In this issue of Blood, Cunningham et al report on the largest prospective, single-center natural history study of familial platelet disorder with associated myeloid malignancies (FPDMM), which is caused by deleterious RUNX1 germ line variants.1 Their findings expand our current understanding of this rare disease, providing further groundwork for future growth in the field and evidence-based care.
Since Song et al described the causative association of heterozygous RUNX1 germ line variants with FPDMM in 1999,2 roughly 200 families have been reported worldwide.3 Individuals affected with autosomal dominantly inherited FPDMM classically present with mild to moderate thrombocytopenia, have platelet aggregation defects that may lead to bleeding, and have an estimated 30% to 40% lifetime probability of developing a hematological malignancy, mainly myelodysplastic syndrome or acute myeloid leukemia. Myeloid neoplasms with germ line RUNX1 mutation are one of the first recognized by the World Health Organization as a leukemia-predisposing disease. Malignancies are frequently preceded by clonal hematopoiesis and can manifest from early childhood to late adulthood.
Twenty years after Song’s initial report,2 Liu et al at the US National Institutes of Health (NIH) launched the Natural History Study of Hematologic and Premalignant Conditions Associated with RUNX1 Mutation. Study participants had annual visits to the NIH Clinical Center, with history and physical examination supplemented as needed by additional studies (eg, immunological, pulmonary, dermatological, gastroenterological) as well as peripheral blood and bone marrow investigations. Samples from index patients and familial controls were collected and stored, providing a valuable resource for current and future studies. In this issue of Blood, the authors report data from the first 111 index patients and 85 familial controls enrolled between 2019 and 2021, representing 45 unrelated families with 39 unique RUNX1 germ line variants. Causative germ line variants as well as the reported clinical phenotype mirrored known allelic heterogeneity and previous clinical findings. Briefly, nonsense or frameshift variants, partial/whole deletions of the RUNX1 locus, or missense variants of the runt homology domain of RUNX1 were identified as the most frequent causative genetic alteration in FPDMM. Most, but not all, patients (91%) had thrombocytopenia, but all tested index individuals (n = 18) had abnormal platelet aggregometry response. Nonspecific baseline dysmegakaryopoiesis was present in nonmalignant bone marrow in almost all adult patients (23/24, 96%) and in two-thirds of the pediatric samples (14/21, 67%). In contrast, hypocellular bone marrow was seen in most pediatric samples (17/21, 81%) but only in one-half of the adult specimens (13/24, 54%). Expansive phenotyping confirmed a strong atopic phenotype in a subset of patients, and comparisons with disease frequencies to the All of Us database expanded the spectrum of FPDMM-associated symptoms to include gastrointestinal tract motility issues. Results of additional genetic investigations have been published on a preprint server.4 In summary, the current study of the national history study confirms previous findings (eg, Brown et al, Churpek et al, Rio-Machin et al, and Decker at al3,5-7). No groundbreaking novel insights were reported after 3 years of enrollment. However, for this longitudinal natural history study of a rare disease with a broad clinical phenotype, the goal is to provide a better understanding of underlying pathophysiology over time, help optimize clinical care, and inform future research directions and care for people with FPDMM.
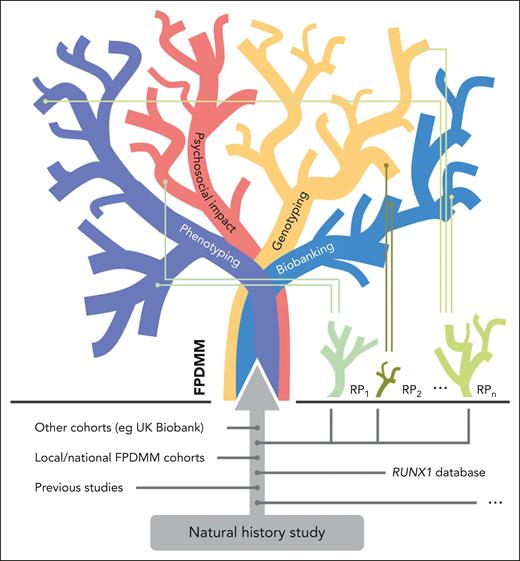
The natural history study has unique potential, but it also faces several challenges. This study is going to create a massive data pool with linked biomaterial, including peripheral blood, bone marrow, and skin biopsies. Altogether, this provides a unique chance to further characterize the natural course of the germ line RUNX1 mutation–associated disease. Future research projects will branch from it, using its valuable clinical data and biomaterials. Thus, the study is fueling a self-feeding FPDMM knowledge tree (see figure) that will expand beyond our present knowledge hubs (eg, the international RUNX1 database8). As discussed by the authors, there are several limitations to be overcome. These include biases due to ascertainment of the most severely affected individuals, the impact of socioeconomic capabilities/limitations on enrollment, and country-of-origin restrictions impacting participation. These issues can be partially solved by cascade testing in families, reimbursement for travel expenses, and increasing awareness for the disease in the general public and among physicians, especially those dealing with immune thrombocytopenia or benign hematological issues in general. From my perspective, the challenges are, firstly, the successful linkage of additional independent data sets and external biobanking hubs linking together into a network and, secondly, enabling enrollment of individuals proven to have FPDMM found incidentally or uncovered in the context of national cohorts (eg, UK Biobank). These measures will significantly enhance the growth of the FPDMM knowledge tree and further minimize ascertainment biases. All rare diseases face challenges that are best addressed by an international network of dedicated scientists, physicians, and patient representatives. The network has to define scientific, legal, and ethical frameworks for the integration of independent data and biomaterial without significantly minimizing the data quality achieved by single-center studies. Data and biomaterial need to be handled following FAIR principles9 (ie, findable, accessible, interoperable, and reusable). Implementation of patient self-reporting tools will allow effective documentation of medical histories and foster our understanding of psychosocial issues linked to FPDMM.
Knowledge tree of FPDMM. In decades to come, the FPDMM knowledge tree will be fed by the natural history study. The tree branches out in main categories such as phenotyping, psychosocial impact, genotyping, and biobanking, which all include various sub-branches. To integrate already existing data sets as well as future studies and to allow international data sharing and sample access, global scientific, legal, and ethical frameworks need to be defined to integrate these data sets and provide biomaterials for research projects. In a positive feedback loop, the growing knowledge will influence and foster future research projects (RP), illustrated as small plants below the tree.
Knowledge tree of FPDMM. In decades to come, the FPDMM knowledge tree will be fed by the natural history study. The tree branches out in main categories such as phenotyping, psychosocial impact, genotyping, and biobanking, which all include various sub-branches. To integrate already existing data sets as well as future studies and to allow international data sharing and sample access, global scientific, legal, and ethical frameworks need to be defined to integrate these data sets and provide biomaterials for research projects. In a positive feedback loop, the growing knowledge will influence and foster future research projects (RP), illustrated as small plants below the tree.
The Natural History Study of Hematologic and Premalignant Conditions Associated with RUNX1 Mutation has unique potential, but it will take a global team effort to understand the complete spectrum of FPDMM. That is a prerequisite for optimization of personalized risk-adapted genetic counseling, cancer surveillance, treatment options, or even future cure.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal