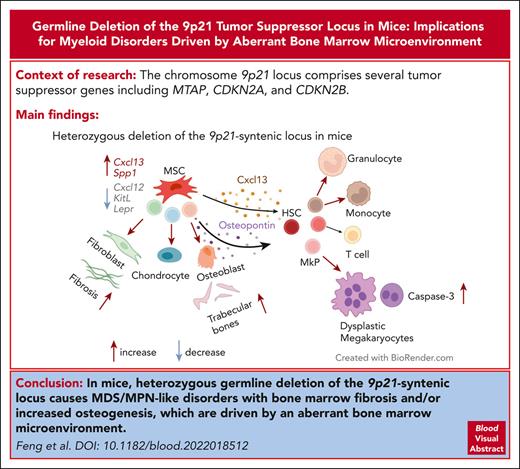
Key Points
Loss of the 9p21-syntenic locus in mice causes MDS/MPN–like disease with BM fibrosis and/or ossification.
The disease is driven by the aberrant BM niche producing less CXCL12 and more CXCL13 and osteopontin.
Abstract
The chromosome 9p21 locus comprises several tumor suppressor genes including MTAP, CDKN2A, and CDKN2B, and its homo- or heterozygous deletion is associated with reduced survival in multiple cancer types. We report that mice with germ line monoallelic deletion or induced biallelic deletion of the 9p21-syntenic locus (9p21s) developed a fatal myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN)-like disease associated with aberrant trabecular bone formation and/or fibrosis in the bone marrow (BM). Reciprocal BM transfers and conditional targeting of 9p21s suggested that the disease originates in the BM stroma. Single-cell analysis of 9p21s-deficient BM stroma revealed the expansion of chondrocyte and osteogenic precursors, reflected in increased osteogenic differentiation in vitro. It also showed reduced expression of factors maintaining hematopoietic stem/progenitor cells, including Cxcl12. Accordingly, 9p21s-deficient mice showed reduced levels of circulating Cxcl12 and concomitant upregulation of the profibrotic chemokine Cxcl13 and the osteogenesis- and fibrosis-related multifunctional glycoprotein osteopontin/Spp1. Our study highlights the potential of mutations in the BM microenvironment to drive MDS/MPN–like disease.
Introduction
Genetic abnormalities including the loss of tumor suppressors, activation of signaling pathways, or epigenetic dysregulation are at the core of hematologic disorders such as myelodysplastic syndromes (MDS), myeloproliferative neoplasms (MPNs), and acute myeloid leukemia (AML).1,2 The chronic myeloid neoplasms MDS and MPN are heterogeneous clonal diseases that arise from hematopoietic stem cells (HSCs) in the bone marrow (BM). MDS is characterized by myeloid cell dysplasia, ineffective hematopoiesis, and peripheral blood cytopenia. MPNs are generally distinguished from MDS by the absence of morphologic dysplasia and the overproduction of differentiated hematopoietic cells. Some myeloid neoplasms display features of both MDS (morphologic dysplasia) and MPN (elevated peripheral blood cell counts or BM fibrosis [BMF]) and are termed MDS/MPN overlap syndromes.
In addition to hematopoietic cell–intrinsic abnormalities, recent evidence suggests a potential role of cell-extrinsic abnormalities within the BM microenvironment in hematologic malignancies.3,4 BMF including reticulin and/or collagen I/III fibers is a common occurrence in several myeloid malignancies, including MDS/MPN disorders and AML.5-7 Multiple studies have demonstrated that patients with MDS with BMF have lower overall survival, higher rate of progression to AML, and lower survival after HSC transplantation. This is particularly relevant in low-grade MDS,8 in which reduced patient survival is essentially confined to the BMF group. Although BMF represents a common and particularly dangerous feature of myeloid malignancies, its pathogenesis and specific role in the disease remain unexplored.
The human 9p21 locus (the syntenic locus at murine chromosome 4qC4) represents a cluster of several genes relevant for carcinogenesis: MTAP, CDKN2A, CDKN2B, and CDKN2BAS (ANRIL). A combined deletion of these genes has been observed in multiple cancer types, including leukemia,9 melanoma,10 mesothelioma,11 and glioblastoma.12 Classical tumor suppressors CDKN2A and CDKN2B encode cyclin-dependent kinase inhibitors that oppose proliferation and facilitate senescence and apoptosis.13CDKN2BAS (AK148321 in the mouse) has been shown to regulate CDKN2A/CDKN2B via epigenetic mechanisms.14MTAP encodes the enzyme methylthioadenosine phosphorylase, a regulator of polyamine metabolism. Reexpression of MTAP in MTAP-deficient tumor cell lines impairs their tumorigenicity, suggesting that the loss of MTAP is a functional oncogenic event.15-17 Germ line deletion of Mtap in mice causes early embryonic lethality, whereas haploinsufficiency accelerates Myc-driven lymphomas.18,19 However, the consequences of combined loss of the entire 9p21 locus with its effect on cancer and/or fibrosis have never been explored in vivo.
To examine the role of the 9p21 locus in tumor suppression, we generated a mouse strain that allows conditional deletion of the entire 9p21-syntenic locus (9p21s). We found that mice with heterozygous loss of 9p21s developed a fatal MDS/MPN–like disease with fibrosis and/or aberrant osteogenesis, which was driven entirely by the abnormal BM microenvironment. The deletion of 9p21 altered the lineage spectrum of mesenchymal stem cell (MSC) differentiation, leading to the remodeling of the BM microenvironment, reduction of the key stroma-derived factor Cxcl12, and induction of a profibrotic chemokine Cxcl13 and osteogenesis- and fibrosis-related multifunctional glycoprotein osteopontin (OPN)/Spp1. Our findings demonstrate the ability of aberrant BM stroma to drive MDS/MPN with fibrosis and establish a mouse model for its experimental study.
Methods
Animals
Pdzk1ip1-CreER mice described in previous studies,20Cx3cr1CreER-YFP mice obtained from The Jackson Laboratory, and R26CreER mice were crossed with 9p21sflox mice. B6.129-Flt3tm1Dgg/J (Flt3ITD/+) mice and B6.Cg-Tg(Sp7-tTA,tetO-EGFP/cre)1Amc/J mice (Osx1-Cre) were obtained from The Jackson Laboratory. The Flt3ITD/+ mice were crossed with 9p21s+/− mice, and the Osx1-Cre mice were crossed with 9p21sfl/fl mice. C57BL/6 (CD45.2) mice and B6.SJL-PtprcaPepcb/BoyCrl (Ly5.1, CD45.1) mice were purchased from Charles River for transplantation.
All animal maintenance and experimentation were performed per the investigators’ protocols approved by the institutional animal care and use committees of New York University School of Medicine. Mice were monitored and euthanized per the humane end points listed in our animal protocol, which include weight loss of >20%, abnormal appearance or behavior, hunched posture, loss of grooming abilities, lack of food consumption, or visible breathing difficulty. For Cre recombinase induction, tamoxifen was administrated to every mouse at a dose of 5 mg by gavage every day for 3 days. Cre recombination using Osx1-Cre were conducted following the instruction of The Jackson Laboratory (JAX006361).
Generation of 9p21sflox and 9p21s+/− mice
To perform conditional deletion of the ∼400 kb region containing Mtap, Cdkn2a, Cdkn2b, and Cdkn2bas (AK148321) from murine chromosome 4q, we inserted LoxP (locus of x-over, P1) sites at both ends of the region using 2 targeting rounds in murine embryonic stem cells (ESCs).21 To allow screening for ESC clones in which the gene targeting occurred on the same chromosome, an enhanced green fluorescent protein (eGFP) mini gene and a phosphoglycerate kinase 1 (PGK) promoter were involved in 5′-targeting and 3′-targeting vectors,22 respectively. In the absence of Cre, the targeted allele expressed wild-type (WT) levels of Mtap_Cdkn2a/2b/2bas. However, Cre-mediated recombination resulted in the deletion of the LoxP-flanked region. ESC clones with both LoxP sites inserted on the same chromosome were used to produce mice with the conditional LoxP-flanked (floxed) allele of 9p21s (9p21sflox). The allele was crossed with ubiquitously expressed tamoxifen-inducible Cre recombinase allele (R26CreER) for global inducible deletion of 9p21s. The allele was also crossed with a germ line Cre deleter to produce mice with a single germ line null allele (9p21s+/−). The GFP expression was detected in ESC clones by microscopy but not in mice. The GFP analysis was not performed in all mice for this study.
Additional methods
Additional methods, including cell processing and flow cytometry, transplantation assays, enzyme-linked immunosorbent assay (ELISA), quantitative real-time polymerase chain reaction (qRT-PCR) analysis, Genomic Data Commons (GDC) data portal and cBioPortal database analysis, microcomputed tomography (micro-CT) analysis, colony-forming unit assay, osteogenic differentiation of BM stromal cells, histology analysis and immunohistochemistry analysis, isolation and staining of primary stromal cells for cellular indexing of transcriptomes and epitopes sequencing (CITE-seq), isolation of BM stromal cells–derived-osteogenic cells for bulk RNA-sequencing (RNA-seq) analysis, CITE-seq data analysis, bulk RNA-seq data analysis, human MDS patient analysis, and statistical analysis, are provided in supplemental Methods, available on the Blood website.
Results
Frequent loss of the 9p21 region in human cancers
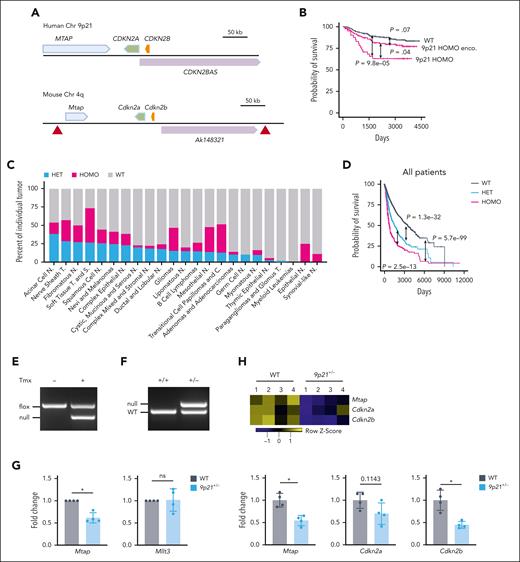
To systematically define the role of the 9p21 region (Figure 1A) in human cancers, we analyzed the deletion of 9p21 in the samples from multiple human tumor types in the cBioPortal database for pediatric patients with acute lymphoid leukemia (phase 2, TRARGET, 2018) (a total of 530 cases selected) and in the NCI GDC data portal (a total of 7622 cases selected).23 In 530 pediatric acute lymphoid leukemia cases, we observed that 41 of 530 cases had a homozygous (HOMO) deletion of the 9p21 locus containing MTAP, CDKN2A, CDKN2B, and CDKN2BAS, whereas 180 of 530 cases had the 9p21 HOMO deletion encompassed (Figure 1B; supplemental Figure 1A). Both types of deletions significantly reduced the survival rates of patients, with the exact 9p21 locus deletion having a particularly worse impact on survival.
Deletion of the 9p21 region in human cancers and genetic targeting of the 9p21s region in mice. (A) Schematic representation of the human 9p21 locus and the syntenic murine locus (9p21s). Genes and their 5′-3′ orientation are indicated by thick arrows (blue, green, and orange). A noncoding RNA is shown by a purple arrow. Red triangles indicate the positions of LoxP sites inserted into the conditional allele. (B) Kaplan-Meier plots of 9p21 locus homozygous (HOMO) deletion, 9p21 HOMO deletion encompassed (enco) and non-9p21 locus deletion–related WT in pediatric acute lymphoid leukemia (ALL). P value shows log-rank test result. (C) Graphical representation of the frequency of the of HET and HOMO 9p21 locus deletions enco in different human cancers. (D) Kaplan-Meier plots of patients with 9p21 deletions in total human cancers. P value shows log-rank test result. (E-F) Cre recombination of the targeted allele. Deleted allele was detected via genomic DNA PCR of BM cells from (E) R26CreER9p21fl/fl mice or (F) Prm-Cre–mediated 9p21+/− mice. (G) Mtap and Mllt3 expressions were confirmed via qRT-PCR. Symbols represent the populations including BM cells, spleen cells, B cells, and T cells from the same WT mouse or 9p21+/− mouse. (H) The haplodeficiency of Mtap, Cdkn2a, and Cdkn2b was demonstrated via bulk RNA-seq of cultured BM stromal cells. Symbols represent individual mice (WT mice, n = 4; 9p21+/− mice, n = 4). Data represent the mean ± standard deviation (SD). Statistical significance was defined using Mann-Whitney test. ∗P < .05 in panels G-H. C, carcinomas; N, neoplasms; ns, not significant; S, sarcomas; T, tumors.
Deletion of the 9p21 region in human cancers and genetic targeting of the 9p21s region in mice. (A) Schematic representation of the human 9p21 locus and the syntenic murine locus (9p21s). Genes and their 5′-3′ orientation are indicated by thick arrows (blue, green, and orange). A noncoding RNA is shown by a purple arrow. Red triangles indicate the positions of LoxP sites inserted into the conditional allele. (B) Kaplan-Meier plots of 9p21 locus homozygous (HOMO) deletion, 9p21 HOMO deletion encompassed (enco) and non-9p21 locus deletion–related WT in pediatric acute lymphoid leukemia (ALL). P value shows log-rank test result. (C) Graphical representation of the frequency of the of HET and HOMO 9p21 locus deletions enco in different human cancers. (D) Kaplan-Meier plots of patients with 9p21 deletions in total human cancers. P value shows log-rank test result. (E-F) Cre recombination of the targeted allele. Deleted allele was detected via genomic DNA PCR of BM cells from (E) R26CreER9p21fl/fl mice or (F) Prm-Cre–mediated 9p21+/− mice. (G) Mtap and Mllt3 expressions were confirmed via qRT-PCR. Symbols represent the populations including BM cells, spleen cells, B cells, and T cells from the same WT mouse or 9p21+/− mouse. (H) The haplodeficiency of Mtap, Cdkn2a, and Cdkn2b was demonstrated via bulk RNA-seq of cultured BM stromal cells. Symbols represent individual mice (WT mice, n = 4; 9p21+/− mice, n = 4). Data represent the mean ± standard deviation (SD). Statistical significance was defined using Mann-Whitney test. ∗P < .05 in panels G-H. C, carcinomas; N, neoplasms; ns, not significant; S, sarcomas; T, tumors.
Furthermore, we analyzed data from 7622 various cancer cases obtained from the GDC data portal. We found that the proportion of the 9p21 locus heterozygous (HET) encompassed accounted for ∼14% (1072) of cases, whereas the 9p21 locus HOMO deletions encompassed accounted for ∼10% (762) of cases. These results suggest that 9p21 locus deletions are relatively common in various cancers and could potentially contribute to poor patient outcomes. In 4 out of 23 human cancers (acinar cells neoplasms, nerve sheath tumors, fibromatous neoplasms, and soft tissue tumors and sarcomas), the combined frequency of HOMO and HET deletions of the entire fragment was >50% (Figure 1C). Moreover, HOMO or HET deletions of the entire locus significantly shortened the survival of patients for all cancer types (Figure 1D) as well as that in 5 specific types of cancer (adenoma and adenocarcinomas, ductal and lobular neoplasms, gliomas, lipomatous neoplasms, and mesothelial neoplasms; supplemental Figure 1B). Notably, the latter 2 types showed no difference between the effect of HOMO vs HET deletions. Thus, the loss of the entire 9p21 locus is highly prevalent in human cancers and adversely affects survival even when it is monoallelic.
Genetic targeting of the 9p21s region in mice
To model the loss of the entire locus in mice, we flanked an ∼400 kb region of the mouse genome that is syntenic to 9p21 (9p21s) with loxP sites (floxed) (Figure 1A). The resulting conditional null 9p21s allele (9p21sflox) was crossed with a tamoxifen-inducible Cre recombinase (R26CreER) for global inducible deletion of 9p21s. The allele was also crossed with a germ line Cre deleter to produce heterozygous mice with a single germ line null allele (9p21s+/−). Correct deletion of 9p21s in the resulting animals was confirmed via genomic PCR in R26CreER9p21sflox/flox mice (Figure 1E; supplemental Table 1) and 9p21s+/− mice (Figure 1F; supplemental Table 1). The RT-PCR analysis in 9p21s+/− hematopoietic cells showed that the expression levels of Mtap were reduced by half, unlike the neighboring Mllt3 gene (Figure 1G; supplemental Table 1). The reduction of Mtap as well as of Cdkn2a and Cdkn2b was further confirmed via the bulk RNA-seq of 9p21s+/− stromal cells (Figure 1H), whereas the neighboring genes were not affected (supplemental Figure 2A). Thus, the resulting animals provide a model for germ line monoallelic (9p21s+/−) or inducible biallelic (R26CreER9p21sflox/flox) loss of the entire 9p21-syntenic region.
Deletion of the 9p21s region causes fatal MDS/MPN–like disease
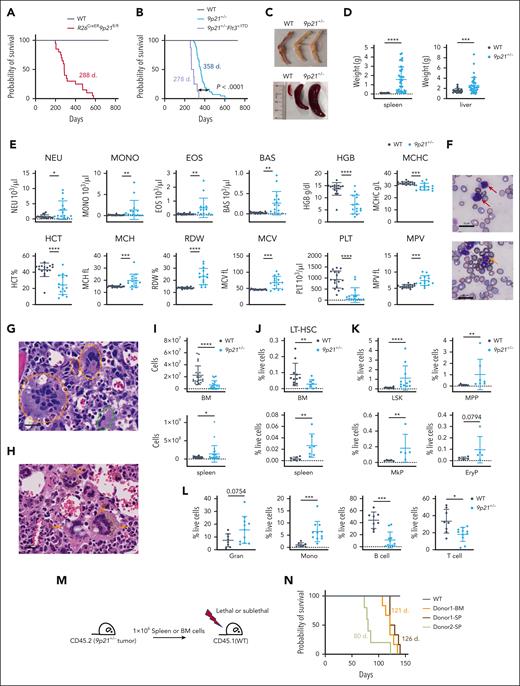
Adult R26CreER9p21sflox/flox mice were treated with tamoxifen to induce global deletion of 9p21s. All mice (n = 20), except WT tamoxifen-treated controls (n = 12), became sick and died with a median survival of ∼9.5 months after treatment (Figure 2A). The sick R26CreER9p21sflox/flox mice displayed splenomegaly and hepatomegaly (supplemental Figure 2B), and the majority of them had hypocellular BM (supplemental Figure 2C). Most of these mice exhibited myeloid disorders, such as expanded dysplastic megakaryocytes with hyperlobulated nuclei in the BM and immature myeloid cells in peripheral blood (supplemental Figure 2D). However, we have also observed severe fibrosis or necrosis in the liver of some R26CreER9p21sflox/flox mice. (supplemental Figure 2E). We then analyzed 9p21s+/− mice with germ line monoallelic loss of 9p21s and observed that these mice (n = 43) but not WT littermates (n = 23) similarly became sick at a median age of 12 months (Figure 2B). No liver fibrosis or necrosis was observed in these mice; however, sick 9p21s+/− animals uniformly displayed anemic BM, splenomegaly, and hepatomegaly (Figure 2C-D). Furthermore, they demonstrated severe macrocytic anemia, thrombocytopenia, and leukocytosis (Figure 2E); the presence of immature myeloid cells and dysplastic neutrophils in the blood (Figure 2F); and BM abnormalities including aberrant megakaryocytes with hyperlobulated nuclei (Figure 2G) and abnormal hemosiderin deposits (Figure 2H). BM cellularity of sick 9p21s+/− mice was decreased, whereas the spleen cellularity was increased compared with controls (Figure 2I). The frequency of long-term HSCs was reduced in the BM but elevated in the spleens of 9p21s+/− mice (Figure 2J; supplemental Table 2). The HSC/progenitors enriched–lineage− Sca-1+ c-Kit+24 population (LSK), multipotent progenitors (MPP),25 megakaryocyte progenitors (MkPs), and erythroid progenitors (EryP) were similarly expanded in 9p21s+/− spleens (Figure 2K; supplemental Table 2), revealing prominent extramedullary hematopoiesis. The proportion of myeloid cells, including granulocytes and monocytes, was increased, whereas lymphoid cells, such as T cells and B cells, showed a significant decrease in the spleen (Figure 2L; supplemental Table 2). Reduced frequency of B cells was also found in the BM (supplemental Figure 2H). Accordingly, splenic histology showed abnormal white and red pulp architecture in 9p21s+/− mice (supplemental Figure 2I). Moreover, the colony-forming assay of sick 9p21s+/− BM cells revealed a temporary increase in colonies during the first round of replating but no significant difference during the second round (supplemental Figure 2J). This indicates that the hematopoietic progenitors did not fully transform for clonogenic growth. Collectively, the observed phenotypes included features of both MDS and MPN and, thus, may be best described as MDS/MPN–like disease.
Deletion of the 9p21s region causes MDS/MPN-like disease. (A) Kaplan-Meier plots of R26CreER9p21fl/fl mice (n = 20) and WT mice (n = 12). (B) Kaplan-Meier plots of 9p21+/− mice with and without Flt3 mutant (9p21+/− mice, n = 43; 9p21+/−Flt3+/ITD mice, n = 8; WT mice, n = 23). P value shows log-rank test result. (C) Representative pale bones and splenomegaly in 9p21+/− tumor mice compared with that in WT mice. (D) Quantification of the spleen and liver weight. Symbols represent individual mice (WT mice, n = 22; 9p21+/− tumor mice, n = 38). (E) Complete blood count analysis of 9p21+/− tumor mice and WT mice. Symbols represent individual mice (9p21+/− tumor mice, n = 15; WT mice, n = 17). (F) May-Grünwald-Giemsa staining of peripheral blood smear. Red arrows indicate representative immature myeloid cells; the orange arrow indicates dysplastic neutrophil; white stars indicate dysplastic red blood cells. (G-H) Hematoxylin and eosin staining of BM sections. Dotted orange circles and a dotted green circle indicate multinucleated and small binucleated megakaryocytes, respectively (G); orange arrows indicate accumulated hemosiderin in abnormal megakaryocytes (H). (I) Absolute numbers of hematopoietic cells in the BM and spleen were analyzed via flow cytometry. Symbols represent individual mice (WT mice, n = 24; 9p21+/− tumor mice; n = 24). (J-L) The frequency of hematopoietic populations was analyzed via flow cytometry. (J) The frequency of long-term HSC (LT-HSC) in mice BM and spleen. Symbols represent individual mice BM (WT mice, n = 14; 9p21+/− tumor mice, n = 13) and spleens (WT mice, n = 10; 9p21+/− tumor mice, n = 11). (K) The frequencies of hematopoietic progenitors in mice spleens. Symbols represent individual mice. LSK (WT mice, n = 14; 9p21+/− tumor mice, n = 13); MPP (WT mice, n = 11; 9p21+/− tumor mice, n = 9); MkP (WT mice, n = 9; 9p21+/− tumor mice, n = 7); and EryP (WT mice, n = 5; 9p21+/− tumor mice, n = 5). (L) The frequencies of hematopoietic lineages in the spleens of WT and 9p21+/− tumor mice. Symbols represent individual mice. WT mice, n = 11; 9p21+/− tumor mice, n = 9. (M) Schematic representation of 9p21+/− MDS/MPN mouse BM and spleen cells transplantation experiment approach. (N) Kaplan-Meier plots of 9p21+/− MDS/MPN mouse BM and spleen cell–derived recipients. WT BM–derived recipients (lethal irradiation, n = 5); 9p21+/− donor 1 BM–derived recipients (lethal irradiation, n = 6), and spleen cell–derived recipients (lethal irradiation, n = 6); and 9p21+/− donor 2 spleen–derived recipients (sublethal irradiation, n = 6). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001 in panels D-E,I-L. EryP, erythroid progenitors; Gran, granulocyte; LSK, lineage− Sca1+cKit+ population; MPP, multipotent progenitors; Mono, monocyte.
Deletion of the 9p21s region causes MDS/MPN-like disease. (A) Kaplan-Meier plots of R26CreER9p21fl/fl mice (n = 20) and WT mice (n = 12). (B) Kaplan-Meier plots of 9p21+/− mice with and without Flt3 mutant (9p21+/− mice, n = 43; 9p21+/−Flt3+/ITD mice, n = 8; WT mice, n = 23). P value shows log-rank test result. (C) Representative pale bones and splenomegaly in 9p21+/− tumor mice compared with that in WT mice. (D) Quantification of the spleen and liver weight. Symbols represent individual mice (WT mice, n = 22; 9p21+/− tumor mice, n = 38). (E) Complete blood count analysis of 9p21+/− tumor mice and WT mice. Symbols represent individual mice (9p21+/− tumor mice, n = 15; WT mice, n = 17). (F) May-Grünwald-Giemsa staining of peripheral blood smear. Red arrows indicate representative immature myeloid cells; the orange arrow indicates dysplastic neutrophil; white stars indicate dysplastic red blood cells. (G-H) Hematoxylin and eosin staining of BM sections. Dotted orange circles and a dotted green circle indicate multinucleated and small binucleated megakaryocytes, respectively (G); orange arrows indicate accumulated hemosiderin in abnormal megakaryocytes (H). (I) Absolute numbers of hematopoietic cells in the BM and spleen were analyzed via flow cytometry. Symbols represent individual mice (WT mice, n = 24; 9p21+/− tumor mice; n = 24). (J-L) The frequency of hematopoietic populations was analyzed via flow cytometry. (J) The frequency of long-term HSC (LT-HSC) in mice BM and spleen. Symbols represent individual mice BM (WT mice, n = 14; 9p21+/− tumor mice, n = 13) and spleens (WT mice, n = 10; 9p21+/− tumor mice, n = 11). (K) The frequencies of hematopoietic progenitors in mice spleens. Symbols represent individual mice. LSK (WT mice, n = 14; 9p21+/− tumor mice, n = 13); MPP (WT mice, n = 11; 9p21+/− tumor mice, n = 9); MkP (WT mice, n = 9; 9p21+/− tumor mice, n = 7); and EryP (WT mice, n = 5; 9p21+/− tumor mice, n = 5). (L) The frequencies of hematopoietic lineages in the spleens of WT and 9p21+/− tumor mice. Symbols represent individual mice. WT mice, n = 11; 9p21+/− tumor mice, n = 9. (M) Schematic representation of 9p21+/− MDS/MPN mouse BM and spleen cells transplantation experiment approach. (N) Kaplan-Meier plots of 9p21+/− MDS/MPN mouse BM and spleen cell–derived recipients. WT BM–derived recipients (lethal irradiation, n = 5); 9p21+/− donor 1 BM–derived recipients (lethal irradiation, n = 6), and spleen cell–derived recipients (lethal irradiation, n = 6); and 9p21+/− donor 2 spleen–derived recipients (sublethal irradiation, n = 6). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001 in panels D-E,I-L. EryP, erythroid progenitors; Gran, granulocyte; LSK, lineage− Sca1+cKit+ population; MPP, multipotent progenitors; Mono, monocyte.
To prove that the hematopoietic abnormalities observed were indeed causing the disease, we transferred BM cells or spleen cells from 2 sick 9p21s+/− mice (CD45.2) into lethally or sublethally irradiated WT (CD45.1) mice (Figure 2M). Blood cell tracing of donor 1–derived recipients at ∼4 to 12 weeks after transplantation showed that transplanted spleen cells and BM cells elicited similar phenotypes, including increased granulocytes and basophils and decreased B cells and platelets (supplemental Figure 3A,C). Additionally, we analyzed sick recipients of the BM or spleen and found that their phenotypes were similar to each other and to that of the primary donor 1 (supplemental Figure 3C,G), with a median survival time of ∼4 months (Figure 2N). The donor 2 spleen–derived group had a shorter median survival time of 80 days (Figure 2N). These results demonstrate that the MDS/MPN–like disease is transplantable from both the spleen and BM.
To investigate the potential cooperation between the cell-extrinsic abnormality in 9p21s mice and a cell-intrinsic leukemogenic mutation, we crossed 9p21s+/− mice with the heterozygous knock-in allele of FLT3 internal tandem duplication (Flt3ITD), which by itself causes mild myeloproliferation but no apparent disease. The resulting 9p21s+/−Flt3ITD/+ mice showed a significant acceleration of MDS/MPN–like disease and succumbed with a median survival of 278 days (Figure 2B; supplemental Figure 2F-G). However, the double-mutant mice did not develop AML, which suggests that the 2 mutations alone may not be sufficient to fully drive leukemogenesis but may work in cooperation with other genetic or environmental factors. Thus, the global inducible or monoallelic germ line deletion of the 9p21-syntenic locus causes multiple hematopoietic abnormalities consistent with MDS/MPN overlap syndromes.
MDS/MPN–like disease in 9p21s+/− mice is driven by the BM microenvironment
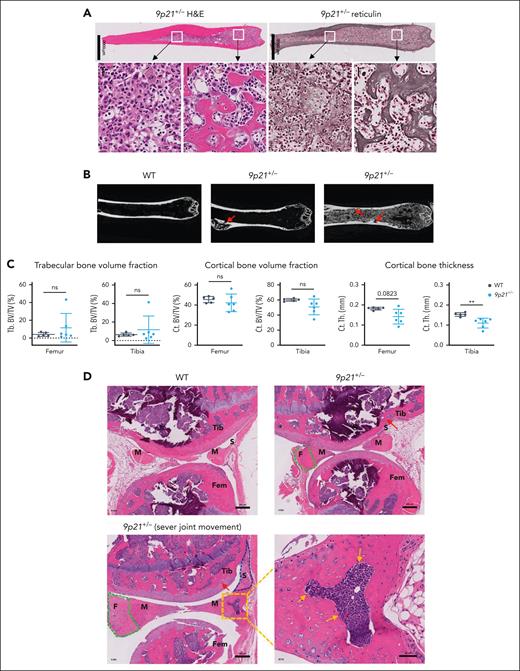
We noticed that a fraction of sick 9p21s+/− mice showed excessive trabecular bone in the BM, and all mice displayed BMF, including reticulin fibrosis and/or collagen fibrosis (Figure 3A; supplemental Figure 4A). We examined trabecular and cortical morphology of the femurs and tibia via micro-CT in 6 randomly selected 9p21s+/− sick mice and observed greatly expanded trabeculae in 2 of 6 femurs (Figure 3B). Micro-CT analysis revealed a tendency toward increased trabecular bone and decreased cortical bone volumes in the tibia and femurs of 9p21s+/− mice, with a significant thinning of the cortical bone in the tibia (Figure 3C). This phenotypic heterogeneity renders the quantification of fibrosis unfeasible; consequently, no notable differences in bone volumes were detected. Trabecular bones in 9p21s+/− mice were lined with tartrate-resistant acid phosphatase (TRAP)-positive osteoclasts (supplemental Figure 4B), likely as a secondary response to excessive bone formation. We also noted that a fraction of sick 9p21s+/− mice had impaired movement of the joints; the affected knee joints showed enlargement and fibrous region expansion in the meniscus as well as thickening of the synovium (Figure 3D). Collectively, the deletion of 9p21s leads to MDS/MPN–like disease with BM atrophy and extramedullary hematopoiesis, which is associated with aberrant osteogenesis, fibrogenesis, and chondrogenesis in the long bones and joints.
The disorders of the BM environment in 9p21s+/− MDS/MPN–like mice. (A) Hematoxylin and eosin and reticulin staining of serial femur bone sections from a representative 9p21+/− MDS/MPN mouse. (B) Micro-CT images of the femurs in 9p21+/− tumor and WT mice. Red arrows show the abnormal cortical bone in 9p21+/− tumor mice. (C) Quantification of trabecular bones and cortical bones volume in femur and tibia total bone volume. Symbols represent individual mice (WT mice, n = 5; 9p21+/− tumor mice, n = 6). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗∗P < .01. (D) Hematoxylin and eosin staining of knee joints from 9p21+/− tumor and WT mice. Green dotted lines indicate increased fibrous regions in meniscus; gold arrows indicate bone formation in meniscus; a blue dotted line indicates thicker synovium; red arrows indicate osteophyte in cartilage. Ct. BV/TV, cortical bone volume fraction; Ct.Th, cortical bone thickness; F, fibrous region; Fem, femur; M, meniscus; S, synovium; Tb. BV/TV, trabecular bone volume fraction; Tib, tibia.
The disorders of the BM environment in 9p21s+/− MDS/MPN–like mice. (A) Hematoxylin and eosin and reticulin staining of serial femur bone sections from a representative 9p21+/− MDS/MPN mouse. (B) Micro-CT images of the femurs in 9p21+/− tumor and WT mice. Red arrows show the abnormal cortical bone in 9p21+/− tumor mice. (C) Quantification of trabecular bones and cortical bones volume in femur and tibia total bone volume. Symbols represent individual mice (WT mice, n = 5; 9p21+/− tumor mice, n = 6). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗∗P < .01. (D) Hematoxylin and eosin staining of knee joints from 9p21+/− tumor and WT mice. Green dotted lines indicate increased fibrous regions in meniscus; gold arrows indicate bone formation in meniscus; a blue dotted line indicates thicker synovium; red arrows indicate osteophyte in cartilage. Ct. BV/TV, cortical bone volume fraction; Ct.Th, cortical bone thickness; F, fibrous region; Fem, femur; M, meniscus; S, synovium; Tb. BV/TV, trabecular bone volume fraction; Tib, tibia.
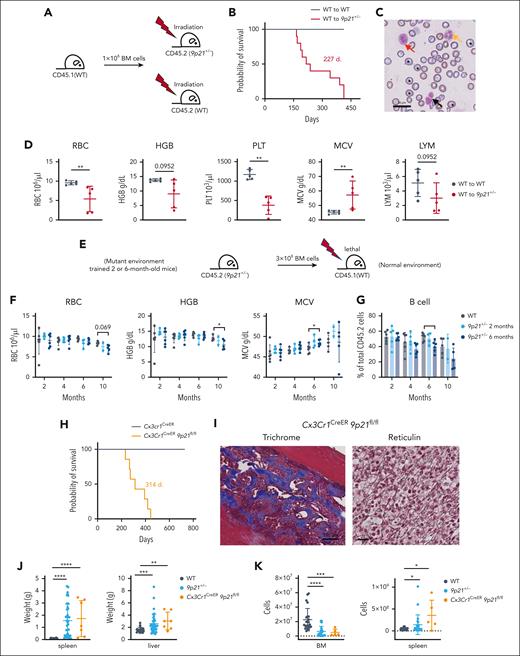
To test whether the abnormal BM microenvironment might contribute to the MDS/MPN–like disease, we transferred 8- or 10-week-old WT BM (CD45.1) into lethally irradiated 8- or 12-week-old WT (CD45.2) or 9p21s+/− mice (CD45.2) to generate BM chimeras (Figure 4A). Despite near-complete donor chimerism (data not shown), 9p21s+/− recipients developed the same hematologic disease with 100% mortality (10 of 10) (Figure 4B), displaying multilineage dysplasia (Figure 4C), anemia, thrombocytopenia, abnormal neutrophils, and mild lymphopenia (Figure 4D; supplemental Figure 5A). We also performed reciprocal transplantation of BM cells from 9p21s+/− mice at 2 or 6 months of age into irradiated WT recipients (Figure 4E). No abnormalities were observed in the recipients of the 2-month-old 9p21s+/− BM cells, and only minor abnormalities such as slightly reduced hemoglobin and B-cell fraction were observed in the recipients of the 6-month-old 9p21s+/− BM cells (Figure 4F-G; supplemental Figure 5B-C); the latter likely reflects the remodeling of hematopoiesis in the older 9p21s+/− donor mice. Importantly, no signs of MDS/MPN and no morbidity or mortality were detected in any recipient of 9p21s+/− BM cells, suggesting that the disease is driven by the BM microenvironment rather than hematopoietic cells.
MDS/MPN–like disease in 9p21s+/− mice is driven by the BM microenvironment. (A) Schematic representation of reciprocal transplantation experiment approach. The CD45.1 congenic WT mice BM cells were transplanted to lethal irradiated WT mice (CD45.2) and 9p21+/− mice (CD45.2). (B) Kaplan-Meier plots of WT mice (WT to WT mice, n = 10) and 9p21+/− tumor mice (WT to 9p21+/− mice, n = 9). (C) May-Grünwald-Giemsa staining of peripheral blood smear from a representative sick 9p21+/− mouse. The orange arrow indicates abnormal neutrophil; the red arrow indicates immature myeloid cell; the black arrow indicates immature erythroid cells; and black stars indicate large red blood cells. (D) Complete blood count analysis of mice that had received transplantation with CD45.1 congenic WT mice BM cells. Symbols represent individual mice (WT to WT mice, n = 5; WT to 9p21+/− tumor mice, n = 5). (E) Schematic representation of 9p21+/− BM cells (2- and 6-month-old mice) and WT BM cells (2-month-old mice) transplantation experiment approach. (F) Complete blood count analysis of WT (CD45.2) donor– and 9p21+/− (CD45.2) donor–derived CD45.1 congenic mice peripheral blood at different time points. Symbols represent individual mice (WT to WT mice, n = 6; 2-month-old 9p21+/− to WT mice, n = 5; and 6-month-old 9p21+/− to WT mice, n = 6). (G) The frequency of B cells was analyzed via flow cytometry in 9p21+/− BM cells and WT BM cells transplantation recipient peripheral blood. Symbols represent individual mice (WT to WT mice, n = 6; 2-month-old 9p21+/− to WT mice, n = 5; and 6-month-old 9p21+/− to WT mice, n = 6). (H) Kaplan-Meier plots of Cx3Cr1CreER (n = 7) and Cx3Cr1CreER9p21fl/fl mice (n = 7). (I) Trichrome Masson and reticulin staining of Cx3Cr1CreER9p21fl/fl mice BM. (J) Quantification of the spleen and liver weights. Symbols represent individual mice (WT mice, n = 22; 9p21+/− tumor mice, n = 38; and Cx3Cr1CreER9p21fl/fl tumor mice, n = 7). (K) Complete blood count analysis of mice. Symbols represent individual mice (9p21+/− tumor mice, n = 15; WT mice, n = 17; and Cx3Cr1CreER9p21fl/fl tumor mice, n = 5). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 in panels D,F-G,J-K.
MDS/MPN–like disease in 9p21s+/− mice is driven by the BM microenvironment. (A) Schematic representation of reciprocal transplantation experiment approach. The CD45.1 congenic WT mice BM cells were transplanted to lethal irradiated WT mice (CD45.2) and 9p21+/− mice (CD45.2). (B) Kaplan-Meier plots of WT mice (WT to WT mice, n = 10) and 9p21+/− tumor mice (WT to 9p21+/− mice, n = 9). (C) May-Grünwald-Giemsa staining of peripheral blood smear from a representative sick 9p21+/− mouse. The orange arrow indicates abnormal neutrophil; the red arrow indicates immature myeloid cell; the black arrow indicates immature erythroid cells; and black stars indicate large red blood cells. (D) Complete blood count analysis of mice that had received transplantation with CD45.1 congenic WT mice BM cells. Symbols represent individual mice (WT to WT mice, n = 5; WT to 9p21+/− tumor mice, n = 5). (E) Schematic representation of 9p21+/− BM cells (2- and 6-month-old mice) and WT BM cells (2-month-old mice) transplantation experiment approach. (F) Complete blood count analysis of WT (CD45.2) donor– and 9p21+/− (CD45.2) donor–derived CD45.1 congenic mice peripheral blood at different time points. Symbols represent individual mice (WT to WT mice, n = 6; 2-month-old 9p21+/− to WT mice, n = 5; and 6-month-old 9p21+/− to WT mice, n = 6). (G) The frequency of B cells was analyzed via flow cytometry in 9p21+/− BM cells and WT BM cells transplantation recipient peripheral blood. Symbols represent individual mice (WT to WT mice, n = 6; 2-month-old 9p21+/− to WT mice, n = 5; and 6-month-old 9p21+/− to WT mice, n = 6). (H) Kaplan-Meier plots of Cx3Cr1CreER (n = 7) and Cx3Cr1CreER9p21fl/fl mice (n = 7). (I) Trichrome Masson and reticulin staining of Cx3Cr1CreER9p21fl/fl mice BM. (J) Quantification of the spleen and liver weights. Symbols represent individual mice (WT mice, n = 22; 9p21+/− tumor mice, n = 38; and Cx3Cr1CreER9p21fl/fl tumor mice, n = 7). (K) Complete blood count analysis of mice. Symbols represent individual mice (9p21+/− tumor mice, n = 15; WT mice, n = 17; and Cx3Cr1CreER9p21fl/fl tumor mice, n = 5). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 in panels D,F-G,J-K.
To further dissect the origin of MDS/MPN in the stromal vs hematopoietic compartment, we used conditional targeting of the 9p21sflox allele. Firstly, we crossed it with the Pdzk1ip1-CreER deleter strain that mediates tamoxifen-inducible Cre recombination in HSCs.20 The administration of tamoxifen to the resulting Pdzk1ip1-CreER 9p21sflox/+ or 9p21sflox/flox mice resulted in efficient recombination of the conditional allele (supplemental Figure 5D-E). However, the mice did not show any signs of disease or reduced survival for up to 2 years (supplemental Figure 5F) or any hematopoietic abnormalities (supplemental Figure 5G). We then crossed 9p21sflox mice with the Cx3cr1CreER strain, which is commonly used to target genes in Cx3cr1-expressing monocytes and hematopoietic progenitors.26 However, Cx3cr1 is also expressed in osteogenic cells,27 and, indeed, the Cx3cr1CreER strain mediated efficient recombination in osteoblast cells (supplemental Figure 5H). The administration of tamoxifen would be expected to permanently delete 9p21s in long-lived cells such as osteoblasts, whereas short-lived cells such as monocytes would quickly turn over26 (supplemental Figure 5I). We found that all tamoxifen-treated 9p21sflox/floxCx3cr1CreER mice succumbed to MDS/MPN–like disease with a median survival of 314 days (Figure 4H). The BM of these mice showed excessive trabecular osteogenesis and/or reticulin fibrosis (Figure 4I). These mice also exhibited hypocellular BM, splenomegaly, and hepatomegaly, similar to 9p21s+/− mice (Figure 4J-K). These data support the notion that the deletion of 9p21s in the stromal rather than hematopoietic cells is necessary and sufficient for MDS/MPN–like disease.
Single-cell analysis of the disease-promoting 9p21s+/− microenvironment
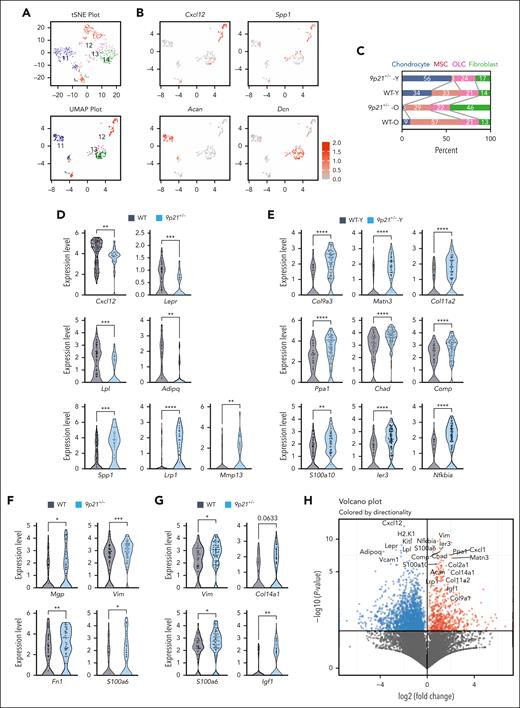
To characterize the MDS/MPN–driving stromal compartment in an unbiased manner, we sorted primary stromal cells by excluding hematopoietic lineage–positive cells, CD71+ erythroid progenitors. The CD31+ endothelial cells are more numerous, and their yield has been variable; therefore, they were excluded to allow better capture of other stromal cells. These Lin− CD71− CD31− populations were isolated from the BM of 4 individual mice: 1 10-month-old 9p21s+/− mouse with restricted joint mobility but no overt MDS/MPN (young or 9p21+/−; Y); 1 16-month-old 9p21s+/− mouse with overt MDS/MPN (old or 9p21+/−; O); and 2 age-matched Y and O WT mice (WT-Y and WT-O, respectively). The samples were stained with oligonucleotide-conjugated antibodies (“hashtags”), and sorted cells were pooled, analyzed via single-cell RNA-seq, and assigned to individual samples based on their hashtags, per the CITE-seq protocol.28 Our stringent filtering criteria retained 825 cells from 9p21s+/− mice and 3426 cells from control WT mice, revealing 14 distinct clusters based on differentially expressed genes (supplemental Figure 6A-C; supplemental Table 3). Despite the intended enrichment of stromal cells, their relative scarcity resulted in the majority of identified clusters (3621 cells, clusters 1-10) corresponding to hematopoietic cells, including SiglecH+ plasmacytoid dendritic cells (cl. 1), hemoglobin-positive erythroid cells (cl. 2), among others (supplemental Figure 6D-E). The remaining clusters 11 to 14 (630 cells) corresponded to stromal cells; notably, their combined frequency was higher in both old and young 9p21s+/− mice, consistent with the depletion of hematopoietic cells (supplemental Figure 6D). The identity of stromal cell clusters was assigned by unbiased matching to a published integrated analysis of the BM stromal compartment.29 The clusters corresponded to MSCs (cluster 12, Cxcl12+) and mesenchymal lineages classified as osteolineage cells (cluster 13, Spp1+), chondrocytes (cluster 11, Acan+), and fibroblasts (cluster 14, Dcn+) (Figure 5A-B; supplemental Figure 7A).
Single-cell analysis of the primary BM microenvironment in sick 9p21s+/− mice. (A) t-Distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) showing 4 niche clusters 11 to 14 in sick 9p21+/− mice (n = 2) and WT mice (n = 2). (B) Expression level of population marker genes. Acan (cluster 11 = chondrocyte); Cxcl12 (cluster 12 = MSC); Spp1 (cluster 13 = osteolineage cell, osteolineage); and Dcn (cluster 14 = fibroblast). (C) The frequency of clusters in total niche cells. (D) Gene expression changes in the MSC population. Symbols represent single cells (WT cells, n = 134; 9p21+/− cells, n = 41). (E) Gene expression changes in the chondrocyte population. Symbols represent single cells (WT-Y cells, n = 67; 9p21+/−-Y cells, n = 105). (F) Gene expression changes in osteolineage cell population. Symbols represent single cells (WT cells, n = 66; 9p21+/− cells, n = 71). (G) Gene expression changes in fibroblast population. Symbols represent single cells (WT cells, n = 43; 9p21+/− cells, n = 88). (H) Volcano plot showing the upregulated and downregulated gene expression in sick 9p21+/− mice total niche cells (pooled clusters 11-14) compared with WT mice total niche cells. The significantly differentially expressed genes were labeled. Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 in panels D-G. 9p21+/− -O, 16-month-old sick mouse; 9p21+/− -Y, 10-month-old sick mouse; WT-O, 16-month-old WT mouse; WT-Y, 10-month-old WT mouse.
Single-cell analysis of the primary BM microenvironment in sick 9p21s+/− mice. (A) t-Distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) showing 4 niche clusters 11 to 14 in sick 9p21+/− mice (n = 2) and WT mice (n = 2). (B) Expression level of population marker genes. Acan (cluster 11 = chondrocyte); Cxcl12 (cluster 12 = MSC); Spp1 (cluster 13 = osteolineage cell, osteolineage); and Dcn (cluster 14 = fibroblast). (C) The frequency of clusters in total niche cells. (D) Gene expression changes in the MSC population. Symbols represent single cells (WT cells, n = 134; 9p21+/− cells, n = 41). (E) Gene expression changes in the chondrocyte population. Symbols represent single cells (WT-Y cells, n = 67; 9p21+/−-Y cells, n = 105). (F) Gene expression changes in osteolineage cell population. Symbols represent single cells (WT cells, n = 66; 9p21+/− cells, n = 71). (G) Gene expression changes in fibroblast population. Symbols represent single cells (WT cells, n = 43; 9p21+/− cells, n = 88). (H) Volcano plot showing the upregulated and downregulated gene expression in sick 9p21+/− mice total niche cells (pooled clusters 11-14) compared with WT mice total niche cells. The significantly differentially expressed genes were labeled. Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 in panels D-G. 9p21+/− -O, 16-month-old sick mouse; 9p21+/− -Y, 10-month-old sick mouse; WT-O, 16-month-old WT mouse; WT-Y, 10-month-old WT mouse.
The relative abundance of stromal cell clusters was different between 9p21s mice and their matching controls, including decreased MSC cluster in both 9p21s+/− mice, and expanded chondrocyte and fibroblast clusters in 9p21s+/−-Y and 9p21s+/−-O mice, respectively (Figure 5C). To detect subpopulations in the individual clusters whose abundance differs between 9p21+/− and WT mice, we performed an integrated analysis of our BM microenvironment data set with the aforementioned published integrated analysis data set (supplemental Figure 7B-C). The analysis revealed that osteo-primed mesenchymal progenitor levels were increased in both 9p21s+/−-Y and 9p21s+/−-O mice compared with those in WT mice (supplemental Figure 8A-B). To explore the heterogeneity within the individual subpopulation, we applied the original subpopulations from 3 published data sets for the integrated analysis.24,25,29,30 We observed abundant fibroblast progenitors (fibroblasts-1-09) expressing mesenchymal stroma markers (Ly6a, Pdgfra, Thy1, and Cd44)24 in the 9p21s+/− fibroblast cluster (supplemental Figure 8C), and osteoarthritis-related hypertrophic chondrocytes (hondrocytes-hypertrophic-02)24 among 9p21s+/−-Y chondrocytes, respectively (supplemental Figure 8D). These data demonstrate shifts of osteo-primed MSC and fibroblast progenitors in both 9p21s+/− mice, suggesting the possibility that these stem and progenitor cells may contribute to the production of excessive trabecular bone and BMF, respectively; furthermore, the abundant osteoarthritis-related hypertrophic chondrocytes in the 9p21s+/− mouse with the knee problem may be associated with joint inflammation.
Within the MSC cluster, the expression of HSC niche genes Cxcl12 and Lepr was significantly decreased in 9p21s+/− mice (Figure 5D). Similarly, the expression of adipogenic markers Lpl and Adipoq was reduced, whereas the expression of osteogenic markers Spp1, Lrp1, and Mmp13 were increased in MSCs (Figure 5D). Cells in the chondrocyte cluster showed a significant upregulation of ossification-related genes (Matn3, Col9a3, Chad, Col11a2, Comp, Ppa1, and S100a10) and cartilage metabolism/NF-κB–related genes (Ier3 and Nfkbia; Figure 5E). The osteolineage cell cluster showed upregulation of osteogenesis-related genes (Mgp and Fn1) and fibrosis-related genes (S100a6 and Vim) (Figure 5F), whereas fibroblasts exhibited upregulation of these fibrosis-related genes as well as extracellular matrix (ECM)-related genes (Col14a1 and Igf1; Figure 5G). Finally, we compared gene expression between WT and 9p21s+/− cells within all stromal clusters from 11 to 14, yielding 328 upregulated genes and 1085 downregulated genes (Figure 5H; supplemental Table 4). Among the most significantly downregulated genes were Cxcl12 and Kitl, encoding the 2 stroma-derived factors Cxcl12 and Kit ligand (also known as stem cell factor [SCF]) that are critical for HSC maintenance.31 Conversely, upregulated genes encoded multiple ECM proteins such as collagens and vimentin. Disease analysis of the upregulated genes with Enrichr32 (Jensen diseases database) yielded growth disorders in bone, cartilage, and collagen-related connected tissue (supplemental Table 5). Moreover, the upregulated genes in BioPlanet database analysis with Enrichr identified several crucial signaling pathways including transforming growth factor β, Oncostatin M, and ECM regulatory networks were responsible for the regulation of 9p21 deletion–mediated fibrosis (supplemental Table 5). These results collectively suggest that the BM stroma in 9p21s haplodeficient mice harbors a contracted MSC compartment that is primed toward osteogenic, chondrogenic, and fibrogenic differentiation at the expense of functional support of hematopoiesis.
To further explore the potential cross talk between the aberrant 9p21s+/− stroma and hematopoietic cells, we took advantage of the serendipitous presence of the latter in our CITE-seq samples. In particular, cluster 7 was identified as a MkP based on the expression of megakaryocyte/platelet-specific genes Pf4, Plek, Itga2b, and Serpinb1 and of the cell proliferation signature (supplemental Figure 9A-B). Cells in this cluster were abundant in both 9p21s+/− samples (supplemental Figure 9C) and showed upregulation of 285 genes compared with that in WT MkPs (supplemental Table 6); Enrichr analysis of these genes revealed enrichment of genes associated with chronic myelogenous leukemia (supplemental Figure 9D; supplemental Table 7). The aberrant gene expression in MkPs corresponded to the dysplasia and apoptosis in megakaryocytes within the BM of sick 9p21s+/− mice, consistent with thrombocytopenia in these animals (supplemental Figure 9E). We then performed CellPhoneDB analysis of cell-to-cell communication mediated by ligand-receptor pairs33 between each niche cell population and MkPs, or within 2 niche cell populations. This analysis suggested a potential interaction between the Spp1 product OPN, which is elevated in 9p21s+/− MSCs, and its receptors CD44 and integrin α4β1 on MkPs (supplemental Figure 9F; supplemental Table 8). OPN is highly expressed in the BM stroma and affects HSC aging–associated lineage skewing.34 OPN protein levels in patients with MPNs (primary myelofibrosis [PMF]) correlate with more severe fibrosis and shorter overall survival.35 These results suggest possible pathways through which the aberrant stromal compartment may affect hematopoietic differentiation to promote the MDS/MPN phenotype.
Increased osteogenesis and aberrant chemokine production in the 9p21s+/− BM stroma
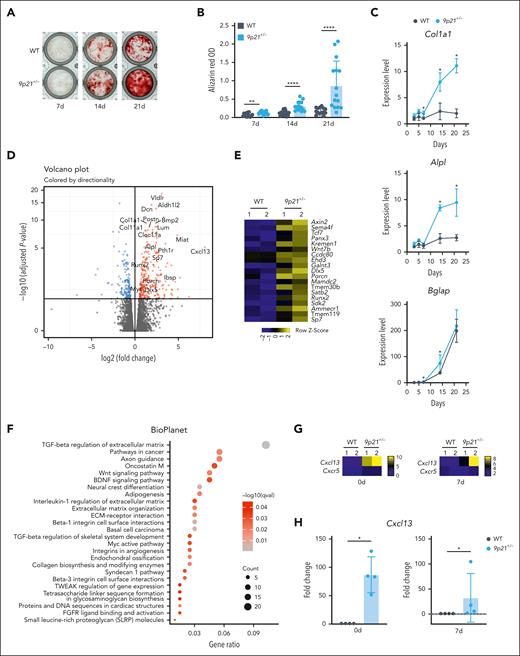
Together with excessive trabecular bone formation and BMF, the single-cell analysis of 9p21s-haplodeficient BM stroma suggested a bias toward osteogenic, chondrogenic, and fibrogenic differentiations. It is important to note that these cells also play a crucial role in the basic maturation process of cells of bone origin. To test this notion, we cultured primary BM stromal cells in the conditions that promote osteogenic differentiation in vitro.36 We found that stromal cultures from sick 9p21s+/− mice showed enhanced Alizarin Red staining (indicative of osteogenesis and calcium deposition) between 7 and 21 days (Figure 6A-B). Accordingly, RT-PCR analysis of 9p21s+/− cultures showed upregulation of the collagen type I gene (Col1a1) at all time points as well as osteogenic differentiation markers Bglap (at 7-14 days) and Alpl (at 14-21 days) (Figure 6C). We then performed global RNA-seq on day 7 of cultures derived from 3 separate sick 9p21s+/− mice and age-matched WT mice. Differential gene expression helped confirm the upregulation of osteogenic differentiation markers including Col1a1 and Alpl, osteogenic mediators such as Bmp2, and transcriptional master regulators of osteogenesis such as Sp7 (Osterix) and Runx2 (Figure 6D-E) in 9p21s+/− cultures. The pathway enrichment analysis of upregulated genes revealed transforming growth factor β–dependent regulation of the ECM as the most enriched pathway (Figure 6F; supplemental Tables 9 and 10). Furthermore, our in vitro osteogenic differentiation of Cx3cr1CreER-YFP9p21fl/fl BM-derived stromal cells showed a near-complete expression of the Cre-linked YFP reporter in osteoblast cells at day 8 after differentiation (supplemental Figure 10A), and calcium deposition was increased in Cx3cr1CreER-YFP9p21fl/fl compared with R26CreER mice (supplemental Figure 10B). We also used the osteogenic-specific Cre line Osx1, which induces Cre activity in embryonic and early postnatal osteogenic cells under no tamoxifen induction. Genomic DNA qPCR analysis showed ∼50% Cre efficiency in BM-derived stromal cells of the Osx1-Cre 9p21fl/+ cohort, corresponding to ∼25% deletion of the total 9p21 ortholog locus (supplemental Figure 10C). After 18 months of complete blood count and fluorescence-activated cell sorting lineage tracing, we did not observe any mice that developed MDS/MPN disease in the Osx1-Cre 9p21fl/+ cohort (supplemental Figure 10D-E). Critically, although homozygous deletion of Mtap causes embryonic lethality, we obtained 2 Osx1-Cre 9p21fl/fl mice that might not have had sufficient Cre efficiency and escaped the lethal crisis. These 2 mice developed the same MDS/MPN disease with excessive trabecular bones (supplemental Figure 10F-H). These data directly confirm that MDS/MPN–like diseases in 9p21s+/− mice are associated with increased osteogenic differentiation of the BM stroma.
Osteogenic differentiation of cultured 9p21s+/− MDS/MPN–like mice BM stromal cells. (A) Alizarin Red staining of osteogenic differentiation of BM stromal cells at 7, 14, and 21 days. (B) Quantification of Alizarin Red staining. Results were carried out in triplicate wells from individual mice (WT mice, n = 5; 9p21+/− tumor mice, n = 5). Symbols represent individual wells. (C) Expression of osteogenic marker genes in the differentiated BM stromal cells at different time points was confirmed via qRT-PCR. Symbols represent the average gene expression of 2 groups (WT mice group, n = 4; and 9p21+/− mice group, n = 4). (D) Volcano plot showing the upregulated and downregulated genes expression of cultured 9p21+/− tumor mice BM stromal cells compared with cultured WT mice BM stromal cells at 7 days after osteogenic differentiation. The significantly differentially expressed genes are labeled. (E) Heatmap showing increased levels of osteoblast signaling pathway–related genes in 9p21+/− tumor mice (n = 2) and WT mice (n = 2) BM stromal cells at 7 days after osteogenic differentiation. (F) Signaling pathway enrichment analysis with Enrichr for significant upregulated genes in 9p21+/− tumor mice BM stromal cells at 7 days after osteogenic differentiation. Gene count, the number of genes enriched in a Gene Ontology (GO) term; gene ratio, the ratio of genes counted in total of 199 upregulated genes. (G) Heatmap showing log2 Cxcl13 and Cxcr5 gene expression in 9p21+/− tumor mice (n = 2) and WT mice (n = 2) BM stromal cells at 0 and 7 days after osteogenic differentiation. (H) Cxcl13 gene expression change in BM stromal cells at 0 and 7 days after osteogenic differentiation were confirmed via qRT-PCR. Symbols represent individual mice (WT mice, n = 4; 9p21+/− tumor mice, n = 4). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗P < .05; ∗∗P < .01; ∗∗∗∗P <.0001 in panels B-C,H.
Osteogenic differentiation of cultured 9p21s+/− MDS/MPN–like mice BM stromal cells. (A) Alizarin Red staining of osteogenic differentiation of BM stromal cells at 7, 14, and 21 days. (B) Quantification of Alizarin Red staining. Results were carried out in triplicate wells from individual mice (WT mice, n = 5; 9p21+/− tumor mice, n = 5). Symbols represent individual wells. (C) Expression of osteogenic marker genes in the differentiated BM stromal cells at different time points was confirmed via qRT-PCR. Symbols represent the average gene expression of 2 groups (WT mice group, n = 4; and 9p21+/− mice group, n = 4). (D) Volcano plot showing the upregulated and downregulated genes expression of cultured 9p21+/− tumor mice BM stromal cells compared with cultured WT mice BM stromal cells at 7 days after osteogenic differentiation. The significantly differentially expressed genes are labeled. (E) Heatmap showing increased levels of osteoblast signaling pathway–related genes in 9p21+/− tumor mice (n = 2) and WT mice (n = 2) BM stromal cells at 7 days after osteogenic differentiation. (F) Signaling pathway enrichment analysis with Enrichr for significant upregulated genes in 9p21+/− tumor mice BM stromal cells at 7 days after osteogenic differentiation. Gene count, the number of genes enriched in a Gene Ontology (GO) term; gene ratio, the ratio of genes counted in total of 199 upregulated genes. (G) Heatmap showing log2 Cxcl13 and Cxcr5 gene expression in 9p21+/− tumor mice (n = 2) and WT mice (n = 2) BM stromal cells at 0 and 7 days after osteogenic differentiation. (H) Cxcl13 gene expression change in BM stromal cells at 0 and 7 days after osteogenic differentiation were confirmed via qRT-PCR. Symbols represent individual mice (WT mice, n = 4; 9p21+/− tumor mice, n = 4). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as follows: ∗P < .05; ∗∗P < .01; ∗∗∗∗P <.0001 in panels B-C,H.
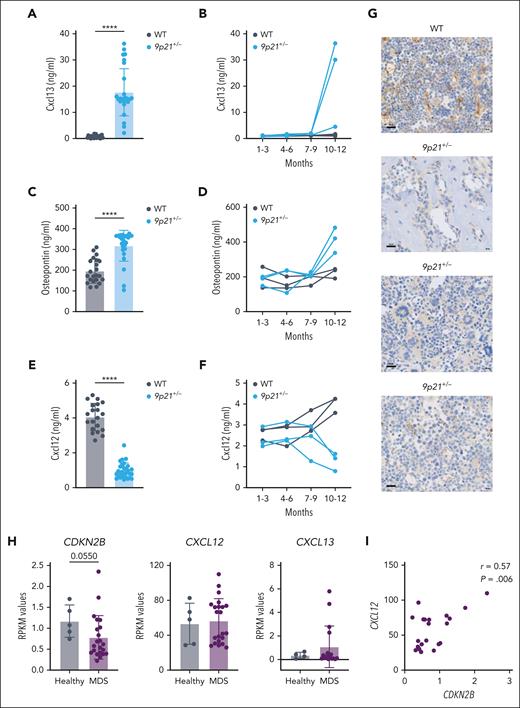
Differential expression analysis of BM stromal cultures revealed Cxcl13 as the most upregulated gene in 9p21s+/− stromal cells (Figure 6D). Cxcl13 encodes a chemokine that regulates the migration and function of B lymphocytes37 and has not been implicated in myeloid malignancy; however, its overexpression in stromal cells is thought to facilitate fibrotic diseases.38 The induction of Cxcl13 expression without change in expression of its receptor Cxcr5 (Figure 6G) was confirmed via qRT-PCR in 9p21s+/− stromal cells at the onset of (day 0) and during (day 7) osteogenic differentiation (Figure 6H). The observed induction of Cxcl13 and OPN, along with the loss of Cxcl12, in the BM stroma from sick 9p21s+/− mice suggested a global switch in the expression of these chemokines during the disease. Indeed, Cxcl13 was virtually undetectable in the sera of control WT mice but was present at high levels (∼20 ng/mL) in the sera of 9p21s+/− mice with MDS/MPN–like disease (Figure 7A). The upregulation of Cxcl13 and OPN occurred concomitantly with the disease between 10 and 12 months of age (Figure 7B). The increase of OPN pattern along with Cxcl13 was very similar (Figure 7C-D). In contrast, the serum levels of Cxcl12 were reduced, on average, fourfold in sick 9p21s+/− mice (Figure 7E), and the trend toward reduction, although not significant, became apparent before the onset of the disease (Figure 7F). Accordingly, the loss of Cxcl12 protein expression in the BM stroma of 9p21s+/− mice was confirmed via immunochemistry (Figure 7G). Furthermore, we performed a gene expression analysis on bone chips collected from patients with MDS, which revealed a marginally significant decrease in CDKN2B expression compared with that in healthy individuals (Figure 7H). Notably, we observed a strong correlation between the decrease in CDKN2B expression and CXCL12 levels in the bone of patients with MDS (Figure 7I). Among the 17 patients with MDS that we analyzed, we found that 4 had high levels of CXCL13 expression (Figure 7H). Future studies with larger cohorts of patients with MDS are needed to confirm and extend these observations. Collectively, these data show that BM stromal cells in 9p21s+/− mice lose the expression of Cxcl12 while upregulating the related factors Cxcl13 and Spp1. Given that the former is critical for HSC/progenitor maintenance39 and the latter factors promote fibrosis,38 this switch likely contributes to the ability of the abnormal BM stroma to drive MDS/MPN–like disease.
Two CXCL chemokines and osteopontin/Spp1 are involved in the pathogenesis of MDS/MPN-like disease in 9p21+/− mice. (A) Cxcl13 protein level in mice serum was measured via enzyme-linked immunosorbent assay (ELISA). Symbols represent individual mice (WT mice, n = 23; 9p21+/− tumor mice, n = 27). (B) Cxcl13 protein level in mice serum was measured via ELISA at different time points of tumorigenesis. Symbols represent individual mice (WT mice, n = 3; 9p21+/− mice, n = 3). (C) Osteopontin protein level in mice serum was measured via ELISA. Symbols represent individual mice (WT mice, n = 22; 9p21+/− tumor mice, n = 25). (D) Osteopontin protein level in mice serum was measured via ELISA at different time points of tumorigenesis. Symbols represent individual mice (WT mice, n = 3; 9p21+/− mice, n = 3). (E) Cxcl12 protein level in mice serum was measured via ELISA. Symbols represent individual mice (WT mice, n = 27; 9p21+/− tumor mice, n = 36). (F) Cxcl12 protein level in mice serum was measured by ELISA at different time points of tumorigenesis. Symbols represent individual mice (WT mice, n = 3; 9p21+/− mice, n = 3). (G) Immunohistochemical staining of Cxcl12 in mice BM. The images represent individual mice. The results represent a total of 5 9p21+/− MDS/MPN mice and 6 WT mice. (H) Gene expression changes in the bone of patients with MDS. Symbols represent individual human samples. CXCL12 and CDKN2A (samples from healthy individuals, n = 5; samples from patients with MDS, n = 22); CXCL13 (samples from healthy individuals, n = 5; samples from patients with MDS, n = 17). RPKM, the reads per kilobase per million value. (I) Positive correlation between CXCL12 and CDKN2B in the bone of patients with MDS. Symbols represent individual patients with MDS (n = 22). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as ∗∗∗∗P < .0001 in panels A,C,E.
Two CXCL chemokines and osteopontin/Spp1 are involved in the pathogenesis of MDS/MPN-like disease in 9p21+/− mice. (A) Cxcl13 protein level in mice serum was measured via enzyme-linked immunosorbent assay (ELISA). Symbols represent individual mice (WT mice, n = 23; 9p21+/− tumor mice, n = 27). (B) Cxcl13 protein level in mice serum was measured via ELISA at different time points of tumorigenesis. Symbols represent individual mice (WT mice, n = 3; 9p21+/− mice, n = 3). (C) Osteopontin protein level in mice serum was measured via ELISA. Symbols represent individual mice (WT mice, n = 22; 9p21+/− tumor mice, n = 25). (D) Osteopontin protein level in mice serum was measured via ELISA at different time points of tumorigenesis. Symbols represent individual mice (WT mice, n = 3; 9p21+/− mice, n = 3). (E) Cxcl12 protein level in mice serum was measured via ELISA. Symbols represent individual mice (WT mice, n = 27; 9p21+/− tumor mice, n = 36). (F) Cxcl12 protein level in mice serum was measured by ELISA at different time points of tumorigenesis. Symbols represent individual mice (WT mice, n = 3; 9p21+/− mice, n = 3). (G) Immunohistochemical staining of Cxcl12 in mice BM. The images represent individual mice. The results represent a total of 5 9p21+/− MDS/MPN mice and 6 WT mice. (H) Gene expression changes in the bone of patients with MDS. Symbols represent individual human samples. CXCL12 and CDKN2A (samples from healthy individuals, n = 5; samples from patients with MDS, n = 22); CXCL13 (samples from healthy individuals, n = 5; samples from patients with MDS, n = 17). RPKM, the reads per kilobase per million value. (I) Positive correlation between CXCL12 and CDKN2B in the bone of patients with MDS. Symbols represent individual patients with MDS (n = 22). Data represent the mean ± SD. Statistical significance was defined using Mann-Whitney test and is shown as ∗∗∗∗P < .0001 in panels A,C,E.
Discussion
Our study of 9p21s+/− mice provides a novel mouse model that develops an MDS/MPN–like disease driven by the BM microenvironment. Previous studies as well as this study on the cBioPortal database and GDC data portal samples have demonstrated that the deletion of the entire 9p21 locus including CDKN2A/CDKN2B, CDKN2BAS, and MTAP occurs commonly in various diseases including leukemias.9,40,41 Furthermore, deletion of the 9p21 locus has been associated with the emergence of tumor subclones in the prolonged culture of telomerized human BM stromal cells.42 Notably, the loss of genes within the 9p21 locus is also associated with fibrosis and bone diseases. The loss of CDKN2B can promote lung fibrosis through increasing fibroblast differentiation43; downregulation of the p16 protein encoded by CDKN2A reportedly correlates with cirrhosis in humans,44 and downregulation of MTAP45 or epigenetic silencing CDKN2BAS (ANRIL)46 was shown to accelerate liver fibrosis. The MTAP gene is commonly deleted in osteosarcoma, and its loss is frequently associated with the deletion of CDKN2A.47 Genomic loss of 1 or 2 of cdkn2 locus–related genes (CDKN2A and CDKN2B) can be considered as the key mediators of murine MSCs transformation in osteosarcoma.21,48 Here, we found that the deletion of the entire 9p21-syntenic locus can promote the differentiation of multiple BM stromal lineages, including fibrogenesis, osteogenesis, and chondrogenesis, while inhibiting adipogenesis. In our MDS/MPN mice, this aberrant stromal progenitor differentiation manifests as BMF, increase in trabecular bone formation, and restriction in joint mobility. Our reciprocal BM marrow transplantations prove that the BM microenvironment affected by 9p21 deletion cannot support normal hematopoiesis, causing the occurrence of myeloid disorders. Thus, to the best of our knowledge, this study is the first to report that haplodeficiency of the 9p21 locus causes an aberrant BM microenvironment in mice, contributing to the development of MDS/MPN–like disease. Furthermore, our findings demonstrate reduced expression of the CDKN2B in the bones of patients with MDS, suggesting a potential role for the human 9p21 locus in the BM microenvironment and pathogenesis of the disease.
BMF is associated with poor prognosis and increased risk of AML in patients with MDS.7,49 However, the mechanisms by which mutations in the BM microenvironment contribute to the development of MDS and the role of BMF in this process are still poorly understood. Patients with MDS with deletions in chromosomes 5q, 7q, or −7 have an increased risk of developing fibrosis during follow-up, but it is unclear whether chromosomal instability precedes BM microenvironment or hematopoietic disorders.50 Studies in mice have shown that germ line deletion of the retinoic acid receptor subtype γ gene or conditional knock out of the retinoblastoma protein leads to BM microenvironment failure in the development of PMF.51,52 Recent evidence suggests that cell-extrinsic abnormalities within the BM microenvironment play a significant role in driving myeloid leukemia,3 including abnormalities in Dicer1,53 β-catenin,54 or Ptpn11.3,55 However, there is limited understanding of how the BM microenvironment contributes to the development of full-blown disease in pre-MDS/MPNs mice. Our established mouse model serves as a valuable tool for investigating the pathogenesis of MDS/MPN with BMF and/or excess trabecular bones originating from the BM microenvironment. Through this model, dysregulated pathways in stromal differentiation and stromal-hematopoietic cross talk can be identified. Notably, the transplantability of MDS/MPN disease in 9p21 mice underscores the potential of this model in deciphering the underlying mechanisms of MDS and in developing novel therapies and treatments for patients with MDS-BMF. Moreover, the lack of progression to AML in 9p21 mice with the leukemogenic mutation Flt3-ITD highlights the significance of considering both cell-extrinsic and -intrinsic abnormalities in comprehending the development and progression of hematologic malignancies.
Normal differentiation of MSCs results in a BM microenvironment that supports normal HSC function. However, our observations in 9p21s+/− mice suggest that alterations in MSC differentiation may trigger a complex phenotype of MDS/MPN overlap disease. Specifically, we found reductions in stem cell–related genes (eg, Cxcl12 and Lepr) and adipogenesis-related genes (eg, Adipoq and Lpl), along with increases in osteogenesis-related genes (eg, Spp1, Lrp1, and Mmp13), suggesting the preferential differentiation of MSCs into osteogenic but not adipogenic cells. Fibroblast progenitors also persisted in the older 9p21s+/− mice and were likely to differentiate into fibroblasts or other mesenchymal-like stromal cells.24 Notably, the amount of Cxcl12 protein secreted from 9p21s+/− MSCs was decreased in the BM, thus weakening the core role of the CXCL12-CXCR4 axis in the BM microenvironment and hematopoietic system. Furthermore, the reduction of Kit (encoding Kit ligand/SCF) in stromal cells may also affect hematopoiesis. In PMF, downregulation of the expression of key HSC supporting factors (Cxcl12 and SCF) and upregulation of genes associated with osteogenesis and fibrosis in Lerp+ mesenchymal stromal linage cells can cause fibrogenic conversion.56 In the context of 9p21s deletion, MSCs or osteogenic cells may secrete the profibrosis factor Cxcl13 and osteogenesis- and fibrosis-related Spp1, leading to BMF and/or excessive trabecular bone and even the formation of fibrotic solid tumors outside of the BM (data not shown). Notably, our findings revealed a correlation between CDKN2B and CXCL12 in the bone of individuals with human MDS, indicating that the 9p21 locus may be involved in the differentiation of human MSCs. However, it is important to acknowledge that these data reflect only a limited number of samples and should be replicated in a larger collection of samples from patients with MDS in the future, when they become available. Therefore, our hypothesis is that the deletion of the 9p21 locus leads to an abnormal priming of the MSC compartment toward osteogenic, chondrogenic, and fibrogenic differentiation. This results in an imbalance of secreted stromal and hematopoietic differentiation factors that ultimately negatively affects the functional support of hematopoiesis.
In summary, our study describes a model in which an aberrant BM microenvironment drives the development of MDS/MPN–like disease with BMF and/or increased osteogenesis. Because fibrosis is common in human MDS, our study suggests that it may represent a causal factor of MDS rather than a secondary effect of hematopoietic abnormalities. Conversely, MDS in older human patients is weakly associated with osteoporosis rather than increased osteogenesis.57 This association can be secondary to the age dependency of both conditions, and/or driven by the primary hematopoietic defects such as Dnmt3a mutations, which may activate osteoclastogenesis.58 In contrast, PMF shows a strong association with osteosclerosis.59 Thus, increased osteogenesis in our mouse model may better represent the latter condition, in which abnormalities of hematopoietic and stromal cells create a vicious cycle to amplify one another. Future studies of the primary BM stromal compartment of patients with MDS or MPN should further elucidate its role in the development of these diseases.
Acknowledgments
The authors thank Masayuki Yazawa and Nicholas Adams for discussions throughout this project; Chyuan-Sheng Lin for ES targeting; Ulf Klein for providing targeting vectors; Azra Raza for samples from patients with MDS or AML; and Roshan P. Shah for samples from healthy patients. The authors also thank Justin Mehl, Christopher Beam, and Joseph David for technical support. Furthermore, the authors acknowledge the usage of resources provided by the Cytometry & Cell Sorting Laboratory, the Genome Technology Core (GTC), the Applied Bioinformatics Facility Laboratories (ABL), and the Experimental Pathology Core by the NYU Grossman School of Medicine, and the micro-CT Core of the NYU College of Dentistry.
The Cytometry & Cell Sorting Laboratory, the GTC, and the ABL are shared resources partially supported by National Institutes of Health (NIH) National Cancer Institute (NCI) grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center. The micro-CT Core is supported by NIH Office of the Director grant S10-OD010751. S.K. is supported by NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases grant AR054447 and the Edward P. Evans Foundation for MDS Research. NIH NCI grant P30CA013696 to the Herbert Irving Comprehensive Cancer Center at Columbia University. B.R. is supported by the Discovery Research Grant from the Edward P. Evans Foundation.
Authorship
Contribution: J.F. conceptualized and supervised the study, performed all experiments, analyzed data, and wrote the manuscript; P.-F.H., R.L., Y.W., and J.P. performed experiments; E.E. and A.K.-J. performed computational analysis; C.Z.L., A.A.A., and S.K. interpreted data; A.T. supervised computational analysis; and B.R. conceptualized and supervised the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: B.R. is an adviser for Related Sciences and a cofounder of Danger Bio, which are not related to this work. The remaining authors declare no competing financial interests.
Correspondence: Jue Feng, Department of Pathology, New York University Grossman School of Medicine, 435 E 30th St, SB423C, New York, NY 10016; e-mail: jue.feng@nyulangone.org; and Boris Reizis, Department of Pathology, New York University Grossman School of Medicine, 435 E 30th St, SB413, New York, NY 10016; e-mail: boris.reizis@nyulangone.org.
References
Author notes
CITE-seq data and bulk RNA-seq data have been deposited at Gene Expression Omnibus database and are publicly available as of the date of publication (accession number GSE211779).
Further information on resources and reagents are available on request from the corresponding author, Jue Feng (jue.feng@nyulangone.org).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal