG mice were injected with 1 × 105 cells, after 5 days antibiotic selection, via tail vein injection. Before transplantation, mice were conditioned with sublethal radiation (2.5 Gy). The log-rank (Mantel-Cox) test was used to compare survival curves. (G) Pathways enrichment analysis (Fisher exact test) of control, PHF8, and F279S cells (n = 3 biological replicates per group). Significantly regulated proteins were compared with the unchanged proteome in the data set based on the gene ontology (GO), KEGG, and keyword terms. (H) Bar charts represent colony numbers of primary mHSPCs cotransduced with MLL-fusions or CDX2 and sgRNAs for specific CRISPR-Cas9–mediated knockout of PHF8 (sgRNA 1, sgRNA 2) or nontargeting control (control). Error bars indicate SDs of 3 independent experiments. Significance was tested using Tukey multiple comparison test (2-way ANOVA, ns). (I) Bar charts represent colony numbers of human AML cells transduced with lenti-CRISPRv2 nontargeting (control) or lenti-CRISPRv2 guide targeting endogenous PHF8 (sgRNA 1 or sgRNA 3). Error bars indicate SDs of 4 independent experiments. Significance was tested using Tukey multiple comparison test (2-way ANOVA, ns); (lower panel) immunoblot analyses showing PHF8 levels. (J) Profile plot showing significant increases of phosphorylation in human THP1 AML cells at defined phosphosites of endogenous PHF8 upon 12 hours of ATRA treatment compared with controls. Each data point is the averaged median of biological quadruplicate of z scored log 2 phosphosite intensities; significance was tested using multiple sample t test (1-way ANOVA, permutation-based false discovery rate [FDR] <0.05). (K) Violin plots depict numbers of colonies of human AML cells treated with 500 nM OKA (OKA 20 minutes incubation) or/and ATRA 10–8 M. Significance was tested using the Tukey multiple comparison test (2-way ANOVA, ∗P < .05; ∗∗P < .0085, ns). (L) Immunoblot of endogenous total PHF8 protein levels. (M) Western blot analyses (upper panel) of PHF8 levels before and after knocking out in AML cells; (lower panel) typical INT-stained colony pictures. KEGG, Kyoto Encyclopedia of Genes and Genomes; KG-1, Koeffler and Golde 1; MLL, mixed lineage leukemia; sgRNA, single guide RNA.
Epigenetic activity of phosphorylated PHF8 counteracts differentiation block via induction of cell-intrinsic immune response in AML. (A-C) Bar charts represent colony numbers in a serial replating assay and typical morphology of cells from fourth round of replating; primary mHSPCs were cotransduced with the indicated AML oncogenes or oncofusions as well as with wt PHF8 (PHF8), PHF8-F279S (F279S), or empty vector (control). Error bars indicate standard deviation (SD) of 3 independent experiments. Significance was tested using the Tukey multiple comparison test (2-way analysis of variance [ANOVA], ∗P < .04; ∗∗∗∗P < .000, nonsignificant [ns]). (D) Bar charts depict colony numbers of human AML cells transduced with PHF8 vs F279S vs Control. Error bars indicate SD of at least 4 independent experiments. Significance was tested using the Tukey multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001; ∗∗∗P < .0004; ∗∗P < .006; ∗P < .04). (E) Bar charts represent early apoptosis as percentage of Annexin-V–positive cells from flow cytometry analysis. Error bars indicate SD of 4 independent experiments. Sidak multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001 each cell group). (F) Kaplan-Meier curves for disease-free survival (DFS) of NSG mice (Charles River Laboratories) that received transplantation with human THP1 cells transduced with indicated constructs (n = 10 mice per group). Six-to-8-week-old NSG mice were injected with 1 × 105 cells, after 5 days antibiotic selection, via tail vein injection. Before transplantation, mice were conditioned with sublethal radiation (2.5 Gy). The log-rank (Mantel-Cox) test was used to compare survival curves. (G) Pathways enrichment analysis (Fisher exact test) of control, PHF8, and F279S cells (n = 3 biological replicates per group). Significantly regulated proteins were compared with the unchanged proteome in the data set based on the gene ontology (GO), KEGG, and keyword terms. (H) Bar charts represent colony numbers of primary mHSPCs cotransduced with MLL-fusions or CDX2 and sgRNAs for specific CRISPR-Cas9–mediated knockout of PHF8 (sgRNA 1, sgRNA 2) or nontargeting control (control). Error bars indicate SDs of 3 independent experiments. Significance was tested using Tukey multiple comparison test (2-way ANOVA, ns). (I) Bar charts represent colony numbers of human AML cells transduced with lenti-CRISPRv2 nontargeting (control) or lenti-CRISPRv2 guide targeting endogenous PHF8 (sgRNA 1 or sgRNA 3). Error bars indicate SDs of 4 independent experiments. Significance was tested using Tukey multiple comparison test (2-way ANOVA, ns); (lower panel) immunoblot analyses showing PHF8 levels. (J) Profile plot showing significant increases of phosphorylation in human THP1 AML cells at defined phosphosites of endogenous PHF8 upon 12 hours of ATRA treatment compared with controls. Each data point is the averaged median of biological quadruplicate of z scored log 2 phosphosite intensities; significance was tested using multiple sample t test (1-way ANOVA, permutation-based false discovery rate [FDR] <0.05). (K) Violin plots depict numbers of colonies of human AML cells treated with 500 nM OKA (OKA 20 minutes incubation) or/and ATRA 10–8 M. Significance was tested using the Tukey multiple comparison test (2-way ANOVA, ∗P < .05; ∗∗P < .0085, ns). (L) Immunoblot of endogenous total PHF8 protein levels. (M) Western blot analyses (upper panel) of PHF8 levels before and after knocking out in AML cells; (lower panel) typical INT-stained colony pictures. KEGG, Kyoto Encyclopedia of Genes and Genomes; KG-1, Koeffler and Golde 1; MLL, mixed lineage leukemia; sgRNA, single guide RNA.
Although ATRA treatment increased PHF8 phosphorylation in AML cells (Figure 1J), the potent protein phosphatase inhibitor OKA has been shown to inhibit PHF8 dephosphorylation.11 We demonstrated that cell lines with comparably high expression of PHF8 were very susceptible to combinatorial treatment with OKA/ATRA, resulting in a significant reduction of colony formation upon low-dose drug exposure. In contrast, KASUMI-1 cells with low PHF8 levels showed only a moderate response, and MOLM-13 cells with undetectable expression of PHF8 were entirely resistant to this therapy (Figure 1K-L). Furthermore, KO of PHF8 abolished the sensitivity of AML cells to OKA/ATRA (Figure 1M; supplemental Figure 1M), confirming the requirement of PHF8 for AML blockage by this treatment. Thus, the antileukemic activity of PHF8 across different AML subtypes largely depends on its phosphorylation status. Our data strongly suggest that pharmacological induction of phospho-PHF8 is a promising novel approach to overcome ATRA resistance in AML.
Site-specific phosphorylation of PHF8 blocks growth of AML in vitro and in vivo
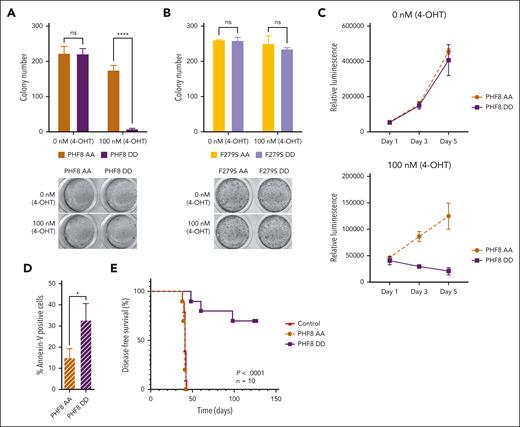
Next, using an inducible expression system that mimics constitutively phosphorylated PHF8 (PHF8 DD and F279S DD) or the nonphosphorylated protein (PHF8 AA and F279S AA),11,23 we demonstrated that only PHF8 DD compromised colony formation of leukemia cells (Figure 2A; supplemental Figure 2A). In cells carrying the catalytically inactive PHF8 mutants (F279S AA and F279S DD), the phosphorylation status did not induce any functional changes (Figure 2B). Cells expressing PHF8 DD showed significantly reduced proliferation and increased apoptosis compared with cells expressing PHF8 AA (Figure 2C-D), whereas there were no changes in total PHF8 protein levels or cell cycle (supplemental Figure 2B-C). Moreover, mice that received transplantation with human AML THP1-PHF8 DD cells showed significantly improved disease-free survival compared with mice that received transplantation with THP1-PHF8 AA or THP1 control cells (Figure 2E; supplemental Figure 2D). Thus, the antileukemic activity of PHF8 critically depends on both, its demethylase activity as well as the phosphorylation status of the protein.
Serine phosphorylation of the protein orchestrates PHF8 functions in AML. (A) Colony numbers of human THP1 AML cells transduced with ER-fused PHF8 AA (mimicking nonphosphorylated form) or PHF8 DD (mimicking phosphorylated form), allowing for an inducible expression in the presence of 4-OHT. Error bars indicate SD of 4 independent experiments. Sidak multiple comparison test (2-way ANOVA, ∗∗∗∗P < .000, ns); (lower panel) typical INT-stained colonies. (B) Number of colonies using equivalent constructs with an additional enzymatic dead F279S mutation. Error bars indicate standard deviation of 4 independent experiments (2-way ANOVA, ns); (lower panel) typical INT-stained colonies. (C) Proliferation assays, 1500 to 2000 cells were seeded in a 96-well plate and in 100 μL of medium per well. The CellTiter-Glo luminescent assay kit was used to determine the number of viable cells in culture. (D) Bar charts represent percentage of Annexin-V–positive cells analyzed by flow cytometry after 4-OHT induction. Pair t test (∗P < .05). (E) Kaplan-Meier curves for DFS of NSG mice that received transplantation via tail vein injection with 1 × 105 human THP1 cells, after 5 days antibiotic selection, transduced with indicated constructs (n = 10 mice per group). Before transplantation, mice were conditioned with sublethal radiation (2.5 Gy). The log-rank (Mantel-Cox) test was used to compare survival curves. 4-OHT, 4-hydroxytamoxifen.
Serine phosphorylation of the protein orchestrates PHF8 functions in AML. (A) Colony numbers of human THP1 AML cells transduced with ER-fused PHF8 AA (mimicking nonphosphorylated form) or PHF8 DD (mimicking phosphorylated form), allowing for an inducible expression in the presence of 4-OHT. Error bars indicate SD of 4 independent experiments. Sidak multiple comparison test (2-way ANOVA, ∗∗∗∗P < .000, ns); (lower panel) typical INT-stained colonies. (B) Number of colonies using equivalent constructs with an additional enzymatic dead F279S mutation. Error bars indicate standard deviation of 4 independent experiments (2-way ANOVA, ns); (lower panel) typical INT-stained colonies. (C) Proliferation assays, 1500 to 2000 cells were seeded in a 96-well plate and in 100 μL of medium per well. The CellTiter-Glo luminescent assay kit was used to determine the number of viable cells in culture. (D) Bar charts represent percentage of Annexin-V–positive cells analyzed by flow cytometry after 4-OHT induction. Pair t test (∗P < .05). (E) Kaplan-Meier curves for DFS of NSG mice that received transplantation via tail vein injection with 1 × 105 human THP1 cells, after 5 days antibiotic selection, transduced with indicated constructs (n = 10 mice per group). Before transplantation, mice were conditioned with sublethal radiation (2.5 Gy). The log-rank (Mantel-Cox) test was used to compare survival curves. 4-OHT, 4-hydroxytamoxifen.
The histone demethylase activity of phosphorylated PHF8 triggers IFN-I response–mediated apoptosis
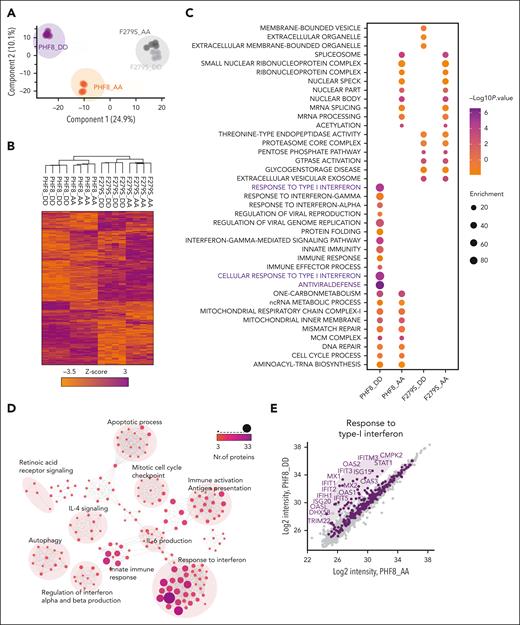
We further evaluated the molecular mechanisms underlying growth arrest of THP1-PHF8 DD cells via deep-proteome quantification analysis. Principal component analysis (PCA) and heat map profiling revealed that PHF8 DD cells contrasted significantly from PHF8 AA cells and from samples carrying catalytically deficient PHF8 (F279S) (Figure 3A). PHF8 DD cells presented a unique cluster dominated by highly expressed proteins (Figure 3B). Analysis of specific biological features revealed antiviral defense and IFN-I response as the most predominant pathways activated by PHF8 DD than by nonphosphorylated PHF8 (PHF8 AA) or the catalytically inactive mutants (F279S DD and F279S AA) that presented common profiles enriched with pathways of low significance (Figure 3C; supplemental Figure 3A). Accordingly, network analysis of highly expressed proteins in PHF8 DD cells revealed functionally interconnected clusters related to IFN response, interleukin-6 production, innate immune response, immune activation, and induction of apoptosis (Figure 3D; supplemental Figure 3B-C).
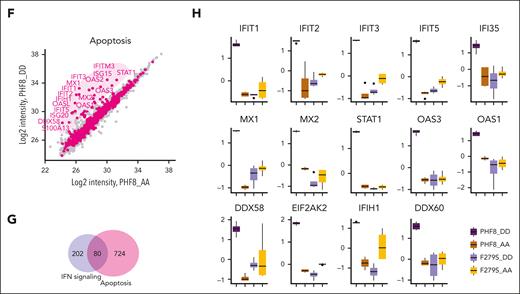
Epigenetic activity of phosphorylated PHF8 triggers IFN-I response–mediated apoptosis. (A-H) Deep-proteome quantification analysis of the indicated samples; (A) PCA of all quantified proteins of the samples in the proteome data set (A-H, n = 4 biological replicates per group). (B) Unsupervised hierarchical clustering of differentially regulated proteins between the sample groups as heat map. The color code indicates z scored log 2 protein intensities of multiple sample t test (1-way ANOVA, permutation-based FDR <0.05, significant). (C) Significantly enriched GO terms and pathways. Enrichment analysis (Fisher exact test) of differentially regulated proteins in each of the sample groups. Regulated proteins were compared to the unchanged proteome in the data set based on the GO, KEGG, and keyword terms. (D) Network analysis of enriched pathways for proteins significantly upregulated in PHF8 DD cells. The size and color of nodes indicate number of proteins participating in each pathway. The network analysis was performed in Cytoscape version 3.9.1. (E) Scatterplot comparing the quantified log 2 protein intensities in PHF8 AA vs PHF8 DD cells (data obtained by fractionation proteome experiment, n = 8 fractions per sample). Purple dots indicate proteins participating in response to IFN-I (GO term). The marked protein names represent significant regulations (permutation-based FDR, 0.05). (F) Scatterplot comparing the quantified log 2 protein intensities in PHF8 AA vs PHF8 DD cells (data obtained by fractionation proteome experiment, n = 8 fractions per sample). Pink dots indicate proteins related with apoptotic pathways (GO term). The marked protein names represent significant regulations (permutation-based FDR, 0.05). (G) Venn diagram showing the number of shared proteins between pathways in response to IFN-I and apoptosis in PHF8 DD cells (GO term). (H) Box plot representation of selected significantly regulated proteins participating in response to cell-intrinsic immune response and IFN-I (n = 4 biological replicates per group).
Epigenetic activity of phosphorylated PHF8 triggers IFN-I response–mediated apoptosis. (A-H) Deep-proteome quantification analysis of the indicated samples; (A) PCA of all quantified proteins of the samples in the proteome data set (A-H, n = 4 biological replicates per group). (B) Unsupervised hierarchical clustering of differentially regulated proteins between the sample groups as heat map. The color code indicates z scored log 2 protein intensities of multiple sample t test (1-way ANOVA, permutation-based FDR <0.05, significant). (C) Significantly enriched GO terms and pathways. Enrichment analysis (Fisher exact test) of differentially regulated proteins in each of the sample groups. Regulated proteins were compared to the unchanged proteome in the data set based on the GO, KEGG, and keyword terms. (D) Network analysis of enriched pathways for proteins significantly upregulated in PHF8 DD cells. The size and color of nodes indicate number of proteins participating in each pathway. The network analysis was performed in Cytoscape version 3.9.1. (E) Scatterplot comparing the quantified log 2 protein intensities in PHF8 AA vs PHF8 DD cells (data obtained by fractionation proteome experiment, n = 8 fractions per sample). Purple dots indicate proteins participating in response to IFN-I (GO term). The marked protein names represent significant regulations (permutation-based FDR, 0.05). (F) Scatterplot comparing the quantified log 2 protein intensities in PHF8 AA vs PHF8 DD cells (data obtained by fractionation proteome experiment, n = 8 fractions per sample). Pink dots indicate proteins related with apoptotic pathways (GO term). The marked protein names represent significant regulations (permutation-based FDR, 0.05). (G) Venn diagram showing the number of shared proteins between pathways in response to IFN-I and apoptosis in PHF8 DD cells (GO term). (H) Box plot representation of selected significantly regulated proteins participating in response to cell-intrinsic immune response and IFN-I (n = 4 biological replicates per group).
Further peptide fractionation experiments and direct comparison of the proteomes of PHF8 DD and PHF8 AA cells ultimately highlighted the most significantly upregulated proteins (>log 2) in PHF8 DD cells as part of antiviral response, IFN-I response (Figure 3E), and related apoptotic processes (Figure 3F). Specifically, 80 proteins were directly associated and shared with IFN signaling–related apoptotic pathways (Figure 3G). Among the most significantly upregulated proteins in PHF8 DD cells were the double-stranded (dsRNA) pattern recognition receptors, PKR (EIF2AK2), RIG-I (DDX58), MDA5 (IFIH1), and DDX60 as well as members of the IFIT family, MX1, MX2, STAT1, and components of the oligoadenylate synthetase family, all of them part of the antiviral signature via IFN-I responses and functionally related with induction of apoptosis or differentiation (Figure 3H; supplemental Figure 3D).25 Consistently, our transcriptome analysis of PHF8 DD vs PHF8 AA cells showed differentially upregulated genes from the apoptotic FAS-associated death domain (FADD)-caspase-8 signaling pathway that is typically activated upon IFN-I response–mediated late apoptosis (supplemental Figure 3E).26,27
Together, these results consistently demonstrated that the major antileukemic effects of constitutively phosphorylated PHF8 correlate with the same IFN-I response signaling pathways activated during cell-intrinsic immune response and terminate in its associated late apoptosis pathway.
Epigenetic activity of phospho-PHF8 confers direct activation of key initiators of cell-intrinsic immune response
After identification of the unique activated cluster of PHF8 DD cells linked to cell-intrinsic immune response, we aimed to pinpoint its direct targets, specifically initiators of the IFN-I response. To this end, a new set of RNAseq experiments and genome wide chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) were performed to identify genes activated and the genomic sites occupied by PHF8 depending on its phosphorylation status (PHF8 AA vs PHF8 DD). The profiles of the ChIP-seq tracks showed significantly higher occupancy of PHF8 DD in promoter regions (Figure 4A). As a result of the correlation of the most significantly upregulated genes with PHF8 occupancy at the promoter site, 437 genes were identified (Figure 4B; supplemental Table 1). These genes fell into 5 functional clusters in which transcription and RNA binding had the highest significance (supplemental Figure 4A). Among the most significantly upregulated genes, 16 have functions related to induction of cell intrinsic immune response (Figure 4C). Network analysis28 revealed experimental functional interconnection among 5 of these genes: cytidine/uridine monophosphate kinase 2 (CMPK2),29interferon-induced protein with tetratricopeptide repeats 5 (IFIT5),30tripartite motif 25 (TRIM25),31transporter associated with antigen processing (TAP1),32 and Moloney leukemia virus 10 (MOV10)33 (Figure 4D). We confirmed PHF8 as the common direct activator of all 5 genes (supplemental Figure 4B) and showed that phospho-PHF8 is the transcriptional activator of CMPK2, IFIT5, TRIM25, TAP1, and MOV10 by specific demethylation of the H3K9me2 repressive mark in PHF8 DD vs PHF8 AA (Figure 4E; supplemental Figure 4C).
Phospho-PHF8 directly activates key initiators of cell-intrinsic immune response. (A) Profile plot of PHF8 genome occupancy (ChIP-seq) in THP1 PHF8 AA compared with THP1 PHF8 DD cells. (B) Volcano graph depicts all regulated genes in THP1 PHF8 DD cells. Differential expression is represented as LogFC in (x-axis) vs PHF8 ChIP-seq MACS analysis presented as peakScore in (y-axis). Upregulated genes with significance higher than log 0.5 (>log 0.5) are colored in violet. (C) Scatterplot of the highly significantly (>log 0.5) upregulated genes in the transcriptome of PHF8 DD cells (logFC) and PHF8 DD targets resulting from PHF8 ChIP sequencing (peakScore). (D) Protein network analysis of the correlation of 16 highly significantly (>log 0.5) upregulated genes (cell intrinsic immune response inducers) in the transcriptome of PHF8 DD cells and PHF8 DD targets resulting from ChIP sequencing. Pink color represents higher confidence based on experimental evidence. The protein-protein interaction network was established using STRING database and visualized in Cytoscape version 3.9.1. (E) ChIP-qPCR analysis of histone mark H3K9me2 on the targeted promoter of the indicated genes in THP1 PHF8 AA and THP1 PHF8 DD cells. ChIP signals are presented as percentage of input. Error bars indicate SD of 3 independent experiments. Sidak multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001). (F) RT-qPCR time-course analysis (left) of the expression of specified human genes. The relative expression of each gene was independently analyzed in 2 cell groups: PHF8 AA and PHF8 DD. Each group consisted of noninduced cells and cells induced with tamoxifen (4-OHT). Noninduced samples within each group were used as references for assessing the relative expression of the genes at each time point. Error bars indicate SD of 3 independent experiments. Bar charts (right) of RT-qPCR analysis at 24 hours after 4-OHT induction. Expression levels are relative only to the PHF8 AA 0mM (4-OHT) cells. Error bars indicate SD of 3 independent experiments; Tukey multiple comparison test (2-way ANOVA, ∗∗P < .0023; ∗∗∗∗P < .0001). (G) RT-qPCR time-course analysis of the expression of RIG-I and IFN-β human genes. The relative expression of each gene was independently analyzed in 2 cell groups: PHF8 AA and PHF8 DD. Each group consisted of noninduced cells and cells induced with tamoxifen (4-OHT). Noninduced samples within each group were used as references for assessing the relative expression of the genes at each time point. Error bars indicate SD of 3 independent experiments; Tukey multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001). (H-I) ChIP-qPCR analysis of (left) PHF8 or (middle) histone mark H3K9me2 on CMPK2 or IFIT5 targeted promoter in THP1 cells (Control) or THP1 cells after 12 hours of OKA/ATRA treatment. ChIP signals are presented as percentage of input. Error bars indicate SD of 3 independent experiments; Sidak multiple comparison test (2-way ANOVA, ∗∗P < .023; ∗∗∗P < .0009; ∗∗∗∗P < .0001). RT-qPCR time-course analysis (right) of the expression of CMPK2 or IFIT5. The relative expression of each gene was independently analyzed in nontreated and treated groups of cells. Nontreated samples were used as references for assessing the relative expression of the genes at each time point. Error bars indicate SD of 3 independent experiments; Tukey multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001). (J-K) Bar charts represent normalized colony numbers of human THP1 AML cells cotransduced with nontargeting empty vector control (Control empty) or lenti-CRISPR-V2 guide targeting the indicated endogenous genes (CMPK2 sgRNA 7, CMPK2 sgRNA 9 or IFIT5 sgRNA 1, IFIT5 sgRNA 2) as well as PHF8 vs empty vector (Control), respectively. Nontargeting empty vector control cells were taken as the reference for normalizing the colony numbers in each group. Error bars indicate SD of 3 independent experiments; Tukey multiple comparison test (2-way ANOVA, ∗∗P < .0033; ∗∗∗P < .0007, ns); (right panels) representative western blot analyses of the indicated proteins. MACS, Model based analysis for ChIP-seq; RT-qPCR, Real time quantitative PCR.
Phospho-PHF8 directly activates key initiators of cell-intrinsic immune response. (A) Profile plot of PHF8 genome occupancy (ChIP-seq) in THP1 PHF8 AA compared with THP1 PHF8 DD cells. (B) Volcano graph depicts all regulated genes in THP1 PHF8 DD cells. Differential expression is represented as LogFC in (x-axis) vs PHF8 ChIP-seq MACS analysis presented as peakScore in (y-axis). Upregulated genes with significance higher than log 0.5 (>log 0.5) are colored in violet. (C) Scatterplot of the highly significantly (>log 0.5) upregulated genes in the transcriptome of PHF8 DD cells (logFC) and PHF8 DD targets resulting from PHF8 ChIP sequencing (peakScore). (D) Protein network analysis of the correlation of 16 highly significantly (>log 0.5) upregulated genes (cell intrinsic immune response inducers) in the transcriptome of PHF8 DD cells and PHF8 DD targets resulting from ChIP sequencing. Pink color represents higher confidence based on experimental evidence. The protein-protein interaction network was established using STRING database and visualized in Cytoscape version 3.9.1. (E) ChIP-qPCR analysis of histone mark H3K9me2 on the targeted promoter of the indicated genes in THP1 PHF8 AA and THP1 PHF8 DD cells. ChIP signals are presented as percentage of input. Error bars indicate SD of 3 independent experiments. Sidak multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001). (F) RT-qPCR time-course analysis (left) of the expression of specified human genes. The relative expression of each gene was independently analyzed in 2 cell groups: PHF8 AA and PHF8 DD. Each group consisted of noninduced cells and cells induced with tamoxifen (4-OHT). Noninduced samples within each group were used as references for assessing the relative expression of the genes at each time point. Error bars indicate SD of 3 independent experiments. Bar charts (right) of RT-qPCR analysis at 24 hours after 4-OHT induction. Expression levels are relative only to the PHF8 AA 0mM (4-OHT) cells. Error bars indicate SD of 3 independent experiments; Tukey multiple comparison test (2-way ANOVA, ∗∗P < .0023; ∗∗∗∗P < .0001). (G) RT-qPCR time-course analysis of the expression of RIG-I and IFN-β human genes. The relative expression of each gene was independently analyzed in 2 cell groups: PHF8 AA and PHF8 DD. Each group consisted of noninduced cells and cells induced with tamoxifen (4-OHT). Noninduced samples within each group were used as references for assessing the relative expression of the genes at each time point. Error bars indicate SD of 3 independent experiments; Tukey multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001). (H-I) ChIP-qPCR analysis of (left) PHF8 or (middle) histone mark H3K9me2 on CMPK2 or IFIT5 targeted promoter in THP1 cells (Control) or THP1 cells after 12 hours of OKA/ATRA treatment. ChIP signals are presented as percentage of input. Error bars indicate SD of 3 independent experiments; Sidak multiple comparison test (2-way ANOVA, ∗∗P < .023; ∗∗∗P < .0009; ∗∗∗∗P < .0001). RT-qPCR time-course analysis (right) of the expression of CMPK2 or IFIT5. The relative expression of each gene was independently analyzed in nontreated and treated groups of cells. Nontreated samples were used as references for assessing the relative expression of the genes at each time point. Error bars indicate SD of 3 independent experiments; Tukey multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001). (J-K) Bar charts represent normalized colony numbers of human THP1 AML cells cotransduced with nontargeting empty vector control (Control empty) or lenti-CRISPR-V2 guide targeting the indicated endogenous genes (CMPK2 sgRNA 7, CMPK2 sgRNA 9 or IFIT5 sgRNA 1, IFIT5 sgRNA 2) as well as PHF8 vs empty vector (Control), respectively. Nontargeting empty vector control cells were taken as the reference for normalizing the colony numbers in each group. Error bars indicate SD of 3 independent experiments; Tukey multiple comparison test (2-way ANOVA, ∗∗P < .0033; ∗∗∗P < .0007, ns); (right panels) representative western blot analyses of the indicated proteins. MACS, Model based analysis for ChIP-seq; RT-qPCR, Real time quantitative PCR.
Time course experiments comparing gene expression levels of our 5 targets, after induction of PHF8 AA or PHF8 DD, revealed a significant increase in CMPK2, IFIT5, and TRIM25 expression levels at earlier time points only in PHF8 DD cells, whereas in the same cells, TAP1 and MOV10 expression increased slowly and reached their peak much later (Figure 4F). Significantly increased expression of CMPK2, IFIT5, and TRIM25 occurred earlier than the increased expression of IFNβ, indicating their role in the initiation of cell-intrinsic immune response (Figure 4F-G). In fact, IFIT5 is a RNA binding protein that recognizes endogenous RNAs such as 5’triphosphate-RNA, which is also recognized by RIG-I,34,35 a fundamental RNA sensor and initiator of the antiviral response that triggers IFNβ. Consistently, in cells expressing PHF8 DD, both RNA-binding proteins displayed peak expression levels at earlier time points (Figure 4F-G). Furthermore, TRIM25, a key activator of RIG-I, was also directly upregulated by PHF8 DD at the same time points (Figure 4F).
The most notable directly upregulated genes within these clusters were CMPK2 and IFIT5 (Figure 4F). Consistently in THP1 cells, increased phosphorylation of endogenous PHF8 upon OKA/ATRA treatment results in increased expression of CMPK2 and IFIT5 (Figure 4H-I).
Knockout of CMPK2 or that of IFIT5 reversed the effect of PHF8, confirming the crucial role of these 2 genes in PHF8-mediated growth impairment of human AML (Figure 4J-K; supplemental Figure 4D-F).
Thus, our comparative multiomics and functional analyses confirmed that phospho-PHF8 directly induces gene expression signatures of functionally interconnected clusters. The most prominently upregulated genes among these clusters were associated with cell-intrinsic immune response, most eminently CMPK2 and IFIT5.
Pharmacological support of PHF8 phosphorylation orchestrates cell-intrinsic immune response in human AML
Next, we investigated whether the cell-intrinsic immune response triggered by PHF8 phosphorylation could be pharmacologically induced in AML cells. We performed a gene set enrichment analysis36 correlation comparing the published transcriptome data of AML cells treated with high ATRA concentration of 10–6 M (THP1-ATRA)37 and the transcriptome of AML cells carrying our PHF8 phospho mutants. Indeed, the gene signature of THP1-ATRA cells significantly overlapped with THP1-PHF8 DD cells and contrasted with the THP1-PHF8 AA signature (Figure 5A). Thus, we wondered whether simultaneous induction of PHF8 and ATRA treatment could enhance AML destruction synergistically. Indeed, increased expression of PHF8 sensitized AML cells to very low concentrations of ATRA (10–8 M) (Figure 5B-C). Phosphoproteome analysis of AML cells expressing PHF8 under different ATRA concentrations identified 15 phospho-residues with increased phosphorylation upon ATRA treatment, with a further increase at higher ATRA concentrations (supplemental Figure 5A). These results indicate that low levels of ATRA are sufficient to trigger site-specific phosphorylation of PHF8. Thus, these joint forces within the same pathway decisively enhanced the effect, resulting in wide elimination of AML cells.
Pharmacological mediated phosphorylation of PHF8 orchestrates cell-intrinsic immune response in human AML. (A) Barcode plot showing GSEA of ATRA gene signature. (B) Bar charts illustrating the impact of ATRA treatment on human AML cells. It displays normalized colony numbers of THP1 cells transduced with empty vector (EV) (control) or PHF8 wt (PHF8) and treated with ATRA (12 hours) at the indicated concentration (10–8 M). Nontreated cells were taken as the reference for each group. Error bars are representative of 4 independent experiments. Tukey multiple comparison test (2-way ANOVA, ∗P < .01; ∗∗∗∗P < .0001). (C) Bar charts display normalized percentage of Annexin-V–positive cells after flow cytometry analysis. Nontreated cells were taken as the reference for each group. Error bars indicate standard deviation of 4 independent experiments. Tukey multiple comparison test (2-way ANOVA, ∗P < .03; ∗∗∗P < .0005; ∗∗∗∗P < .0001). (D) Scatterplot showing enriched GO terms of significantly regulated phosphoproteins in THP1 PHF8 cells upon 12 hours of ATRA treatment (10–8 M). Changes of phosphoproteins were compared with the unchanged phosphoproteome in the data set based on GO, KEGG, and keyword terms; y-axis shows the negative log of P value obtained from the Fisher exact test. (E) Dot plot showing changes in phosphosites of displayed proteins (12 hours of ATRA) in THP1 cells transduced with EV or PHF8 (PHF8). Red color and asterisk highlight specific IFIT5 targets. (ANOVA test permutation-based FDR <0.05). Sizes and colors of the dots are proportional to the averaged phosphosite intensity, z score (log2 intensity; n = 4 biological replicates per group). (F) Representative western blot analyses of time-course experiments (n = 3) after treatment with indicated ATRA concentration. Actin was used as a loading control for total extract and Lamin B1 for nuclear extract of THP1 cells expressing PHF8. (G) Western blot analysis of NF-κB (p65) protein levels (12 hours of ATRA) in nuclear (Lamin B1 loading control) and cytosolic (tubulin loading control) extracts of THP1 cells (n = 3). Cells were transduced with EV (control) or PHF8 wt (PHF8). (H) Phosphoproteome map of early IFN-I response and apoptosis in PHF8 AML cells after ATRA stimulation. Significantly regulated phosphoresidues are depicted on proteins participating in the process (n = 4 biological replicates per group; 1-way ANOVA, permutation-based FDR <0.05). Arrows indicate protein-protein interactions and phosphorylation events curated from experimentally defined databases. GSEA, gene set enrichment analysis; RELA, REL-associated; S, serine; T, threonine; Y, tyrosine.
Pharmacological mediated phosphorylation of PHF8 orchestrates cell-intrinsic immune response in human AML. (A) Barcode plot showing GSEA of ATRA gene signature. (B) Bar charts illustrating the impact of ATRA treatment on human AML cells. It displays normalized colony numbers of THP1 cells transduced with empty vector (EV) (control) or PHF8 wt (PHF8) and treated with ATRA (12 hours) at the indicated concentration (10–8 M). Nontreated cells were taken as the reference for each group. Error bars are representative of 4 independent experiments. Tukey multiple comparison test (2-way ANOVA, ∗P < .01; ∗∗∗∗P < .0001). (C) Bar charts display normalized percentage of Annexin-V–positive cells after flow cytometry analysis. Nontreated cells were taken as the reference for each group. Error bars indicate standard deviation of 4 independent experiments. Tukey multiple comparison test (2-way ANOVA, ∗P < .03; ∗∗∗P < .0005; ∗∗∗∗P < .0001). (D) Scatterplot showing enriched GO terms of significantly regulated phosphoproteins in THP1 PHF8 cells upon 12 hours of ATRA treatment (10–8 M). Changes of phosphoproteins were compared with the unchanged phosphoproteome in the data set based on GO, KEGG, and keyword terms; y-axis shows the negative log of P value obtained from the Fisher exact test. (E) Dot plot showing changes in phosphosites of displayed proteins (12 hours of ATRA) in THP1 cells transduced with EV or PHF8 (PHF8). Red color and asterisk highlight specific IFIT5 targets. (ANOVA test permutation-based FDR <0.05). Sizes and colors of the dots are proportional to the averaged phosphosite intensity, z score (log2 intensity; n = 4 biological replicates per group). (F) Representative western blot analyses of time-course experiments (n = 3) after treatment with indicated ATRA concentration. Actin was used as a loading control for total extract and Lamin B1 for nuclear extract of THP1 cells expressing PHF8. (G) Western blot analysis of NF-κB (p65) protein levels (12 hours of ATRA) in nuclear (Lamin B1 loading control) and cytosolic (tubulin loading control) extracts of THP1 cells (n = 3). Cells were transduced with EV (control) or PHF8 wt (PHF8). (H) Phosphoproteome map of early IFN-I response and apoptosis in PHF8 AML cells after ATRA stimulation. Significantly regulated phosphoresidues are depicted on proteins participating in the process (n = 4 biological replicates per group; 1-way ANOVA, permutation-based FDR <0.05). Arrows indicate protein-protein interactions and phosphorylation events curated from experimentally defined databases. GSEA, gene set enrichment analysis; RELA, REL-associated; S, serine; T, threonine; Y, tyrosine.
Phosphoproteome analysis of specific biological features, comparing THP1 cells expressing PHF8 (PHF8) or THP1 empty vector control after 12 hours of exposure to different ATRA concentrations, revealed significantly enriched phosphorylation levels on proteins and specific kinase motifs involved in the IFN-I response pathways (Figure 5D; supplemental Figure 5B-E).
IFIT5 not only functions as an RNA-binding protein but also as a positive regulator of IFN-I response, pulsing NF-κB activation via increase of phosphorylation and activation of IκB kinase (IKK).38 Consistently, our comprehensive phosphoproteome analyses demonstrated increased phosphorylation of known IFIT5 targets, TAK1(MAP3K7) at its functionally relevant residue S439, and IKKB (IKBKB) at active residues S672 and S679 (Figure 5E). Additionally, our data revealed full activation of the pathway downstream of IKK, including phosphorylation of the NF-κB inhibitor IκB-epsilon at S157 and IκB-beta at S183 as degradation signals (Figure 5E).
The phosphoproteome data show increased phosphorylation of the NF-κB subunit p65 at Ser45, a critical phosphorylation site known to disrupt DNA binding and transcriptional activity39 (Figure 5E). Additionally, we observed phosphorylation of GSK3β at Tyr216, which facilitates the phosphorylation and subsequent degradation of NF-κB subunits,40 including Ser903 and Ser907, which prime NFKB1 for Skp, cullin, F-box (SCF) containing complex, a cluster of NF-κB–specific proteasome proteins. Notably, the SCF complex was directly upregulated by phospho-PHF8 (Figure 5E). These results suggest an association between ATRA-induced phosphorylation of PHF8 and the degradation of NF-κB.
Time-course experiments showed IFIT5 expression peaking within the first 15 minutes of ATRA treatment (Figure 5F). NF-κB activation analysis revealed increasing nuclear levels of NF-κB subunit p65 reaching a maximum at 60 minutes of ATRA treatment in AML cells with ectopic PHF8 expression. This finding is consistent with the role of IFIT5 in early activation of NF-κB during early IFN-I response. Nuclear NF-κB subunit p65 levels likewise decreased gradually within the next hours and vanished entirely after 12 hours of ATRA treatment (Figure 5G). This indicates a tight control of NF-κB activity (expression) in the nucleus in the presence of ATRA and PHF8.
These results emphasize that pharmacological activation of PHF8 phosphorylation triggers early IFIT5 expression, subsequently leading to a cascade of phosphorylation-mediated activation and inhibition of key regulators involved in the IFN response (Figure 5H).
A distinct PHF8/IFN-I response protein signature is frequent in patients with AML and provides novel therapeutic opportunities
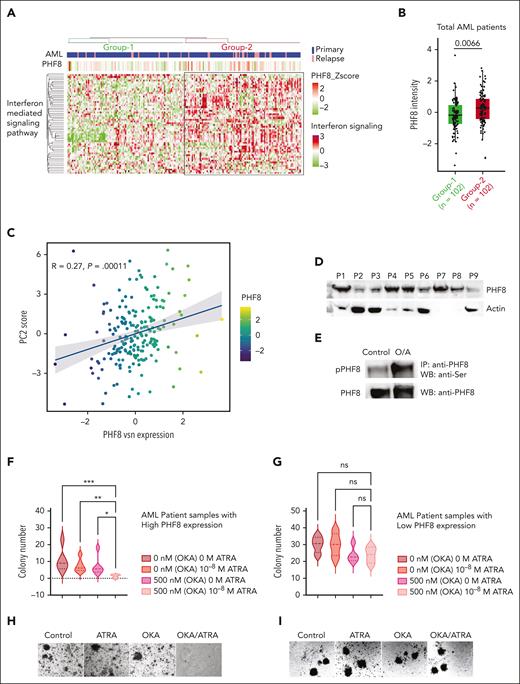
To consolidate the finding that IFN signaling pathway proteins are regulated by PHF8, we performed unsupervised enrichment analysis of our proteome data set from bone marrow biopsies of 177 patients with uniformly treated AML16 and from proteome of patients with relapsed AML, totaling 204 patient samples. This analysis identified 2 major groups of IFN signaling protein expression patterns (Figure 6A; supplemental Table 2). Based on this unbiased grouping of samples, we identified a positive correlation of elevated IFN signaling protein cluster sample group associated with significantly higher PHF8 expression levels (Figure 6A-B). Clinical characteristics did not show relevant differences regarding commonly defined AML subtypes (supplemental Figure 6A) or survival between the 2 groups (supplemental Figure 6B-D). PCA showed a clear dichotomization of the AML cohort with significantly higher expression of PHF8 in cluster 2 (Figure 6C; supplemental Figure 6E), whereas PCA of CD34+ hematopoietic progenitor cells from healthy volunteers did not show clustering (supplemental Figure 6F). Combined OKA/ATRA treatment increased PHF8 phosphorylation, reducing clonogenic growth in samples from patients with AML with high PHF8 levels, whereas single-agent ATRA or OKA had no significant impact. No change in clonogenic growth was observed in samples with low PHF8 levels (Figure 6D-I). Thus, we identified 2 AML proteome subgroups (high-PHF8/IFN-I and low-PHF8/IFN-I) based on the IFN signaling cluster. Both subtypes account for about half the population of patients with AML and reflect specific biological signatures spanning boundaries of common genetic AML subtypes and, most likely, also distinct susceptibility to ATRA/phospho-PHF8-based immunotherapeutic approaches.
PHF8/IFN-I response protein signature is frequent in patients with AML and provides novel opportunities for future targeted therapies. (A) Unsupervised hierarchical clustering of samples from patients with AML (n = 204 samples) based on proteins participating in GO term IFN-I–mediated signaling pathway. The clustering identifies 2 major sample groups (group-1 and group-2) as highlighted in the heat map. Annotations at the top of the heat map show patient disease status and PHF8 protein expression. (B) Box plot showing the differential expression of PHF8 in AML samples based on IFN protein expression cluster (group-1 = 102 patient samples vs group-2 = 102 patient samples). Two sample t test, permutation-based FDR <0.05 represented as significant. (C) Dot plot for PHF8 expression vs PC2 scores of 204 samples from patients with AML (primary, n = 177; relapsed AML, n = 27) based on the IFN-I signature subproteome. (D) Immunoblot analysis of total PHF8 levels of 9 samples (P1-P9) from patients with AML. Actin was used as loading control. (E) Immunoprecipitated PHF8 from cells from patients with primary AML expressing high PHF8 levels, untreated (control) or under combinatorial treatment with OKA/ATRA (O/A), and immunoblotted for phospho-Ser (upper panel) as well as for total PHF8 protein after membrane stripping (lower panel). (F) Violin plot depicting total numbers of colonies of 5 samples from patients with primary AML with high expression of PHF8 (n = 5). Significance was tested using the Tukey multiple comparison test (2-way ANOVA, ∗P < .02; ∗∗P < .008; ∗∗∗P < .0002). (G) Samples from patients with AML with low expression of PHF8 (n = 4), treated with the indicated concentrations of ATRA and OKA (OKA 20 minutes incubation). Tukey multiple comparison test, ns. (H-I) Colony morphology of primary human AML cells expressing high (H) or low (I) PHF8 levels. Cells were treated with ATRA (10–8 M) or/and OKA (500 nM, 20 minutes incubation). IP, Immunoprecipitation; WB, western blot.
PHF8/IFN-I response protein signature is frequent in patients with AML and provides novel opportunities for future targeted therapies. (A) Unsupervised hierarchical clustering of samples from patients with AML (n = 204 samples) based on proteins participating in GO term IFN-I–mediated signaling pathway. The clustering identifies 2 major sample groups (group-1 and group-2) as highlighted in the heat map. Annotations at the top of the heat map show patient disease status and PHF8 protein expression. (B) Box plot showing the differential expression of PHF8 in AML samples based on IFN protein expression cluster (group-1 = 102 patient samples vs group-2 = 102 patient samples). Two sample t test, permutation-based FDR <0.05 represented as significant. (C) Dot plot for PHF8 expression vs PC2 scores of 204 samples from patients with AML (primary, n = 177; relapsed AML, n = 27) based on the IFN-I signature subproteome. (D) Immunoblot analysis of total PHF8 levels of 9 samples (P1-P9) from patients with AML. Actin was used as loading control. (E) Immunoprecipitated PHF8 from cells from patients with primary AML expressing high PHF8 levels, untreated (control) or under combinatorial treatment with OKA/ATRA (O/A), and immunoblotted for phospho-Ser (upper panel) as well as for total PHF8 protein after membrane stripping (lower panel). (F) Violin plot depicting total numbers of colonies of 5 samples from patients with primary AML with high expression of PHF8 (n = 5). Significance was tested using the Tukey multiple comparison test (2-way ANOVA, ∗P < .02; ∗∗P < .008; ∗∗∗P < .0002). (G) Samples from patients with AML with low expression of PHF8 (n = 4), treated with the indicated concentrations of ATRA and OKA (OKA 20 minutes incubation). Tukey multiple comparison test, ns. (H-I) Colony morphology of primary human AML cells expressing high (H) or low (I) PHF8 levels. Cells were treated with ATRA (10–8 M) or/and OKA (500 nM, 20 minutes incubation). IP, Immunoprecipitation; WB, western blot.
Discussion
The reversibility of epigenetic marks that control the cell-intrinsic immune response provides a promising platform for personalized cancer immunotherapies. Our study reveals PHF8 as the first epigenetic regulator driving cell-intrinsic immune response, promoting differentiation and apoptosis in a wide range of AML subtypes. Targeting PHF8 holds therapeutic potential for AML and provides a new direction for the development of epigenetic-based immunotherapies.
Through systematic multiomics and functional analyses, we reveal that the collaborative effects of catalytic activity and site-specific phosphorylation of PHF8 drive the acute IFN-I response as the primary mechanism underlying the antileukemic activity of the protein in AML.
Histone H3K9me2 is an epigenetic signature in IFN response associated with suppression of IFN and ISG.7 Active IFN and ISG promoters exhibit reduced levels of H3K9me2 and enriched H3K4me3 marks.7 Given its ability to target H3K9me2 and its distinct characteristics, such as a more flexible interdomain structure, PHF8 represents an ideal candidate for activating the IFN response more efficiently than other KDMs such as LSD1 or KDM7A.14,41 We provide evidence that phospho-PHF8 specifically targets the repressive H3K9me2 mark, leading to the upregulation of IFIT5 and CMPK2.
Our findings shed light on the role of PHF8 in coordinating the RNA sensing machinery for initiation of cell-intrinsic immune response. We observed a concomitant upregulation of IFIT5, RIG-I, and TRIM25 after PHF8 phosphorylation. IFIT5 binds to viral RNA and endogenous 5′-phosphate RNAs,34,35 suggesting its involvement in triggering an innate immune reaction upon detection of endogenous RNA molecules. The simultaneous upregulation of RIG-I, a critical sensor for viral RNA also binding 5′-phosphatase RNAs,42 supports the notion of a coordinated response to RNA patterns. Importantly, this observation aligns with previous reports indicating the colocalization of both proteins with mitochondria, in which they synergistically interact with mitochondrial antiviral-signaling protein to enhance the antiviral response.30,34 TRIM25 serves as a RING-finger E3 ubiquitin ligase crucial for the ubiquitination, stabilization, and activation of RIG-I, thereby facilitating the production of IFN-β.31 All 3 proteins exhibit upregulation during ATRA-induced cell differentiation,43 strongly implying a synergistic interplay between the effects of ATRA and PHF8 in initiation of the IFN-I response via the TRIM25-RIG-I-IFIT5 axis. Previous studies have emphasized the importance of dsRNA recognition proteins as key inducers of FADD and caspase-8, which are essential initiators of IFN-I–induced apoptosis.26,27 Our findings provide compelling evidence that phosphorylated PHF8 induces the upregulation of the entire machinery involved in IFN-induced apoptosis and differentiation.
Furthermore, the progressive upregulation of CMPK2, a mitochondrial protein involved in nucleotide metabolism, cellular homeostasis, and antiviral response,29,44 after PHF8 phosphorylation suggests its involvement in cellular energetics and metabolism, reflecting a coordinated adaptation of the cell to mounting antiviral responses. Further investigation of the molecular mechanisms underlying the regulation of CMPK2 by PHF8 will yield valuable insights into the intricate network of immune regulatory pathways and their coordinated role in antiviral defense.
We unveil a novel multifunctional role of IFIT5 as a positive regulator of the early IFN-I response. Consistent with its ability to facilitate NF-κB activation through interactions with IKK and TAK1,38 we demonstrate increased phosphorylation of active residues in TAK1 and IKK, indicating that IFIT5 effectively enhances IKK phosphorylation. Moreover, we observe temporal regulation of IFIT5 and nuclear NF-κB subunit p65 upon pharmacological induction of PHF8 phosphorylation, suggesting a coordinated control of NF-κB activity in the presence of ATRA and PHF8. These mechanistic insights provide a deeper understanding of the regulatory network underlying NF-κB activation and highlight the interplay between IFIT5, PHF8, and these signaling molecules.
Our study also implicates that phosphorylated PHF8 stimulates NF-κB inhibition and induces its degradation through distinct mechanisms. Specifically, we observe phosphorylation of the NF-κB subunit p65 at Ser45, consistent with previous studies demonstrating its disruptive effect on DNA binding and transcriptional activity.39 Additionally, we identify phosphorylation events on NFKB1 and GSK3β, contributing to the degradation of NF-κB subunits and their recognition by the SCF complex, a cluster of NF-κB–specific proteasome proteins.45 These findings emphasize the multifaceted role of PHF8 in regulating NF-κB activity and degradation.
Finally, our unsupervised analysis of proteome data from patients with AML reveals the presence of a PHF8/IFN-I signature in ∼50% of all AML cases. Importantly, this clustering is absent in CD34+ hematopoietic progenitor cells from healthy volunteers. These findings, combined with the significant response of primary patient cells with high expression of PHF8 to phospho-PHF8/ATRA treatment, suggest the susceptibility of a substantial proportion of patients with AML to regimens triggering the phospho-PHF8/IFN-I pathway. Our study provides a novel strategy promoting differentiation and apoptosis via induction of cell-intrinsic immune response and holds promise as a targeted approach for future AML therapies.
Acknowledgments
The authors thank the MedK program of the Medical Faculty of Münster University for support of Franca Seifert; Igor Paron and Mario Oroshi, Department of Proteomics and Signal Transduction at the Max Planck Institute for Biochemistry for excellent mass spectrometry and technical support; Stefan Froehling, Emily Bernstein, Hugh Brady, and Kristian Helin for CDX2, CBX7, E2A-HLF, and PHF8 constructs respectively; and all members of the Mikesch-Arteaga laboratory for constructive discussion.
This work was funded by The Deutsche Forschungsgemeinschaft (no. AR 969/2-1). The project was also supported by IZKF Münster (no. Art1/019/18) and Innovative Medizinische Forschung Universität Münster (no. AR 12 13 11).
Authorship
Contribution: J.-H.M. and M.F.A. conceptualized the study; E.F.F., A.K.J., J.-H.M., and M.F.A. prepared the methodology; E.F.F., F.S., V.A., and M.F.A. provided validation; C.W., J.M.F., L.A., M.D., M.M., A.K.J., and M.F.A. conducted formal analysis; E.F.F., F.S., V.A., A.K.J., J.-H.M., and M.F.A. conducted investigation; W.H., S.W., H.S., T.O., G.L., C.M.-T., C.S., O.H., M.M., J.-H.M., and M.F.A. acquired resources; J.-H.M. and M.F.A. wrote the original draft; all authors wrote, reviewed, and edited the manuscript; J.M.F., T.H., A.K.J., and M.F.A. performed visualization; J.-H.M. and M.F.A. supervised the project; and M.F.A. administered the project and acquired funding.
Conflict-of-interest disclosure: T.O. received research funding from Gilead (related to this work) and Merck KGaA (not related to this work). T.O. is a consultant for Roche and Merck KGaA (both not related to this work). The remaining authors declare no competing financial interests.
Correspondence: Maria Francisca Arteaga, Department of Medicine A, University Hospital Muenster, Albert-Schweitzer-Campus 1, 48149 Muenster, Germany; email: marifrancis.arteaga@uni-muenster.de.
References
Author notes
A.K.J., J.-H.M., and M.F.A. are joint senior authors and jointly supervised this work.
Proteomic data sets that were generated during this project are accessible via the Proteomics Identifications Database (PRIDE) for the AML cell lines’ proteome (accession number PXD035635) and the discovery cohort proteome of 177 patients with AML (accession number PXD023201).
Raw and processed genomic data of RNA sequencing and ChIP sequencing are available in the Gene Expression Omnibus database (accession number GSE211643).
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Maria Francisca Arteaga (marifrancis.arteaga@uni-muenster.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


![Epigenetic activity of phosphorylated PHF8 counteracts differentiation block via induction of cell-intrinsic immune response in AML. (A-C) Bar charts represent colony numbers in a serial replating assay and typical morphology of cells from fourth round of replating; primary mHSPCs were cotransduced with the indicated AML oncogenes or oncofusions as well as with wt PHF8 (PHF8), PHF8-F279S (F279S), or empty vector (control). Error bars indicate standard deviation (SD) of 3 independent experiments. Significance was tested using the Tukey multiple comparison test (2-way analysis of variance [ANOVA], ∗P < .04; ∗∗∗∗P < .000, nonsignificant [ns]). (D) Bar charts depict colony numbers of human AML cells transduced with PHF8 vs F279S vs Control. Error bars indicate SD of at least 4 independent experiments. Significance was tested using the Tukey multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001; ∗∗∗P < .0004; ∗∗P < .006; ∗P < .04). (E) Bar charts represent early apoptosis as percentage of Annexin-V–positive cells from flow cytometry analysis. Error bars indicate SD of 4 independent experiments. Sidak multiple comparison test (2-way ANOVA, ∗∗∗∗P < .0001 each cell group). (F) Kaplan-Meier curves for disease-free survival (DFS) of NSG mice (Charles River Laboratories) that received transplantation with human THP1 cells transduced with indicated constructs (n = 10 mice per group). Six-to-8-week-old NSG mice were injected with 1 × 105 cells, after 5 days antibiotic selection, via tail vein injection. Before transplantation, mice were conditioned with sublethal radiation (2.5 Gy). The log-rank (Mantel-Cox) test was used to compare survival curves. (G) Pathways enrichment analysis (Fisher exact test) of control, PHF8, and F279S cells (n = 3 biological replicates per group). Significantly regulated proteins were compared with the unchanged proteome in the data set based on the gene ontology (GO), KEGG, and keyword terms. (H) Bar charts represent colony numbers of primary mHSPCs cotransduced with MLL-fusions or CDX2 and sgRNAs for specific CRISPR-Cas9–mediated knockout of PHF8 (sgRNA 1, sgRNA 2) or nontargeting control (control). Error bars indicate SDs of 3 independent experiments. Significance was tested using Tukey multiple comparison test (2-way ANOVA, ns). (I) Bar charts represent colony numbers of human AML cells transduced with lenti-CRISPRv2 nontargeting (control) or lenti-CRISPRv2 guide targeting endogenous PHF8 (sgRNA 1 or sgRNA 3). Error bars indicate SDs of 4 independent experiments. Significance was tested using Tukey multiple comparison test (2-way ANOVA, ns); (lower panel) immunoblot analyses showing PHF8 levels. (J) Profile plot showing significant increases of phosphorylation in human THP1 AML cells at defined phosphosites of endogenous PHF8 upon 12 hours of ATRA treatment compared with controls. Each data point is the averaged median of biological quadruplicate of z scored log 2 phosphosite intensities; significance was tested using multiple sample t test (1-way ANOVA, permutation-based false discovery rate [FDR] <0.05). (K) Violin plots depict numbers of colonies of human AML cells treated with 500 nM OKA (OKA 20 minutes incubation) or/and ATRA 10–8 M. Significance was tested using the Tukey multiple comparison test (2-way ANOVA, ∗P < .05; ∗∗P < .0085, ns). (L) Immunoblot of endogenous total PHF8 protein levels. (M) Western blot analyses (upper panel) of PHF8 levels before and after knocking out in AML cells; (lower panel) typical INT-stained colony pictures. KEGG, Kyoto Encyclopedia of Genes and Genomes; KG-1, Koeffler and Golde 1; MLL, mixed lineage leukemia; sgRNA, single guide RNA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/22/10.1182_blood.2023021640/2/m_blood_bld-2023-021640-gr1b.jpeg?Expires=1769092643&Signature=3KkHUgMxYNd4n05JYSoc3Y67M7i1UA6vSQsEIZiFXCgMBcATFRy5su~kp65yznMch2fn9n6rqhJtiBUxNklD39F3mHvM59GjAQhGdCLUA-FEDwtWouELGlxEYg643vLztQyBNBtspVqi2EL6Uck2KxXZg9P5A~Cjzi14q3KZsT1oUEYJtS6jVW~-VoMI2YF~NrKmx5CAezvSSAwKWBqaQ-0Dq6S~PWYOqoDk38kj-8ZcbsSD7D5vKVy07ivNeE6IqLVfapO6MTRPptqgX6bVpMleYecpp4X1QpeqEsFpClGHjiRcSS7uPYIPlXyvWTbOOiG0ApJiiB~w5hv71G91Tw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal