Notch might overcome its inherent weakness as a transactivator in T-ALL by cobinding powerful RNA synthesis machinery.
The CDC73-induced DNA repair program co-opted by Notch is more highly expressed in T-ALL than in other tumors and mitigates genotoxic stress.
Visual Abstract
Activated Notch signaling is highly prevalent in T-cell acute lymphoblastic leukemia (T-ALL), but pan-Notch inhibitors showed excessive toxicity in clinical trials. To find alternative ways to target Notch signals, we investigated cell division cycle 73 (Cdc73), which is a Notch cofactor and key component of the RNA polymerase–associated transcriptional machinery, an emerging target in T-ALL. Although we confirmed previous work that CDC73 interacts with NOTCH1, we also found that the interaction in T-ALL was context-dependent and facilitated by the transcription factor ETS1. Using mouse models, we showed that Cdc73 is important for Notch-induced T-cell development and T-ALL maintenance. Mechanistically, chromatin and nascent gene expression profiling showed that Cdc73 intersects with Ets1 and Notch at chromatin within enhancers to activate expression of known T-ALL oncogenes through its enhancer functions. Cdc73 also intersects with these factors within promoters to activate transcription of genes that are important for DNA repair and oxidative phosphorylation through its gene body functions. Consistently, Cdc73 deletion induced DNA damage and apoptosis and impaired mitochondrial function. The CDC73-induced DNA repair expression program co-opted by NOTCH1 is more highly expressed in T-ALL than in any other cancer. These data suggest that Cdc73 might induce a gene expression program that was eventually intersected and hijacked by oncogenic Notch to augment proliferation and mitigate the genotoxic and metabolic stresses of elevated Notch signaling. Our report supports studying factors such as CDC73 that intersect with Notch to derive a basic scientific understanding on how to combat Notch-dependent cancers without directly targeting the Notch complex.
Introduction
Activation of the Notch signaling pathway plays essential roles in the development and homeostasis of diverse tissues, including multiple stages of T-cell fate specification and development.1 In normal cells, the Notch1-4 receptors are activated by ligands. Next, gamma-secretase cleaves Notch, which releases intracellular Notch (ICN1-4). ICN translocates to the nucleus, where it binds its DNA-binding cofactor recombination signal binding protein for immunoglobulin kappa J region (RBPJ) to induce transcription.2 In T-cell acute lymphoblastic leukemia (T-ALL), Notch signaling is activated constitutively to supraphysiological levels through NOTCH1 mutations that occur in ∼60% of cases.3 Gamma-secretase inhibitors inhibit Notch activation in both normal and cancer cells. Unfortunately, early clinical studies reported excessive toxicities with continuous gamma-secretase inhibitor dosing.4,5
The polymerase-associated factor 1 complex (Paf1C) was previously linked to the Notch pathway, as Drosophila with mutations in Rtf1 (a Paf1C subunit) or Bre1 (a direct Paf1C cofactor) show notched wings and impaired transcription of Notch target genes.6,7 Paf1C acts as a scaffold that physically links transcriptional machinery (eg, RNA polymerase II) to chromatin-modifying enzymes and promotes messenger RNA (mRNA) synthesis from initiation and elongation to termination and processing.8-10 In this role, Paf1C is estimated to promote the mRNA synthesis of ∼15% to 20% of the most highly expressed genes.11,12 Paf1C also has enhancer functions, regulating eRNA synthesis, and H3K27ac deposition.13,14 CDC73 is a subunit of Paf1C that was previously shown to interact with NOTCH1 in breast cancer cells.15 However, the precise link between CDC73 and NOTCH1 and its relevance for Notch-driven cancers remains unclear. In this context, we proposed and experimentally tested the role of CDC73 in Notch-induced T-ALL. Our results show that CDC73 intersects Notch-regulated pathways, particularly at known T-ALL drivers, DNA repair genes, and oxidative phosphorylation (OXPHOS) genes, and identify CDC73 as a new dependency in this cancer.
Materials and methods
Mice
C57/BL6 mice between 4 and 8 weeks old and LckCre mice were purchased from Taconic. Rosa26CreERT2Cdc73f/f mice were generated by crossing Cdc73f/f mice16 with Rosa26CreERT2 mice (Jackson). Mouse experiments were performed according to National Institutes of Health guidelines and approved protocols from the institutional animal care and use committee at the University of Michigan.
Cell lines
CUTLL1 cells were obtained from Adolfo Ferrando and Andrew Weng. All other cell lines, including SUP-T1, LOUCY, HPB-ALL, CEM, THP-6, and OP9-DL4 cells, were obtained as previously published.17
PDX experiments
Patient-derived xenograft 3 (PDX3; M71) and PDX4 (2583AB) were obtained from Andrew Weng and Moshe Talpaz, respectively. PDXs were expanded by injection into NOD-scid-IL2γnull mice.
Constructs
Antibodies and primers
Antibodies and primers are listed in supplemental Table 2, available on the Blood website.
Bone marrow transplantation
Flow cytometry
Flow cytometry and cell sorting was performed as previously described.17 Mitochondrial potential staining was done using tetramethylrhodamine methyl ester assay (Thermo Fisher).
Bru-seq, BruUV-seq, and ChIP-seq library preparation and sequencing
Bromouridine (Bru)-seq and BruUV-seq were performed as previously described.22-24 chromatin immunoprecipitation (ChIP)-polymerase chain reaction and -sequencing (ChIP-seq) library preparation and sequencing were performed as previously described17 with the following additions. All reactions were followed by a solid-phase reversible immobilization (SPRI) bead cleanup using AMPure or SPRISelect beads. Size selection for the library was performed using AMPure or SPRISelect beads. Paired-end sequencing was performed using Nova-seq or Next-seq with ∼20 million reads per sample by the University of Michigan Advanced Genomics Core.
Results
CDC73 interacts with NOTCH1 and ETS1 in T-ALL cells
To confirm the CDC73-NOTCH1 interaction in T-ALL cells, we performed coimmunoprecipitation (co-IP) assays. We detected endogenous CDC73 with Flag-NOTCH1 pulldown (supplemental Figure 1A). Because we previously showed that the transcription factor ETS1 nucleates multiple transcription factors at chromatin, including NOTCH1,17 we wondered whether CDC73 had a stronger interaction with ETS1 than with NOTCH1. Consistently, ETS1 interacted with CDC73 in forward and reciprocal co-IP assays with Flag-tagged proteins (supplemental Figure 1A-B) and endogenous proteins (supplemental Figure 1C). To define the genomic locations of CDC73-associated complexes, we performed CDC73 ChIP-seq in a control cell line and ETS1 knockdown in the human T-ALL cell line THP-6.17 The strongest CDC73 signals were associated with the most highly transcribed genes (supplemental Figure 1D) and active chromatin (supplemental Figure 1E). Consistent with our co-IP data, CDC73 peaks overlapped more frequently with ETS1 peaks (56%) than with RBPJ/Notch peaks (40%) (supplemental Figure 1F). Further, motif analysis showed that CDC73 peaks were more frequently associated with ETS motifs than any other motif (supplemental Figure 1G). ETS1 knockdown attenuated CDC73 peaks that overlapped with dynamic ETS1 peaks (defined as ETS1 peaks significantly diminished by ETS1 knockdown; supplemental Figure 1H-I) relative to all CDC73 peaks. Thus, the NOTCH1-CDC73 interaction appears to be dependent, at least partially, on an ETS1 context.
Cdc73 is important for Notch-dependent T-cell development
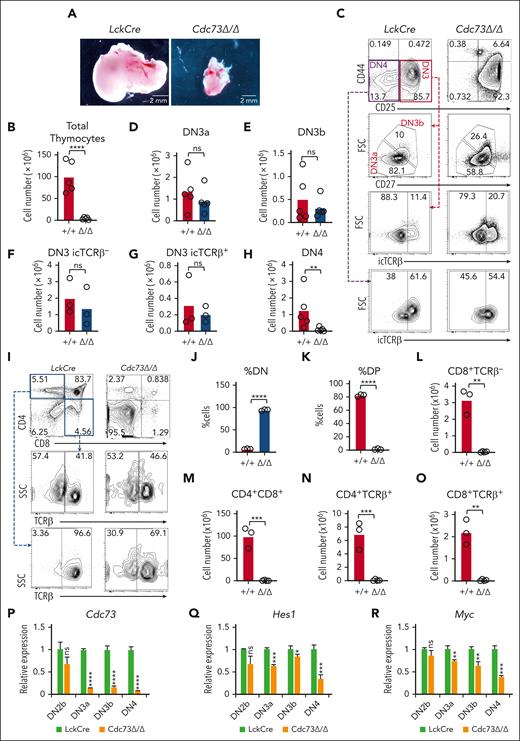
To investigate the importance of Cdc73 for Notch functions in T cells, we first studied its effect on T-cell development, which is driven primarily by Notch signals.25 Murine Notch-dependent T-cell development proceeds in the thymus through a series of stages, from the double-negative (DN) stages (DN1-DN4) to the immature single-positive and CD4+CD8+ double-positive (DP) stages, and then to the single-positive CD4+ or CD8+ stages. Like Notch1 and Ets1, Cdc73 and other Paf1C members are expressed throughout T-cell development (supplemental Figure 2A-B). To test whether Cdc73 might have similar functions as Notch1 and Ets1 in early T cells, we crossed LckCre mice with Cdc73f/f mice16 to generate LckCre Cdc73f/f mice (Cdc73Δ/Δ mice). Like Notch-deficient and Ets1-deficient mice,26-29Cdc73Δ/Δ mice showed severely impaired thymopoiesis (Figure 1A-B), with a significant loss of early T cells by the DN4 stage (Figure 1C-H) and impaired DN-to-DP cell transition (Figure 1I-O). Expression analysis of sorted DN subsets showed that Cdc73 deletion was detected at the DN3a stage but was strongest at the DN4 stage (Figure 1P). This effect is consistent with DN4 showing the most significant loss of cell number (Figure 1H) and the most significant loss of expression of the classic Notch target genes Hes1 and Myc (Figure 1Q-R). Thus, like Notch1 and Ets1, Cdc73 promotes the DN-to-DP transition.
Cdc73 is important for Notch-dependent T-cell development. (A-H) Representative images of thymuses (A); absolute thymocyte counts (B); representative flow cytometric profiles of DN subsets (C); and absolute numbers of DN3a (D), DN3b (E), DN3 icTCRβ− (F), DN3 icTCRβ+ (G), and DN4 (H) subsets in LckCre control and LckCre Cdc73f/f(Cdc73Δ/Δ) mice. (I-O) Representative flow cytometric profiles of CD4/CD8 subsets (I); %DN (J); %DP (K); and absolute numbers of immature single-positive (ISP) (L), DP (M), CD4 single-positive (SP) (N), and CD8 SP (O) thymic subsets in LckCre control and Cdc73Δ/Δ mice. DN3a = Lineage−CD44−CD25+FSCloCD27−; DN3b = Lineage−CD44−CD25+FSChiCD27+; DN4 = Lineage−CD44−CD25−; ISP = CD8+TCRb−; DP = CD4+CD8+; CD4 SP = CD4+TCRb+; And CD8 SP = CD8+TCRb+. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. (P-R) Relative expression of Cdc73 (P), Hes1 (Q), and Myc (R) in sorted thymic subsets from LckCre mice (green) and Cdc73Δ/Δ mice (orange). FSC, forward scatter; icTCR, intracellular T-cell receptor; ns, not significant.
Cdc73 is important for Notch-dependent T-cell development. (A-H) Representative images of thymuses (A); absolute thymocyte counts (B); representative flow cytometric profiles of DN subsets (C); and absolute numbers of DN3a (D), DN3b (E), DN3 icTCRβ− (F), DN3 icTCRβ+ (G), and DN4 (H) subsets in LckCre control and LckCre Cdc73f/f(Cdc73Δ/Δ) mice. (I-O) Representative flow cytometric profiles of CD4/CD8 subsets (I); %DN (J); %DP (K); and absolute numbers of immature single-positive (ISP) (L), DP (M), CD4 single-positive (SP) (N), and CD8 SP (O) thymic subsets in LckCre control and Cdc73Δ/Δ mice. DN3a = Lineage−CD44−CD25+FSCloCD27−; DN3b = Lineage−CD44−CD25+FSChiCD27+; DN4 = Lineage−CD44−CD25−; ISP = CD8+TCRb−; DP = CD4+CD8+; CD4 SP = CD4+TCRb+; And CD8 SP = CD8+TCRb+. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. (P-R) Relative expression of Cdc73 (P), Hes1 (Q), and Myc (R) in sorted thymic subsets from LckCre mice (green) and Cdc73Δ/Δ mice (orange). FSC, forward scatter; icTCR, intracellular T-cell receptor; ns, not significant.
Cdc73 is important for murine Notch-induced T-ALL maintenance
Next, we wondered whether the dependence of T-cell precursors on Cdc73 would be conserved after they transform in leukemia. To test this possibility, we used a well-established murine model of Notch-induced T-ALL.20,21 We transduced bone marrow stem and progenitor cells from Rosa26CreERT2 or Rosa26CreERT2Cdc73f/f mice with an activated Notch1 allele (ΔE/Notch1).30,31 We transplanted these cells into recipient mice to generate primary tumors (Figure 2A; supplemental Figure 3). Next, we transplanted different primary tumors into secondary recipients, which were injected with tamoxifen to delete Cdc73. In contrast to the control T-ALL mice (Figure 2B), Cdc73f/f T-ALL mice treated with tamoxifen showed reduced blast or white blood cell counts of 700-fold or 11-fold, respectively (Figure 2C). Tamoxifen did not change the median survival relative to vehicle in control T-ALL mice (Figure 2D) but prolonged survival by >200% or 84% in Cdc73f/f T-ALL mice (Figure 2E). To test whether Cdc73 is important in T-ALLs in which Notch is secondarily activated, we transplanted CD45.2+Lmo2-tg Rosa26CreERT2Cdc73f/f tumors (supplemental Figure 4A) into CD45.1+ congenic wild-type recipients, which were injected with tamoxifen to delete Cdc73. In this model, the Lmo2 transgene is the primary driver, and Notch activation is secondary.32 Some Lmo2-tg mice develop Notch-inactive immature T-ALL, which were excluded by quantitative polymerase chain reaction for Notch target genes (supplemental Figure 4B). This model is particularly relevant for studying ETS1-associated factors because mutations create ETS1 binding sites that activate LMO2 in human T-ALL.33 Mice treated with tamoxifen showed 80-fold reduced donor-derived peripheral blasts and prolonged median survival by >200% compared with control mice (supplemental Figure 4C-H). Thus, Cdc73 is important for the maintenance of primarily and secondarily Notch-activated T-ALL.
Cdc73 is important for Notch-induced T-ALL maintenance. (A) Experimental strategy to study dependence of Notch-induced T-ALL maintenance on Cdc73. (B-E) Mice were injected with 2 different primary ΔE/Notch1-induced Rosa26CreERT2 control T-ALL tumors (B,D) or 2 different primary ΔE/Notch1-induced Rosa26CreERT2Cdc73f/f murine T-ALL tumors (C,E). Numbers indicate tumor IDs. Peripheral blood green fluorescent protein–positive (GFP+) or white blood cell (WBC) counts for panels B-C at 2.5 weeks after transplant and survival for panels D-E were measured. (F) Western blot showing CDC73 knockdown in shRNA-transduced CEM cells. Numbers indicate relative band intensity. (G) Fold expansion (day 9 cell count / day 0 cell count) of Notch1-activated T-ALL cells transduced with 2 independent shCDC73. (H-I) Viability of conventional T-ALL PDX cells transduced with shCDC73 in OP9-DL4 stromal cell culture. N = 3. (J) Leukemia-free survival of NOD-scid-IL2γnull (NSG) mice injected with PDX4 cells transduced with shCDC73 that were passaged in NSG mice for 24 weeks. N = 5. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. cKO, comditional knockout; Tam, 25 mg/kg tamoxifen; WT, wild-type.
Cdc73 is important for Notch-induced T-ALL maintenance. (A) Experimental strategy to study dependence of Notch-induced T-ALL maintenance on Cdc73. (B-E) Mice were injected with 2 different primary ΔE/Notch1-induced Rosa26CreERT2 control T-ALL tumors (B,D) or 2 different primary ΔE/Notch1-induced Rosa26CreERT2Cdc73f/f murine T-ALL tumors (C,E). Numbers indicate tumor IDs. Peripheral blood green fluorescent protein–positive (GFP+) or white blood cell (WBC) counts for panels B-C at 2.5 weeks after transplant and survival for panels D-E were measured. (F) Western blot showing CDC73 knockdown in shRNA-transduced CEM cells. Numbers indicate relative band intensity. (G) Fold expansion (day 9 cell count / day 0 cell count) of Notch1-activated T-ALL cells transduced with 2 independent shCDC73. (H-I) Viability of conventional T-ALL PDX cells transduced with shCDC73 in OP9-DL4 stromal cell culture. N = 3. (J) Leukemia-free survival of NOD-scid-IL2γnull (NSG) mice injected with PDX4 cells transduced with shCDC73 that were passaged in NSG mice for 24 weeks. N = 5. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. cKO, comditional knockout; Tam, 25 mg/kg tamoxifen; WT, wild-type.
CDC73 is important for human NOTCH1–activated T-ALL propagation and maintenance
In a clinically annotated cohort of pediatric T-ALL, CDC73 is expressed across all oncogenomic and developmental subgroups (supplemental Figure 5A-B). CDC73 expression is not associated with white blood cell (supplemental Figure 5C) or survival (supplemental Figure 5D). To test the functional importance of CDC73 for human T-ALL cell proliferation, we transduced CDC73 shRNAs into human NOTCH1–activated T-ALL cells with effective suppression of the CDC73 protein (Figure 2F). CDC73 knockdown reduced proliferation of NOTCH1-activated T-ALL cell lines by twofold to ninefold, including Notch-independent T-ALLs CEM and THP-6 that are ETS1-dependent17 (Figure 2G). To test the antitumor effects of CDC73 inactivation in nonimmortalized human T-ALL cells, we took advantage of the success of shRNA protocols in knocking down gene expression in Notch-activated PDXs.17,34CDC73 knockdown reduced the viability of PDX cells by fivefold or sixfold (Figure 2H-I). Transplantation of these cells into immunodeficient mice showed that CDC73 knockdown significantly prolonged survival (Figure 2J). Taken together, these results demonstrate a strong and highly prevalent CDC73 dependency in human NOTCH1–activated T-ALL.
Cdc73 promotes gene expression in DNA repair and OXPHOS pathways
Next, we sought to understand the underlying mechanism that explains why Cdc73 is required for Notch1-activated T-ALL maintenance. Toward this goal, we generated 2 independent Notch-induced CreERT2Cdc73f/f T-ALL cell lines (969 and 970) from the tumors described in Figure 2C. We confirmed that 4-hydroxytamoxifen (OHT) induced Cdc73 deletion in these cells (supplemental Figure 6A) and impaired cell proliferation (Figure 3A). In contrast, OHT treatment had no effect on control CreERT2 cells. Next, we performed Bru-seq22,24 in these cells to measure nascent mRNA transcripts after Cdc73 deletion. We identified 1062 differentially expressed genes (Cdc73 target genes) shared between the 2 Cdc73f/f T-ALL cells but not by control cells (Figure 3B; supplemental Figure 6B). Consistent with previous studies, we did not observe large-scale downregulation of gene expression, which indicates that Cdc73 regulates specific genes.
Cdc73 shares ETS1 and Notch-driven pathways. (A) Rosa26CreERT2Cdc73f/f T-ALL cells (969 and 970) and control Rosa26CreERT2 T-ALL cells derived from tumors in Figure 2C were treated with 3 nM OHT to delete Cdc73 and measured for growth. Fold expansion = day 3 or day 6 cell count / day 0 cell count. (B) Venn diagram showing 1062 Cdc73 target genes shared by both Cdc73f/f T-ALL cell lines from panel A. Target genes defined as fold change (FC) > 1.5; Padj < .05 in Bru-seq counts at 30 hours after OHT addition in both Cdc73f/f cell lines but not in controls. (C-D) GSEA using the top 258 ETS1-induced gene list17 of Cdc73-induced genes in 969 (C) and 970 (D) cells. (E-H) GSEA analyses showing the top 6 Hallmark pathways enriched for Cdc73- (E-F), ETS1- (G) and Notch- (H) induced target genes. ETS1 and Notch target genes were previously described in human THP-6 T-ALL cells.17 Pathways shared across the 4 analyses are highlighted in blue. (I-L) GSEA using the MSigDB C2 Kauffman_DNA_repair gene list of Cdc73-induced genes (I-J), ETS1-induced genes (K), and Notch-induced genes (L). (M-N) Venn diagram showing overlap of Cdc73-induced and ETS1-induced (M) or Notch-induced (N) genes in the Kauffman_DNA_repair gene list. (O-T) Volcano plots of significance vs Bru-seq (O-P,S) or RNA-seq (Q-R,T) Log2FC data showing the control vs OHT (Cdc73Δ/Δ) comparison and highlighting important genes in nonhomologous end-joining (NHEJ; O,Q), homologous recombination (HR; P,R), and OXPHOS (S-T) pathways in 969 T-ALL cells (O-P,S) and AML cells (Q-R,T) on background of all genes (gray) giving average RPKM > 0.8. RNA-seq analysis of Control and Cdc73-deleted AML cells were obtained from.35 ∗∗P < .01; ∗∗∗∗P < .0001. DMSO, dimethyl sulfoxide; EtOH, ethyl alcohol; GSI, gamma secretase inhibitor; RPKM, reads per kilobase per million.
Cdc73 shares ETS1 and Notch-driven pathways. (A) Rosa26CreERT2Cdc73f/f T-ALL cells (969 and 970) and control Rosa26CreERT2 T-ALL cells derived from tumors in Figure 2C were treated with 3 nM OHT to delete Cdc73 and measured for growth. Fold expansion = day 3 or day 6 cell count / day 0 cell count. (B) Venn diagram showing 1062 Cdc73 target genes shared by both Cdc73f/f T-ALL cell lines from panel A. Target genes defined as fold change (FC) > 1.5; Padj < .05 in Bru-seq counts at 30 hours after OHT addition in both Cdc73f/f cell lines but not in controls. (C-D) GSEA using the top 258 ETS1-induced gene list17 of Cdc73-induced genes in 969 (C) and 970 (D) cells. (E-H) GSEA analyses showing the top 6 Hallmark pathways enriched for Cdc73- (E-F), ETS1- (G) and Notch- (H) induced target genes. ETS1 and Notch target genes were previously described in human THP-6 T-ALL cells.17 Pathways shared across the 4 analyses are highlighted in blue. (I-L) GSEA using the MSigDB C2 Kauffman_DNA_repair gene list of Cdc73-induced genes (I-J), ETS1-induced genes (K), and Notch-induced genes (L). (M-N) Venn diagram showing overlap of Cdc73-induced and ETS1-induced (M) or Notch-induced (N) genes in the Kauffman_DNA_repair gene list. (O-T) Volcano plots of significance vs Bru-seq (O-P,S) or RNA-seq (Q-R,T) Log2FC data showing the control vs OHT (Cdc73Δ/Δ) comparison and highlighting important genes in nonhomologous end-joining (NHEJ; O,Q), homologous recombination (HR; P,R), and OXPHOS (S-T) pathways in 969 T-ALL cells (O-P,S) and AML cells (Q-R,T) on background of all genes (gray) giving average RPKM > 0.8. RNA-seq analysis of Control and Cdc73-deleted AML cells were obtained from.35 ∗∗P < .01; ∗∗∗∗P < .0001. DMSO, dimethyl sulfoxide; EtOH, ethyl alcohol; GSI, gamma secretase inhibitor; RPKM, reads per kilobase per million.
Gene set enrichment analysis (GSEA) using a list of high-confidence direct NOTCH1 target genes in T-ALL36 showed no significant enrichment for Cdc73-regulated target genes (supplemental Figure 6C-D). However, Cdc73 deletion significantly impaired the expression of 25% of these genes in both Cdc73f/f T-ALL cell lines at q < 0.05 (P = .0219, Fisher exact test; supplemental Figure 6E). Three of the Cdc73-induced Notch signature target genes (HES1, IL7R, and SHQ1) were previously reported to have oncogenic function in T-ALL.36-43 For GSEA, all genes were weighted based on fold change, whereas only genes meeting statistical criteria were included for the Fisher exact test enrichment analysis. Statistical results between these 2 methods can diverge when the statistical significance is modest. In contrast, GSEA using a list of high-confidence, direct ETS1 target genes in T-ALL17 showed strongly significant enrichment for Cdc73-regulated genes in both 969 cells (Cdc73f/f T-ALL cell line; normal enrichment score [NES], 3.26; false discovery rate [FDR] < 0.001) and 970 cells (Cdc73f/f T-ALL cell line; NES, 2.75; FDR < 0.001; Figure 3C-D). Cdc73 deletion impaired the expression of 22% of ETS1 signature genes at q < 0.05 (P = .0003, Fisher exact test). These data are consistent with our protein-protein interaction and ChIP-seq data showing a stronger interaction of CDC73 with ETS1 than with NOTCH1. Hallmark GSEA analysis identified DNA repair and OXPHOS gene signatures as the top 2 shared pathways enriched for Cdc73-induced, ETS1-induced, and Notch-induced genes (highlighted in blue in Figure 3E-H). In contrast to ETS1 and Notch, Cdc73-induced genes were not enriched for MYC pathway genes. Thus, Cdc73 shares essential roles in T-cell development and leukemogenesis with Notch1 and Ets1 but has only partially overlapping functions with these factors in regulating gene expression.
The most enriched DNA repair signature for Cdc73-induced genes in MSigDB was Kauffman_DNA_repair_genes, which gave an FDR < 0.001 for Cdc73-induced genes (NES, 4-4.54; Figure 3I-J), ETS1-induced genes (NES, 3.94; Figure 3K), and Notch-induced genes (NES, 3.2; Figure 3L). Analysis of core enrichment genes showed that ETS1 and Notch1 induce large fractions of Cdc73-induced DNA repair genes (58% in Figure 3M and 58% in Figure 3N, respectively). Cdc73 deletion generally downregulated important DNA repair genes in multiple pathways, such as nonhomologous end joining (Figure 3O; supplemental Figure 6F) and homologous recombination (Figure 3P; supplemental Figure 6G). To test whether Cdc73 functions apply to Notch-independent and nonlymphoid contexts, we interrogated our gene expression datasets of murine mixed-lineage leukemia-to-mixed-lineage leukemia traslocated to, 3 super elongation complex subunit translocation (MLL)-AF9–driven Cdc73f/f acute myeloid leukemia (AML) cells.35 Accordingly, Cdc73 deletion in these cells led to the general downregulation of the same DNA repair genes (Figure 3Q-R). Thus, Cdc73 is not cell-specific or Notch-specific but happens to converge on Notch target genes in DNA repair.
OXPHOS is a synthetic lethal vulnerability in T-ALL, in part, because Notch promotes high anabolic demand.40,44-48 Of the OXPHOS gene lists in MSigDB, the hallmark list gave the highest enrichment with FDR < 0.001 for Cdc73-induced genes (NES, 3.52-3.83; supplemental Figure 6H-I), ETS1-induced genes (NES, 5.15; supplemental Figure 6J), and Notch-induced genes (NES, 4.63; supplemental Figure 6K). Analysis of core enrichment genes showed that ETS1 and Notch induce large fractions of Cdc73-induced OXPHOS genes (60% in supplemental Figure 6L and 62% in supplemental Figure 6M, respectively). Cdc73 deletion generally downregulated important OXPHOS pathway genes (Figure 3S; supplemental Figure 6N). These same genes were also generally downregulated upon Cdc73 deletion in AML cells (Figure 3T).35 Thus, Cdc73 is not cell-specific or Notch-specific but happens to converge on Notch target genes in OXHPHOS. Expression analysis of sorted DN subsets showed loss of important DNA repair and OXPHOS genes, particularly at the DN4 stage, when the strongest Cdc73 deletion was achieved (supplemental Figure 6O). Taken together, like ETS1 and Notch, Cdc73 promotes the expression of genes that are important for DNA repair and OXPHOS.
Cdc73 is important for genome integrity
DepMap analysis of several cancer types showed that T-ALL cells express the highest levels of DNA repair target genes that are shared between Cdc73, Notch, and Ets1 (leftmost group in supplemental Figure 7A). Given these data, we wondered whether Cdc73 helps preserve the genome integrity in T-ALL. To test this, we treated Cdc73f/f T-ALL cell lines with OHT and measured levels of γH2AX, a marker of DNA double-strand breaks. Consistently, OHT treatment of both Cdc73f/f T-ALL cell lines, but not the control cell line, effectively suppressed Cdc73 expression and increased gamma-H2A histone family X (γH2AX) levels (Figure 4A). Further, CDC73 knockdown had similar effects in SUP-T1 and CUTLL1, 2 human Notch-induced T-ALL cell lines (Figure 4B). Because γH2AX signals cluster at the promoters of genes regardless of expression level in T-ALL cell lines,49 we examined the localization of γH2AX at the promoters of a panel of genes. Consistently, quantitative chromatin immunoprecipitation (qChIP) showed that Cdc73 deletion induced γH2AX localization to chromatin (supplemental Figure 7B). In contrast, OHT had no effect on control cells (supplemental Figure 7C). To confirm that Cdc73 deletion led to spontaneous DNA damage, we performed blinded assays to assess chromosomal damage in metaphase spreads. Consistently, we observed increased chromosomal abnormalities in Cdc73f/f T-ALL metaphases upon treatment with OHT (Figure 4C-E). As expected, DNA damage upon OHT treatment led to a small but significant increase in apoptosis (approximately fourfold; Figure 4F-G). These data suggest that Cdc73 promotes DNA repair globally through gene expression, which might help protect T-ALL cells from chromosomal damage.
Cdc73 is important for genome integrity. (A) Western blot for γH2AX in Rosa26CreERT2Cdc73f/f T-ALL cells (969 and 970) and control Rosa26CreERT2 T-ALL cells treated with OHT for 30 hours to delete Cdc73. Numbers represent band intensities normalized to β-actin loading control. (B) Western blot for γH2AX in human NOTCH1-induced human T-ALL cell lines (SUP-T1; CUTLL1) transduced with shCDC73. (C-E) Representative metaphase spreads (C) and quantification of metaphase abnormalities in aggregate (D) or per replicate (E) in blinded analyses of Cdc73f/f T-ALL cells treated with OHT for 30 hours. White arrows represent gaps; orange arrow represents break. (F-G) Representative Annexin V/7-AAD flow cytometric plots (F) and Annexin V+/7-AAD− scatterplot (G) of Cdc73f/f T-ALL cells treated with vehicle (Control) or OHT for 30 hours (Cdc73Δ/Δ). 7-AAD, 7-aminoactinomycin D.
Cdc73 is important for genome integrity. (A) Western blot for γH2AX in Rosa26CreERT2Cdc73f/f T-ALL cells (969 and 970) and control Rosa26CreERT2 T-ALL cells treated with OHT for 30 hours to delete Cdc73. Numbers represent band intensities normalized to β-actin loading control. (B) Western blot for γH2AX in human NOTCH1-induced human T-ALL cell lines (SUP-T1; CUTLL1) transduced with shCDC73. (C-E) Representative metaphase spreads (C) and quantification of metaphase abnormalities in aggregate (D) or per replicate (E) in blinded analyses of Cdc73f/f T-ALL cells treated with OHT for 30 hours. White arrows represent gaps; orange arrow represents break. (F-G) Representative Annexin V/7-AAD flow cytometric plots (F) and Annexin V+/7-AAD− scatterplot (G) of Cdc73f/f T-ALL cells treated with vehicle (Control) or OHT for 30 hours (Cdc73Δ/Δ). 7-AAD, 7-aminoactinomycin D.
Cdc73 is important for OXPHOS
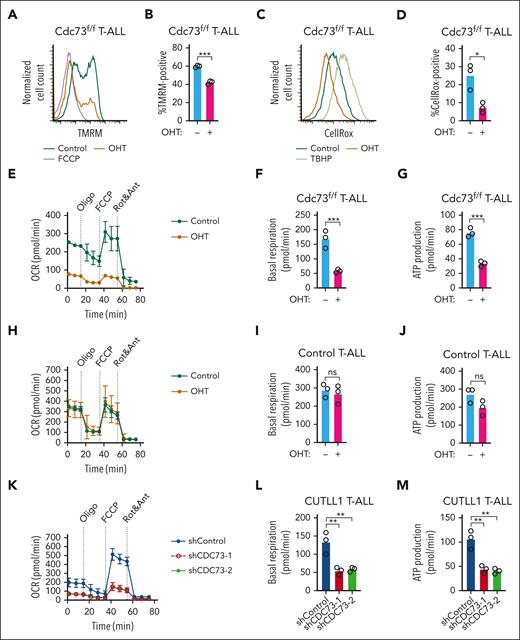
Supraphysiological Notch signals in T-ALL have been previously shown to directly and indirectly induce OXPHOS genes, such as electron transport complex I.44,48 DepMap analysis of several cancer types showed that T-ALL cells express average levels of OXPHOS target genes shared between Cdc73, Notch, and Ets1 (leftmost group in supplemental Figure 8). Because Cdc73 promotes the expression of genes important for the electron transport chain, we considered the possibility that Cdc73 helps maintain membrane potential to protect T-ALL cells from metabolic stress. To test this possibility, we treated Cdc73f/f T-ALL cell lines with OHT and measured the mitochondrial membrane potential of live cells by the tetramethylrhodamine methyl ester assay (Figure 5A-B). Cdc73 deletion reduced membrane potential to nearly the same degree as carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP,) a mitochondrial uncoupler. Based on our gene expression analysis showing that Cdc73 regulates complex I, we predicted that Cdc73 deletion would reduce reactive oxygen species. Consistently, the CellROX assay showed that OHT treatment reduced reactive oxygen species production (Figure 5C-D). Our gene expression analyses also predicted impaired oxygen consumption. Consistently, the mitochondrial stress test showed that Cdc73 deletion reduced basal respiration and mitochondrial adenosine triphosphate production rates of live cells (Figure 5E-G). In contrast, treating control T-ALL cells with OHT had no significant effect (Figure 5H-J). CDC73 knockdown in live human Notch-induced T-ALL cells also impaired oxygen consumption rates (Figure 5K-M). These data suggest that Cdc73 promotes OXPHOS, which might help protect T-ALL cells from the metabolic stresses of supraphysiological Notch signaling.
Cdc73 is important for OXPHOS. (A-B) Representative flow cytometric histogram (A) and mean florescence intensity scatterplot (N = 3) (B) showing mitochondrial membrane potentials measured by tetramethylrhodamine methyl ester (TMRM) assay on live 4′,6-diamidino-2-phenylindole (DAPI)-negative Rosa26CreERT2Cdc73f/f T-ALL cells (970) treated with OHT for 30 hours to delete Cdc73. FCCP was added as a positive control. (C-D) Representative flow cytometric histogram (C) and mean fluorescence intensity scatterplot (N = 3) (D) showing mitochondrial reactive oxygen species (ROS) production measured by the CellROX assay on live DAPI-negative 970 cells treated with OHT for 30 hours to delete Cdc73. TBHP was added as a positive control. (E-M) Seahorse XFe96 instrument measurements of real-time oxygen consumption rate (OCR) normalized to live cell number and protein concentration under basal conditions or in response to the indicated mitochondrial inhibitors (E,H,K) and scatterplots of basal (F,I,L) and adenosine triphosphate (ATP) production (G,J,M) respiration phases of 970 cells treated with OHT for 30 hours to delete Cdc73 (E-G), control Rosa26CreERT2 cells treated with OHT for 30 hours (H-J), and CUTLL1 cells at 4 days after transduction with 2 independent shCDC73 (K-M).
Cdc73 is important for OXPHOS. (A-B) Representative flow cytometric histogram (A) and mean florescence intensity scatterplot (N = 3) (B) showing mitochondrial membrane potentials measured by tetramethylrhodamine methyl ester (TMRM) assay on live 4′,6-diamidino-2-phenylindole (DAPI)-negative Rosa26CreERT2Cdc73f/f T-ALL cells (970) treated with OHT for 30 hours to delete Cdc73. FCCP was added as a positive control. (C-D) Representative flow cytometric histogram (C) and mean fluorescence intensity scatterplot (N = 3) (D) showing mitochondrial reactive oxygen species (ROS) production measured by the CellROX assay on live DAPI-negative 970 cells treated with OHT for 30 hours to delete Cdc73. TBHP was added as a positive control. (E-M) Seahorse XFe96 instrument measurements of real-time oxygen consumption rate (OCR) normalized to live cell number and protein concentration under basal conditions or in response to the indicated mitochondrial inhibitors (E,H,K) and scatterplots of basal (F,I,L) and adenosine triphosphate (ATP) production (G,J,M) respiration phases of 970 cells treated with OHT for 30 hours to delete Cdc73 (E-G), control Rosa26CreERT2 cells treated with OHT for 30 hours (H-J), and CUTLL1 cells at 4 days after transduction with 2 independent shCDC73 (K-M).
Cdc73 promotes expression of oncogenes but not DNA repair and OXPHOS genes through enhancers
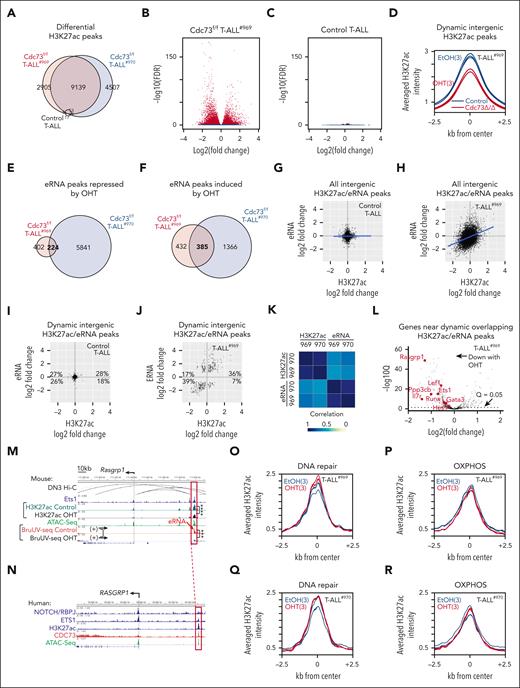
Consistent with a possible role in regulating enhancer activity, we observed strong CDC73 ChIP-seq signals at intergenic regulatory elements relative to promoters (supplemental Figure 9A). To test this further, we performed H3K27ac ChIP-seq in our CreERT2Cdc73f/f T-ALL cell lines (969 and 970) after OHT treatment. Consistently, ChIP-seq showed differential H3K27ac signals upon Cdc73 deletion at FDR < 0.05 (Figure 6A-B; supplemental Figure 9B). In contrast, OHT had little effect on control cells (Figure 6C). We intersected the data sets to obtain 9139 dynamic H3K27ac peaks, which were defined based on differential H3K27ac signals at FDR < 0.05 in the same direction for both CreERT2Cdc73f/f T-ALL cell lines but not the control CreERT2 T-ALL cell line. In general, Cdc73 deletion impaired H3K27ac signals at dynamic intergenic H3K27ac peaks (Figure 6D; supplemental Figure 9C).
Cdc73 promotes expression of oncogenes but not DNA repair and OXPHOS genes through enhancers. (A) Venn diagram of differential H3K27ac (FDR < 0.05) in Rosa26CreERT2Cdc73f/f T-ALL cells (969 and 970) and control Rosa26CreERT2 T-ALL cells upon treatment with 6 nM OHT for 30 hours. (B-C) Volcano plots of significance vs log2(OHT/control) H3K27ac ChIP-seq signals of 969 (B) and control (C) cells in panel A. (D) Metagene plot of dynamic intergenic H3K27ac signals (defined as FDR < 0.05 in 969 and 970 cells but not in control cells) in 969 cells. (E-F) Venn diagram of differential eRNAs in Cdc73f/f T-ALL cells (969 and 970) that were repressed (E) or induced (F) upon treatment with 6 nM OHT for 30 hours. eRNAs were defined as intergenic BruUV-seq peaks or intragenic peaks that were antisense in direction relative to mRNAs. No differential eRNAs were identified after OHT treatment of control T-ALL cells. (G-J) BruUV-Seq log2(OHT/control) vs H3K27ac log2(OHT/Control) scatterplots of all overlapping intergenic peaks (G-H) or overlapping dynamic intergenic peaks (I-J) in control T-ALL cells (G,I) and Cdc73f/f T-ALL cells (969) (H,J). Overlapping dynamic peaks were defined as giving q < 0.05 and FDR < 0.05 in the same direction for the BruUV-Seq and H3K27ac comparisons respectively in both Cdc73f/f T-ALL cells but not in control T-ALL cells. (K) Spearman correlation coefficient analysis of eRNA and H3K27ac log2(OHT/control) from panel J and supplemental Figure 7G in 969 and 970 Cdc73f/f T-ALL cells. (L) Volcano plot of significance vs Bru-seq log2(OHT/control) of genes nearest overlapping OHT-downregulated dynamic intergenic BruUV-Seq and H3K27ac peaks in 969 Cdc73f/f T-ALL cells. (M-N) Display tracks of indicated ChIP-seq and assay for transposase-accessible chromatin (ATAC)-seq data sets at the Rasgrp1 locus in mouse 969 cells (M) or human THP-6 cells (N) showing nearest mouse-human homologous enhancers in red boxes that contain overlapping dynamic intergenic eRNA and H3K27ac peaks. Ets1 ChIP-seq (GSM461516); ATAC-seq (GSM2461649); DN3 Hi-C (GSE79422) analyzed in.50 (O-R) Metagene plots of H3K27ac signals at nonpromoter H2K27ac peaks nearest DNA repair (O,Q) and OXPHOS genes (P,R) from supplemental Table 1 in 969 cells (O-P) and 970 cells (Q-R). ∗∗∗FDR < 0.001; ∗∗∗∗FDR < 0.0001.
Cdc73 promotes expression of oncogenes but not DNA repair and OXPHOS genes through enhancers. (A) Venn diagram of differential H3K27ac (FDR < 0.05) in Rosa26CreERT2Cdc73f/f T-ALL cells (969 and 970) and control Rosa26CreERT2 T-ALL cells upon treatment with 6 nM OHT for 30 hours. (B-C) Volcano plots of significance vs log2(OHT/control) H3K27ac ChIP-seq signals of 969 (B) and control (C) cells in panel A. (D) Metagene plot of dynamic intergenic H3K27ac signals (defined as FDR < 0.05 in 969 and 970 cells but not in control cells) in 969 cells. (E-F) Venn diagram of differential eRNAs in Cdc73f/f T-ALL cells (969 and 970) that were repressed (E) or induced (F) upon treatment with 6 nM OHT for 30 hours. eRNAs were defined as intergenic BruUV-seq peaks or intragenic peaks that were antisense in direction relative to mRNAs. No differential eRNAs were identified after OHT treatment of control T-ALL cells. (G-J) BruUV-Seq log2(OHT/control) vs H3K27ac log2(OHT/Control) scatterplots of all overlapping intergenic peaks (G-H) or overlapping dynamic intergenic peaks (I-J) in control T-ALL cells (G,I) and Cdc73f/f T-ALL cells (969) (H,J). Overlapping dynamic peaks were defined as giving q < 0.05 and FDR < 0.05 in the same direction for the BruUV-Seq and H3K27ac comparisons respectively in both Cdc73f/f T-ALL cells but not in control T-ALL cells. (K) Spearman correlation coefficient analysis of eRNA and H3K27ac log2(OHT/control) from panel J and supplemental Figure 7G in 969 and 970 Cdc73f/f T-ALL cells. (L) Volcano plot of significance vs Bru-seq log2(OHT/control) of genes nearest overlapping OHT-downregulated dynamic intergenic BruUV-Seq and H3K27ac peaks in 969 Cdc73f/f T-ALL cells. (M-N) Display tracks of indicated ChIP-seq and assay for transposase-accessible chromatin (ATAC)-seq data sets at the Rasgrp1 locus in mouse 969 cells (M) or human THP-6 cells (N) showing nearest mouse-human homologous enhancers in red boxes that contain overlapping dynamic intergenic eRNA and H3K27ac peaks. Ets1 ChIP-seq (GSM461516); ATAC-seq (GSM2461649); DN3 Hi-C (GSE79422) analyzed in.50 (O-R) Metagene plots of H3K27ac signals at nonpromoter H2K27ac peaks nearest DNA repair (O,Q) and OXPHOS genes (P,R) from supplemental Table 1 in 969 cells (O-P) and 970 cells (Q-R). ∗∗∗FDR < 0.001; ∗∗∗∗FDR < 0.0001.
Because Paf1C regulates enhancer activity through enhancer ribonucleic acid (eRNA) synthesis,13,14 we next sought to identify enhancers directly regulated by Cdc73 by integrating our dynamic H3K27ac data set with differential analysis of eRNA expression. Toward this goal, we performed BruUV-seq on the CreERT2Cdc73f/f T-ALL cell lines (969 and 970) and the control CreERT2 T-ALL cell line. BruUV-seq is a nascent RNA technique that detects and quantifies rapidly degraded RNAs such as eRNAs and is performed using living cells.23 We identified 609 differential eRNAs at q < 0.05 upon Cdc73 deletion that were shared by both Cdc73f/f T-ALL cell lines but not by the control cell line (Figure 6E-F; supplemental Figure 9D-E). Next, we integrated the H3K27ac and BruUV-seq data sets to identify intergenic H3K27ac and eRNA peaks that overlapped in both T-ALL cell lines. There was no correlation between changes in eRNA and the H3K27ac signal in control cells (Figure 6G). In contrast, there was a modest correlation in Cdc73f/f T-ALL cells (Figure 6H; supplemental Figure 9F). To determine the genomic locations where Cdc73 might have strong direct effects on enhancers, we defined dynamic eRNA peaks as showing q < 0.05 in the same direction for the BruUV-seq comparison in both Cdc73f/f T-ALL cells but not in control T-ALL cells. Changes in dynamic intergenic H3K27ac signals showed no correlation in control cells (Figure 6I) but a strong correlation in both Cdc73f/f T-ALL cells with changes in dynamic intergenic eRNA signals (r > 0.66; Figure 6J-K; supplemental Figure 9G).
Next, we sought to identify direct target genes of Cdc73 by associating overlapping dynamic intergenic H3K27ac/eRNA peaks that diminish upon Cdc73 deletion with the nearest expressed genes. We found that OHT-downregulated H3K27ac/eRNA peaks were associated with OHT-downregulated expression of several known genes that are important for maintaining T-ALL proliferation, such as Il7r, Ets1, Rasgrp1, and Lef136,39,51-56 (Figure 6L-N; supplemental Figure 9H-N). Importantly, we consistently observed CDC73 occupancy at homologous human enhancers (Figure 6N; supplemental Figure 9J,L,N). Analysis of publicly available high-throughput chromosome conformation capture technique (Hi-C) data sets of DN3 cells confirmed interactions of these enhancers with respective promoters, except for the previously described Notch-dependent Il7r enhancer,36 possibly because DN3 cells express low Il7r. Interestingly, we did not observe an association of overlapping dynamic intergenic H3K27ac/eRNA peaks with core enrichment genes from GSEA pathway analyses in DNA repair or OXPHOS (supplemental Table 1). Furthermore, we did not observe any differences in H3K27ac signal changes upon Cdc73 deletion at nonpromoter peaks nearest DNA repair or OXPHOS genes relative to all genes (Figure 6O-R; supplemental Figure 9O-P). Taken together, these data suggest that Cdc73 activates enhancers that induce important T-ALL driver genes but not DNA repair or OXPHOS genes.
Cdc73 promotes DNA repair and OXPHOS gene expression through gene body functions
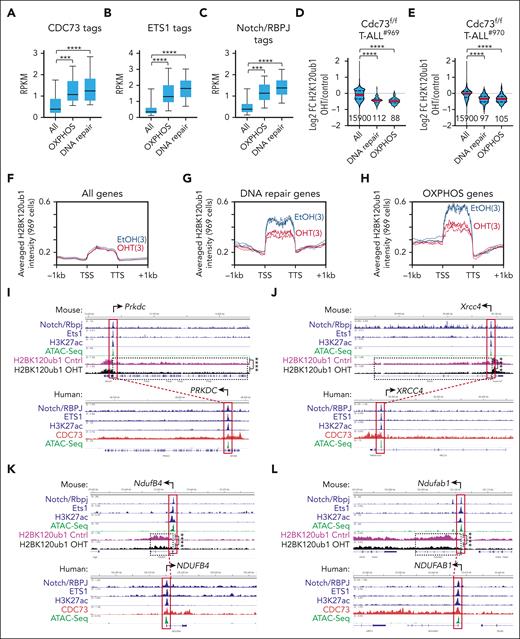
Because Cdc73 did not appear to induce expression of DNA repair and OXPHOS genes through enhancers, we considered the possibility that Cdc73 induces these genes through its well-established functions in promoting mRNA synthesis. To test this, we first counted CDC73 tags between the transcriptional start site and transcriptional termination site of GSEA core enrichment genes in DNA repair or OXPHOS (Figure 7A). Consistently, CDC73 tags were generally more abundant at DNA repair and OXPHOS genes relative to all genes. ETS1 and RBPJ/Notch tags were also more abundant at the promoters of these genes (Figure 7B-C). This observation is consistent with our ChIP-seq and gene expression analyses showing that CDC73 intersects with the ETS1 and Notch pathways. Next, we performed H2BK120ub1 ChIP-seq on our CreERT2Cdc73f/f T-ALL cell lines upon treatment with OHT to delete Cdc73. We chose H2BK120ub1 because one of the best-established mRNA functions of Paf1C is to promote transcriptional elongation by recruiting the Bre1-Rad6 E3 ubiquitin ligase complex to monoubiquitinate H2BK120.57-61
Cdc73 promotes DNA repair and OXPHOS gene expression through its gene body functions. (A-C) Box and whisker plots of CDC73 (A), ETS1 (B), and Notch/RBPJ (C) tag counts in human THP-6 cells at gene bodies (A) and promoters (B-C) of all genes, OXPHOS genes, and DNA repair genes shared by the GSEA enrichment cores in Cdc73f/f 969 and 970 T-ALL cells (Figure 3I; supplemental Figure 4G; supplemental Table 1). (D-E). Violin plots showing H2BK120ub1 ChIP-seq Log2FC in 969 cells (D) and 970 cells (E) for all genes, DNA repair genes, and OXPHOS genes in the GSEA enrichment cores of 969 and 970 cells (supplemental Table 1). OHT was added for 30 hours to delete Cdc73. (F-H) Metagene plots of H2BK120ub1 signals in EtOH-treated (blue) and OHT-treated (red) 969 cells at all genes (F), DNA repair genes (G), and OXPHOS genes (H) in core enrichment genes of GSEA analyses (supplemental Table 1). (I-L) Display tracks of indicated ChIP-seq and ATAC-seq datasets at important DNA repair genes (I-J) and OXPHOS genes (K-L) in mouse 969 cells (top) or human THP-6 cells (bottom) showing representative tracks and FDR values upon OHT addition (Cdc73 deletion) of H2BK120ub1 signals between transcriptional start site (TSS) and transcriptional termination site (TTS). ∗∗∗FDR < 0.001; ∗∗∗∗FDR < 0.0001. ATAC-seq (GSM2461649). Ets1 ChIP-seq (GSM2461515).
Cdc73 promotes DNA repair and OXPHOS gene expression through its gene body functions. (A-C) Box and whisker plots of CDC73 (A), ETS1 (B), and Notch/RBPJ (C) tag counts in human THP-6 cells at gene bodies (A) and promoters (B-C) of all genes, OXPHOS genes, and DNA repair genes shared by the GSEA enrichment cores in Cdc73f/f 969 and 970 T-ALL cells (Figure 3I; supplemental Figure 4G; supplemental Table 1). (D-E). Violin plots showing H2BK120ub1 ChIP-seq Log2FC in 969 cells (D) and 970 cells (E) for all genes, DNA repair genes, and OXPHOS genes in the GSEA enrichment cores of 969 and 970 cells (supplemental Table 1). OHT was added for 30 hours to delete Cdc73. (F-H) Metagene plots of H2BK120ub1 signals in EtOH-treated (blue) and OHT-treated (red) 969 cells at all genes (F), DNA repair genes (G), and OXPHOS genes (H) in core enrichment genes of GSEA analyses (supplemental Table 1). (I-L) Display tracks of indicated ChIP-seq and ATAC-seq datasets at important DNA repair genes (I-J) and OXPHOS genes (K-L) in mouse 969 cells (top) or human THP-6 cells (bottom) showing representative tracks and FDR values upon OHT addition (Cdc73 deletion) of H2BK120ub1 signals between transcriptional start site (TSS) and transcriptional termination site (TTS). ∗∗∗FDR < 0.001; ∗∗∗∗FDR < 0.0001. ATAC-seq (GSM2461649). Ets1 ChIP-seq (GSM2461515).
Consistent with this function, Cdc73 deletion reduced most of the H2BK120ub1 signal in mouse genes ranked in the top tercile of CDC73 signal in homologous human genes (supplemental Figure 10A,D) relative to the middle tercile (supplemental Figure 10B,E) and bottom tercile (supplemental Figure 10C,F) in both Cdc73f/f cell lines. Cdc73 deletion generally caused downregulation of H2BK120ub1 signals in DNA repair and OXPHOS genes relative to all genes in both Cdc73f/f cell lines (Figure 7D-H; supplemental Figure 10G-I). For example, we observed significant downregulation (FDR < 0.01) of H2BK120ub1 counts in important DNA repair genes, such as Prkdc, Xrcc4, Rad50, and Atr (Figure 7I-J; supplemental Figure 10J-K) and important OXPHOS genes, such as NdufB4, Ndufab1, Etfa, and Ndufa6 (Figure 7K-L; supplemental Figure 10L-M). Importantly, broad CDC73 signals were consistently observed across the gene bodies of the homologous human genes (Figure 7I-L, bottom; supplemental Figure 10J-M, bottom). In accordance with our analyses of ETS1 and RBPJ/Notch binding in Figure 7B-C, we observed the occupancy of these factors at the promoters of the mouse genes and human homologs. These data suggest that CDC73 might intersect NOTCH1 and ETS1 occupancy at DNA repair and OXPHOS genes and directly induce mRNA synthesis by promoting H2BK120 monoubiquitination.
CDC73 protects T-ALL cells from excessive oncogenic stress
Because our model is that CDC73 promotes cellular pathways that protect cells from oncogenic activation (supplemental Figure 11), we predict that raising oncogenic stress would be particularly lethal to cells lacking Cdc73. To test this prediction, we transduced the oncogenes from supplemental Figure 11 (activated Myc and AKT [to activate mTOR]) and controls into Cdc73f/f T-ALL cells (supplemental Figure 12). Next, we added vehicle or OHT to delete Cdc73 (supplemental Figure 12A). Consistently, enforced expression of Myc (supplemental Figure 12C) and AK mouse transforming (AKT) (albeit modestly; supplemental Figure 12D) did not “rescue” cells from Cdc73 deficiency but had the opposite effect, significantly impairing fitness (supplemental Figure 12G). Transduction of the control vector had no effect (supplemental Figure 12B). In contrast, enforced expression of our rescue controls, CDC73 and the antiapoptotic gene BCL2 (supplemental Figure 12E-F), rescued cells from Cdc73 deficiency, significantly increasing fitness (supplemental Figure 12G). These data suggest that Cdc73-deficient T-ALL cells lack fitness in the context of enforced expression of oncogenes. Next, we wondered whether Cdc73 is important for protecting T-ALL cells from the DNA damage of excessive oncogenic activity. Consistently, Cdc73 deletion in MYC-transduced cells induced ∼1.7-fold and ∼27-fold increases in γH2AX levels relative to MYC-transduced Cdc73+/+ cells and vector-transduced Cdc73+/+ cells, respectively (supplemental Figure 12H-I). These data suggest that Cdc73 might protect T-ALL cells from oncogenic activity, in particular the Notch downstream target Myc.
Discussion
Notch signaling controls developmentally important activities in diverse tissues, raising barriers to developing anti-Notch therapies that directly target Notch. Emerging evidence originating in Drosophila highlights an alternative strategy for targeting transcriptional regulators that cobind Notch-occupied regulatory elements.62,63 Previous work showed that NOTCH1 bound CDC73 in breast cancer cells and relied on Paf1C to induce Notch target gene expression for Drosophila wing development.6,7,15 Here, we showed that this interaction is context-dependent. RBPJ/Notch co-occupied only a subset of CDC73-bound elements, which were highly enriched for ETS motifs and ETS1 occupancy. Further, ETS1 interacted with CDC73, and ETS1 knockdown generally impaired CDC73 binding to chromatin, to which ETS1 was also bound. Thus, the chromatin context (here, ETS1 in T-ALL) might restrict the NOTCH1-CDC73 interaction. We are mindful that CDC73 binds chromatin in the absence of NOTCH1 or ETS1. Cdc73 was previously shown to interact with non-Notch oncogenic transcription factors such as β-catenin and Gli115 that were previously implicated in driving the proliferation of Notch-active and -inactive T-ALL cells.64-69 Thus, CDC73 might be recruited separately from Notch into other large, highly active transcription factor complexes to drive cell proliferation.
Ccd73 acts as a tumor suppressor70-72 or an oncogene.35,73-76 In T-ALL, we showed that Cdc73 has an oncogenic role. Consistent with our chromatin profiling, gene expression analyses showed that Notch and Ets1 converge on a subset of Cdc73-induced pathways, notably boosting ∼60% of Cdc73 target genes in DNA repair and OXPHOS pathways through gene body functions and key oncogenes such as Il7r and Lef1 through enhancer functions. Although we have not identified the most important Cdc73 target genes, the shared DNA repair gene program is strikingly more highly expressed in T-ALL than in any other cancer. Further, Cdc73 deletion increased chromosomal damage, induced γH2AX, and impaired oxygen consumption. Cdc73 also appears to have similar functions in the Notch-independent and nonlymphoid context of AML cells, which die when Cdc73 is deleted.76 Thus, Cdc73 is not specific to Notch or T-ALL cells but happens to converge on Notch target genes in DNA repair and OXPHOS. Our data extend previous studies77-82 and suggest that Cdc73 promoted a gene expression program important for DNA repair and mitochondrial function, which Notch intersected and co-opted to mitigate the genotoxic and metabolic stresses of elevated Notch signaling (supplemental Figure 11). Previous studies by others showed divergent roles of Paf1C in regulating enhancer activity in colon cancer cells and embryonic stem cells.13,14 Consistent with the view of Paf1C as a transcriptional activator,8,11 our analysis showed that Cdc73 promotes eRNA synthesis and H3K27ac signals at CDC73-bound enhancers associated with known T-ALL drivers. In contrast, H2BK120ub1 profiling showed that Cdc73 primarily induces DNA repair and OXPHOS genes by promoting mRNA synthesis.
Our investigation seems to imply that DNA repair, OXPHOS, and CDC73 are therapeutic targets in T-ALL. However, a drug targeting all CDC73 functions would likely be toxic. Cdc73 interacts with pathways such as Wnt, Hedgehog, and Notch15; causes severe intestinal toxicity in adult mice when deleted15,16; and is essential for Notch-independent cells16 and hematopoietic stem cells.35 Thus, extensive structure-function and CDC73 interactome studies are needed to find ways to safely target CDC73 functions, such as through inhibitors of context-dependent protein-protein interactions that are important for cancer functions but not for essential homeostasis. One possibility to explore is the CDC73-ETS1 interaction because we showed that postnatal deletion of Ets1 in mice did not significantly affect survival or overall health.17 Furthermore, our study supports the notion that T-ALL cells are a cancer type that is sensitive to inhibition of general transcriptional machinery, which had once been considered to be too toxic to block given its vital functions in normal cells.83-89 Previously, we showed that leukemia-associated NOTCH1 alleles are weak transactivators.90 Here, we suggest that oncogenic Notch might overcome its inherent weakness through local intersection at chromatin with cobinding transcription factors and powerful RNA synthesis machinery, which induce the most highly active chromatin and expressed genes. Our report shows that studying intersecting pathways might reveal clues on how to combat Notch-dependent cancers without directly targeting the Notch complex.
Acknowledgments
The authors thank Qing Li, David Ferguson, Shannon Carty, Michelle Paulsen, Arvind Rao, Rohan Kogdule, Amparo Serna Alarcon, Ran Yan, Cher Sha, Morgan Jones, Koral Campbell, Andrea Hartlerod, Courtney Hames, and Luis Correa for their thoughtful input and technical assistance during this project. The authors thank Andrew Weng, Malathi Kandarpa, and Moshe Talpaz for PDX and primary T-ALL samples. The authors thank Utpal Davé for the Lmo2-tg mice. The authors thank Daniela Salgado Figueroa and Katia Georgopoulos for Hi-C contact maps of GSE79422.50
This work was supported by funding from the National Institutes of Health (NIH), National Cancer Institute grants 1F31CA260929 [A.F.M.] and R01CA27611701 [M.Y.C.]; NIH, National Institute of Allergy and Infectious Diseases grant R01AI136941 [M.Y.C.]; NIH, National Institute of General Medical Sciences grants T32GM007315 [A.F.M.], and R01GM101171 [D.B.L.]; University of Michigan Rackham Graduate School; Michigan Medicine Rogel Cancer Center; Rally Foundation for Childhood Cancer Research; and Alex’s Lemonade Stand Foundation.
Authorship
Contribution: M.Y.C. and A.F.M. conceptualized the study; A.F.M., C.M., K.L., S.L., N.A.J., E.C., B.M., A.C.M., F.A., N.K., G.B., Y.E.L., and Q.W. performed investigation; M.Y.C., A.F.M., C.M., K.L., S.L., N.A.J., E.C., S.K., D.B.L., M.L., A.C.M., J.S., K.L., R.J.H.R., and A.G.M. visualized the data; A.F.M., E.C., B.M., and N.A.D. performed formal analysis; A.C.M., N.K., K.L., E.C., B.M., M.Y.C., and R.J.H.R. curated the data; M.Y.C. and A.F.M. acquired funds and wrote the original draft; M.Y.C, A.F.M., R.J.H.R., D.B.L., M.L., S.K., B.M., E.C., N.K., K.L., N.A.J., J.S., C.M., and A.G.M. reviewed and edited the manuscript; and M.Y.C. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark Y. Chiang, University of Michigan School of Medicine, 109 Zina Pitcher Pl, Room 2043 BSRB, Ann Arbor, MI 48109-2200; email: markchia@med.umich.edu.
References
Author notes
High-throughput sequencing data, results, and statistics were deposited in the Gene Expression Omnibus (GEO) database (accession number GSE221345). The data are accessible to reviewers using the following token: unejosqivlwdvkj. The publicly available next-generation sequencing data sets used in this study can be found in the GEO database (accession numbers GSE79422, GSE117749, GSE93755, GSE138516, GSE109125, GSE22601, and GSE138659).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal