In this issue of Blood, Pagani et al1 demonstrate that the residual BCR::ABL1 DNA level detected in granulocytes and T cells can be used as a predictive measure for the successful discontinuation of tyrosine kinase inhibitors (TKIs) in patients with chronic myeloid leukemia (CML). Based on these results, a sensitive prognostic model capable of identifying the probability of recurrence at 60 months was designed.1 Do we finally have in our hands a tool that can indicate the potential of success after discontinuation of TKIs?
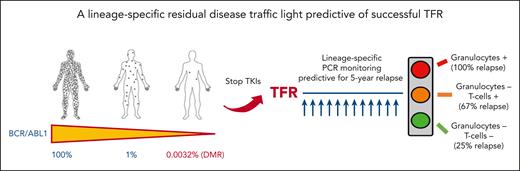
Over the past 15 years, the outcome of patients with CML has improved dramatically following the introduction of TKIs, with overall survival now approaching that of healthy people.2 The use of second-generation TKIs as first-line treatment has resulted in faster achievement of molecular response and increased the rate of deep molecular response (DMR, or MR4 and MR4.5 corresponding to 0.01% and 0.0032% BCR::ABL1 ratio in International Scale [IS], respectively).3 The concept of treatment-free remission (TFR) has been introduced as a possible goal for patients in the chronic phase with a long-lasting DMR. The first French trial, the STIM study, showed that patients with long-term MR4.5 can safely stop treatment, with approximately 40% of them remaining in remission (ie, no molecular detection of disease) without therapy.4 Several studies and real-world evidence have shown that the percentage of patients who maintained the remission after discontinuation increases to 50% through 60% if the loss of major molecular response (0.1% in IS) was considered as the threshold to resume the treatment.5 Prognostic factors for a successful discontinuation have been extensively studied, but only the duration of treatment for more than 5 years and the duration of a DMR for more than 3 years have been internationally confirmed as the optimal prerequisites for effective long-lasting discontinuation.3,5 In recent years, the search for new methodologies capable of increasing the sensitivity of real-time quantitative polymerase chain reaction monitoring has been explored. The use of digital droplet polymerase chain reaction (ddPCR) seems to provide a valid tool for the subgroup of patients attempting TFR, with a better sensitivity and specificity.6 DMR confirmed by ddPCR appears to increase the likelihood of success after discontinuation.7 Shorter BCR::ABL1 halving time after the start of first-line treatment was also found to correlate with the likelihood of success, as was reported in an earlier study by the same group.8 Currently, there is still an ongoing discussion about the appropriate cutoff to use for ddPCR to define the increased risk of recurrence after discontinuation. Indeed, in this study, Pagani et al sought to dissect the value of residual disease by sorting the different leukocyte fractions followed by a DNA-based patient-specific PCR in the different leukocyte lineages. In a prospective comparison of patients who relapsed vs those who maintained remission, BCR::ABL1 DNA was significantly detected in granulocytes and T cells but not in monocytes, B cells, or natural killer cells. Three groups of patients were defined based on the detection of BCR::ABL1 in granulocytes and/or T cells, with different probability of recurrence at 60 months, in a model providing an accuracy of 77% (see figure). In multivariate analysis, detection of residual disease in granulocytes remained the only prognostic factor that could identify patients at risk of relapse in 2 different models that included T lymphocytes and halving time.
A lineage-specific residual disease traffic light predictive of successful TFR.
A lineage-specific residual disease traffic light predictive of successful TFR.
The results of this study expanded on previous reports by the same group on measurable residual disease (MRD) in patients attempting TFR,9 specifically that finding of BCR::ABL1+ lymphocytes at presentation and detectable disease in this subset of cells in TFR denoted the persistence of a multipotent progenitor. Sorting different subsets of leukocytes seems to increase the predictive accuracy in defining the risk of relapse. The increased sensitivity of the method requires that laboratories are familiar with the use of DNA-based patient-specific PCR. As with ddPCR, this is not yet currently used by all centers and will require centralized laboratory analysis of samples for patients attempting TFR in specific laboratories.
There is now a biological traffic light for predicting successful TFR.10 Patients with BCR::ABL1+ granulocytes have a red light, with residual disease still present. In these patients, a proactive switch to improve the depth of molecular response or a prolonged treatment with the same TKI should be required before considering discontinuing the therapy. Considering the poor results obtained in patients attempting a second round of discontinuation after the first TFR failure, a "red light" should help mitigate clinical failure. What remains to be done in the near future? In laboratories that can perform these new monitoring methods, an initial evaluation should better identify patients for whom therapy may be suspended safely, identifying the best timing for TFR, while also providing monitoring that can identify an early recurrence. In the near future, these biological data could be combined with new prognostic factors (eg, immunological, next generation sequencing) to better shape a withdrawal strategy.
Conflict-of-interest disclosure: The author received honoraria from Novartis, Pfizer, BMS, Incyte, AOP, and AbbVie.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal