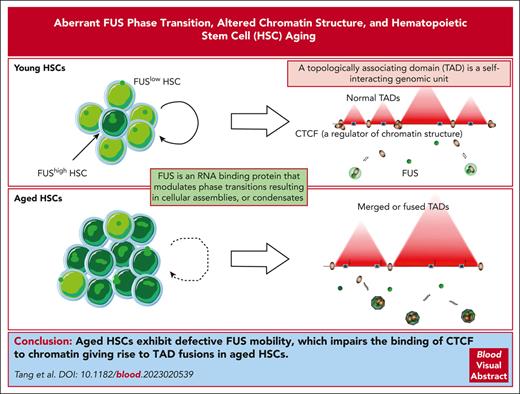
FUShigh HSCs exhibit compromised FUS mobility and resemble aged HSCs both functionally and transcriptionally.
Aberrant FUS condensates diminish the binding of CTCF with chromatin and incite TAD-fusion events in aged HSCs.
Visual Abstract
Aged hematopoietic stem cells (HSCs) exhibit compromised reconstitution capacity. The molecular mechanisms behind this phenomenon are not fully understood. Here, we observed that the expression of FUS is increased in aged HSCs, and enforced FUS recapitulates the phenotype of aged HSCs through arginine-glycine-glycine–mediated aberrant FUS phase transition. By using Fus-gfp mice, we observed that FUShigh HSCs exhibit compromised FUS mobility and resemble aged HSCs both functionally and transcriptionally. The percentage of FUShigh HSCs is increased upon physiological aging and replication stress, and FUSlow HSCs of aged mice exhibit youthful function. Mechanistically, FUShigh HSCs exhibit a different global chromatin organization compared with FUSlow HSCs, which is observed in aged HSCs. Many topologically associating domains (TADs) are merged in aged HSCs because of the compromised binding of CCCTC-binding factor with chromatin, which is invoked by aberrant FUS condensates. It is notable that the transcriptional alteration between FUShigh and FUSlow HSCs originates from the merged TADs and is enriched in HSC aging-related genes. Collectively, this study reveals for the first time that aberrant FUS mobility promotes HSC aging by altering chromatin structure.
Introduction
Aged hematopoietic stem cell (HSC) exhibits impaired reconstitution capacity and differentiation bias toward myeloid lineage, wherein the molecular mechanism remains largely unknown.1,2 Several studies have shown that aged HSCs exhibit epigenetic drift, which results in transcriptome alteration and compromised function.3-7 Genome organization is the process of compacting the long linear genomic DNA into a 3-dimensional (3D) micrometer-sized nucleus, which is regulated by a variety of molecular mechanisms.8 Phase separation, which provides a venue for biochemical reactions through the dynamic changes of membrane-less compartments,9 has been shown to be involved in the regulation of chromatin structure.10-12 The internal connection between them has not been investigated.
Here, we investigate the functional role of FUS in HSC aging and, to our knowledge, for the first time demonstrate that aberrant FUS condensate promotes HSC aging by reorganizing chromatin structure. We also point out that this mechanism might be generalized to aging and aging-related diseases.
Methods
Mice
To generate Fus reporter mice (Fus-gfp mice), the TAG stop codon was replaced with the 3xEAAAK-EGFP cassette.
Flow cytometric analysis and cell sorting
Peripheral blood (PB) cells and bone marrow (BM) cells were harvested and stained with antibodies as mentioned in supplemental Table 1, available on the Blood website.
Lentivirus production
CDS were cloned into the pRRL-PPT-SF-newMCS-IRES2-EGFP vector, and Fus shRNAs were cloned into the SF-LV-miRE-mCherry vector. Lentiviruses were produced in 293T cells.
Transplantation experiment
HSCs (CD45.2) were injected into lethally irradiated recipient mice (CD45.1/2 or CD45.2) together with competitor cells (CD45.1).
Western blotting
Samples were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transferring onto a polyvinylidene fluoride membrane. Membranes were blocked and then probed with the indicated primary antibodies. All antibodies are listed in supplemental Table 2.
Reverse transcriptase polymerase chain reaction
Freshly isolated HSCs were lysed in Trizol. RNA was extracted by phenol-chloroform and reversely transcribed to complementary DNA by PrimeScript RT reagent Kit (Takara). PowerUp SYBR Green mix was used for reverse transcriptase polymerase chain reaction.
RNA-sequencing
The transcriptome of HSCs was conducted by the Illumina Hiseq platform with a 150-bp paired-end (Berry Genomics, Beijing, China).
tagHi-C assay
CUT&Tag
The CUT&Tag was performed according to the protocol of CUT&Tag Assay Kit (TD903, Vazyme).
CUT&RUN
Coimmunoprecipitation
293T cells transferred by SFB-tagged and Myc-tagged plasmids were collected, and the lysis supernatant was incubated with S-protein agarose beads. Beads were collected and washed with NETN buffer for 3 times. Then, 2× loading buffer was added to the beads and boiled for western blot.
DNA pull-down
293T cells transferred with SFB-tagged and Myc-tagged plasmids were collected, and the supernatant was incubated with streptavidin sepharose beads and biotin M1 motif. Beads were collected for western blot as described above.
Statistical analysis
Statistical analysis was made using the Prism software. Data are shown as mean ± standard error of the mean. The student t tests (2-tailed unpaired) were used for comparisons, and the Wilcoxon rank-sum tests were used for Hi-C comparisons. All experiments were repeated 2 or 3 times independently.
Results
Aging-increased FUS impairs HSCs
We observed that FUS protein, not Fus mRNA, is significantly increased in aged HSCs but not ST-HSC, MPP, common myeloid progenitor (CMP), GMP, or megakaryocyte-erythrocyte progenitor (MEP) cells (Figure 1A; supplemental Figure 1A-B). To confirm this observation, we found that the protein level of FUS is indeed increased in aged HSCs in a published proteomics database17 (supplemental Figure 1C).
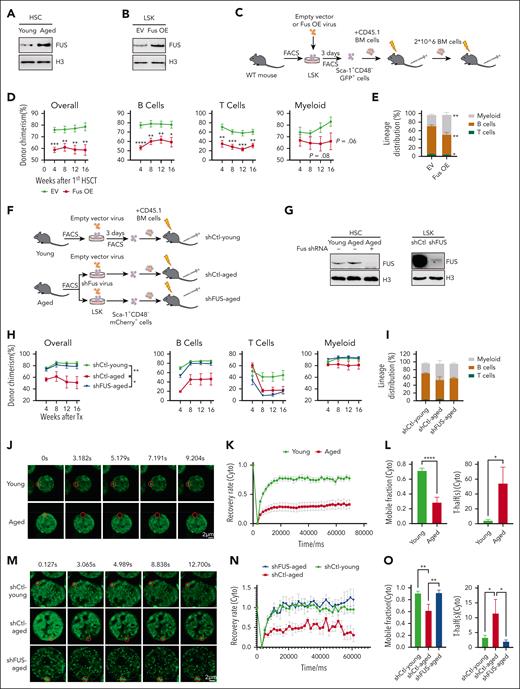
Aged hematopoietic stem cells exhibit compromised mobility of FUS. (A) Representative western blot showing the expression of FUS in young and aged HSCs. Freshly isolated 20 000 CD34– LSK (Lineage− Sca-1+ c-Kit+, hematopoietic stem and progenitor cell-enriched population) cells from young (2 months) and aged (24-28 months) wild-type (WT) mice were lysed in 2 × sodium dodecyl sulfate loading buffer. Lysis was completed by sonication, denatured by boiling, and subjected to a western blot assay to detect FUS. (B) Representative western blot showing the expression of FUS in FUS-overexpressed LSK cells. Freshly isolated LSK cells from young (2 months) WT mice were infected by FUS-carrying lentivirus (Fus OE) and empty vector (EV), and 3 days later, 105 GFP+ cells were purified for western blot assay with indicated antibodies. (C) Experimental design of a competitive transplantation assay for FUS-overexpressed HSCs. Freshly isolated 105 LSK cells from young WT mice (2 months) were infected by FUS-carrying lentivirus (Fus OE). Three days later, 2000 GFP+ Sca-1+CD48– cells were purified and transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitor cells (CD45.1). Secondary transplantation was performed by using 2 × 106 total BM cells from the primary recipients. Chimera in peripheral blood was evaluated every 4 weeks until the 16th week. (D) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in peripheral blood of primary recipients at indicated time points. Refer to supplemental Figure 1E. N = 8 recipients per group, data are shown as mean ± SEM. (E) The histogram depicts the lineage distribution of donor-derived peripheral blood cells at the 16th week. N = 8 recipients per group, data are shown as mean ± SEM. Refer to supplemental Figure 1F. (F) Experimental design of competitive transplantation assay. Freshly isolated 105 LSK cells from young (2 months) and aged (18 months) WT mice were infected by shFus-carrying lentivirus, which is labeled by mCherry fluorescence. Three days later, 2000 mCherry+Sca-1+CD48– cells were purified and transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitor cells (CD45.1). Chimera in peripheral blood was evaluated every 4 weeks until the 16th week. (G) Representative western blot showing the knockdown efficiency of shRNA against Fus. Freshly isolated 2 × 104 HSCs (mCherry+ CD48– LSK) from the recipients of panel F at the end of the 16th week after transplantation (left) and 2 × 104 cultured mCherry+ LSK from young WT mice (right) were lysed for western blot assay with indicated antibodies. (H) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in peripheral blood of primary recipients at indicated timepoints. N = 7 recipients per group, data are shown as mean ± SEM. (I) The histogram depicts the lineage distribution of donor–derived peripheral blood cells at the 16th week. N = 7 recipients per group, data are shown as mean ± SEM. (J) Representative fluorescence recovery after photobleaching (FRAP) images show the fluorescence recovery of FUS condensates in cytoplasm of HSCs (CD34–LSK) from young (2 months) and aged (18 months) Fus-gfp mice after 0.5 mM ARS treatment for 6 hours. Scale bar represents 2 μm. Refer to supplemental Video 1. (K) The line plot depicts the recovery rate of FUS condensates in young (N = 21) and aged (N = 26) HSCs of panel J at indicated time points. Data are shown as mean ± SEM. (L) The histograms depict the mobile fraction (71.5% vs 28.7%) and T-half (4.3% vs 54.4%) of young and aged HSCs. T-half is the half-maximal recovery time. Data are shown as mean ± SEM. (M) Representative FRAP images show the fluorescence recovery of FUS condensates in the cytoplasm of shCtl-young, shCtl-aged, and shFUS-aged LSK cells after 0.5 mM ARS treatment for 6 hours. (N) The line plot depicts the recovery rate of FUS condensates in shCtl-young (N = 21), shCtl-aged (N = 15), and shFUS-aged (N = 13) LSK cells of panel M at indicated time points. (O) The histograms depict mobile fraction and T-half of shCtl-young, shCtl-aged, and shFUS-aged LSK cells. Data are shown as mean ± SEM. SEM, standard error of the mean. Freshly isolated 2 × 104 HSCs (mCherry+ CD48– LSK) from the recipients of panel F at the end of the 16th week after transplantation (left) and 2 × 104 cultured mCherry+ LSK from young WT mice (right) were lysed for western blot assay with indicated antibodies. Freshly isolated 2 × 104 HSCs (mCherry+ CD48– LSK) from the recipients of panel F at the end of the 16th week after transplantation (left) and 2 × 104 cultured mCherry+ LSK from young WT mice (right) were lysed for western blot assay with indicated antibodies. GFP, green fluorescent protein.
Aged hematopoietic stem cells exhibit compromised mobility of FUS. (A) Representative western blot showing the expression of FUS in young and aged HSCs. Freshly isolated 20 000 CD34– LSK (Lineage− Sca-1+ c-Kit+, hematopoietic stem and progenitor cell-enriched population) cells from young (2 months) and aged (24-28 months) wild-type (WT) mice were lysed in 2 × sodium dodecyl sulfate loading buffer. Lysis was completed by sonication, denatured by boiling, and subjected to a western blot assay to detect FUS. (B) Representative western blot showing the expression of FUS in FUS-overexpressed LSK cells. Freshly isolated LSK cells from young (2 months) WT mice were infected by FUS-carrying lentivirus (Fus OE) and empty vector (EV), and 3 days later, 105 GFP+ cells were purified for western blot assay with indicated antibodies. (C) Experimental design of a competitive transplantation assay for FUS-overexpressed HSCs. Freshly isolated 105 LSK cells from young WT mice (2 months) were infected by FUS-carrying lentivirus (Fus OE). Three days later, 2000 GFP+ Sca-1+CD48– cells were purified and transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitor cells (CD45.1). Secondary transplantation was performed by using 2 × 106 total BM cells from the primary recipients. Chimera in peripheral blood was evaluated every 4 weeks until the 16th week. (D) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in peripheral blood of primary recipients at indicated time points. Refer to supplemental Figure 1E. N = 8 recipients per group, data are shown as mean ± SEM. (E) The histogram depicts the lineage distribution of donor-derived peripheral blood cells at the 16th week. N = 8 recipients per group, data are shown as mean ± SEM. Refer to supplemental Figure 1F. (F) Experimental design of competitive transplantation assay. Freshly isolated 105 LSK cells from young (2 months) and aged (18 months) WT mice were infected by shFus-carrying lentivirus, which is labeled by mCherry fluorescence. Three days later, 2000 mCherry+Sca-1+CD48– cells were purified and transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitor cells (CD45.1). Chimera in peripheral blood was evaluated every 4 weeks until the 16th week. (G) Representative western blot showing the knockdown efficiency of shRNA against Fus. Freshly isolated 2 × 104 HSCs (mCherry+ CD48– LSK) from the recipients of panel F at the end of the 16th week after transplantation (left) and 2 × 104 cultured mCherry+ LSK from young WT mice (right) were lysed for western blot assay with indicated antibodies. (H) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in peripheral blood of primary recipients at indicated timepoints. N = 7 recipients per group, data are shown as mean ± SEM. (I) The histogram depicts the lineage distribution of donor–derived peripheral blood cells at the 16th week. N = 7 recipients per group, data are shown as mean ± SEM. (J) Representative fluorescence recovery after photobleaching (FRAP) images show the fluorescence recovery of FUS condensates in cytoplasm of HSCs (CD34–LSK) from young (2 months) and aged (18 months) Fus-gfp mice after 0.5 mM ARS treatment for 6 hours. Scale bar represents 2 μm. Refer to supplemental Video 1. (K) The line plot depicts the recovery rate of FUS condensates in young (N = 21) and aged (N = 26) HSCs of panel J at indicated time points. Data are shown as mean ± SEM. (L) The histograms depict the mobile fraction (71.5% vs 28.7%) and T-half (4.3% vs 54.4%) of young and aged HSCs. T-half is the half-maximal recovery time. Data are shown as mean ± SEM. (M) Representative FRAP images show the fluorescence recovery of FUS condensates in the cytoplasm of shCtl-young, shCtl-aged, and shFUS-aged LSK cells after 0.5 mM ARS treatment for 6 hours. (N) The line plot depicts the recovery rate of FUS condensates in shCtl-young (N = 21), shCtl-aged (N = 15), and shFUS-aged (N = 13) LSK cells of panel M at indicated time points. (O) The histograms depict mobile fraction and T-half of shCtl-young, shCtl-aged, and shFUS-aged LSK cells. Data are shown as mean ± SEM. SEM, standard error of the mean. Freshly isolated 2 × 104 HSCs (mCherry+ CD48– LSK) from the recipients of panel F at the end of the 16th week after transplantation (left) and 2 × 104 cultured mCherry+ LSK from young WT mice (right) were lysed for western blot assay with indicated antibodies. Freshly isolated 2 × 104 HSCs (mCherry+ CD48– LSK) from the recipients of panel F at the end of the 16th week after transplantation (left) and 2 × 104 cultured mCherry+ LSK from young WT mice (right) were lysed for western blot assay with indicated antibodies. GFP, green fluorescent protein.
To determine whether the increase of FUS promotes HSC aging, we cloned the complementary DNA of mouse FUS into a lentiviral vector, which exhibits comparable expression of FUS to aged HSCs (Figure 1B; supplemental Figure 1D). Competitive transplantation assay shows that enforced FUS significantly impairs the reconstitution capacity of HSCs and promotes HSC differentiation bias toward myeloid lineage (Figure 1C-E; supplemental Figure 1E-F). The secondary transplantation result reveals that the FUS-carrying cells were almost undetectable at the end of the 16th week (supplemental Figure 1G), indicating that enforced FUS severely impairs the self-renewal capacity of HSCs.
To further validate this observation from the opposite direction, we conducted a competitive transplantation assay by knocking down Fus in young and aged hematopoietic stem and progenitor cells (HSPCs) with an efficient shRNA (Figures 1F-G). The PB and BM analyses reveal that knockdown of FUS ameliorates the reconstitution capacity of aged HSCs and has no effect on young HSCs (Figure 1H, supplemental Figure 1H-J), but does not rescue the differentiation skewing (Figure 1I; supplemental Figure 1K). These data suggest that aging-increased FUS is a pro-aging factor in HSCs.
Aged HSCs exhibit aberrant phase transition of FUS
Because abnormal aggregation of FUS protein has been identified in multiple neurodegenerative diseases,18,19 we generated Fus-gfp mice to investigate whether FUS protein in aged HSCs exhibits aberrant mobility (supplemental Figure 1L; see details in Methods). The immunofluorescence result shows that FUS extensively colocalized with green fluorescent protein (GFP) in non-DAPI (4′,6-diamidino-2-phenylindole)–dense regions (supplemental Figures 1M-N), suggesting that the fusion of GFP at the C-terminal of FUS does not perturb the location of endogenous FUS. It is notable that we observed that the GFP signal is significantly increased in HSCs of aged Fus-gfp mouse (supplemental Figure 1O), indicating that FUS is increased upon aging, which is consistent with western blotting result (Figure 1A). Lineage composition and hematopoietic reconstitution of Fus-gfp mice are comparable to those of the control (supplemental Figure 1P, 1-1A-C), indicating that it is an ideal model to investigate the behavior of FUS in HSCs.
Time-lapse imaging reveals that FUS forms dynamic and reversible assemblies upon sodium arsenite (ARS) treatment in HSPCs, which are featured as undergoing frequent fusion and fission events (cytoplasm, supplemental Figure 1-1D; nucleoplasm, supplemental Figure 1-1E). The fluorescence recovery after photobleaching (FRAP) assay reveals that FUS exchanges rapidly for recovery within 30 seconds (supplemental Figure 1-1F). These data suggest that FUS undergoes liquid-liquid phase separation (LLPS) in HSPCs. To further investigate whether aged HSCs exhibit abnormal dynamic behavior of FUS, the FRAP assay shows that the recovery rate of FUS condensates of aged HSCs is significantly compromised compared with young HSCs (cytoplasm: Figures 1J-K; supplemental Video 1; nucleoplasm: supplemental Figures 1-1G and 1-1H; supplemental Video 2). In addition, the mobile fraction is drastically reduced and the half-maximal recovery time (T-half) is prolonged in aged HSCs (Figure 1L; supplemental Figure 1-1I), suggesting that the mobility of FUS in aged HSCs is significantly compromised. Notably, knockdown of FUS in aged HSCs rescues the dynamic behavior of FUS (cytoplasm: Figure 1M-O; nucleoplasm: supplemental Figures 1-1J-L).
To examine the influence of enforced FUS on its mobility in young HSCs, the FRAP assay shows that enforced FUS indeed compromises the dynamic behavior of FUS condensates (cytoplasm: supplemental Figures 1-1M-O; nucleoplasm: supplemental Figures 1-1P-R). These data indicate that the expression level of FUS in HSCs is the key determinant of its dynamic behavior.
FUShigh HSCs exhibit aberrant FUS phase transition and increase with aging
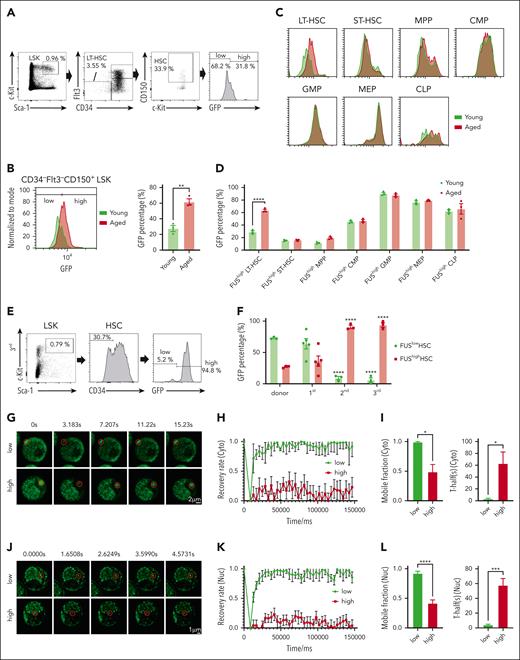
To determine whether the increase of FUS in aged HSCs is homogenous or heterogeneous, we observed that HSCs (CD34– Flt3– CD150+ LSK) are fractionated into 2 subpopulations: FUShigh and FUSlow HSCs (Figure 2A) and the percentage of FUShigh HSCs is significantly increased with aging (Figure 2B). Further analysis reveals that the percentage of FUShigh cells exhibits a conspicuous increase in LT-HSC but not in ST-HSC, MPP, CMP, GMP, MEP, or common lymphoid progenitor (CLP) with aging (Figures 2C-D; supplemental Figure 2A). These results indicate that FUS is heterogeneously expressed in HSCs, and the increase of FUS with aging attributes to the increase of FUShigh HSCs.
FUShigh HSCs exhibit compromised mobility of FUS. (A) Representative FACS plots showing the percentage of FUSlow and FUShigh HSCs (CD34–Flt3–CD150+LSK) in a young (2 months) Fus-gfp mouse. (B) The FACS plot and histogram show the GFP expression in HSCs (CD34–Flt3–CD150+LSK) between young (2 months) and aged (17-18 months) Fus-gfp mice. Data are shown as mean ± SEM, N = 3 mice per group. (C) The FACS plot shows the GFP expression in HSPCs, including CD34–Flt3–LSK (LT-HSC), CD34+Flt3–LSK (ST-HSC), CD34+Flt3+LSK (MPP), CD34+CD16/32– Lineage–Sca-1–c-Kit+ (CMP), CD34+CD16/32+Lineage–Sca-1–c-Kit+ (GMP), CD34–CD16/32– Lineage–Sca-1–c-Kit+ (MEP), and CD127+Flt3+Lineage–Sca-1loc-Kitlo (CLP). (D) The histogram depicts the percentage of FUShigh HSPCs between young (2 months) and aged (17-18 months) Fus-gfp mice. Data are shown as mean ± SEM, N = 3 mice per group. (E) Representative FACS plots showing the percentage of FUSlow and FUShigh HSCs (CD34–LSK) at the end of the third round of transplantation. (F) The histogram depicts the percentage of FUSlow and FUShigh HSCs (CD34–LSK) at the end of the indicated round of transplantation. Data are shown as mean ± SEM, N = 3 to 5 mice per group. (G) Representative FRAP images show the fluorescence recovery of FUS condensates in the cytoplasm of FUSlow and FUShigh LSK cells of 2-month-old Fus-gfp mice upon 0.5 mM ARS treatment for 6 hours. Scale bar represents 2 μm. See also supplemental Video 3. (H) The line plot shows the recovery rate of FUS condensates in FUSlow LSK cells (N = 6) and FUShigh LSK cells (N = 12) of panel G at indicated time points. Data are shown as mean ± SEM. (I) The histograms depict the mobile fraction (99.2% vs 48.9%) and T-half (3.383% vs 63.064%) of FUSlow and FUShigh LSK cells. Data are shown as mean ± SEM. (J) Representative FRAP images show the fluorescence recovery of FUS condensates in the nucleoplasm of FUSlow and FUShigh LSK cells of 2-month-old Fus-gfp mice upon 0.5 mM ARS treatment for 4 hours. Scale bar represents 1 μm. Refer to supplemental Video 4. (K) The line plot shows the recovery rate of FUS condensates in FUSlow LSK cells (N = 19) and FUShigh LSK cells (N = 47) of panel J at indicated time points. Data are shown as mean ± SEM. (L) The histograms depict the mobile fraction (92.2% vs 41.6%) and T-half (4.264% vs 57.8%) of FUSlow and FUShigh LSK cells. Data are shown as mean ± SEM.
FUShigh HSCs exhibit compromised mobility of FUS. (A) Representative FACS plots showing the percentage of FUSlow and FUShigh HSCs (CD34–Flt3–CD150+LSK) in a young (2 months) Fus-gfp mouse. (B) The FACS plot and histogram show the GFP expression in HSCs (CD34–Flt3–CD150+LSK) between young (2 months) and aged (17-18 months) Fus-gfp mice. Data are shown as mean ± SEM, N = 3 mice per group. (C) The FACS plot shows the GFP expression in HSPCs, including CD34–Flt3–LSK (LT-HSC), CD34+Flt3–LSK (ST-HSC), CD34+Flt3+LSK (MPP), CD34+CD16/32– Lineage–Sca-1–c-Kit+ (CMP), CD34+CD16/32+Lineage–Sca-1–c-Kit+ (GMP), CD34–CD16/32– Lineage–Sca-1–c-Kit+ (MEP), and CD127+Flt3+Lineage–Sca-1loc-Kitlo (CLP). (D) The histogram depicts the percentage of FUShigh HSPCs between young (2 months) and aged (17-18 months) Fus-gfp mice. Data are shown as mean ± SEM, N = 3 mice per group. (E) Representative FACS plots showing the percentage of FUSlow and FUShigh HSCs (CD34–LSK) at the end of the third round of transplantation. (F) The histogram depicts the percentage of FUSlow and FUShigh HSCs (CD34–LSK) at the end of the indicated round of transplantation. Data are shown as mean ± SEM, N = 3 to 5 mice per group. (G) Representative FRAP images show the fluorescence recovery of FUS condensates in the cytoplasm of FUSlow and FUShigh LSK cells of 2-month-old Fus-gfp mice upon 0.5 mM ARS treatment for 6 hours. Scale bar represents 2 μm. See also supplemental Video 3. (H) The line plot shows the recovery rate of FUS condensates in FUSlow LSK cells (N = 6) and FUShigh LSK cells (N = 12) of panel G at indicated time points. Data are shown as mean ± SEM. (I) The histograms depict the mobile fraction (99.2% vs 48.9%) and T-half (3.383% vs 63.064%) of FUSlow and FUShigh LSK cells. Data are shown as mean ± SEM. (J) Representative FRAP images show the fluorescence recovery of FUS condensates in the nucleoplasm of FUSlow and FUShigh LSK cells of 2-month-old Fus-gfp mice upon 0.5 mM ARS treatment for 4 hours. Scale bar represents 1 μm. Refer to supplemental Video 4. (K) The line plot shows the recovery rate of FUS condensates in FUSlow LSK cells (N = 19) and FUShigh LSK cells (N = 47) of panel J at indicated time points. Data are shown as mean ± SEM. (L) The histograms depict the mobile fraction (92.2% vs 41.6%) and T-half (4.264% vs 57.8%) of FUSlow and FUShigh LSK cells. Data are shown as mean ± SEM.
To further confirm this observation, we performed a serial transplantation assay by using FUSlow HSCs (supplemental Figure 2B), which is a classical approach to recapitulate the aging process by inducing replication stress.20-22 The result reveals that the percentage of FUShigh HSCs is increased at the end of the first transplantation (36.1%), further increased at the end of the second transplantation (90.8%), and kept stable at the end of the third transplantation (94.0%) (Figure 2E-F; supplemental Figure 2C).
To investigate the dynamic behavior of FUS condensates in these 2 populations, we performed FRAP assays for FUShigh and FUSlow HSPCs from young Fus-gfp mice. The result shows that FUShigh HSPCs exhibit compromised mobility of FUS condensates compared with FUSlow cells (cytoplasm: Figure 2G-I; supplemental Video 3; nucleoplasm: Figure 2J-L; supplemental Video 4).
Briefly, these results reveal that FUS is heterogeneously expressed in HSCs, and FUShigh HSPCs display compromised mobility of FUS condensates. The percentage of FUShigh HSCs is significantly increased in response to physiological aging and serial transplantation–induced replication stress.
FUShigh HSCs resemble aged HSCs
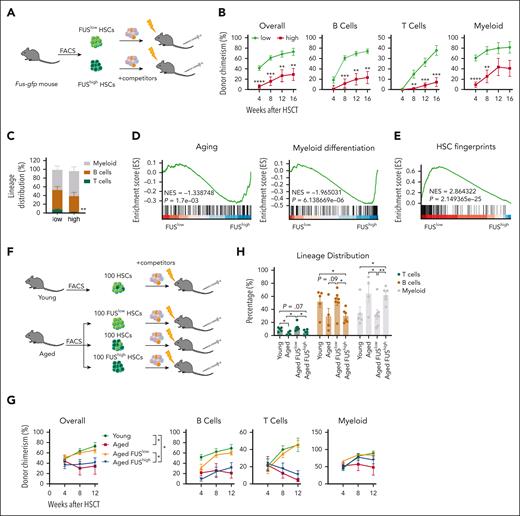
We conducted competitive transplantation assays for FUShigh and FUSlow HSCs (Figure 3A), and observed that the reconstitution capacity of FUShigh HSCs is impaired, including myeloid, B, and T cells (Figure 3B). In addition, FUShigh HSCs exhibit differentiation bias toward myeloid lineage (Figure 3C), which recapitulates the phenotype of aged HSCs. In a biological repeated experiment (supplemental Figure 3A), we observed a significant decrease in donor-derived HSPCs in the FUShigh group (supplemental Figure 3B-C), indicating that the impaired reconstitution of FUShigh HSCs is due to compromised proliferation capacity.
FUShigh HSCs resemble aged HSCs. (A) Experimental design of a competitive transplantation assay for FUSlow and FUShigh HSCs. 35 freshly isolated FUShigh and FUSlow HSCs (CD34–CD150+LSK) from young (2-4 months old) Fus-gfp mice were transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitors (CD45.1). The chimera in peripheral blood was evaluated every 4 weeks until week 16. (B) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in the peripheral blood of the recipients at the indicated time points. N = 7 recipients per group, data are shown as mean ± SEM. (C) The histogram depicts the lineage distribution of donor-derived peripheral blood cells at the 16th week after transplantation. N = 7 recipients per group, data are shown as mean ± SEM. (D-E) These figures show the GSEA of aging-related genes and myeloid differentiation–related genes (D) and HSC fingerprint genes between FUSlow and FUShigh HSCs (CD34–CD150+LSK). NES, normalized enrichment score. |NES| > 0.3 and P < .05 represent significant difference. The genes are listed in supplemental Table 2. (F) Experimental design of competitive transplantation assay for young, aged, aged FUSlow, and aged FUShigh HSCs (CD34–LSK). A total of 100 freshly isolated HSCs from 3 young (2-month-old) Fus-gfp mice and 100 FUShigh HSCs, 100 FUSlow HSCs, 100 unfractionated HSCs from 2 aged (18-month-old) Fus-gfp mice were transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitors (CD45.1). The chimera in peripheral blood was evaluated every 4 weeks until week 12. The experiment was repeated 2 times. (G) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in peripheral blood of the recipients at indicated time points. N = 4-7 recipients per group, data are shown as mean ± SEM. (H) The histogram depicts the lineage distribution of donor-derived peripheral blood cells at week 12 after transplantation. N = 4-7 recipients per group, data are shown as mean ± SEM.
FUShigh HSCs resemble aged HSCs. (A) Experimental design of a competitive transplantation assay for FUSlow and FUShigh HSCs. 35 freshly isolated FUShigh and FUSlow HSCs (CD34–CD150+LSK) from young (2-4 months old) Fus-gfp mice were transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitors (CD45.1). The chimera in peripheral blood was evaluated every 4 weeks until week 16. (B) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in the peripheral blood of the recipients at the indicated time points. N = 7 recipients per group, data are shown as mean ± SEM. (C) The histogram depicts the lineage distribution of donor-derived peripheral blood cells at the 16th week after transplantation. N = 7 recipients per group, data are shown as mean ± SEM. (D-E) These figures show the GSEA of aging-related genes and myeloid differentiation–related genes (D) and HSC fingerprint genes between FUSlow and FUShigh HSCs (CD34–CD150+LSK). NES, normalized enrichment score. |NES| > 0.3 and P < .05 represent significant difference. The genes are listed in supplemental Table 2. (F) Experimental design of competitive transplantation assay for young, aged, aged FUSlow, and aged FUShigh HSCs (CD34–LSK). A total of 100 freshly isolated HSCs from 3 young (2-month-old) Fus-gfp mice and 100 FUShigh HSCs, 100 FUSlow HSCs, 100 unfractionated HSCs from 2 aged (18-month-old) Fus-gfp mice were transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitors (CD45.1). The chimera in peripheral blood was evaluated every 4 weeks until week 12. The experiment was repeated 2 times. (G) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in peripheral blood of the recipients at indicated time points. N = 4-7 recipients per group, data are shown as mean ± SEM. (H) The histogram depicts the lineage distribution of donor-derived peripheral blood cells at week 12 after transplantation. N = 4-7 recipients per group, data are shown as mean ± SEM.
We then performed RNA sequencing to identify the transcriptional difference between FUShigh and FUSlow HSCs, and the resemblance between young and aged HSCs. Principle component analysis (PCA) shows that FUShigh HSCs are close to aged HSCs, and FUSlow HSCs are close to young HSCs (supplemental Figure 3-1A). The differentially regulated genes between FUShigh and FUSlow HSCs on the PC1 axis are mainly enriched in hemopoiesis and myeloid differentiation pathways (supplemental Figure 3-1B; supplemental Table 1). Gene set enrichment analysis (GSEA) reveals that aging, myeloid differentiation, DNA repair, DNA damage response, cell cycle, and DNA replication–related genes are enriched in FUShigh HSCs (Figure 3D; supplemental Figure 3-1C), whereas HSC fingerprint genes and autophagy-related genes are enriched in FUSlow HSCs (Figure 3E; supplemental Figure 3-1D; supplemental Table 2). In addition, dGSE analysis reveals that the canonical HSC aging genes23 are enriched in both aged HSCs and FUShigh HSCs (supplemental Figure 3-1E; supplemental Table 2).
To further investigate whether knockdown of FUS ameliorates aged HSCs at the transcriptomic level, we conducted RNA sequencing for shFUS-carrying and shCtl-carrying HSCs freshly isolated from the recipients (Figure 1F). The result shows that knockdown of FUS reforms the transcriptional profile of aged HSCs on the dimension of PC1 axis (supplemental Figure 3-1F). Enrichment analysis reveals that knockdown of FUS extricates aged HSCs from aging-related genes, whereas it does not influence the myeloid differentiation–related genes (supplemental Figure 3-1G), which is consistent with the result that knockdown of FUS does not rescue the differentiation skewing (Figure 1I).
In brief, the above data show that FUShigh HSCs resemble aged HSCs both functionally and transcriptionally. Knockdown of FUS reforms the transcriptional alteration of aged HSCs.
FUSlow HSCs of aged mice exhibit youthful function
Because FUShigh HSCs of young mice exhibit compromised reconstitution capacity, we then wondered whether the FUSlow HSCs of aged mice exhibited rejuvenated function. The result of the competitive transplantation assay shows that the reconstitution capacity of FUShigh HSCs and unfractionated HSCs of aged mice is comparable (Figures 3F-G), and both of them exhibit differentiation bias toward myeloid lineages (Figure 3H). It is notable that the reconstitution capacity of FUSlow HSCs from aged mice is comparable to those of young HSCs (Figure 3G), and they exhibit balanced differentiation potential from B/T/myeloid lineages (Figure 3H). This result suggests that the expression of FUS is inversely proportional to the function of HSC, and FUSlow HSCs of aged mice exhibit youthful function.
To further investigate the effect of FUS expression on HSC, we conducted a competitive transplantation assay by knocking down Fus in aged FUSlow and FUShigh HSPCs. The PB and BM analyses reveal that knockdown of FUS improves the reconstitution capacity of aged FUSlow HSCs but does not rescue the reconstitution and differentiation skewing of aged FUShigh HSCs (supplemental Figures 3-1H-K). These results indicate that the expression level of FUS controls the function of HSCs until HSCs are severely impaired.
Arginine-glycine-glycine domain mediates FUS dynamics in HSCs and enforced FUS impairs HSC via arginine-glycine-glycine domain
Given that FUS undergoes phase transition in HSCs, we then set out to identify the key domain by generating FUS variants: FUSΔLC, FUSΔRGG1, FUSΔRGG2, FUSΔRGG3, FUSΔRGG123, and FUSΔNLS, wherein the indicated domains have been proven to modulate FUS mobility (Figure 4A).24 Western blot assays reveal that these FUS variants are efficiently expressed (supplemental Figure 4A).
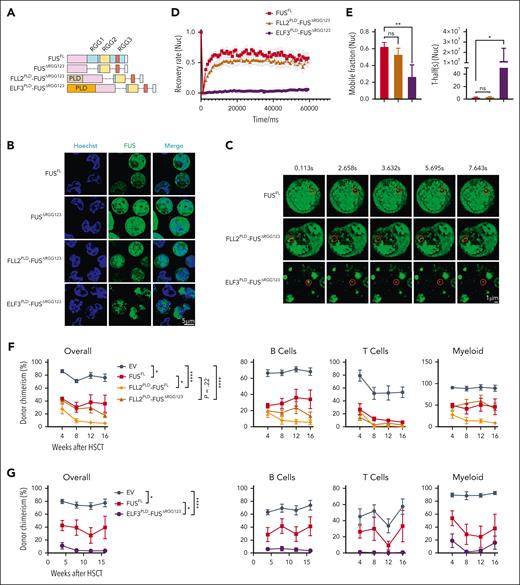
RGG domain mediates FUS mobility and its functional role in HSCs. (A) This schematic diagram shows the components of each FUS variants, including FUSΔLC, FUSΔRGG1, FUSΔRGG2, FUSΔRGG3, FUSΔRGG123, and FUSΔNLS. (B-C) Freshly isolated LSK cells from 2-month-old young WT mice were infected by lentivirus carrying FUS and FUS variants, and 3 days later, GFP+ cells were purified and treated with 0.5 mM ARS. (B) These representative images show the FUS condensates in the cytoplasm of indicated groups upon 0.5 mM ARS treatment for 1 hour. Scale bar represents 5 μm. (C) These representative images show the FUS condensates in the nucleoplasm of the indicated groups upon 0.5 mM ARS treatment for 4 hours. Scale bar represents 5 μm. (D) This histogram shows the quantification of the percentage of cells with FUS condensates in panel B. N = 89-313 cells per group. Data are shown as mean ± SEM. (E) This histogram shows the quantification of the FUS condensate numbers per cell in panel C. N = 74-256 cells per group. Data are shown as mean ± SEM. (F) These representative images exhibit the droplets formed by FUS variants at the protein concentration of 200 nM. The scale bar indicates 20 μm. (G) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in the peripheral blood of the recipients at the indicated time points. Freshly isolated 105 LSK cells from 2-month-old young WT mice were infected by lentivirus carrying FUS and FUS variants. Three days later, 2000 GFP+Sca-1+CD48– cells were purified and transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitor cells (CD45.1). Chimera in peripheral blood was evaluated every 4 weeks until the 16th week. N = 4-7 recipients per group; data are shown as mean ± SEM.
RGG domain mediates FUS mobility and its functional role in HSCs. (A) This schematic diagram shows the components of each FUS variants, including FUSΔLC, FUSΔRGG1, FUSΔRGG2, FUSΔRGG3, FUSΔRGG123, and FUSΔNLS. (B-C) Freshly isolated LSK cells from 2-month-old young WT mice were infected by lentivirus carrying FUS and FUS variants, and 3 days later, GFP+ cells were purified and treated with 0.5 mM ARS. (B) These representative images show the FUS condensates in the cytoplasm of indicated groups upon 0.5 mM ARS treatment for 1 hour. Scale bar represents 5 μm. (C) These representative images show the FUS condensates in the nucleoplasm of the indicated groups upon 0.5 mM ARS treatment for 4 hours. Scale bar represents 5 μm. (D) This histogram shows the quantification of the percentage of cells with FUS condensates in panel B. N = 89-313 cells per group. Data are shown as mean ± SEM. (E) This histogram shows the quantification of the FUS condensate numbers per cell in panel C. N = 74-256 cells per group. Data are shown as mean ± SEM. (F) These representative images exhibit the droplets formed by FUS variants at the protein concentration of 200 nM. The scale bar indicates 20 μm. (G) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in the peripheral blood of the recipients at the indicated time points. Freshly isolated 105 LSK cells from 2-month-old young WT mice were infected by lentivirus carrying FUS and FUS variants. Three days later, 2000 GFP+Sca-1+CD48– cells were purified and transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitor cells (CD45.1). Chimera in peripheral blood was evaluated every 4 weeks until the 16th week. N = 4-7 recipients per group; data are shown as mean ± SEM.
We treated HSPCs carrying FUS variants by sodium arsenite (ARS) to induce FUS condensates. The result shows that the FUS condensates are still generated in FUSΔLC and FUSΔNLS groups, but are significantly compromised in arginine-glycine-glycine (RGG)-truncated groups (cytoplasm, Figures 4B,D; nucleoplasm, Figures 4C,E), indicating that the RGG domain is essential for FUS to form condensates in HSPCs. To further confirm this observation, we purified protein of FUS variants, including FUSΔRGG1, FUSΔRGG2, and FUSΔRGG3. In vitro assays reveal that RGG-truncated groups exhibit significantly fewer condensates than full-length FUS protein (FUSFL) at the same concentration (Figure 4F), demonstrating that the RGG domain indeed mediates FUS condensate formation.
We next set out to investigate whether the RGG domain mediates the inhibitory function of enforced FUS on HSCs in vivo. We conducted a competitive transplantation assay and observed that enforced FUS impairs the reconstitution capacity of HSCs, whereas the reconstitution capacity is restored in the FUSΔRGG1, FUSΔRGG2, FUSΔRGG3, and FUSΔRGG123 groups, including myeloid, T, and B cells (Figure 4G). The result suggests that the impairment of enforced FUS on the reconstitution capacity of HSCs is mediated by the RGG domain and perhaps the RGG–mediated phase transition.
Heterografting restores the ability of FUS to form condensates
To rule out the influence of the potential non-LLPS function of the RGG domain on HSCs, we generated FLL2PLD-FUSΔRGG123 and ELF3PLD-FUSΔRGG123 by complementing FUSΔRGG123 with the PLD domain of Arabidopsis ELF3 and FLL2, which are in vivo regulators of phase separation (Figure 5A).25,26 Western blot assay reveals that these variants are efficiently expressed (supplemental Figure 4B). Imaging analysis reveals that the cells transfected by FUSΔRGG123 cannot form droplets, whereas the cells transfected by ELF3PLD-FUSΔRGG123 and FLL2PLD-FUSΔRGG123 form condensates efficiently in the nucleoplasm (Figure 5B) and cytoplasm (supplemental Figure 5A), indicating that the PLD domain of Arabidopsis ELF3 and FLL2 indeed restores the ability of FUS to form condensates.
Aberrant phase transition of FUS impairs HSCs. (A) This schematic diagram shows the components of each FUS variants, including FUSΔRGG123, FLL2PLD-FUSΔRGG123, and ELF3PLD-FUSΔRGG123. (B-C) Freshly isolated 105 LSK cells from 2-month-old young WT mice were infected by lentivirus carrying FUSFL, FUSΔRGG123, FLL2PLD-FUSΔRGG123, ELF3PLD-FUSΔRGG123, and 3 days later, GFP+ cells were purified and treated with 0.5 mM ARS. (B) These representative images show the FUS condensates in the nucleoplasm of indicated groups upon 0.5 mM ARS treatment for 4 hours, Scale bar represents 5 μm. (C) Representative FRAP images show the fluorescence recovery of FUS condensates in the nucleoplasm of the indicated groups in response to 0.5 mM ARS treatment for 4 hours. Scale bar represents 1 μm. (D) The line plot shows the recovery rate of FUS condensates in panel C at indicated time points. N = 9-32 condensates per group. (E) The histograms depict the mobile fraction and T-half of FRAP assay of panel C. Data are shown as mean ± SEM. (F-G) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in the peripheral blood of the recipients at the indicated time points. Freshly isolated 105 LSK cells from 2-month-old young WT mice were infected by lentivirus carrying FUS and indicated FUS variants. Three days later, 2000 GFP+Sca-1+CD48– cells were purified and transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitor cells (CD45.1). Chimera in peripheral blood was evaluated every 4 weeks until the 16th week. N = 5-6 (F) and N = 6-9 (G) recipients per group, data are shown as mean ± SEM.
Aberrant phase transition of FUS impairs HSCs. (A) This schematic diagram shows the components of each FUS variants, including FUSΔRGG123, FLL2PLD-FUSΔRGG123, and ELF3PLD-FUSΔRGG123. (B-C) Freshly isolated 105 LSK cells from 2-month-old young WT mice were infected by lentivirus carrying FUSFL, FUSΔRGG123, FLL2PLD-FUSΔRGG123, ELF3PLD-FUSΔRGG123, and 3 days later, GFP+ cells were purified and treated with 0.5 mM ARS. (B) These representative images show the FUS condensates in the nucleoplasm of indicated groups upon 0.5 mM ARS treatment for 4 hours, Scale bar represents 5 μm. (C) Representative FRAP images show the fluorescence recovery of FUS condensates in the nucleoplasm of the indicated groups in response to 0.5 mM ARS treatment for 4 hours. Scale bar represents 1 μm. (D) The line plot shows the recovery rate of FUS condensates in panel C at indicated time points. N = 9-32 condensates per group. (E) The histograms depict the mobile fraction and T-half of FRAP assay of panel C. Data are shown as mean ± SEM. (F-G) These line plots depict the percentage of donor-derived cells (overall, B cell, T cell, myeloid cell) in the peripheral blood of the recipients at the indicated time points. Freshly isolated 105 LSK cells from 2-month-old young WT mice were infected by lentivirus carrying FUS and indicated FUS variants. Three days later, 2000 GFP+Sca-1+CD48– cells were purified and transplanted into lethally irradiated recipients (CD45.2) together with 3.5 × 105 competitor cells (CD45.1). Chimera in peripheral blood was evaluated every 4 weeks until the 16th week. N = 5-6 (F) and N = 6-9 (G) recipients per group, data are shown as mean ± SEM.
To further investigate the dynamic behavior of these variants, we performed a FRAP assay and observed that FLL2PLD-FUSΔRGG123 recovers moderately within 60 seconds in the nucleoplasm (Figures 5C-D), which exhibits a mild difference with FUSFL in mobile fraction (53% vs 63%) and T-half (3.43s vs 2.68s) (Figure 5E). Meanwhile, ELF3PLD-FUSΔRGG123 exhibits weak recovery in both the nucleoplasm (26.8%) (Figures 5C-E) and cytoplasm (28.6%) (supplemental Figures 5B-D). The result shows that the mobility of FLL2PLD-FUSΔRGG123 is comparable with FUS, whereas the mobility of ELF3PLD-FUSΔRGG123 is significantly compromised.
To examine the effect of additional PLD domain grafting on the mobility of FUS, we generated FLL2PLD-FUSFL and ELF3PLD-FUSFL by grafting the PLD domains of FLL2 and ELF3 on FUSFL (supplemental Figures 4B and 5E). Imaging analysis reveals that the cells transfected by FLL2PLD-FUSFL exhibit more condensates, whereas the cells transfected by ELF3PLD-FUSFL form FUS aggregation (supplemental Figure 5F-G).
To further investigate the dynamic behavior of FUS variants, we performed a FRAP assay on HSPCs carrying ELF3PLD-FUSFL and FLL2PLD-FUSFL, respectively. The result shows that additional FLL2PLD grafting indeed compromises the mobility of FUSFL (59% vs 69% in the nucleoplasm and 49% vs 63% in the cytoplasm), and ELF3PLD grafting results in the formation of FUS aggregation (cytoplasm, supplemental Figure 5H-J; nucleoplasm, supplemental Figure 5K-M).
Enforced chimeric FUS impairs the reconstitution capacity of HSCs
To further test whether enforced FUS variants impair the reconstitution capacity of HSCs in vivo, we conducted a competitive transplantation assay and observed that enforced FLL2PLD-FUSΔRGG123 exhibits comparable chimera with FUSFL (Figure 5F), which is consistent with the result that the mobility of FLL2PLD-FUSΔRGG123 exhibits no difference with FUSFL (Figure 5E). We observed that enforced FLL2PLD-FUS significantly impairs the reconstitution capacity of HSCs (Figure 5F), which is consistent with the result that the mobility of FLL2PLD-FUS is compromised (supplemental Figure 5I,L).
Moreover, we performed the same assay for ELF3PLD-FUSΔRGG123 and ELF3PLD-FUSFL. The result reveals that enforced ELF3PLD-FUSΔRGG123 and ELF3PLD-FUSFL severely impair HSCs, whereas Arabidopsis ELF3PLD domain exhibits no inhibitory effect on the reconstitution capacity of HSCs (Figure 5G; supplemental Figure 5N).
The above results show that compromised mobility of FUS induced by heterografting of Arabidopsis FLL2PLD and ELF3PLD domains indeed impairs HSCs. The worse the dynamic behavior of FUS is, the more it impairs HSCs.
FUShigh HSCs exhibit global chromatin reorganization
Previous studies have shown that senescent cells undergo multiscale, 3-dimensional genome reorganization.27 Combining with the results that FUShigh HSCs resemble aged HSCs functionally and transcriptionally (Figure 3G; supplemental Figure 3-1A), we tentatively assume that the destructive role of enforced FUS on HSCs may be achieved by altering chromatin structure. To test this hypothesis, we reanalyzed the RNA-sequencing data of FUShigh and FUSlow HSCs and observed that the upregulated genes are enriched for nuclear division and chromosome segregation signaling (supplemental Figure 6A), which is confirmed by GSEA analysis (supplemental Figure 6B; supplemental Table 2). To further test this hypothesis, we profiled genome-wide chromatin interactions for FUShigh, FUSlow, and aged HSCs. The result exhibits sufficient sequencing depth and high reproducibility (supplemental Figure 6C-D). Analysis of FUShigh and FUSlow, young and aged HSCs reveals a different global chromatin organization (the Hi-C data of young HSC is obtained from a published data set13).
On the genomic level, we observed that the interaction matrixes of FUShigh HSCs exhibit enhanced proximal interactions and weakened distal interactions compared with FUSlow HSCs (Figure 6A). Similarly, aged HSCs exhibit the same alteration (supplemental Figure 6E). The mean intrachromosomal contact probability shows that FUShigh HSCs and aged HSCs display increased contact probabilities at <6 Mb and decreased contact probabilities at >6 Mb distances (Figure 6B). To further exhibit the difference in contact probability between FUShigh and FUSlow, young and aged HSCs, we observed that PvD is significantly higher in FUShigh and aged HSCs (Figure 6C), indicating that the chromosome structure is reorganized.
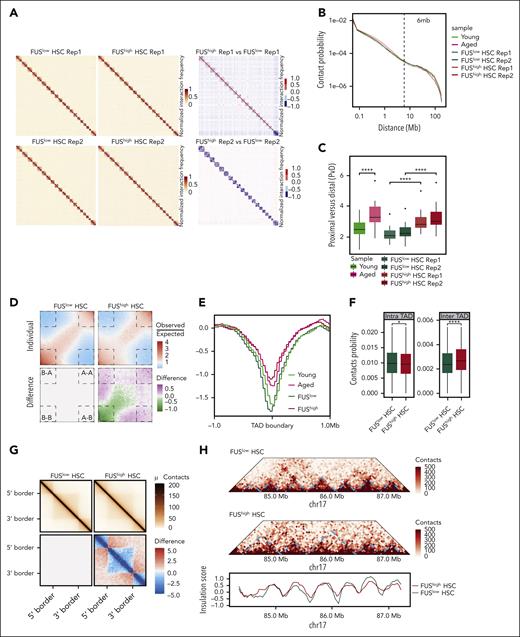
FUShigh HSCs exhibit global chromatin reorganization. (A) Heatmaps showing the normalized Hi-C interaction frequencies of the whole genome (1 Mb bin) of FUSlow and FUShigh HSCs (CD34–LSK) from young (2-4 months old) Fus-gfp mice (2 biological replicates for each group) (left) and differential interactions between them (right). (B) The decaying curve shows the genome-wide contact probability for interactions within individual chromosomes by genomic distance. The dotted line represents 6 Mb. (C) The boxplot showing the ratio of proximal and distal contacts (PvD) for young, aged, FUSlow, and FUShigh HSCs. (D) Saddle plots of Hi-C data binned at 100 kb resolution showing the compartmental interactions for FUSlow HSCs (top left) and FUShigh HSCs (top right), and the differential interactions between them (bottom right). The differentially interacting compartments are marked by the dashed lines. (E) These curves show the insulation score around TAD boundaries in young, aged, FUSlow, and FUShigh HSCs (±1 Mb). (F) These boxplots show the contact probability of intra-TAD and inter-TAD interactions for FUSlow and FUShigh HSCs. (G) Aggregation analysis for TADs showing the interactions for FUSlow (top left) and FUShigh HSCs (top right) and differential interactions between them (bottom right), wherein blue denotes loss of interactions and red denotes gain of interactions in FUShigh HSCs. (H) Example of TAD fusion and insulation score distribution for chromosome 17 (84-88 Mb) in FUSlow and FUShigh HSCs.
FUShigh HSCs exhibit global chromatin reorganization. (A) Heatmaps showing the normalized Hi-C interaction frequencies of the whole genome (1 Mb bin) of FUSlow and FUShigh HSCs (CD34–LSK) from young (2-4 months old) Fus-gfp mice (2 biological replicates for each group) (left) and differential interactions between them (right). (B) The decaying curve shows the genome-wide contact probability for interactions within individual chromosomes by genomic distance. The dotted line represents 6 Mb. (C) The boxplot showing the ratio of proximal and distal contacts (PvD) for young, aged, FUSlow, and FUShigh HSCs. (D) Saddle plots of Hi-C data binned at 100 kb resolution showing the compartmental interactions for FUSlow HSCs (top left) and FUShigh HSCs (top right), and the differential interactions between them (bottom right). The differentially interacting compartments are marked by the dashed lines. (E) These curves show the insulation score around TAD boundaries in young, aged, FUSlow, and FUShigh HSCs (±1 Mb). (F) These boxplots show the contact probability of intra-TAD and inter-TAD interactions for FUSlow and FUShigh HSCs. (G) Aggregation analysis for TADs showing the interactions for FUSlow (top left) and FUShigh HSCs (top right) and differential interactions between them (bottom right), wherein blue denotes loss of interactions and red denotes gain of interactions in FUShigh HSCs. (H) Example of TAD fusion and insulation score distribution for chromosome 17 (84-88 Mb) in FUSlow and FUShigh HSCs.
At compartment scale, we observed that the interaction between A and B compartments is strengthened in FUShigh HSCs (AA and AB) (Figure 6D) and aged HSCs (AA) (supplemental Figure 6F), whereas BB is compromised in both FUShigh and aged HSCs (supplemental Figure 6G).
Substantial TAD fusions emerge in aged and FUShigh HSCs
At the topologically associating domain (TAD) scale, the contact probability of the TAD border is increased in aged HSCs (supplemental Figure 6-1A). Further analysis reveals that the insulation score is significantly increased and 250 TADs exhibit contact changes with an insulation score change greater than 0.5 in aged HSCs (Figure 6E; supplemental Figure 6-1B; supplemental Table 3), indicating the insulation of TAD of aged HSCs is dampened. Consistently, the insulation score is increased in FUShigh HSCs and 603 merged TADs are observed (Figure 6E; supplemental Figure 6-1C; supplemental Table 3). Given that aged HSCs are composed of FUShigh and FUSlow HSCs, it is reasonable that fewer merged TADs are observed in aged HSCs. We next explored TAD changes between FUShigh and FUShigh HSCs in details.
The contact probability of the intra-TAD score of FUShigh HSCs is significantly reduced (Figure 6F), indicating that the TADs are decompacted. The inter-TAD score is increased in FUShigh HSCs (Figures 6F-G), indicating the interaction between adjacent TADs is strengthened. The contact probability among 10 adjacent TADs shows that the interactions between a TAD and its 2 neighboring TADs are enhanced (supplemental Figure 6-1D). A representative example exhibits TADs at chr17: 85 Mb-87 Mb of FUSlow HSCs are merged with FUShigh HSCs because of the blurred boundary between them (Figure 6H).
In brief, the above data reveal that the interaction within TADs of FUShigh HSCs is compromised, whereas the interaction between adjacent TADs is strengthened, which is correlated with the blurred boundaries of TADs in FUShigh HSCs.
Aberrant FUS condensates compromise the binding of CCCTC-binding factor with its motif
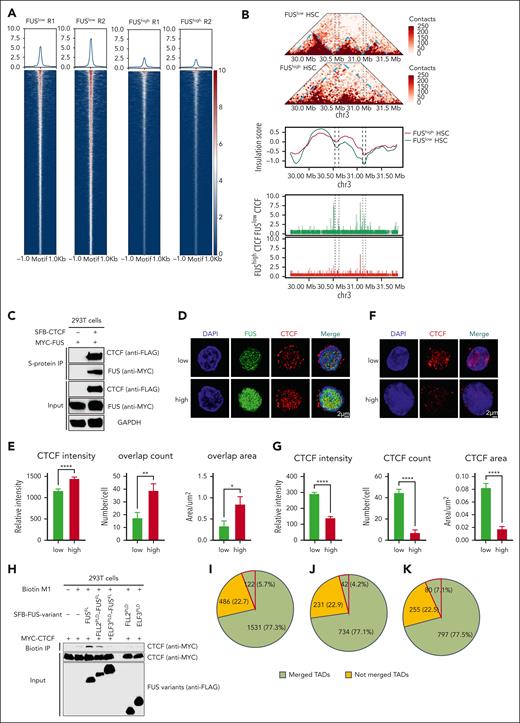
Given that CCCTC-binding factor (CTCF) is one of the most important insulators separating TADs,28,29 we profiled CTCF binding sites for young and aged HSCs, FUShigh and FUSlow HSPCs. The result shows that the binding intensity of CTCF is significantly compromised in FUShigh HSPCs (Figure 7A) and aged HSCs (supplemental Figure 7A). A representative example exhibits TADs at chr3: 30 Mb-31.5 Mb of FUSlow HSCs merge into 1 TAD in FUShigh HSCs because of acute reduction of CTCF binding (Figure 7B).
Aberrant FUS condensates limit the binding of CCCTC-binding factor with chromatin. (A) Heatmaps showing the CCCTC-binding factor (CTCF) peaks around the CTCF motif in FUSlow and FUShigh LSK cells (±1 kb). Freshly isolated 5 × 104 LSK from young (2-4 months) Fus-gfp mice are subjected to a CTCF CUT&Tag assay. (B) Representative example showing the merged TAD, insulation score, and CTCF peaks at chr3: 30-31.5 Mb of FUSlow and FUShigh HSCs. (C) Representative western blot showing the interaction of FUS and CTCF. HEK293T cells were cotransfected with plasmids encoding SFB-tagged FUS and Myc-tagged CTCF. Lysates were subjected to immunoprecipitation by using anti-S-protein agarose beads for 3 hours. Input and copurified proteins were blotted by probing with corresponding antibodies. (D) Representative images showing the immunofluorescence staining of CTCF (red) in FUSlow and FUShigh LSK cells from a 2-month-old Fus-gfp mouse. DAPI (blue) was used for nuclei counterstaining. Scale bar represents 2 μm. (E) These histograms depict the CTCF intensity, overlap count, and overlap area between FUS and CTCF in FUSlow and FUShigh LSK cells. Data are shown as mean ± SEM, N = 32-40 cells per group. (F) Representative images showing immunofluorescence staining of CTCF (red) in CSK buffer–preextracted FUSlow and FUShigh LSK cells from young (2-4 months old) Fus-gfp mice. DAPI (blue) was used for nuclei counterstaining. Scale bar represents 2 μm. (G) These histograms depict the intensity, count, and area of CTCF condensates in FUSlow and FUShigh LSK cells preextracted with CSK buffer. Data are shown as mean ± SEM, N = 33-34 cells per group. (H) Representative western blot showing the detection of CTCF binding to the biotinylated M1 motifs in the presence FUSFL or variants by DNA pull-down assay. HEK293T cells were cotransfected with plasmids encoding SFB-tagged FUSFL or FUS variants and Myc-tagged CTCF. A total of 2 μg biotinylated M1 motifs were added to lysates and subjected to immunoprecipitation by using streptavidin agarose beads for 3 hours. Input and copurified proteins were blotted by probing with corresponding antibodies. (I-K) These pie charts show the distribution of DEG (I), upregulated genes (J), and downregulated genes (K) in FUShigh HSCs compared with FUSlow HSCs. The green background represents the genes that originated from merged TADs, wherein the red lines indicate the genes that have been reported to modulate HSC function.
Aberrant FUS condensates limit the binding of CCCTC-binding factor with chromatin. (A) Heatmaps showing the CCCTC-binding factor (CTCF) peaks around the CTCF motif in FUSlow and FUShigh LSK cells (±1 kb). Freshly isolated 5 × 104 LSK from young (2-4 months) Fus-gfp mice are subjected to a CTCF CUT&Tag assay. (B) Representative example showing the merged TAD, insulation score, and CTCF peaks at chr3: 30-31.5 Mb of FUSlow and FUShigh HSCs. (C) Representative western blot showing the interaction of FUS and CTCF. HEK293T cells were cotransfected with plasmids encoding SFB-tagged FUS and Myc-tagged CTCF. Lysates were subjected to immunoprecipitation by using anti-S-protein agarose beads for 3 hours. Input and copurified proteins were blotted by probing with corresponding antibodies. (D) Representative images showing the immunofluorescence staining of CTCF (red) in FUSlow and FUShigh LSK cells from a 2-month-old Fus-gfp mouse. DAPI (blue) was used for nuclei counterstaining. Scale bar represents 2 μm. (E) These histograms depict the CTCF intensity, overlap count, and overlap area between FUS and CTCF in FUSlow and FUShigh LSK cells. Data are shown as mean ± SEM, N = 32-40 cells per group. (F) Representative images showing immunofluorescence staining of CTCF (red) in CSK buffer–preextracted FUSlow and FUShigh LSK cells from young (2-4 months old) Fus-gfp mice. DAPI (blue) was used for nuclei counterstaining. Scale bar represents 2 μm. (G) These histograms depict the intensity, count, and area of CTCF condensates in FUSlow and FUShigh LSK cells preextracted with CSK buffer. Data are shown as mean ± SEM, N = 33-34 cells per group. (H) Representative western blot showing the detection of CTCF binding to the biotinylated M1 motifs in the presence FUSFL or variants by DNA pull-down assay. HEK293T cells were cotransfected with plasmids encoding SFB-tagged FUSFL or FUS variants and Myc-tagged CTCF. A total of 2 μg biotinylated M1 motifs were added to lysates and subjected to immunoprecipitation by using streptavidin agarose beads for 3 hours. Input and copurified proteins were blotted by probing with corresponding antibodies. (I-K) These pie charts show the distribution of DEG (I), upregulated genes (J), and downregulated genes (K) in FUShigh HSCs compared with FUSlow HSCs. The green background represents the genes that originated from merged TADs, wherein the red lines indicate the genes that have been reported to modulate HSC function.
Coimmunoprecipitation assay demonstrates that FUS exhibits strong interaction with CTCF (Figure 7C). Immunofluorescence assay shows that CTCF exhibits increased overlap with FUS in FUShigh HSPCs (Figure 7D-E), aged HSCs (supplemental Figure 7B-C), and HSPCs with enforced FUS (supplemental Figure 7D-E). Given that CTCF forms TADs boundary by binding with its motif, we then performed a preextraction procedure to leave the CTCF stably associated with chromatin intact. The result exhibits a reduced amount of chromatin-associated CTCF (Figure 7F-G), indicating that the binding of CTCF with chromatin is compromised in FUShigh HSPCs.
We next investigated the influence of FUS mobility on the binding of CTCF with its motif by performing a DNA pull-down assay. The result shows that FLL2PLD-FUSFL and ELF3PLD-FUSFL compromise it (Figure 7H), indicating that the binding of CTCF with DNA decreases as long as the mobility of FUS is deteriorated.
To confirm the observation, we performed a preextraction immunofluorescence assay for CTCF in aged HSPCs carrying Fus shRNA. The result shows that CTCF intensity is significantly increased compared with the control group (supplemental Figure 7F-G), indicating that the binding of CTCF with chromatin is increased in response to FUS reduction. Consistent with this result, we observed that the binding of CTCF with its motif is strengthened in response to Fus reduction (supplemental Figure 7H).
Briefly, these results show that aberrant phase–separated FUS limits the binding of CTCF with chromatin and furthermore results in the compromise of TAD insulation in aged HSCs.
Most differentially expressed genes (DEG) between FUShigh and FUSlow HSCs locate in merged TADs
Previous studies have shown that TADs’ fusion leads to the alteration of genes within them.30,31 We then wondered whether the fusion of TADs in FUShigh HSCs results in transcriptome alteration. Firstly, we observed that 77.3% of the DEG (Figure 7I), including 77.1% of the upregulated genes (Figure 7J) and 77.5% of the downregulated genes (Figure 7K) between FUShigh and FUSlow HSCs, are located in merged TADs (hereafter, the DEG inside merged TADs are named DEGsTAD, the upregulated genes inside merged TADs are named UPGsTAD, and the downregulated genes inside merged TADs are named DWGsTAD). The dGSE and GSEA of DEGs show that the genes involved in HSC aging, myeloid differentiation, chromosome segregation, DNA repair, DNA damage response, DNA replication, and cell cycle, but not HSC fingerprints or autophagy, are enriched in FUShigh HSCs (supplemental Figure 7-1A). Furthermore, gene ontology (GO) analysis reveals that genes within the differential CTCF binding region between FUSlow and FUShigh HSPCs are enriched in B-cell differentiation and the cell cycle pathway (supplemental Figure 7-1A).
Except for the pathway, chromosome segregation, GO analysis, and dGSE analysis reveal that the UPGsTAD are enriched in the DNA damage response pathway, including DNA repair, DNA replication, and double-strand break repair, but not in HSC aging genes (supplemental Figure 7-2A,B), whereas the DWGsTAD are enriched in inflammatory signaling and HSC aging (supplemental Figure 7-2C,D). We conducted a conjoint analysis by using the database23 and Top20 FUShigh HSC-specific aging signature genes are listed in supplemental Table 3, including Mt1 and CD86. Furthermore, it is notable that 122 genes within the DEGsTAD (42 are upregulated and 80 are downregulated) have been demonstrated to modulate HSC function (supplemental Figure 7-2E; supplemental Table 3). For example, the gene Tgfbi is downregulated in FUShigh HSCs (supplemental Figure 7-2F) and a previous study reported that dysfunction of Tgfbi impairs the reconstitution capacity of HSCs.32 By exploring the proteomic database,17 we observed that TGFBI is indeed decreased in aged HSCs (supplemental Figure 7-2G).
Taken together, our study demonstrates that aged HSCs exhibit aberrant FUS mobility, which promotes HSC aging by blurring the boundary of TADs and then altering the expression of HSC function–related signaling.
Discussion
Biomolecular condensates are an epoch-making biological phenomenon discovered in the last few decades. To our knowledge, our study demonstrates for the first time that the aberrant phase transition of FUS promotes HSC aging by altering chromatin structure and further points out that aberrant biomolecular condensate assembly may be one of the driving forces behind aging and aging-related diseases.
Acknowledgments
The authors thank Hong Zhang of Institute of Biophysics, Chinese Academy of Sciences and Pilong Li and Yi Lin of Tsinghua University for detailed discussion. They thank Qianwen Sun of Tsinghua University for offering Arabidopsis complementary DNA and Zhangbin Bao of Tsinghua University for offering purified protein of FUSFL, FUSΔRGG1, FUSΔRGG2, and FUSΔRGG3. The authors also thank the Tsinghua-Peking Center for Life Sciences and the China Telecom Corporation Limited for facility and financial support.
This work was supported by grant numbers Z200022, 82250002, 92249305, 2018YFA0800200 and 81870118 to J.W., and 81890991 and 20222000298 to M.S. from the National Key R&D Program of China or the Beijing Municipal Science & Technology Commission or the National Natural Science Foundation of China and CapitalBio Technology Co, Ltd, and HH23KYZX0002 from Haihe Laboratory of Cell Ecosystem Innovation Fund.
Authorship
Contribution: J.W. and M.S. contributed in conceptualization; B.T., X.W., H.H., R.C., G.Q., Y.Y., Z.X., L.W., and Q.D. contributed in methodology; B.T., X.W., H.H., R.C., G.Q., Y.Y., Z.X., L.W., Q.D., and J.Y. contributed in investigation; B.T., X.W., L.W., and Q.D. performed the formal analysis; J.W., J.Y., and M.S. contributed in resources; J.W. and M.S. contributed in writing; J.W. and M.S. contributed in funding and acquisition; and J.W., M.S., and M.Q.Z. supervised the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael Q. Zhang, The Ministry of Education Key Laboratory of Bioinformatics, Bioinformatics Division and Center for Synthetic and Systems Biology, Beijing National Research Center for Information Science and Technology, School of Medicine, Tsinghua University, Beijing, China; email: michael.zhang@utdallas.edu; Minglei Shi, The Ministry of Education Key Laboratory of Bioinformatics, Bioinformatics Division and Center for Synthetic and Systems Biology, Beijing National Research Center for Information Science and Technology, School of Medicine, Tsinghua University, Beijing, China; email: shiml79@tsinghua.edu.cn; and Jianwei Wang, School of Pharmaceutical Sciences, Tsinghua University, Haidian District, Beijing 100084, China;.
References
Author notes
The Gene Expression Omnibus accession number for the data reported in this manuscript is GSE209715.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal