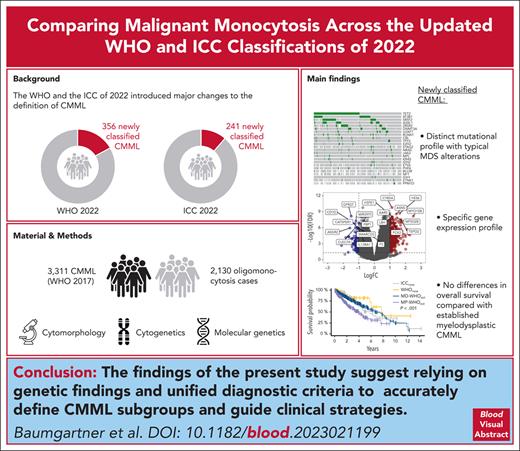
When reclassifying 2130 oligomonocytosis cases using the WHO and ICC 2022 classifications, 356 and 241 cases are newly classified as CMML.
Compared with MD-CMML, newly classified CMML cases show distinct mutational and transcriptional profiles but comparable overall survival.
Visual Abstract
The World Health Organization (WHO) classification of hematolymphoid tumors and the International Consensus Classification (ICC) of 2022 introduced major changes to the definition of chronic myelomonocytic leukemia (CMML). To assess its qualitative and quantitative implications for patient care, we started with 3311 established CMML cases (according to WHO 2017 criteria) and included 2130 oligomonocytosis cases fulfilling the new CMML diagnostic criteria. Applying both 2022 classification systems, 356 and 241 of oligomonocytosis cases were newly classified as myelodysplastic (MD)-CMML (WHO and ICC 2022, respectively), most of which were diagnosed as myelodysplastic syndrome (MDS) according to the WHO 2017 classification. Importantly, 1.5 times more oligomonocytosis cases were classified as CMML according to WHO 2022 than based on ICC, because of different diagnostic criteria. Genetic analyses of the newly classified CMML cases showed a distinct mutational profile with strong enrichment of MDS-typical alterations, resulting in a transcriptional subgroup separated from established MD and myeloproliferative CMML. Despite a different cytogenetic, molecular, immunophenotypic, and transcriptional landscape, no differences in overall survival were found between newly classified and established MD-CMML cases. To the best of our knowledge, this study represents the most comprehensive analysis of routine CMML cases to date, both in terms of clinical characterization and transcriptomic analysis, placing newly classified CMML cases on a disease continuum between MDS and previously established CMML.
Introduction
For the past half century, chronic myelomonocytic leukemia (CMML) was defined as a myelodysplastic (MD)/myeloproliferative (MP) neoplasm with the hallmark characteristic of peripheral blood (PB) monocytosis ≥1 × 109/L, in conjunction with MD and/or MP bone marrow (BM) features.1-3
Because of constant technological advancement and a rapidly evolving knowledge base, preexisting conceptions are being challenged, making the continuous adaptation of disease definitions necessary. In this regard, the fifth edition of the World Health Organization (WHO) classification and the International Consensus Classification (ICC) introduced major common changes to the diagnostic criteria of CMML.4,5 First, in the presence of additional CMML features, the cutoff for absolute monocytosis was lowered to ≥0.5 × 109/L absolute monocyte count in both classifications, thereby incorporating cases formerly referred to as oligomonocytic (OM)-CMML into the diagnosis of CMML.6-9 Second, the CMML-0 subgroup was eliminated because of its limited clinical relevance.10,11 Finally, in the current classifications, the diagnostic focus is shifted onto recurrent molecular aberrations, thus replacing solely clinical criteria such as persisting monocytosis for ≥3 months in the absence of alternative diagnoses.
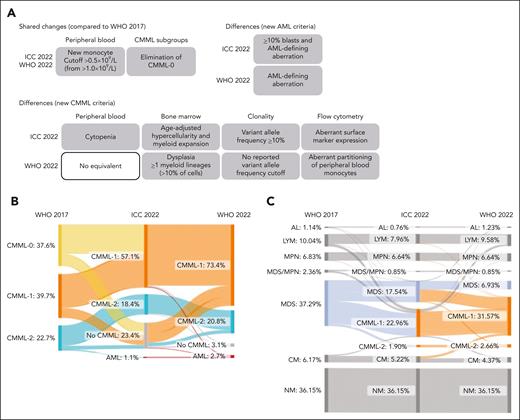
However, the updated classification systems rely on partly different diagnostic criteria, thus introducing 2 discordant definitions of CMML.12 Specifically, the main differences lie in the requirements for PB cytopenia, BM, and flow cytometric features, as well as variant allele frequency (VAF) cutoff values to define clonality through mutations (Figure 1A).4,5
Reclassification of established CMML and monocytosis cases based on the updated WHO22 and ICC22 classifications. (A) Schematic depiction of the main differences and changes between the WHO17, the ICC22, and the WHO22 classifications. (B) Diagnosis update of 1279 established CMML cases from the WHO17 to the ICC22 and the WHO22 classifications. “No CMML” stands for alternative diagnosis due to CMML criteria not being fulfilled. Notably, when evaluating all cases with absolute monocyte count (AMC) ≥1 × 109/L and ≥10% PB monocytes irrespective of their original diagnosis, only 69 cases (<0.01%) were not diagnosed as CMML at the time of workup. Of these 69 cases, 62 either had a concurrent diagnosis of lymphoma or non-CMML leukemia (n = 12) or received a CMML diagnosis during follow-up (n = 50). (C) Diagnosis update of 1054 monocytosis cases (AMC, 0.5 × 109/L-1 × 109/L and >10%; no prior CMML diagnosis) from the WHO17 to the ICC22, and the WHO22 classifications. AL, acute leukemia; CCUS, clonal cytopenia of unknown significance; CHIP, clonal hematopoiesis of indeterminate potential; LYM, lymphoma; MPN, myeloproliferative neoplasia; NM, nonmalignant.
Reclassification of established CMML and monocytosis cases based on the updated WHO22 and ICC22 classifications. (A) Schematic depiction of the main differences and changes between the WHO17, the ICC22, and the WHO22 classifications. (B) Diagnosis update of 1279 established CMML cases from the WHO17 to the ICC22 and the WHO22 classifications. “No CMML” stands for alternative diagnosis due to CMML criteria not being fulfilled. Notably, when evaluating all cases with absolute monocyte count (AMC) ≥1 × 109/L and ≥10% PB monocytes irrespective of their original diagnosis, only 69 cases (<0.01%) were not diagnosed as CMML at the time of workup. Of these 69 cases, 62 either had a concurrent diagnosis of lymphoma or non-CMML leukemia (n = 12) or received a CMML diagnosis during follow-up (n = 50). (C) Diagnosis update of 1054 monocytosis cases (AMC, 0.5 × 109/L-1 × 109/L and >10%; no prior CMML diagnosis) from the WHO17 to the ICC22, and the WHO22 classifications. AL, acute leukemia; CCUS, clonal cytopenia of unknown significance; CHIP, clonal hematopoiesis of indeterminate potential; LYM, lymphoma; MPN, myeloproliferative neoplasia; NM, nonmalignant.
Currently, clinical implications resulting from the updated, albeit discordant CMML definitions remain widely unexplored. In this work, through the use case of 2 well-characterized routine diagnostic data sets from the Munich Leukemia Laboratory (MLL), we examined the specific changes and differences introduced by the fifth edition of the WHO (WHO22) and ICC 2022 (ICC22) classifications for CMML and their resulting real-world implications for patients, clinicians, and researchers.
Methods
Sample collection, processing, and sequencing
All samples were analyzed at the MLL between August 2005 and October 2022 and investigated by cytomorphology, chromosome banding analysis, flow cytometry, panel sequencing, and, in a subset of 226 cases, by whole-genome (WGS) and whole-transcriptome sequencing (WTS). Fluorescence in situ hybridization (FISH) was performed if required in a clinical setting (supplemental Figure 1A-B, available on the Blood website). Diagnoses were established based on cytomorphology, immunophenotype, cytogenetics, and molecular genetics as previously published.13-15 In-house cytomorphological diagnosis was considered as the time point of first diagnosis. Cases with co-occurring malignancies, incomplete PB reports, or inadequate BM sampling (dysplasia or cellularity not assessable) were excluded from the study (supplemental Figure 2A-B). All patients provided written informed consent in accordance with the Declaration of Helsinki, and the study was approved by the internal review board.
Diagnostic criteria definitions and risk scoring
Primary diagnoses were based on the WHO 2001, 2008, or 2017 classifications and updated to the WHO17, WHO22, and ICC22 classifications for this study (supplemental Figure 3A-C). For risk scoring, both CMML-specific Prognostic Scoring System (CPSS) and clinical/molecular CPSS (CPSS-Mol) were applied.16,17 Because of incomplete clinical information regarding red blood cell transfusion dependency, sex-specific hemoglobin thresholds were used as a surrogate parameter.17,18 For reclassified cases with a prior myelodysplastic syndrome (MDS) diagnosis, Revised International Prognostic Scoring System (IPSS-R), and Molecular International Prognostic Scoring System (IPSS-M) scores were calculated using the “ipssm” package.19,20
Targeted and panel sequencing
Targeted and panel sequencing were performed as previously described.21 Because samples were recruited from routine diagnostics over a period of 17 years, the number of genes sequenced varied between 1 and 78. The number of samples assessed per gene is listed where mutation frequencies are reported.
WGS and WTS
WGS and WTS were performed for 226 patients and 64 controls as previously described.22 WGS was performed from polymerase chain reaction (PCR)–free libraries and 150 bp paired-end sequencing on NovaSeq6000 and HiSeqX instruments to a median depth of 100×. Reads were aligned to the hg19/GRCh37 reference genome using Isaac.23 WTS was performed from total RNA libraries on the same instruments using 100 bp paired-end sequencing to a median of 50 million reads per sample. Reference alignment was performed using spliced transcripts alignment to a reference (STAR),24 and gene counts were estimated using Cufflinks.25 After normalization/filtering with edgeR, differential gene expression of weighted samples was assessed using limma, and gene set enrichment analysis was performed.
Identification of somatic mutations, rearrangements, and chromosomal deletions
Somatic mutations from panel sequencing were called with Pisces26 or SeqPilot (JSI Medisys). Mutations from WGS were identified using Strelka227 and filtered through an in-house pipeline.22 The definition of pathogenicity and VAF for mutations are described in the supplemental Methods. NPM1-mutation status was assessed if required in a clinical setting. Depending on the panel sequenced, different quantitative PCR assays with different sensitivities were used for KIT analysis. KMT2A-partial tandem duplications (PTD) was analyzed by a quantitative PCR assay and FLT3-internal tandem duplications (ITD) by gene scan, as described previously.28 Structural abnormalities were analyzed by cytogenetics and fluorescence in situ hybridization analysis.
Statistical analysis
All statistical analyses were performed inside an R programming environment (version 4.2.1).29 The error bars shown in the figures represent the standard deviation unless specified otherwise. The correlation of continuous to categorical variables was statistically assessed with the Kruskal-Wallis test or the Wilcoxon rank sum test, whereas nominal variables were compared using the χ2 and Fisher exact tests. Unless stated otherwise an (adjusted) P value < .05 was considered significant. Overall survival (OS) was visualized using Kaplan-Meier curves and compared using a two-sided log-rank test.
Results
Diagnosis update of established CMML cases
We retrospectively evaluated 3311 routine CMML cases diagnosed according to the WHO17 classification from August 2005 to October 2022 at the MLL. Only first diagnosis cases with complete PB count information and adequate BM sampling were considered, with the final cohort sizes being 1205 and 966 CMML cases according to the WHO224 and ICC225 classifications (supplemental Figure 3A).
Reclassification compared with WHO17 was assessed for 1279 cases with all data available for WHO22 and ICC22. Reclassification occurred in 41.9% of CMML cases according to the WHO22 classification (ICC22, 50.2%; Figure 1B; supplemental Table 1). This resulted from the following major changes. First, the CMML-0 was incorporated into the CMML-1 subgroup, which increased from 39.7% to 73.4% in the WHO22 classification (ICC22, 57.1%). Additionally, because the WHO17 criterion of persisting monocytosis (in the absence of clonality or dysplasia) was removed in both revised classifications, 3.1% (39 cases; WHO22) and 23.4% (299 cases; ICC22) of cases were no longer classified as CMML. Finally, because of the revised AML diagnostic criteria, 2.7% (35 cases; WHO22) and 1.1% (14 cases; ICC22) of prior CMML cases were reclassified as AML. This discrepancy resulted from the ICC22 requirement of ≥10% blasts in addition to AML-defining aberrations for the diagnosis of AML.
Diagnosis update of OM cases previously not classified as CMML
Considering that the most impactful revision in both classifications was lowering the PB monocytosis threshold from ≥1 × 109/L to ≥0.5 × 109/L for the diagnosis of CMML, we additionally analyzed 2130 well-described OM cases (PB monocytes, 0.5 × 109/L-1.0 × 109/L) diagnosed during the same time interval defined earlier. Applying WHO22 and ICC22 criteria to diagnose OM-CMML led to 1.5 times more cases being classified as CMML based on the WHO22 than with the ICC22 criteria (356 vs 241 cases; Figure 1C; supplemental Figure 3B; supplemental Table 1). This difference originated mainly from differing BM requirements: the ICC22 requires myeloid proliferation and age-adjusted hypercellularity, whereas the WHO22 requires dysplasia in ≥1 myeloid lineage.
A closer inspection of reclassified OM cases revealed the majority of those being previously classified as MDS (WHO22, 89.6%; ICC22, 88.8%; supplemental Figure 4A-C), in accordance with previous reports showing high transformation rates of OM-MDS cases into genuine CMML.30-32 Importantly, when assessing the equivalent ICC22 diagnosis of OM cases exclusively classified as CMML in the WHO22 classification, they fell into either the MDS or clonal monocytosis of undetermined significance (CMUS)/clonal cytopenia and monocytosis of undetermined significance (CCMUS) categories (supplemental Figure 4D-E).
Comparison of established and OM-CMML cases
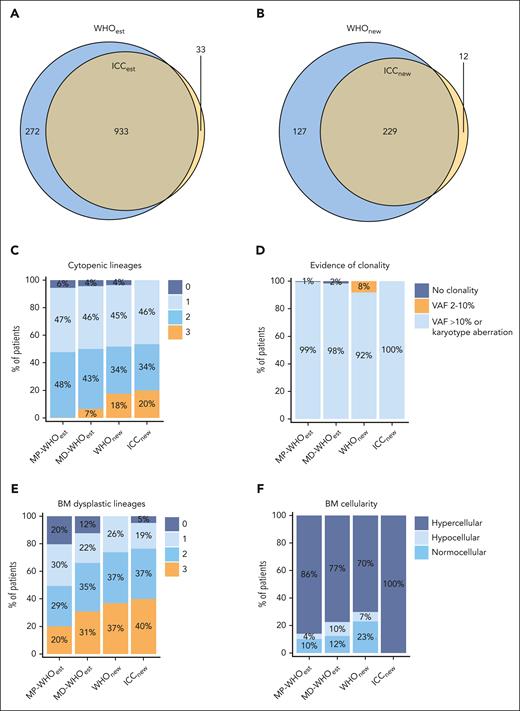
Our next aim was to compare established CMML cases with PB monocytosis ≥1 × 109/L to newly classified CMML cases with PB monocytosis of 0.5 × 109/L to 1.0 × 109/L. In terms of established CMML cases, there was a 75.4% overlap between WHO22 (WHOest, n = 1205) and ICC22 (ICCest, n = 966; Figure 2A). Because of a broader use of the WHO classification in the clinical setting, established WHO22 cases are shown as comparison (WHOest, n = 1205), whereas comparisons with established ICC22 cases (ICCest, n = 966) are listed in the supplemental Tables 2.1 to 2.4. These were subdivided into the MD and MP subgroups, in consideration of reclassified cases categorized exclusively as MD-CMML, known to have distinct clinical and molecular features from MP-CMML.33 In terms of newly classified CMML cases, final cohort sizes were 356 according to WHO22 (WHOnew cohort) and 241 according to ICC22 (ICCnew cohort; supplemental Figure 4A-B). Except for 12 patients, all reclassified ICC22 cases were a subgroup of the reclassified WHO22 cases (Figure 2B).
WHO22 and ICC22 classification criteria analysis for WHOest, WHOnew, and ICCnew cases. (A) Overlap between the reclassified WHOnew and ICCnew cohorts, with the ICCnew cohort being a subgroup of the WHOnew cohort. (B) Overlap between established CMML cases after ICC22 (ICCest) and after WHO22 (WHOest) diagnosis update. (C-F) Classification criteria analysis in the WHOest (subdivided into MD-CMML and MP-CMML), WHOnew, and ICCnew cohorts. (C) Number of cytopenic lineages in the PB. (D) Evidence of clonality. Patients were separated into “no clonality” (maximal VAF <2% of myeloid malignancy associated mutations), VAF between 2% and 10%, and VAF >10% or karyotypical alteration. Only data of patients with ≥10 genes sequenced are shown. (E) Number of dysplastic lineages in the BM. (F) Age-adjusted BM cellularity.
WHO22 and ICC22 classification criteria analysis for WHOest, WHOnew, and ICCnew cases. (A) Overlap between the reclassified WHOnew and ICCnew cohorts, with the ICCnew cohort being a subgroup of the WHOnew cohort. (B) Overlap between established CMML cases after ICC22 (ICCest) and after WHO22 (WHOest) diagnosis update. (C-F) Classification criteria analysis in the WHOest (subdivided into MD-CMML and MP-CMML), WHOnew, and ICCnew cohorts. (C) Number of cytopenic lineages in the PB. (D) Evidence of clonality. Patients were separated into “no clonality” (maximal VAF <2% of myeloid malignancy associated mutations), VAF between 2% and 10%, and VAF >10% or karyotypical alteration. Only data of patients with ≥10 genes sequenced are shown. (E) Number of dysplastic lineages in the BM. (F) Age-adjusted BM cellularity.
WHO22 and ICC22 criteria analysis for newly classified and established CMML cases
We then assessed the differences in the diagnostic criteria of both classification systems. According to the ICC22, cytopenia of ≥1 lineage is required for the diagnosis of CMML. Expectedly, all ICCnew cases presented with cytopenia, in contrast to 94% to 96% in the WHO cohorts (Figure 2C; supplemental Figure 5A; Table 1). Trilineage cytopenia occurred significantly more frequently in the WHOnew and ICCnew cohorts than in the WHOest cohorts (18%-20% vs 0.2%-6.6%, respectively; P < .001). Importantly, this observation held true irrespective of the distinction between MP- and MD-CMML, possibly reflecting a hypoproliferative disease biology of WHOnew and ICCnew cases.
Clinicopathological parameters of WHOest, WHOnew, and ICCnew
| . | MP-WHOest n = 551∗ . | MD-WHOest n = 654∗ . | WHOnew n = 356∗ . | q value† MD-WHOest vs WHOnew . | ICCnew n = 241∗ . | q value† MD-WHOest vs ICCnew . |
|---|---|---|---|---|---|---|
| Age, y | 74.3 ± 8.6; 76.3 (37.8-93.3) | 74.3 ± 9.1; 75.6 (19.9-92.7) | 74.2 ± 9.3; 75.9 (30.8-91.9) | 0.9 | 75.3 ± 8.1; 76.8 (43.8-91.9) | 0.12 |
| Sex | 0.2 | 0.3 | ||||
| Female | 171/551 (31%) | 171/654 (26%) | 107/356 (30%) | 71/241 (29%) | ||
| Male | 380/551 (69%) | 483/654 (74%) | 249/356 (70%) | 170/241 (71%) | ||
| Clinical status | ||||||
| First diagnosis | 551/551 (100%) | 654/654 (100%) | 356/356 (100%) | 241/241 (100%) | ||
| Absolute monocyte count (PB), per nL | 9371.7 ± 12 137.4; 5667.0 (1240.0-95 680.0) | 1971.4 ± 1061.1; 1622.5 (1000.0-8160.0) | 709.3 ± 140.0; 694.5 (500.0-998.4) | <0.001 | 713.9 ± 138.5; 704.0 (500.0-998.4) | <0.001 |
| % Monocytes (PB) | 26.7 ± 13.2; 24.0 (10.0-79.0) | 28.1 ± 11.2; 26.0 (10.0-70.0) | 16.2 ± 6.4; 14.0 (10.0-46.0) | <0.001 | 16.6 ± 6.4; 15.0 (10.0-43.0) | <0.001 |
| White blood count, per nL | 35 358.9 ± 33 350.0; 24 800.0 (13 000.0-370 580.0) | 7171.5 ± 2845.8; 6850.0 (750.0-12 990.0) | 4856.4 ± 1687.5; 4700.0 (1500.0-9960.0) | <0.001 | 4786.7 ± 1704.7; 4600.0 (1500.0-9960.0) | <0.001 |
| Hemoglobin, g/dL | 10.9 ± 2.3; 10.9 (4.0-18.7) | 11.3 ± 2.3; 11.2 (4.3-19.6) | 10.7 ± 2.2; 10.6 (4.3-19.6) | <0.001 | 10.5 ± 2.0; 10.3 (4.3-16.3) | <0.001 |
| HB <9 g/dL (transfusion dependency) | 96/546 (18%) | 95/649 (15%) | 54/349 (15%) | 0.7 | 43/238 (18%) | 0.2 |
| Platelets (per nL) | 168 240.9 ± 196 848.4; 114 500.0 (4000.0-1 809 000.0) | 138 521.6 ± 123 464.7; 108 000.0 (4000.0-1 385 000.0) | 197 412.6 ± 151 556.8; 148 000.0 (4000.0-988 000.0) | <0.001 | 196 071.4 ± 161 420.0; 138 000.0 (4000.0-988 000.0) | <0.001 |
| Absolute neutrophil count (PB, per nL) | 17 228.5 ± 16 322.8; 12 266.5 (1400.0-138 250.0) | 3316.8 ± 2053.3; 2866.5 (180.0-9144.0) | 2509.7 ± 1370.8; 2355.0 (124.8-7669.2) | <0.001 | 2463.8 ± 1412.3; 2172.5 (136.8-7669.2) | <0.001 |
| % Neutrophils (PB) | 52.5 ± 14.8; 54.0 (8.0-82.0) | 42.4 ± 15.6; 43.0 (5.0-79.0) | 48.9 ± 15.2; 50.0 (6.0-80.0) | <0.001 | 48.5 ± 15.4; 49.0 (6.0-80.0) | <0.001 |
| Cytopenic lineages | <0.001 | <0.001 | ||||
| 0 | 31/551 (5.6%) | 28/654 (4.3%) | 13/352 (3.7%) | 0/241 (0%) | ||
| 1 | 257/551 (47%) | 299/654 (46%) | 157/352 (45%) | 112/241 (46%) | ||
| 2 | 262/551 (48%) | 284/654 (43%) | 120/352 (34%) | 81/241 (34%) | ||
| 3 | 1/551 (0.2%) | 43/654 (6.6%) | 62/352 (18%) | 48/241 (20%) | ||
| % blasts (PB) | 1.2 ± 2.2; 0.0 (0.0-12.0) | 0.4 ± 1.4; 0.0 (0.0-14.0) | 0.2 ± 1.1; 0.0 (0.0-11.0) | <0.001 | 0.3 ± 1.2; 0.0 (0.0-11.0) | 0.002 |
| % blasts (BM) | 7.0 ± 5.0; 6.0 (0.0-19.5) | 6.3 ± 4.1; 5.5 (0.0-19.5) | 4.0 ± 3.5; 3.0 (0.0-18.0) | <0.001 | 4.4 ± 3.7; 3.5 (0.0-18.0) | <0.001 |
| Blasts in PB and BM | <0.001 | <0.001 | ||||
| <2% PB blasts and <5% BM blasts | 203/551 (37%) | 258/654 (39%) | 256/356 (72%) | 164/241 (68%) | ||
| ≥2% and <10% PB blasts or ≥5% and <10% BM blasts | 202/551 (37%) | 275/654 (42%) | 72/356 (20%) | 57/241 (24%) | ||
| ≥10% and <20% PB blasts or ≥10% and <20% BM blasts | 146/551 (26%) | 121/654 (19%) | 28/356 (7.9%) | 20/241 (8.3%) | ||
| Percentage of monocytes (BM), % | 15.9 ± 11.6; 13.0 (0.0-77.5) | 13.9 ± 10.3; 12.0 (0.0-77.0) | 4.7 ± 4.5; 3.5 (0.0-38.5) | <0.001 | 4.2 ± 4.1; 3.0 (0.0-31.0) | <0.001 |
| Ringsideroblasts (BM, in %) | 3.7 ± 12.1; 0.0 (0.0-89.0) | 7.7 ± 19.8; 0.0 (0.0-96.0) | 20.6 ± 30.2; 0.0 (0.0-97.0) | <0.001 | 23.6 ± 31.8; 1.0 (0.0-97.0) | <0.001 |
| Esterase positivity (BM, in %) | 19.7 ± 14.7; 20.0 (0.0-80.0) | 16.2 ± 13.3; 15.0 (0.0-70.0) | 6.6 ± 9.9; 0.0 (0.0-60.0) | <0.001 | 7.1 ± 10.5; 0.0 (0.0-60.0) | <0.001 |
| Cellularity (BM) | <0.001 | <0.001 | ||||
| Hypercellular | 474/551 (86%) | 506/654 (77%) | 250/356 (70%) | 241/241 (100%) | ||
| Hypocellular | 21/551 (3.8%) | 67/654 (10%) | 24/356 (6.7%) | 0/241 (0%) | ||
| Normal | 56/551 (10%) | 81/654 (12%) | 82/356 (23%) | 0/241 (0%) | ||
| Dysplastic lineages (BM) | <0.001 | 0.004 | ||||
| 0 | 112/551 (20%) | 81/654 (12%) | 0/356 (0%) | 12/241 (5.0%) | ||
| 1 | 167/551 (30%) | 141/654 (22%) | 93/356 (26%) | 45/241 (19%) | ||
| 2 | 161/551 (29%) | 230/654 (35%) | 132/356 (37%) | 88/241 (37%) | ||
| 3 | 111/551 (20%) | 202/654 (31%) | 131/356 (37%) | 96/241 (40%) | ||
| Karyotype risk score | <0.001 | <0.001 | ||||
| 0 | 431/551 (78%) | 560/654 (86%) | 258/356 (72%) | 177/241 (73%) | ||
| 1 | 54/551 (9.8%) | 45/654 (6.9%) | 47/356 (13%) | 28/241 (12%) | ||
| 2 | 66/551 (12%) | 49/654 (7.5%) | 51/356 (14%) | 36/241 (15%) | ||
| Molecular risk score | 0.077 | 0.11 | ||||
| 0 | 84/390 (22%) | 262/531 (49%) | 143/246 (58%) | 91/158 (58%) | ||
| 1 | 105/390 (27%) | 112/531 (21%) | 44/246 (18%) | 26/158 (16%) | ||
| 2 | 81/390 (21%) | 68/531 (13%) | 32/246 (13%) | 24/158 (15%) | ||
| 3 | 120/390 (31%) | 89/531 (17%) | 27/246 (11%) | 17/158 (11%) | ||
| CPSS-Mol (complete observations) | <0.001 | 0.029 | ||||
| Low | 0/429 (0%) | 113/543 (21%) | 95/256 (37%) | 54/168 (32%) | ||
| Intermediate 1 | 37/429 (8.6%) | 172/543 (32%) | 70/256 (27%) | 46/168 (27%) | ||
| Intermediate 2 | 180/429 (42%) | 191/543 (35%) | 68/256 (27%) | 53/168 (32%) | ||
| High | 212/429 (49%) | 67/543 (12%) | 23/256 (9.0%) | 15/168 (8.9%) | ||
| CPSS (complete observations) | 0.021 | 0.012 | ||||
| Low | 0/548 (0%) | 401/650 (62%) | 198/349 (57%) | 133/238 (56%) | ||
| Intermediate 1 | 282/548 (51%) | 173/650 (27%) | 90/349 (26%) | 61/238 (26%) | ||
| Intermediate 2 | 227/548 (41%) | 69/650 (11%) | 60/349 (17%) | 44/238 (18%) | ||
| High | 39/548 (7.1%) | 7/650 (1.1%) | 1/349 (0.3%) | 0/238 (0%) | ||
| Mean IPSS-M | ||||||
| Very low | 52/356 (15%) | 22/241 (9.1%) | ||||
| Low | 165/356 (46%) | 113/241 (47%) | ||||
| Moderate low | 51/356 (14%) | 42/241 (17%) | ||||
| Moderate high | 32/356 (9.0%) | 21/241 (8.7%) | ||||
| High | 35/356 (9.8%) | 25/241 (10%) | ||||
| Very high | 21/356 (5.9%) | 18/241 (7.5%) | ||||
| IPSS-R | ||||||
| Very low | 77/348 (22%) | 45/237 (19%) | ||||
| Low | 162/348 (47%) | 109/237 (46%) | ||||
| Intermediate | 68/348 (20%) | 53/237 (22%) | ||||
| High | 33/348 (9.5%) | 25/237 (11%) | ||||
| Very high | 8/348 (2.3%) | 5/237 (2.1%) |
| . | MP-WHOest n = 551∗ . | MD-WHOest n = 654∗ . | WHOnew n = 356∗ . | q value† MD-WHOest vs WHOnew . | ICCnew n = 241∗ . | q value† MD-WHOest vs ICCnew . |
|---|---|---|---|---|---|---|
| Age, y | 74.3 ± 8.6; 76.3 (37.8-93.3) | 74.3 ± 9.1; 75.6 (19.9-92.7) | 74.2 ± 9.3; 75.9 (30.8-91.9) | 0.9 | 75.3 ± 8.1; 76.8 (43.8-91.9) | 0.12 |
| Sex | 0.2 | 0.3 | ||||
| Female | 171/551 (31%) | 171/654 (26%) | 107/356 (30%) | 71/241 (29%) | ||
| Male | 380/551 (69%) | 483/654 (74%) | 249/356 (70%) | 170/241 (71%) | ||
| Clinical status | ||||||
| First diagnosis | 551/551 (100%) | 654/654 (100%) | 356/356 (100%) | 241/241 (100%) | ||
| Absolute monocyte count (PB), per nL | 9371.7 ± 12 137.4; 5667.0 (1240.0-95 680.0) | 1971.4 ± 1061.1; 1622.5 (1000.0-8160.0) | 709.3 ± 140.0; 694.5 (500.0-998.4) | <0.001 | 713.9 ± 138.5; 704.0 (500.0-998.4) | <0.001 |
| % Monocytes (PB) | 26.7 ± 13.2; 24.0 (10.0-79.0) | 28.1 ± 11.2; 26.0 (10.0-70.0) | 16.2 ± 6.4; 14.0 (10.0-46.0) | <0.001 | 16.6 ± 6.4; 15.0 (10.0-43.0) | <0.001 |
| White blood count, per nL | 35 358.9 ± 33 350.0; 24 800.0 (13 000.0-370 580.0) | 7171.5 ± 2845.8; 6850.0 (750.0-12 990.0) | 4856.4 ± 1687.5; 4700.0 (1500.0-9960.0) | <0.001 | 4786.7 ± 1704.7; 4600.0 (1500.0-9960.0) | <0.001 |
| Hemoglobin, g/dL | 10.9 ± 2.3; 10.9 (4.0-18.7) | 11.3 ± 2.3; 11.2 (4.3-19.6) | 10.7 ± 2.2; 10.6 (4.3-19.6) | <0.001 | 10.5 ± 2.0; 10.3 (4.3-16.3) | <0.001 |
| HB <9 g/dL (transfusion dependency) | 96/546 (18%) | 95/649 (15%) | 54/349 (15%) | 0.7 | 43/238 (18%) | 0.2 |
| Platelets (per nL) | 168 240.9 ± 196 848.4; 114 500.0 (4000.0-1 809 000.0) | 138 521.6 ± 123 464.7; 108 000.0 (4000.0-1 385 000.0) | 197 412.6 ± 151 556.8; 148 000.0 (4000.0-988 000.0) | <0.001 | 196 071.4 ± 161 420.0; 138 000.0 (4000.0-988 000.0) | <0.001 |
| Absolute neutrophil count (PB, per nL) | 17 228.5 ± 16 322.8; 12 266.5 (1400.0-138 250.0) | 3316.8 ± 2053.3; 2866.5 (180.0-9144.0) | 2509.7 ± 1370.8; 2355.0 (124.8-7669.2) | <0.001 | 2463.8 ± 1412.3; 2172.5 (136.8-7669.2) | <0.001 |
| % Neutrophils (PB) | 52.5 ± 14.8; 54.0 (8.0-82.0) | 42.4 ± 15.6; 43.0 (5.0-79.0) | 48.9 ± 15.2; 50.0 (6.0-80.0) | <0.001 | 48.5 ± 15.4; 49.0 (6.0-80.0) | <0.001 |
| Cytopenic lineages | <0.001 | <0.001 | ||||
| 0 | 31/551 (5.6%) | 28/654 (4.3%) | 13/352 (3.7%) | 0/241 (0%) | ||
| 1 | 257/551 (47%) | 299/654 (46%) | 157/352 (45%) | 112/241 (46%) | ||
| 2 | 262/551 (48%) | 284/654 (43%) | 120/352 (34%) | 81/241 (34%) | ||
| 3 | 1/551 (0.2%) | 43/654 (6.6%) | 62/352 (18%) | 48/241 (20%) | ||
| % blasts (PB) | 1.2 ± 2.2; 0.0 (0.0-12.0) | 0.4 ± 1.4; 0.0 (0.0-14.0) | 0.2 ± 1.1; 0.0 (0.0-11.0) | <0.001 | 0.3 ± 1.2; 0.0 (0.0-11.0) | 0.002 |
| % blasts (BM) | 7.0 ± 5.0; 6.0 (0.0-19.5) | 6.3 ± 4.1; 5.5 (0.0-19.5) | 4.0 ± 3.5; 3.0 (0.0-18.0) | <0.001 | 4.4 ± 3.7; 3.5 (0.0-18.0) | <0.001 |
| Blasts in PB and BM | <0.001 | <0.001 | ||||
| <2% PB blasts and <5% BM blasts | 203/551 (37%) | 258/654 (39%) | 256/356 (72%) | 164/241 (68%) | ||
| ≥2% and <10% PB blasts or ≥5% and <10% BM blasts | 202/551 (37%) | 275/654 (42%) | 72/356 (20%) | 57/241 (24%) | ||
| ≥10% and <20% PB blasts or ≥10% and <20% BM blasts | 146/551 (26%) | 121/654 (19%) | 28/356 (7.9%) | 20/241 (8.3%) | ||
| Percentage of monocytes (BM), % | 15.9 ± 11.6; 13.0 (0.0-77.5) | 13.9 ± 10.3; 12.0 (0.0-77.0) | 4.7 ± 4.5; 3.5 (0.0-38.5) | <0.001 | 4.2 ± 4.1; 3.0 (0.0-31.0) | <0.001 |
| Ringsideroblasts (BM, in %) | 3.7 ± 12.1; 0.0 (0.0-89.0) | 7.7 ± 19.8; 0.0 (0.0-96.0) | 20.6 ± 30.2; 0.0 (0.0-97.0) | <0.001 | 23.6 ± 31.8; 1.0 (0.0-97.0) | <0.001 |
| Esterase positivity (BM, in %) | 19.7 ± 14.7; 20.0 (0.0-80.0) | 16.2 ± 13.3; 15.0 (0.0-70.0) | 6.6 ± 9.9; 0.0 (0.0-60.0) | <0.001 | 7.1 ± 10.5; 0.0 (0.0-60.0) | <0.001 |
| Cellularity (BM) | <0.001 | <0.001 | ||||
| Hypercellular | 474/551 (86%) | 506/654 (77%) | 250/356 (70%) | 241/241 (100%) | ||
| Hypocellular | 21/551 (3.8%) | 67/654 (10%) | 24/356 (6.7%) | 0/241 (0%) | ||
| Normal | 56/551 (10%) | 81/654 (12%) | 82/356 (23%) | 0/241 (0%) | ||
| Dysplastic lineages (BM) | <0.001 | 0.004 | ||||
| 0 | 112/551 (20%) | 81/654 (12%) | 0/356 (0%) | 12/241 (5.0%) | ||
| 1 | 167/551 (30%) | 141/654 (22%) | 93/356 (26%) | 45/241 (19%) | ||
| 2 | 161/551 (29%) | 230/654 (35%) | 132/356 (37%) | 88/241 (37%) | ||
| 3 | 111/551 (20%) | 202/654 (31%) | 131/356 (37%) | 96/241 (40%) | ||
| Karyotype risk score | <0.001 | <0.001 | ||||
| 0 | 431/551 (78%) | 560/654 (86%) | 258/356 (72%) | 177/241 (73%) | ||
| 1 | 54/551 (9.8%) | 45/654 (6.9%) | 47/356 (13%) | 28/241 (12%) | ||
| 2 | 66/551 (12%) | 49/654 (7.5%) | 51/356 (14%) | 36/241 (15%) | ||
| Molecular risk score | 0.077 | 0.11 | ||||
| 0 | 84/390 (22%) | 262/531 (49%) | 143/246 (58%) | 91/158 (58%) | ||
| 1 | 105/390 (27%) | 112/531 (21%) | 44/246 (18%) | 26/158 (16%) | ||
| 2 | 81/390 (21%) | 68/531 (13%) | 32/246 (13%) | 24/158 (15%) | ||
| 3 | 120/390 (31%) | 89/531 (17%) | 27/246 (11%) | 17/158 (11%) | ||
| CPSS-Mol (complete observations) | <0.001 | 0.029 | ||||
| Low | 0/429 (0%) | 113/543 (21%) | 95/256 (37%) | 54/168 (32%) | ||
| Intermediate 1 | 37/429 (8.6%) | 172/543 (32%) | 70/256 (27%) | 46/168 (27%) | ||
| Intermediate 2 | 180/429 (42%) | 191/543 (35%) | 68/256 (27%) | 53/168 (32%) | ||
| High | 212/429 (49%) | 67/543 (12%) | 23/256 (9.0%) | 15/168 (8.9%) | ||
| CPSS (complete observations) | 0.021 | 0.012 | ||||
| Low | 0/548 (0%) | 401/650 (62%) | 198/349 (57%) | 133/238 (56%) | ||
| Intermediate 1 | 282/548 (51%) | 173/650 (27%) | 90/349 (26%) | 61/238 (26%) | ||
| Intermediate 2 | 227/548 (41%) | 69/650 (11%) | 60/349 (17%) | 44/238 (18%) | ||
| High | 39/548 (7.1%) | 7/650 (1.1%) | 1/349 (0.3%) | 0/238 (0%) | ||
| Mean IPSS-M | ||||||
| Very low | 52/356 (15%) | 22/241 (9.1%) | ||||
| Low | 165/356 (46%) | 113/241 (47%) | ||||
| Moderate low | 51/356 (14%) | 42/241 (17%) | ||||
| Moderate high | 32/356 (9.0%) | 21/241 (8.7%) | ||||
| High | 35/356 (9.8%) | 25/241 (10%) | ||||
| Very high | 21/356 (5.9%) | 18/241 (7.5%) | ||||
| IPSS-R | ||||||
| Very low | 77/348 (22%) | 45/237 (19%) | ||||
| Low | 162/348 (47%) | 109/237 (46%) | ||||
| Intermediate | 68/348 (20%) | 53/237 (22%) | ||||
| High | 33/348 (9.5%) | 25/237 (11%) | ||||
| Very high | 8/348 (2.3%) | 5/237 (2.1%) |
BM, bone marrow; HB, hemoglobin; PB, peripheral blood; SD, standard deviation,
n / N (%); mean ± SD; median (range).
Pearson χ2 test; Wilcoxon rank sum test; and Fisher exact test after false discovery rate correction for multiple testing.
Clonality assessment is another major criterion, which differs in the VAF requirements for pathological mutations between classifications. Specifically, the ICC22 requires a VAF ≥10%, whereas the current version of the WHO22 has not established a VAF threshold for the definition of clonality in the absence of aberrant cytogenetics. To permit comparability while still minimizing false positives, a 2% VAF threshold was applied to the WHO22 cohort, analogous to the threshold for defining clonal hematopoiesis of indeterminate potential.5 However, only 2 (∼0.002%) of all established CMML cases had a VAF of 2% to 10% without accompanying karyotypical aberrations, making the VAF threshold distinction negligible in routine diagnostics for cases with PB monocytosis ≥1 × 109/L (Figure 2D; supplemental Figure 5B-C). In contrast, 8% of WHOnew cases presented with a mutation with a VAF of 2% to 10% as the sole evidence of clonality, matching previous reports considering OM-CMML as a precursor of CMML disease.7
According to the WHO22 classification, 1 lineage dysplasia is a prerequisite criterion for OM-CMML (equivalent to the WHOnew cohort) while only being a supporting criterium for CMML cases with PB monocytosis ≥1 × 109/L. Therefore, BM dysplasia was present in all WHOnew, vs 88% of MD-WHOest cases (Figure 2E; supplemental Figure 5D).
Another necessary criterion in the ICC22 classification is CMML-typical BM morphology, defined as a hypercellular BM due to myeloid proliferation, often with increased monocytes.5 Unexpectedly, this criterion applied to only 77% and 86% of MD-WHOest and MP-WHOest cases, respectively, and was lower (70%) in the WHOnew cohort (Figure 2F; supplemental Figure 5E). Interestingly, although hypercellularity was necessarily observed in all ICCnew cases, BM monocyte expansion >10% was present in only ∼40% of those cases (supplemental Figure 5F).
In a final step, we assessed the redundancy (not independently contributing to establishing the diagnosis) of diagnostic criteria for all established CMML cases. This analysis suggested that the WHO22 criterion of dysplasia is largely redundant when evidence of clonality can be sufficiently assessed (supplemental Figure 6A). Furthermore, when examining the ICC22 criteria, both increased blast threshold and abnormal immunophenotyping were redundant (supplemental Figure 6B). However, this assessment was only possible in a fully characterized cohort, whereas redundancy was greatly reduced when including samples with partially available information (supplemental Figure 6C-D).
Differential assessment of clinical and immunophenotypical parameters in established and reclassified cases
An assessment of clinical and immunophenotypical parameters was performed between WHOest and OM-CMML cases. A separate comparison of OM to ICCest cases was not performed, because we did not observe relevant differences in the composition of the WHOest and ICCest cohorts warranting a separate comparison (supplemental Tables 2.1-2.4). WHOnew, compared with MD-WHOest cases, presented with significantly lower white blood cell counts (4.9 × 109/L vs 7.2 × 109/L; q <0.001), absolute neutrophil counts (2.5 × 109/L vs 3.3 × 109/L; q <0.001), and absolute (0.7 × 109/L vs 2.0 × 109/L; q <0.001) and relative PB monocyte counts (16.2% vs 28.1%; q <0.001; Table 1). Conversely, the WHOnew cohort showed significantly higher platelet counts (197.4 × 109/L vs 138.5 × 109/L in MD-WHOest; q <0.001). A small but significant difference in hemoglobin levels between WHOnew and MD-WHOest cases (10.7 vs 11.3 g/dL; q <0.001) did not affect sex-specific hemoglobin thresholds as an indicator of red blood cell transfusion dependency (q = 0.7).17,18 Finally, blast counts were significantly lower in WHOnew cases, both in the PB (0.2% vs 0.4%; q <0.001) and BM (4.0% vs 6.3%; q <0.001). Of note, because the ICCnew cohort was almost exclusively a subgroup of the WHOnew cohort, all observations followed the same trends when comparing the ICCnew with the WHOest or ICCest cohorts.
We further compared the immunophenotypic profiles of MD-WHOest with WHOnew and ICCnew cases. Because this was a retrospective analysis, either PB or BM (or both) were considered, depending on sample availability. MD-WHOest cases showed a significantly higher number of aberrantly expressed surface markers than WHOnew (mean 5.2 vs 3.9 per case, q <0.001) and ICCnew cohorts (mean, 5.2 vs 4.4; q = 0.003). Expression differences in individual markers are depicted in Table 2.
Immunophenotypes of WHOest, WHOnew, and ICCnew
| . | MP-WHOest n = 551∗ . | MD-WHOest n = 654∗ . | WHOnew n = 356∗ . | q value† MD-WHOest vs WHOnew . | ICCnew n = 241∗ . | q value† MD-WHOest vs ICCnew . |
|---|---|---|---|---|---|---|
| Aberrant marker expression | 6.0 ± 2.4; 6.0 (0.0-13.0) | 5.2 ± 2.1; 5.0 (0.0-13.0) | 3.9 ± 2.3; 4.0 (0.0-11.0) | <0.001 | 4.4 ± 2.3; 4.0 (0.0-11.0) | 0.003 |
| ≥3 aberrantly expressed surface markers (ICC22 flow criterium) | 318/366 (87%) | 442/484 (91%) | 210/296 (71%) | <0.001 | 152/199 (76%) | <0.001 |
| Blast (%) | 1.4 ± 2.1; 0.6 (0.0-14.0) | 1.3 ± 2.1; 0.7 (0.0-13.0) | 1.3 ± 2.1; 0.7 (0.0-18.0) | >0.9 | 1.5 ± 2.4; 0.8 (0.0-18.0) | 0.2 |
| Granulocytes (%) | 66.8 ± 14.3; 69.0 (20.0-92.0) | 62.9 ± 14.2; 66.0 (13.0-89.0) | 61.6 ± 15.7; 64.0 (11.0-88.0) | 0.7 | 62.3 ± 15.4; 65.0 (11.0-88.0) | >0.9 |
| Monocytes (%) | 12.9 ± 9.5; 11.0 (0.0-56.0) | 10.7 ± 6.0; 10.0 (0.5-37.0) | 5.9 ± 4.1; 5.0 (0.3-22.0) | <0.001 | 5.9 ± 4.0; 4.0 (0.7-20.0) | <0.001 |
| Erythrocytes (%) | 0.9 ± 1.5; 0.4 (0.0-16.0) | 1.3 ± 2.0; 0.6 (0.0-15.0) | 2.0 ± 2.9; 1.0 (0.0-20.0) | 0.002 | 2.2 ± 3.1; 1.0 (0.0-20.0) | 0.002 |
| Monocyte CD2 | 103/209 (49%) | 139/325 (43%) | 49/194 (25%) | <0.001 | 36/129 (28%) | 0.013 |
| Monocyte CD11b | 27/209 (13%) | 37/325 (11%) | 9/194 (4.6%) | 0.021 | 7/129 (5.4%) | 0.15 |
| Monocyte CD13 | 112/209 (54%) | 165/325 (51%) | 65/194 (34%) | <0.001 | 53/129 (41%) | 0.2 |
| Monocyte CD14 | 61/209 (29%) | 86/325 (26%) | 32/194 (16%) | 0.021 | 27/129 (21%) | 0.4 |
| Monocyte CD33 | 28/209 (13%) | 26/325 (8.0%) | 15/194 (7.7%) | >0.9 | 10/129 (7.8%) | >0.9 |
| Monocyte CD45 | 85/209 (41%) | 95/325 (29%) | 21/194 (11%) | <0.001 | 15/129 (12%) | <0.001 |
| Monocyte CD56 | 133/209 (64%) | 179/325 (55%) | 66/194 (34%) | <0.001 | 46/129 (36%) | 0.001 |
| Monocyte HLA-DR | 96/209 (46%) | 125/325 (38%) | 50/194 (26%) | 0.010 | 37/129 (29%) | 0.15 |
| Granulocyte CD13 | 2/209 (1.0%) | 0/325 (0%) | 0/194 (0%) | 0/129 (0%) | ||
| Granulocyte CD33 | 3/209 (1.4%) | 0/325 (0%) | 1/194 (0.5%) | 0.6 | 1/129 (0.8%) | 0.4 |
| Granulocyte CD56 | 29/209 (14%) | 31/325 (9.5%) | 15/194 (7.7%) | 0.7 | 12/129 (9.3%) | >0.9 |
| Granulocyte CD11b-CD16 | 148/209 (71%) | 238/325 (73%) | 124/194 (64%) | 0.053 | 87/129 (67%) | 0.4 |
| Granulocyte CD13-CD16 | 66/209 (32%) | 88/325 (27%) | 49/194 (25%) | 0.8 | 37/129 (29%) | 0.9 |
| Granulocyte CD11b-CD13 | 45/209 (22%) | 40/325 (12%) | 25/194 (13%) | >0.9 | 23/129 (18%) | 0.2 |
| Granulocyte SSC | 0.068 | 0.9 | ||||
| 0 | 92/209 (44%) | 162/325 (50%) | 118/194 (61%) | 70/129 (54%) | ||
| 1 | 89/209 (43%) | 129/325 (40%) | 64/194 (33%) | 47/129 (36%) | ||
| 2 | 28/209 (13%) | 34/325 (10%) | 12/194 (6.2%) | 12/129 (9.3%) | ||
| Erythrocyte CD71 | 44/209 (21%) | 103/325 (32%) | 88/194 (45%) | 0.006 | 61/129 (47%) | 0.008 |
| Blast CD2 | 2/209 (1.0%) | 0/325 (0%) | 1/194 (0.5%) | 0.6 | 1/129 (0.8%) | 0.4 |
| Blast CD5 | 62/209 (30%) | 51/325 (16%) | 16/194 (8.2%) | 0.032 | 12/129 (9.3%) | 0.2 |
| Blast CD7 | 35/209 (17%) | 29/325 (8.9%) | 18/194 (9.3%) | >0.9 | 16/129 (12%) | 0.4 |
| Blast CD11b | 1/209 (0.5%) | 1/325 (0.3%) | 0/194 (0%) | >0.9 | 0/129 (0%) | >0.9 |
| Blast CD13 | 1/209 (0.5%) | 0/325 (0%) | 2/194 (1.0%) | 0.2 | 1/129 (0.8%) | 0.4 |
| Blast CD33 | 1/209 (0.5%) | 0/325 (0%) | 0/194 (0%) | 0/129 (0%) | ||
| Blast CD34 | 1/209 (0.5%) | 3/325 (0.9%) | 1/194 (0.5%) | >0.9 | 1/129 (0.8%) | >0.9 |
| Blast CD34 strong | 0/209 (0%) | 0/325 (0%) | 0/194 (0%) | 0/129 (0%) | ||
| Blast CD45 | 15/209 (7.2%) | 24/325 (7.4%) | 7/194 (3.6%) | 0.14 | 4/129 (3.1%) | 0.2 |
| Blast CD56 | 14/209 (6.7%) | 21/325 (6.5%) | 2/194 (1.0%) | 0.010 | 2/129 (1.6%) | 0.11 |
| Blast CD117 | 0/209 (0%) | 1/325 (0.3%) | 3/194 (1.5%) | 0.2 | 1/129 (0.8%) | 0.6 |
| Blast HLA-DR | 1/209 (0.5%) | 1/325 (0.3%) | 1/194 (0.5%) | >0.9 | 1/129 (0.8%) | 0.6 |
| Blast HLA-DR strong | 1/209 (0.5%) | 0/325 (0%) | 0/194 (0%) | 0/129 (0%) |
| . | MP-WHOest n = 551∗ . | MD-WHOest n = 654∗ . | WHOnew n = 356∗ . | q value† MD-WHOest vs WHOnew . | ICCnew n = 241∗ . | q value† MD-WHOest vs ICCnew . |
|---|---|---|---|---|---|---|
| Aberrant marker expression | 6.0 ± 2.4; 6.0 (0.0-13.0) | 5.2 ± 2.1; 5.0 (0.0-13.0) | 3.9 ± 2.3; 4.0 (0.0-11.0) | <0.001 | 4.4 ± 2.3; 4.0 (0.0-11.0) | 0.003 |
| ≥3 aberrantly expressed surface markers (ICC22 flow criterium) | 318/366 (87%) | 442/484 (91%) | 210/296 (71%) | <0.001 | 152/199 (76%) | <0.001 |
| Blast (%) | 1.4 ± 2.1; 0.6 (0.0-14.0) | 1.3 ± 2.1; 0.7 (0.0-13.0) | 1.3 ± 2.1; 0.7 (0.0-18.0) | >0.9 | 1.5 ± 2.4; 0.8 (0.0-18.0) | 0.2 |
| Granulocytes (%) | 66.8 ± 14.3; 69.0 (20.0-92.0) | 62.9 ± 14.2; 66.0 (13.0-89.0) | 61.6 ± 15.7; 64.0 (11.0-88.0) | 0.7 | 62.3 ± 15.4; 65.0 (11.0-88.0) | >0.9 |
| Monocytes (%) | 12.9 ± 9.5; 11.0 (0.0-56.0) | 10.7 ± 6.0; 10.0 (0.5-37.0) | 5.9 ± 4.1; 5.0 (0.3-22.0) | <0.001 | 5.9 ± 4.0; 4.0 (0.7-20.0) | <0.001 |
| Erythrocytes (%) | 0.9 ± 1.5; 0.4 (0.0-16.0) | 1.3 ± 2.0; 0.6 (0.0-15.0) | 2.0 ± 2.9; 1.0 (0.0-20.0) | 0.002 | 2.2 ± 3.1; 1.0 (0.0-20.0) | 0.002 |
| Monocyte CD2 | 103/209 (49%) | 139/325 (43%) | 49/194 (25%) | <0.001 | 36/129 (28%) | 0.013 |
| Monocyte CD11b | 27/209 (13%) | 37/325 (11%) | 9/194 (4.6%) | 0.021 | 7/129 (5.4%) | 0.15 |
| Monocyte CD13 | 112/209 (54%) | 165/325 (51%) | 65/194 (34%) | <0.001 | 53/129 (41%) | 0.2 |
| Monocyte CD14 | 61/209 (29%) | 86/325 (26%) | 32/194 (16%) | 0.021 | 27/129 (21%) | 0.4 |
| Monocyte CD33 | 28/209 (13%) | 26/325 (8.0%) | 15/194 (7.7%) | >0.9 | 10/129 (7.8%) | >0.9 |
| Monocyte CD45 | 85/209 (41%) | 95/325 (29%) | 21/194 (11%) | <0.001 | 15/129 (12%) | <0.001 |
| Monocyte CD56 | 133/209 (64%) | 179/325 (55%) | 66/194 (34%) | <0.001 | 46/129 (36%) | 0.001 |
| Monocyte HLA-DR | 96/209 (46%) | 125/325 (38%) | 50/194 (26%) | 0.010 | 37/129 (29%) | 0.15 |
| Granulocyte CD13 | 2/209 (1.0%) | 0/325 (0%) | 0/194 (0%) | 0/129 (0%) | ||
| Granulocyte CD33 | 3/209 (1.4%) | 0/325 (0%) | 1/194 (0.5%) | 0.6 | 1/129 (0.8%) | 0.4 |
| Granulocyte CD56 | 29/209 (14%) | 31/325 (9.5%) | 15/194 (7.7%) | 0.7 | 12/129 (9.3%) | >0.9 |
| Granulocyte CD11b-CD16 | 148/209 (71%) | 238/325 (73%) | 124/194 (64%) | 0.053 | 87/129 (67%) | 0.4 |
| Granulocyte CD13-CD16 | 66/209 (32%) | 88/325 (27%) | 49/194 (25%) | 0.8 | 37/129 (29%) | 0.9 |
| Granulocyte CD11b-CD13 | 45/209 (22%) | 40/325 (12%) | 25/194 (13%) | >0.9 | 23/129 (18%) | 0.2 |
| Granulocyte SSC | 0.068 | 0.9 | ||||
| 0 | 92/209 (44%) | 162/325 (50%) | 118/194 (61%) | 70/129 (54%) | ||
| 1 | 89/209 (43%) | 129/325 (40%) | 64/194 (33%) | 47/129 (36%) | ||
| 2 | 28/209 (13%) | 34/325 (10%) | 12/194 (6.2%) | 12/129 (9.3%) | ||
| Erythrocyte CD71 | 44/209 (21%) | 103/325 (32%) | 88/194 (45%) | 0.006 | 61/129 (47%) | 0.008 |
| Blast CD2 | 2/209 (1.0%) | 0/325 (0%) | 1/194 (0.5%) | 0.6 | 1/129 (0.8%) | 0.4 |
| Blast CD5 | 62/209 (30%) | 51/325 (16%) | 16/194 (8.2%) | 0.032 | 12/129 (9.3%) | 0.2 |
| Blast CD7 | 35/209 (17%) | 29/325 (8.9%) | 18/194 (9.3%) | >0.9 | 16/129 (12%) | 0.4 |
| Blast CD11b | 1/209 (0.5%) | 1/325 (0.3%) | 0/194 (0%) | >0.9 | 0/129 (0%) | >0.9 |
| Blast CD13 | 1/209 (0.5%) | 0/325 (0%) | 2/194 (1.0%) | 0.2 | 1/129 (0.8%) | 0.4 |
| Blast CD33 | 1/209 (0.5%) | 0/325 (0%) | 0/194 (0%) | 0/129 (0%) | ||
| Blast CD34 | 1/209 (0.5%) | 3/325 (0.9%) | 1/194 (0.5%) | >0.9 | 1/129 (0.8%) | >0.9 |
| Blast CD34 strong | 0/209 (0%) | 0/325 (0%) | 0/194 (0%) | 0/129 (0%) | ||
| Blast CD45 | 15/209 (7.2%) | 24/325 (7.4%) | 7/194 (3.6%) | 0.14 | 4/129 (3.1%) | 0.2 |
| Blast CD56 | 14/209 (6.7%) | 21/325 (6.5%) | 2/194 (1.0%) | 0.010 | 2/129 (1.6%) | 0.11 |
| Blast CD117 | 0/209 (0%) | 1/325 (0.3%) | 3/194 (1.5%) | 0.2 | 1/129 (0.8%) | 0.6 |
| Blast HLA-DR | 1/209 (0.5%) | 1/325 (0.3%) | 1/194 (0.5%) | >0.9 | 1/129 (0.8%) | 0.6 |
| Blast HLA-DR strong | 1/209 (0.5%) | 0/325 (0%) | 0/194 (0%) | 0/129 (0%) |
Mean ± standard deviation; median (range); n / N (%).
Wilcoxon rank sum test; Pearson χ2 test after false discovery rate correction for multiple testing.
The genetic landscape of established and newly classified CMML cases
Clonal cytogenetic abnormalities have been described in 20% to 40% of CMML cases.34,35 Karyotype assessment showed a significantly higher number of aberrations in WHOnew vs MD-WHOest cases (0.7 vs 0.4 per case; q <0.001), with a higher rate of complex karyotypes (2.6% vs 1.8%) and a concomitantly lower rate of cases with normal karyotype (60% vs 79%). As in the assessment of clinical and immunophenotypical differences, WHOnew and ICCnew were in alignment regarding their differences to the WHOest cohort. Differences in individual cytogenetic alterations are shown in Table 3. Most notably, deletion 5q, deletion 20q, and chrY loss were significantly more frequent in both OM-CMML cohorts.
Cytogenetic alterations in WHOest, WHOnew, and ICCnew
| . | MP-WHOest n = 551∗ . | MD-WHOest n = 654∗ . | WHOnew n = 356∗ . | q value† MD-WHOest vs WHOnew . | ICCnew N = 241∗ . | q value† MD-WHOest vs ICCnew . |
|---|---|---|---|---|---|---|
| Clinical karyotype | <0.001 | <0.001 | ||||
| Normal karyotype | 401/538 (75%) | 515/649 (79%) | 211/351 (60%) | 146/240 (61%) | ||
| Aberrant1 | 105/538 (20%) | 107/649 (16%) | 105/351 (30%) | 74/240 (31%) | ||
| Aberrant2 | 20/538 (3.7%) | 9/649 (1.4%) | 22/351 (6.3%) | 10/240 (4.2%) | ||
| Aberrant3 | 5/538 (0.9%) | 6/649 (0.9%) | 4/351 (1.1%) | 3/240 (1.3%) | ||
| Complex >3 | 7/538 (1.3%) | 12/649 (1.8%) | 9/351 (2.6%) | 7/240 (2.9%) | ||
| Total aberrations | 0.4 ± 1.0; 0.0 (0.0-10.0) | 0.4 ± 1.1; 0.0 (0.0-13.0) | 0.7 ± 1.5; 0.0 (0.0-15.0) | <0.001 | 0.7 ± 1.7; 0.0 (0.0-15.0) | <0.001 |
| del(5q) | 4/551 (0.7%) | 17/654 (2.6%) | 27/356 (7.6%) | 0.003 | 19/241 (7.9%) | 0.003 |
| chr7 loss | 16/551 (2.9%) | 11/654 (1.7%) | 12/356 (3.4%) | 0.3 | 10/241 (4.1%) | 0.15 |
| del(7q) | 4/551 (0.7%) | 6/654 (0.9%) | 6/356 (1.7%) | 0.6 | 2/241 (0.8%) | >0.9 |
| chr8 gain | 36/551 (6.5%) | 27/654 (4.1%) | 26/356 (7.3%) | 0.13 | 18/241 (7.5%) | 0.2 |
| del(11q) | 4/551 (0.7%) | 1/654 (0.2%) | 5/356 (1.4%) | 0.11 | 3/241 (1.2%) | 0.2 |
| del(12p) | 8/551 (1.5%) | 2/654 (0.3%) | 1/356 (0.3%) | >0.9 | 1/241 (0.4%) | >0.9 |
| chr19 gain | 1/551 (0.2%) | 4/654 (0.6%) | 1/356 (0.3%) | 0.9 | 1/241 (0.4%) | >0.9 |
| del(20q) | 9/551 (1.6%) | 9/654 (1.4%) | 15/356 (4.2%) | 0.036 | 14/241 (5.8%) | 0.003 |
| chr21 gain | 13/551 (2.4%) | 1/654 (0.2%) | 2/356 (0.6%) | 0.5 | 2/241 (0.8%) | 0.4 |
| chrY loss | 18/551 (3.3%) | 45/654 (6.9%) | 49/356 (14%) | 0.003 | 31/241 (13%) | 0.034 |
| . | MP-WHOest n = 551∗ . | MD-WHOest n = 654∗ . | WHOnew n = 356∗ . | q value† MD-WHOest vs WHOnew . | ICCnew N = 241∗ . | q value† MD-WHOest vs ICCnew . |
|---|---|---|---|---|---|---|
| Clinical karyotype | <0.001 | <0.001 | ||||
| Normal karyotype | 401/538 (75%) | 515/649 (79%) | 211/351 (60%) | 146/240 (61%) | ||
| Aberrant1 | 105/538 (20%) | 107/649 (16%) | 105/351 (30%) | 74/240 (31%) | ||
| Aberrant2 | 20/538 (3.7%) | 9/649 (1.4%) | 22/351 (6.3%) | 10/240 (4.2%) | ||
| Aberrant3 | 5/538 (0.9%) | 6/649 (0.9%) | 4/351 (1.1%) | 3/240 (1.3%) | ||
| Complex >3 | 7/538 (1.3%) | 12/649 (1.8%) | 9/351 (2.6%) | 7/240 (2.9%) | ||
| Total aberrations | 0.4 ± 1.0; 0.0 (0.0-10.0) | 0.4 ± 1.1; 0.0 (0.0-13.0) | 0.7 ± 1.5; 0.0 (0.0-15.0) | <0.001 | 0.7 ± 1.7; 0.0 (0.0-15.0) | <0.001 |
| del(5q) | 4/551 (0.7%) | 17/654 (2.6%) | 27/356 (7.6%) | 0.003 | 19/241 (7.9%) | 0.003 |
| chr7 loss | 16/551 (2.9%) | 11/654 (1.7%) | 12/356 (3.4%) | 0.3 | 10/241 (4.1%) | 0.15 |
| del(7q) | 4/551 (0.7%) | 6/654 (0.9%) | 6/356 (1.7%) | 0.6 | 2/241 (0.8%) | >0.9 |
| chr8 gain | 36/551 (6.5%) | 27/654 (4.1%) | 26/356 (7.3%) | 0.13 | 18/241 (7.5%) | 0.2 |
| del(11q) | 4/551 (0.7%) | 1/654 (0.2%) | 5/356 (1.4%) | 0.11 | 3/241 (1.2%) | 0.2 |
| del(12p) | 8/551 (1.5%) | 2/654 (0.3%) | 1/356 (0.3%) | >0.9 | 1/241 (0.4%) | >0.9 |
| chr19 gain | 1/551 (0.2%) | 4/654 (0.6%) | 1/356 (0.3%) | 0.9 | 1/241 (0.4%) | >0.9 |
| del(20q) | 9/551 (1.6%) | 9/654 (1.4%) | 15/356 (4.2%) | 0.036 | 14/241 (5.8%) | 0.003 |
| chr21 gain | 13/551 (2.4%) | 1/654 (0.2%) | 2/356 (0.6%) | 0.5 | 2/241 (0.8%) | 0.4 |
| chrY loss | 18/551 (3.3%) | 45/654 (6.9%) | 49/356 (14%) | 0.003 | 31/241 (13%) | 0.034 |
n / N (%).
Wilcoxon rank sum test and Pearson χ2 test with false discovery rate correction for multiple testing.
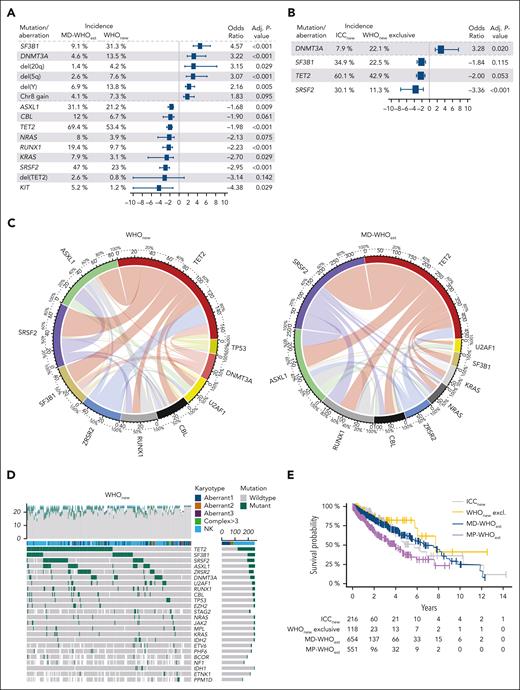
Using targeted next-generation sequencing and covering a mean of 48 genes per sample, we defined the mutational landscape of all cohorts (Figure 3A-D; supplemental Figures 7A-F and 8A-D; Table 4). MP-WHOest presented with the highest number of mutated genes (mean 3.5 per case), followed by MD-WHOest (3.1 per case). The number of mutations did not significantly differ between the WHOnew and ICCnew cohorts (mean, 2.2. vs 2.4 mutations per sample). Importantly, WHOnew cases presented with mutational frequencies closely resembling those of historical MDS cohorts.36 Specifically, mutations occurred more frequently in DNMT3A and SF3B1 while occurring less frequently in ASXL1, RUNX1, SRSF2, and TET2 when compared with the MD-WHOest cohort, aligning with the observation that most WHOnew cases were previously classified as MDS (Figure 3A). Moreover, a significantly lower number of multiple TET2 mutations was observed in patients in WHOnew cohort (30% vs 47.4% in MD-WHOest; q <0.001), previously described as a typical finding in CMML.37 Interestingly, when comparing cases exclusively falling into the WHOnew cohort with combined ICCnew and WHOnew cases, we observed a higher incidence of DNMT3A mutations (odds ratio [OR], 3.2; q = 0.020), whereas SRSF2 (OR, 3.4; q <0.001), SF3B1 (OR, 1.8; q = 0.115) and TET2 (OR, 2.0; q = 0.053) mutations presented with the opposite trend (Figure 3B). Furthermore, MD-WHOest cases showed a significantly higher percentage of KRAS (7.9% vs 3.1%; q = 0.026) mutations and trend toward higher incidences of NRAS (8.0% vs 3.9%; q = 0.065) and CBL (12.0% vs 6.7%; q = 0.053) mutations. The less frequent occurrence of these mutations associated with proliferative features is in line with their lower frequency in OM-CMML cases.38,39
Molecular data analysis of MD-WHOest, WHOnew, and ICCnew cases. (A-B) Forest Plots comparing the incidences and ORs of 15 alterations significantly differing between 654 MD-WHOest and 356 WHOnew cases (A) and 4 alterations significantly differing between 241 ICCnew and 127 WHOnew exclusive cases (B). (C) Circos plots showing the comutational profile of 251 WHOnew (left) and 474 MD-WHOnew (right) cases with >20 genes sequenced. (D) Mutations were identified by NGS in 251 WHOnew patients with >20 genes sequenced. The 24 mutations with highest incidence and respective karyotypes are depicted, ordered from the most to the least frequently mutated, with each column representing a patient and each row representing a gene. The number of mutations identified/genes sequenced per patient is shown as columns in the top row. (E) Kaplan-Meier curves of the ICCnew, WHOnew exclusive, MD-WHOest, and MP-WHOest cohorts. WHOnew and ICCest cases showed a median OS of 7.7 and 5.7 years (P = .520 and P = .796 vs MD-CMMLest, respectively), vs 7.5 years in MD-WHOest and 3.3 years in MP-WHOest. Aberrant1-3, 1-3 cytogenetic aberrancies; Complex >3, complex karyotype with >3 cytogenetic aberrancies; NGS, next-generation sequencing; NK, normal karyotype.
Molecular data analysis of MD-WHOest, WHOnew, and ICCnew cases. (A-B) Forest Plots comparing the incidences and ORs of 15 alterations significantly differing between 654 MD-WHOest and 356 WHOnew cases (A) and 4 alterations significantly differing between 241 ICCnew and 127 WHOnew exclusive cases (B). (C) Circos plots showing the comutational profile of 251 WHOnew (left) and 474 MD-WHOnew (right) cases with >20 genes sequenced. (D) Mutations were identified by NGS in 251 WHOnew patients with >20 genes sequenced. The 24 mutations with highest incidence and respective karyotypes are depicted, ordered from the most to the least frequently mutated, with each column representing a patient and each row representing a gene. The number of mutations identified/genes sequenced per patient is shown as columns in the top row. (E) Kaplan-Meier curves of the ICCnew, WHOnew exclusive, MD-WHOest, and MP-WHOest cohorts. WHOnew and ICCest cases showed a median OS of 7.7 and 5.7 years (P = .520 and P = .796 vs MD-CMMLest, respectively), vs 7.5 years in MD-WHOest and 3.3 years in MP-WHOest. Aberrant1-3, 1-3 cytogenetic aberrancies; Complex >3, complex karyotype with >3 cytogenetic aberrancies; NGS, next-generation sequencing; NK, normal karyotype.
Mutational profiles of WHOest, WHOnew, and ICCnew
| . | MP-WHOest n = 549∗ . | MD-WHOest n = 654∗ . | WHOnew n = 356∗ . | q value† MD-WHOest vs WHOnew . | ICCnew n = 241∗ . | q value† MD-WHOest vs ICCnew . |
|---|---|---|---|---|---|---|
| Assessed genes | 49.9 ± 42.0; 35.0 (1.0-131.0) | 54.8 ± 39.7; 40.0 (1.0-135.0) | 47.0 ± 37.2; 36.0 (1.0-132.0) | <0.001 | 44.8 ± 37.6; 34.0 (1.0-132.0) | <0.001 |
| Mutated genes | 3.5 ± 2.3; 4.0 (0.0-12.0) | 3.1 ± 2.1; 3.0 (0.0-14.0) | 2.2 ± 1.8; 2.0 (0.0-9.0) | <0.001 | 2.4 ± 1.9; 2.0 (0.0-9.0) | <0.001 |
| NGS assessed genes | 46.1 ± 42.3; 32.0 (0.0-129.0) | 50.9 ± 40.2; 35.0 (0.0-130.0) | 46.1 ± 37.4; 35.0 (0.0-130.0) | 0.5 | 43.9 ± 37.9; 34.0 (0.0-130.0) | 0.3 |
| NGS mutated genes | 3.3 ± 2.2; 3.0 (0.0-12.0) | 2.9 ± 1.9; 3.0 (0.0-12.0) | 2.2 ± 1.8; 2.0 (0.0-9.0) | <0.001 | 2.3 ± 1.8; 2.0 (0.0-9.0) | <0.001 |
| Highest VAF | 55.1 ± 19.5; 50.0 (0.0-100.0) | 52.6 ± 19.6; 48.0 (0.0-100.0) | 41.2 ± 21.7; 40.0 (2.1-97.0) | <0.001 | 45.1 ± 20.1; 42.0 (4.0-97.0) | <0.001 |
| VAF cutoffs | <0.001 | 0.3 | ||||
| <2% / no clonality | 35/495 (7.1%) | 47/623 (7.5%) | 42/351 (12%) | 28/238 (12%) | ||
| 2%-10% | 1/495 (0.2%) | 4/623 (0.6%) | 28/351 (8.0%) | 1/238 (0.4%) | ||
| >10% | 459/495 (93%) | 572/623 (92%) | 281/351 (80%) | 209/238 (88%) | ||
| ASXL1 | 260/443 (59%) | 181/582 (31%) | 61/288 (21%) | 0.008 | 40/184 (22%) | 0.082 |
| ASXL2 | 4/229 (1.7%) | 2/312 (0.6%) | 0/154 (0%) | >0.9 | 0/96 (0%) | >0.9 |
| BCOR | 7/275 (2.5%) | 7/360 (1.9%) | 5/195 (2.6%) | 0.8 | 2/126 (1.6%) | >0.9 |
| BRAF | 4/266 (1.5%) | 3/362 (0.8%) | 0/174 (0%) | 0.7 | 0/111 (0%) | >0.9 |
| BRCC3 | 2/214 (0.9%) | 5/295 (1.7%) | 0/159 (0%) | 0.4 | 0/99 (0%) | 0.6 |
| CBL | 105/438 (24%) | 69/574 (12%) | 18/268 (6.7%) | 0.053 | 15/175 (8.6%) | 0.4 |
| CSF3R | 6/356 (1.7%) | 1/492 (0.2%) | 2/227 (0.9%) | 0.4 | 0/144 (0%) | >0.9 |
| CUX1 | 5/168 (3.0%) | 5/231 (2.2%) | 1/111 (0.9%) | 0.8 | 0/68 (0%) | >0.9 |
| DNMT3A | 14/373 (3.8%) | 23/498 (4.6%) | 34/252 (13.5%) | <0.001 | 13/164 (7.9%) | 0.3 |
| ETNK1 | 16/329 (4.9%) | 14/465 (3.0%) | 4/218 (1.8%) | 0.5 | 2/138 (1.4%) | 0.9 |
| ETV6 | 6/278 (2.2%) | 2/367 (0.5%) | 7/247 (2.8%) | 0.086 | 4/157 (2.5%) | 0.2 |
| EZH2 | 45/427 (11%) | 26/568 (4.6%) | 13/285 (4.6%) | >0.9 | 9/183 (4.9%) | >0.9 |
| FLT3-ITD | 12/279 (4.3%) | 0/342 (0%) | 0/198 (0%) | 0/129 (0%) | ||
| IDH1 | 2/381 (0.5%) | 8/508 (1.6%) | 4/261 (1.5%) | >0.9 | 3/169 (1.8%) | >0.9 |
| IDH2 | 13/381 (3.4%) | 21/509 (4.1%) | 7/261 (2.7%) | 0.5 | 6/169 (3.6%) | >0.9 |
| JAK2 | 57/499 (11%) | 23/594 (3.9%) | 12/275 (4.4%) | 0.8 | 12/184 (6.5%) | 0.3 |
| KIT | 22/426 (5.2%) | 29/558 (5.2%) | 3/243 (1.2%) | 0.026 | 2/157 (1.3%) | 0.12 |
| KMT2A-PTD | 5/98 (5.1%) | 5/105 (4.8%) | 1/47 (2.1%) | 0.8 | 1/35 (2.9%) | >0.9 |
| KRAS | 61/439 (14%) | 45/571 (7.9%) | 8/261 (3.1%) | 0.026 | 7/171 (4.1%) | 0.3 |
| MPL | 6/381 (1.6%) | 13/495 (2.6%) | 9/262 (3.4%) | 0.7 | 5/174 (2.9%) | >0.9 |
| NF1 | 25/235 (11%) | 10/316 (3.2%) | 5/171 (2.9%) | >0.9 | 3/109 (2.8%) | >0.9 |
| fish NF1 | 9/551 (1.6%) | 1/654 (0.2%) | 0/356 (0%) | >0.9 | 0/241 (0%) | >0.9 |
| NRAS | 118/454 (26%) | 47/590 (8.0%) | 11/282 (3.9%) | 0.065 | 9/187 (4.8%) | 0.3 |
| PHF6 | 7/291 (2.4%) | 16/393 (4.1%) | 6/227 (2.6%) | 0.5 | 5/142 (3.5%) | >0.9 |
| PPM1D | 1/231 (0.4%) | 2/313 (0.6%) | 4/160 (2.5%) | 0.4 | 0/100 (0%) | >0.9 |
| PTPN11 | 18/280 (6.4%) | 9/381 (2.4%) | 2/227 (0.9%) | 0.4 | 2/146 (1.4%) | >0.9 |
| RUNX1 | 102/449 (23%) | 114/588 (19%) | 30/308 (9.7%) | <0.001 | 24/201 (12%) | 0.082 |
| SETBP1 | 40/378 (11%) | 16/516 (3.1%) | 3/242 (1.2%) | 0.3 | 0/156 (0%) | 0.12 |
| SF3B1 | 18/399 (4.5%) | 50/552 (9.1%) | 99/316 (31%) | <0.001 | 74/212 (35%) | <0.001 |
| SH2B3 | 5/220 (2.3%) | 6/291 (2.1%) | 0/145 (0%) | 0.4 | 0/89 (0%) | 0.6 |
| SRSF2 | 220/434 (51%) | 264/562 (47%) | 59/256 (23%) | <0.001 | 50/166 (30%) | <0.001 |
| STAG2 | 6/231 (2.6%) | 9/311 (2.9%) | 10/217 (4.6%) | 0.5 | 8/139 (5.8%) | 0.4 |
| TET2 | 274/426 (64%) | 388/559 (69%) | 141/264 (53%) | <0.001 | 104/173 (60%) | 0.10 |
| fish TET2 | 9/551 (1.6%) | 17/654 (2.6%) | 3/356 (0.8%) | 0.2 | 2/241 (0.8%) | 0.4 |
| TP53 | 11/345 (3.2%) | 15/488 (3.1%) | 14/284 (4.9%) | 0.4 | 7/185 (3.8%) | >0.9 |
| U2AF1 | 31/384 (8.1%) | 33/524 (6.3%) | 22/267 (8.2%) | 0.5 | 15/173 (8.7%) | 0.5 |
| ZRSR2 | 5/304 (1.6%) | 50/414 (12%) | 36/242 (15%) | 0.5 | 25/155 (16%) | 0.4 |
| . | MP-WHOest n = 549∗ . | MD-WHOest n = 654∗ . | WHOnew n = 356∗ . | q value† MD-WHOest vs WHOnew . | ICCnew n = 241∗ . | q value† MD-WHOest vs ICCnew . |
|---|---|---|---|---|---|---|
| Assessed genes | 49.9 ± 42.0; 35.0 (1.0-131.0) | 54.8 ± 39.7; 40.0 (1.0-135.0) | 47.0 ± 37.2; 36.0 (1.0-132.0) | <0.001 | 44.8 ± 37.6; 34.0 (1.0-132.0) | <0.001 |
| Mutated genes | 3.5 ± 2.3; 4.0 (0.0-12.0) | 3.1 ± 2.1; 3.0 (0.0-14.0) | 2.2 ± 1.8; 2.0 (0.0-9.0) | <0.001 | 2.4 ± 1.9; 2.0 (0.0-9.0) | <0.001 |
| NGS assessed genes | 46.1 ± 42.3; 32.0 (0.0-129.0) | 50.9 ± 40.2; 35.0 (0.0-130.0) | 46.1 ± 37.4; 35.0 (0.0-130.0) | 0.5 | 43.9 ± 37.9; 34.0 (0.0-130.0) | 0.3 |
| NGS mutated genes | 3.3 ± 2.2; 3.0 (0.0-12.0) | 2.9 ± 1.9; 3.0 (0.0-12.0) | 2.2 ± 1.8; 2.0 (0.0-9.0) | <0.001 | 2.3 ± 1.8; 2.0 (0.0-9.0) | <0.001 |
| Highest VAF | 55.1 ± 19.5; 50.0 (0.0-100.0) | 52.6 ± 19.6; 48.0 (0.0-100.0) | 41.2 ± 21.7; 40.0 (2.1-97.0) | <0.001 | 45.1 ± 20.1; 42.0 (4.0-97.0) | <0.001 |
| VAF cutoffs | <0.001 | 0.3 | ||||
| <2% / no clonality | 35/495 (7.1%) | 47/623 (7.5%) | 42/351 (12%) | 28/238 (12%) | ||
| 2%-10% | 1/495 (0.2%) | 4/623 (0.6%) | 28/351 (8.0%) | 1/238 (0.4%) | ||
| >10% | 459/495 (93%) | 572/623 (92%) | 281/351 (80%) | 209/238 (88%) | ||
| ASXL1 | 260/443 (59%) | 181/582 (31%) | 61/288 (21%) | 0.008 | 40/184 (22%) | 0.082 |
| ASXL2 | 4/229 (1.7%) | 2/312 (0.6%) | 0/154 (0%) | >0.9 | 0/96 (0%) | >0.9 |
| BCOR | 7/275 (2.5%) | 7/360 (1.9%) | 5/195 (2.6%) | 0.8 | 2/126 (1.6%) | >0.9 |
| BRAF | 4/266 (1.5%) | 3/362 (0.8%) | 0/174 (0%) | 0.7 | 0/111 (0%) | >0.9 |
| BRCC3 | 2/214 (0.9%) | 5/295 (1.7%) | 0/159 (0%) | 0.4 | 0/99 (0%) | 0.6 |
| CBL | 105/438 (24%) | 69/574 (12%) | 18/268 (6.7%) | 0.053 | 15/175 (8.6%) | 0.4 |
| CSF3R | 6/356 (1.7%) | 1/492 (0.2%) | 2/227 (0.9%) | 0.4 | 0/144 (0%) | >0.9 |
| CUX1 | 5/168 (3.0%) | 5/231 (2.2%) | 1/111 (0.9%) | 0.8 | 0/68 (0%) | >0.9 |
| DNMT3A | 14/373 (3.8%) | 23/498 (4.6%) | 34/252 (13.5%) | <0.001 | 13/164 (7.9%) | 0.3 |
| ETNK1 | 16/329 (4.9%) | 14/465 (3.0%) | 4/218 (1.8%) | 0.5 | 2/138 (1.4%) | 0.9 |
| ETV6 | 6/278 (2.2%) | 2/367 (0.5%) | 7/247 (2.8%) | 0.086 | 4/157 (2.5%) | 0.2 |
| EZH2 | 45/427 (11%) | 26/568 (4.6%) | 13/285 (4.6%) | >0.9 | 9/183 (4.9%) | >0.9 |
| FLT3-ITD | 12/279 (4.3%) | 0/342 (0%) | 0/198 (0%) | 0/129 (0%) | ||
| IDH1 | 2/381 (0.5%) | 8/508 (1.6%) | 4/261 (1.5%) | >0.9 | 3/169 (1.8%) | >0.9 |
| IDH2 | 13/381 (3.4%) | 21/509 (4.1%) | 7/261 (2.7%) | 0.5 | 6/169 (3.6%) | >0.9 |
| JAK2 | 57/499 (11%) | 23/594 (3.9%) | 12/275 (4.4%) | 0.8 | 12/184 (6.5%) | 0.3 |
| KIT | 22/426 (5.2%) | 29/558 (5.2%) | 3/243 (1.2%) | 0.026 | 2/157 (1.3%) | 0.12 |
| KMT2A-PTD | 5/98 (5.1%) | 5/105 (4.8%) | 1/47 (2.1%) | 0.8 | 1/35 (2.9%) | >0.9 |
| KRAS | 61/439 (14%) | 45/571 (7.9%) | 8/261 (3.1%) | 0.026 | 7/171 (4.1%) | 0.3 |
| MPL | 6/381 (1.6%) | 13/495 (2.6%) | 9/262 (3.4%) | 0.7 | 5/174 (2.9%) | >0.9 |
| NF1 | 25/235 (11%) | 10/316 (3.2%) | 5/171 (2.9%) | >0.9 | 3/109 (2.8%) | >0.9 |
| fish NF1 | 9/551 (1.6%) | 1/654 (0.2%) | 0/356 (0%) | >0.9 | 0/241 (0%) | >0.9 |
| NRAS | 118/454 (26%) | 47/590 (8.0%) | 11/282 (3.9%) | 0.065 | 9/187 (4.8%) | 0.3 |
| PHF6 | 7/291 (2.4%) | 16/393 (4.1%) | 6/227 (2.6%) | 0.5 | 5/142 (3.5%) | >0.9 |
| PPM1D | 1/231 (0.4%) | 2/313 (0.6%) | 4/160 (2.5%) | 0.4 | 0/100 (0%) | >0.9 |
| PTPN11 | 18/280 (6.4%) | 9/381 (2.4%) | 2/227 (0.9%) | 0.4 | 2/146 (1.4%) | >0.9 |
| RUNX1 | 102/449 (23%) | 114/588 (19%) | 30/308 (9.7%) | <0.001 | 24/201 (12%) | 0.082 |
| SETBP1 | 40/378 (11%) | 16/516 (3.1%) | 3/242 (1.2%) | 0.3 | 0/156 (0%) | 0.12 |
| SF3B1 | 18/399 (4.5%) | 50/552 (9.1%) | 99/316 (31%) | <0.001 | 74/212 (35%) | <0.001 |
| SH2B3 | 5/220 (2.3%) | 6/291 (2.1%) | 0/145 (0%) | 0.4 | 0/89 (0%) | 0.6 |
| SRSF2 | 220/434 (51%) | 264/562 (47%) | 59/256 (23%) | <0.001 | 50/166 (30%) | <0.001 |
| STAG2 | 6/231 (2.6%) | 9/311 (2.9%) | 10/217 (4.6%) | 0.5 | 8/139 (5.8%) | 0.4 |
| TET2 | 274/426 (64%) | 388/559 (69%) | 141/264 (53%) | <0.001 | 104/173 (60%) | 0.10 |
| fish TET2 | 9/551 (1.6%) | 17/654 (2.6%) | 3/356 (0.8%) | 0.2 | 2/241 (0.8%) | 0.4 |
| TP53 | 11/345 (3.2%) | 15/488 (3.1%) | 14/284 (4.9%) | 0.4 | 7/185 (3.8%) | >0.9 |
| U2AF1 | 31/384 (8.1%) | 33/524 (6.3%) | 22/267 (8.2%) | 0.5 | 15/173 (8.7%) | 0.5 |
| ZRSR2 | 5/304 (1.6%) | 50/414 (12%) | 36/242 (15%) | 0.5 | 25/155 (16%) | 0.4 |
NGS, next-generation sequencing; VAF, variant allele frequency.
Mean ± standard deviation; median (range); n / N (%).
Wilcoxon rank sum test; Fisher exact test; and Pearson χ2 test after false discovery rate correction for multiple testing.
Newly classified CMML cases show an OS comparable with established MD-CMML cases
The prognosis of patients with CMML is defined by their clinical and genomic features, some of which are associated with a higher risk of progression to AML and shorter OS.33 We analyzed survival data from a total of 1565 CMML cases, whereby we confirmed the absence of outcome differences between the CMML-0 and CMML-1 groups, leading to the elimination of CMML-0 in both revised classification systems (supplemental Figure 9A). Additionally, patients in MD-WHOest showed superior outcomes when compared with the MP-WHOest subgroup (median OS, 6.7 vs 4.0 years; P < .001; supplemental Figure 9B).33 This also held true for WHOnew and ICCnew cases, whose OS did not significantly differ from that of the MD-WHOest cohort (Figure 3E).
To identify cytogenetic and molecular drivers of prognosis in our cohorts, we proceeded with calculating the CPSS and CPSSmol risk scores.16,17 Interestingly, WHOnew and ICCnew cases presented with higher cytogenetic risk than MD-WHOest cases (P < .001; Table 1), conflicting with the comparable OS between those cohorts. Intriguingly, this effect was reversed in the CPSSmol risk score, which was significantly lower for patients in WHOnew and ICCnew cohorts, with the low- and high-risk groups showing the most pronounced difference (P < .001 and P = .029, respectively). Given that most WHOnew patients were diagnosed with MDS according to the WHO17 classification, we additionally determined both the IPSS-M and IPSS-R risk scores for cases reclassified from MDS (Table 1; supplemental Table 3). Because of partial availability of molecular information, the median IPSS-M risk was used for comparison. Hereby, prior MDS cases now reclassified as CMML presented with lower risk scores in both the IPSS-M and IPSS-R than the published IPSS-M validation cohort.19,20 These findings suggest a balance of high-risk cytogenetic aberrations and favorable mutational profiles as the molecular basis for the prognosis of WHOnew patients.
Transcriptomic characterization of newly classified and known patients with CMML
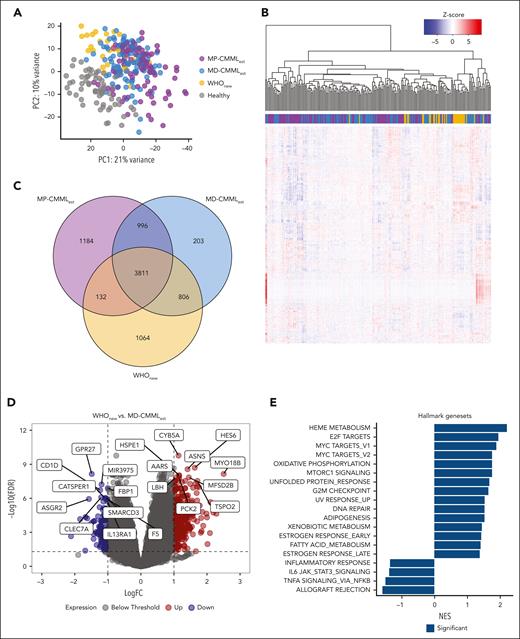
We performed WTS of 22 WHOnew/ICCnew, 11 WHOnew exclusive, and 193 WHOest samples as well as 64 healthy controls. As expected, principal component analysis revealed a separate clustering of healthy and CMML samples, the latter of which formed a transcriptional continuum, with OM-CMML samples partially separating from WHOest samples (Figure 4A). However, no clear separation was observed between the WHOnew exclusive and nonexclusive samples. During unsupervised clustering, WHOnew and ICCnew samples clustered closely, but no clear division was found between the OM and established CMML cohorts (Figure 4B). Approximately half of all differentially expressed genes were shared among the CMML subgroups (62.2%-65.6%) with 1064 genes uniquely differentially expressed in the WHOnew cohort (Figure 4C; supplemental Tables 4-6). Because WHOnew cases present a subgroup of MD-CMML according to the WHO22 classification, we next assessed expression differences between WHOnew and MD-WHOest samples (supplemental Table 7). Here, we found a moderate number of genes differentially expressed (n = 383), with a clear tendency for gene upregulation in the WHOnew cohort (74.4% vs 25.6% downregulated genes; Figure 4D). To identify dysregulated pathways, we performed gene set enrichment analysis for hallmark gene sets (supplemental Table 8).40 Interestingly, among others, the heme metabolism, E2F, and MYC target pathways were significantly enriched in WHOnew compared with MD-WHOest cases (Figure 4E). This was reflected by a strong transcriptional upregulation of genes coding for erythropoiesis associated (MYO18B, HBG1, and HBG2) and proinflammatory proteins (RAG1 and RAG2). In contrast, genes connected to monocytic activity such as the monocytic chemotactic protein CCL741 and the monocyte transporter SLC46A242 were significantly downregulated in WHOnew cases, possibly indicating earlier disease stages. Finally, we did not identify any significant differences between the WHOnew and ICCnew cases, possibly because the ICCnew samples were fully represented within the WHOnew sample cohort. Taken together, our gene expression analysis revealed large transcriptional commonalities between the different CMML cohorts while also highlighting select differences in gene pathway regulation.
Transcriptomic comparison of WHOnew, MD-WHOest, MP-WHOest, and healthy controls. (A) Principal component analysis of all included WHOnew, MD-WHOest, MP-WHOest, and healthy control samples. (B) Unsupervised clustering of 33 WHOnew and 193 MD- and MP-WHOest cases. (C) Overlap between significantly differentially expressed genes in the WHOnew, MD-WHOest, and MP-WHOest cohorts compared against healthy controls. (D) Volcano plot depicting differential gene expression between the WHOnew and MD-WHOest cohorts. The dotted line denotes the FDR cutoff at 0.05. (E) Normalized enrichment score of the 19 significantly enriched hallmark gene sets highlighted in dark blue. FDR, false discovery rate; logFC, log fold change; PC, principal component; NES, normalized enrichment score.
Transcriptomic comparison of WHOnew, MD-WHOest, MP-WHOest, and healthy controls. (A) Principal component analysis of all included WHOnew, MD-WHOest, MP-WHOest, and healthy control samples. (B) Unsupervised clustering of 33 WHOnew and 193 MD- and MP-WHOest cases. (C) Overlap between significantly differentially expressed genes in the WHOnew, MD-WHOest, and MP-WHOest cohorts compared against healthy controls. (D) Volcano plot depicting differential gene expression between the WHOnew and MD-WHOest cohorts. The dotted line denotes the FDR cutoff at 0.05. (E) Normalized enrichment score of the 19 significantly enriched hallmark gene sets highlighted in dark blue. FDR, false discovery rate; logFC, log fold change; PC, principal component; NES, normalized enrichment score.
Discussion
The recently published WHO22 and ICC22 classifications introduced major changes to the diagnostic criteria for CMML, including the reduction of the PB monocyte threshold to include cases previously defined as OM-CMML.4,5,7,8,43 In this study, we systematically assessed their impact on CMML diagnosis. After updating the diagnosis of 1279 established CMML cases to both new classifications, approximately half of the cohort was reclassified. Importantly, due to diverging diagnostic criteria, 21 cases would have been classified as AML according to the WHO22 while still being considered CMML cases according to the ICC22 classification, leaving an open question regarding the different therapeutical implications.33 It should be noted, however, that in the current WHO-HAEM5 BetaBlueBook version,44 AML-defining genetics apply "in the presence of increased blasts," implying a minimum BM blast cutoff of ≥5% (or ≥2% in the PB), which was not mentioned in the publication of the fifth edition of the WHO criteria.4 This discrepancy resulted from an absence of a minimum blast threshold in the presence of AML-defining molecular aberrations in the preliminary WHO22 classification, which, however, may be subject to change in the final classification. Furthermore, a significant proportion of prior OM cases was now classified as CMML, mainly affecting previous MDS cases according to WHO17 and consistent with previous reports of OM-CMML deriving largely from a MD background.43
A critical examination of the, in part diverging, diagnostic criteria of the WHO22 and ICC22 revealed several aspects with critical affecting practicability in routine diagnostics. First and foremost, in our retrospective analysis the necessary BM criterion of age-adjusted hypercellularity excluded 19.9% of cases with a WHO17 CMML diagnosis, adding up to a total of 23.4%, as opposed to only 3.4%, according to the WHO22. Even more critically, hypercellularity was not assessable for ∼450 cases of our initial study cohort, which we excluded from our analysis to avoid biasing the comparability of the remaining diagnostic criteria. In routine diagnostics this would have necessitated repeat sampling, complicating the clinical management of these patients. The hypercellularity requirement also majorly contributed to 1.5 times more cases being classified as CMML according to the WHO22 than according to the ICC22. In contrast, WHO22 has a prerequisite criterion of dysplasia for OM cases, which is only a supporting criterion in cases with PB monocytosis of ≥1 × 109/L and is fulfilled by 95% of ICCnew cases. When assessing the redundancy of diagnostic criteria, we found the criterion of clonality to be the most sensitive in cases with PB monocytosis ≥1 × 109/L for both classifications, whereas the exact VAF threshold distinction was irrelevant in the range of 2% to 10%. In contrast, ∼8% of OM cases newly classified as CMML according to the WHO22 presented with a mutation VAF between 2% and 10% as the sole evidence of clonality. Although the criterion of immunophenotyping for the WHO22 was not retrospectively assessable, our analysis suggests that it is largely redundant because of the high sensitivity of the dysplasia and clonality criteria. It should be critically noted, however, that this assessment was made in a cohort of fully characterized samples, which does not always reflect the reality of routine diagnostics.
An in-depth assessment of reclassified CMML cases showed a hypoproliferative phenotype accompanied by PB cytopenia and reduced blast counts, as previously reported for the OM-CMML entity.7 Karyotyping revealed a significantly higher occurrence of del20q and chrY loss as well as MDS-typical chr5q loss in all reclassified CMML cases. Furthermore, reclassified cases presented fewer mutations normally recurring in CMML, such as in TET2, SRSF2, ASXL1, and RUNX145-48 as well as NRAS, KRAS, and CBL.49-52 Of note, the mutational landscape of reclassified CMML placed those cases between MDS and conventional CMML.53 This is in line with a strong enrichment of MDS-typical alterations in SF3B1 and DNMT3A, as well as del(5q). Importantly, OM cases reclassified according to the ICC22 presented a subgroup of reclassified WHO22 cases with a molecular profile more closely resembling that of established CMML cases. In the transcriptomic analysis of a total of 226 CMML cases, along with 64 healthy control samples, we found transcriptional commonalities between the different CMML cohorts while identifying numerous differentially regulated gene pathways.
From a clinical perspective, conflicting literature exists regarding the prognosis of OM-CMML being considered an early stage of CMML with a more favorable prognosis.6,43 However, inferior OS of OM-CMML cases progressing into overt CMML, not sufficiently accounted for by clinical and molecular phenotypes, was reported.7 In our study, reclassified CMML cases presented with favorable CPSSmol molecular risk scores, not reflected in their comparable OS with MD-CMML cases.17 A possible explanation for this discrepancy may be found in the increased occurrence of higher risk cytogenetics in reclassified CMML cases, comparable with the karyotypic profile of MP-WHOest.
Although the new WHO22 and ICC22 classifications integrate former OM-CMML and established CMML, our study provides evidence for those cases having a distinct clinical and molecular profile, placing them on a continuum between MDS and MD-/MP-CMML. This transitional state should be further evaluated in subsequent studies. Instead of comparing arbitrarily defined PB monocyte thresholds, we suggest relying on genetic findings to define CMML subgroups more accurately and guide clinical strategies, Finally, we underscore the need for unified CMML diagnostic criteria with an emphasis on their practicability in routine diagnostics and clinics.
Acknowledgments
The authors thank all MLL coworkers for their dedicated work and all physicians who provided samples, cared for patients, and collected data.
This work was funded by the Torsten Haferlach Leukämie-Diagnostik Stiftung. F.B., E.A., and P.B. received grant support from the Torsten Haferlach Leukämie-Diagnostik Stiftung.
Authorship
Contribution: F.B. and T.H. designed the study; F.B., S.B., and T.H. interpreted the data and wrote the manuscript; C.H. was responsible for chromosome banding and fluorescence in situ hybridization analyses; F.B., C.B., S.B., E.A., M.T., M.M., M.L., G.H., S.H., H.M., W.W., M.-L.M., N.N., P.B., U.K., W.K., C.H., and T.H. were responsible for molecular and bioinformatic analyses; W.K. was responsible for immunophenotyping; T.H. was responsible for cytomorphologic analyses; and all authors read and contributed to the final version of the manuscript.
Conflict-of-interest disclosure: C.H., W.K., and T.H. declare part ownership of MLL. C.B., M.T., M.M., M.L., G.H., S.H., H.M., W.W., M.-L.M., and N.N. are employed by MLL. The remaining authors declare no competing financial interests.
Correspondence: Torsten Haferlach, MLL Munich Leukemia Laboratory, Max-Lebsche-Platz 31, 81377 Munich, Germany; email: torsten.haferlach@mll.com.
References
Author notes
C.B. and S.B. contributed equally to this work.
Portions of the results were presented by F.B. at the Scientific Workshop on Translational Molecular Diagnostics: Genomic Reclassification of Blood Cancer at the the 64th American Society of Hematology (ASH) Annual Meeting (New Orleans, LA, 9 December 2022).
Sequencing data deposited in the Gene Expression Omnibus database (accession number GSE249547).
Original data are available on request from the corresponding author, Torsten Haferlach (torsten.haferlach@mll.com).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal