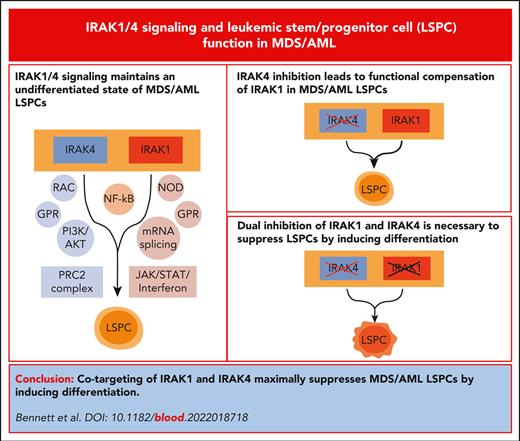
Key Points
Cotargeting of IRAK1 and IRAK4 is required to maximally suppress LSPC function in vitro and in vivo by inducing cellular differentiation.
The dependency of IRAK1 and IRAK4 in MDS/AML is independent of its canonical role using MyD88.
Abstract
Dysregulation of innate immune signaling is a hallmark of hematologic malignancies. Recent therapeutic efforts to subvert aberrant innate immune signaling in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) have focused on the kinase IRAK4. IRAK4 inhibitors have achieved promising, though moderate, responses in preclinical studies and clinical trials for MDS and AML. The reasons underlying the limited responses to IRAK4 inhibitors remain unknown. In this study, we reveal that inhibiting IRAK4 in leukemic cells elicits functional complementation and compensation by its paralog, IRAK1. Using genetic approaches, we demonstrate that cotargeting IRAK1 and IRAK4 is required to suppress leukemic stem/progenitor cell (LSPC) function and induce differentiation in cell lines and patient-derived cells. Although IRAK1 and IRAK4 are presumed to function primarily downstream of the proximal adapter MyD88, we found that complementary and compensatory IRAK1 and IRAK4 dependencies in MDS/AML occur via noncanonical MyD88-independent pathways. Genomic and proteomic analyses revealed that IRAK1 and IRAK4 preserve the undifferentiated state of MDS/AML LSPCs by coordinating a network of pathways, including ones that converge on the polycomb repressive complex 2 complex and JAK-STAT signaling. To translate these findings, we implemented a structure-based design of a potent and selective dual IRAK1 and IRAK4 inhibitor KME-2780. MDS/AML cell lines and patient-derived samples showed significant suppression of LSPCs in xenograft and in vitro studies when treated with KME-2780 as compared with selective IRAK4 inhibitors. Our results provide a mechanistic basis and rationale for cotargeting IRAK1 and IRAK4 for the treatment of cancers, including MDS/AML.
Introduction
Myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) originate in hematopoietic stem and progenitor cells (HSPCs), referred to as leukemia stem/progenitor cells (LSPCs), that acquire genetic and/or epigenetic alterations.1,2 LSPCs replenish the bulk leukemic blasts and contribute to treatment-related relapse.3 Recent therapies targeting LSPCs have shown improved clinical outcomes for patients with high-risk (HR) MDS/AML.4,5 LSPCs share cellular states and transcriptional programs with normal HSPCs, such as ones that maintain self-renewal properties and prevent untimely differentiation. Uncovering signaling dependencies unique to LSPCs is critical to improve the therapeutic responses. It was recently reported that LSPCs from MDS/AML exhibit extensive dysregulated immune and inflammatory pathways.6 Moreover, many genetic alterations in MDS/AML impinge on loll-like receptor (TLR) and interleukin 1 receptor (IL-1R) pathways,7-13 which converge on IL-1R–associated kinases (IRAKs).14 In healthy immune cells, the activation of IL-1R/TLRs leads to the recruitment of MyD88, which then forms a complex with IRAK4.15 IRAK4 subsequently phosphorylates IRAK1 or IRAK2, which then activates IκB kinase (IKK)/NF-κB and MAPKs. Chronic activation of canonical MyD88-IRAK signaling axis is presumed to underlie malignant hematopoiesis, as supported by activating MyD88 mutations in lymphomas and genetic alterations that dysregulate the MyD88-IRAK axis in MDS/AML, including splicing factor mutations that induce hypermorphic IRAK4 isoforms.16-18 Consequently, IRAK4 inhibitors and proteolysis targeting chimeric (PROTAC) small molecule degraders are being assessed in preclinical studies and clinical trials for hematologic malignancies and inflammatory conditions.14 The IRAK4 kinase inhibitor PF-06650833 (zimlovisertib) is being evaluated for chronic inflammatory disorders,19 whereas CA-4948 (emavusertib) is being evaluated for hematologic malignancies, including lymphoma, MDS, and AML.20 The IRAK4 PROTACs are being tested in immunoinflammatory disease and MyD88-mutant lymphomas.21,22 Initial results from the clinical trials in HR-MDS/AML with CA-4948 showed reductions in leukemic blasts and response rates of ∼40%.23 Interestingly, patients with HR-MDS/AML with spliceosome mutations achieved better responses as compared with patients without spliceosome mutations, which supports the findings that mutations in splicing factors induce hypermorphic IRAK4 isoforms and consequently innate immune signaling in MDS/AML.17,18 The ongoing clinical trials and mounting evidence linking IRAK1 and IRAK4 to myeloid malignancies suggest that therapies targeting IRAK4 will likely emerge for MDS/AML. Although these studies propose that IRAK4 is a relevant target in MDS/AML, the magnitude of responses suggest IRAK4 inhibitors as monotherapy will likely be inadequate.23 Herein, we evaluated the mechanistic basis for the limited responses to IRAK4 inhibitors and propose novel IRAK-directed therapeutic strategies for MDS/AML.
Material and methods
See supplemental File, which is available on the Blood website, for additional details.
Compounds and materials
PF-06650833 was purchased from Sigma-Aldrich. CA-4948 and IRAK4 degrader-1 were purchased from MedChemExpress. KME-2780 and KME-3859 were obtained from Kurome Therapeutics.
Clonogenic assays
Clonogenic progenitor frequencies were determined using plating cell lines or patient samples in Methocult H4434 at a density of 500 cells per mL. Plates were imaged after ∼9 to 14 days using the STEMvision counter (StemCell Technologies).
NF-κB reporter
THP1-Blue NF-κB SEAP cells (Invivogen) were plated in triplicates with the indicated inhibitor for 24 hours. The supernatant was added to QuantiBlue Reagent (Invivogen), incubated for 30 minutes, and absorbance was read at 630 nm.
Kinome screens
Dissociation constants were measured using KINOMEscan Profiling (DiscoverX). Kinase inhibition was measured by Reaction Biology.
CRISPR/Cas9 mutant cells
THP1 IRAK4-knockout (IRAK4KO) clone was generated as previously described.17 THP1 IRAK1KO and IRAK1/4-double KO (IRAK1/4dKO) were established from the original wild-type (WT) and IRAK4KO clones. The remaining cell lines and patient-derived samples were generated from parental populations using a guide RNA targeting exon 1 of IRAK4. MyD88KO and TRAF6KO clones were generated from parental populations using synthetic guide RNAs targeting exon 1 of MyD88 andTRAF6, respectively.
Results
IRAK4 inhibition results in compensatory activation of IRAK1 in MDS/AML
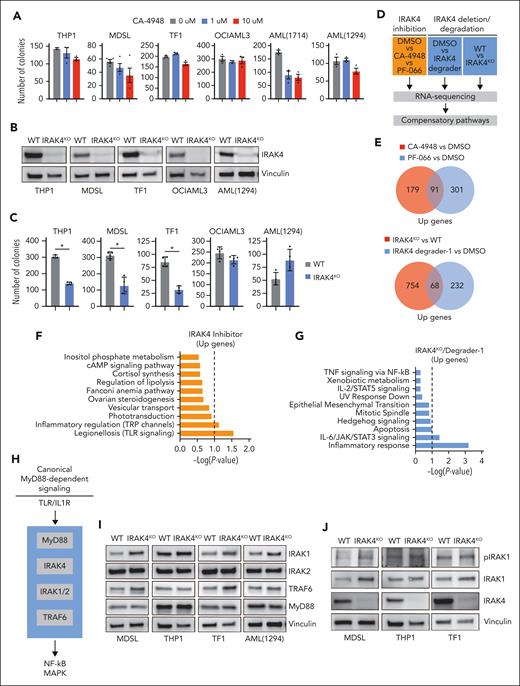
To investigate IRAK4 in MDS/AML, we evaluated the clinical-stage IRAK4 inhibitors CA-4948 and PF-06650833 in cell lines and patient-derived samples (supplemental Table 1). The selected samples form colonies in methylcellulose, an indication of LSPC function24, and express the hypermorphic IRAK4 isoform (IRAK4-L). As expected, both IRAK4 inhibitors suppressed TLR-mediated NF-κB activation in AML cells (supplemental Figure 1A). As reported,17 CA-4948 treatment resulted in a modest inhibition of colony formation in a subset of MDS/AML cell lines and patient-derived samples (Figure 1A). Despite a greater potency in NF-κB assays (supplemental Figure 1A), PF-06650833 was not as effective as CA-4948 at suppressing colony formation (supplemental Figure 1B). This discrepancy may reflect the distinct polypharmacology of the inhibitors. CA-4948 treatment moderately suppressed the proliferation and survival of THP1 cells and had a negligible impact on the growth kinetics of MDSL cells (supplemental Figure 1C), suggesting that IRAK4 inhibition is not cytotoxic. To confirm that the effects observed with the IRAK4 inhibitors were mediated by targeting IRAK4, we generated IRAK4-deficient isogenic cell lines (IRAK4KO) (Figure 1B). The deletion of IRAK4 resulted in a moderate reduction in LSPC colonies (Figure 1C). Collectively, IRAK4 inhibition incompletely suppresses LSPC function, recapitulating the observations from clinical trials for HR-MDS/AML.
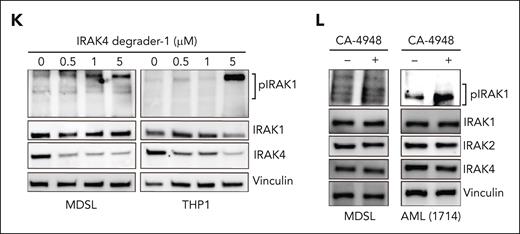
IRAK4 inhibition causes the activation of IRAK1. (A) Colony formation in a panel of MDS/AML cell lines and patient-derived samples treated with the indicated concentrations of CA-4948 (2 independent experiments). (B) Immunoblots for IRAK4 in WT and IRAK4KO AML cell lines and patient-derived samples. (C) Colony formation of WT and IRAK4KO AML cell lines and patient-derived samples. (D) Experimental overview: RNA sequencing was performed using WT and IRAK4KO THP1 cells, and THP1 cells were treated for 24 hours with the indicated inhibitors. Genes upregulated upon IRAK4 deficiency or chemical inhibition were used to annotate compensatory pathways. (E) Venn diagrams of overlapping upregulated genes upon IRAK4 deficiency or IRAK4 chemical inhibition. (F) Pathway enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) data sets using overlapping genes increased upon the treatment with IRAK4 inhibitors. (G) Pathway enrichment of KEGG data sets using overlapping genes increased upon treatment with IRAK4 degrader-1 or after the deletion of IRAK4. (H) Overview of canonical Myd88-dependent signaling: upon TLR ligation, MyD88 nucleates a complex with IRAK4, which signals through IRAK1 and/or IRAK2 and then TRAF6 to activate the NF-κB and MAPK pathways. (I) Immunoblots for IRAK1, IRAK2, TRAF6, and MyD88 in WT and IRAK4KO AML cell lines and patient-derived samples. (J) Immunoblots for phoshpo-IRAK1, total IRAK1, and IRAK4 in WT and IRAK4KO cell lines. (K) Immunoblots for phoshpo-IRAK1, total IRAK1, and IRAK4 in MDSL and THP1 cells treated for 24 hours with IRAK4 degrader-1. (L) Immunoblots for phospho-IRAK1, total IRAK1, IRAK2, and IRAK4 in MDSL and AML (1714) treated for 24 hours with CA-4948 (10 μM). Significance was determined with a Student t test (∗P < .05). Error bars represent the standard deviation.
IRAK4 inhibition causes the activation of IRAK1. (A) Colony formation in a panel of MDS/AML cell lines and patient-derived samples treated with the indicated concentrations of CA-4948 (2 independent experiments). (B) Immunoblots for IRAK4 in WT and IRAK4KO AML cell lines and patient-derived samples. (C) Colony formation of WT and IRAK4KO AML cell lines and patient-derived samples. (D) Experimental overview: RNA sequencing was performed using WT and IRAK4KO THP1 cells, and THP1 cells were treated for 24 hours with the indicated inhibitors. Genes upregulated upon IRAK4 deficiency or chemical inhibition were used to annotate compensatory pathways. (E) Venn diagrams of overlapping upregulated genes upon IRAK4 deficiency or IRAK4 chemical inhibition. (F) Pathway enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) data sets using overlapping genes increased upon the treatment with IRAK4 inhibitors. (G) Pathway enrichment of KEGG data sets using overlapping genes increased upon treatment with IRAK4 degrader-1 or after the deletion of IRAK4. (H) Overview of canonical Myd88-dependent signaling: upon TLR ligation, MyD88 nucleates a complex with IRAK4, which signals through IRAK1 and/or IRAK2 and then TRAF6 to activate the NF-κB and MAPK pathways. (I) Immunoblots for IRAK1, IRAK2, TRAF6, and MyD88 in WT and IRAK4KO AML cell lines and patient-derived samples. (J) Immunoblots for phoshpo-IRAK1, total IRAK1, and IRAK4 in WT and IRAK4KO cell lines. (K) Immunoblots for phoshpo-IRAK1, total IRAK1, and IRAK4 in MDSL and THP1 cells treated for 24 hours with IRAK4 degrader-1. (L) Immunoblots for phospho-IRAK1, total IRAK1, IRAK2, and IRAK4 in MDSL and AML (1714) treated for 24 hours with CA-4948 (10 μM). Significance was determined with a Student t test (∗P < .05). Error bars represent the standard deviation.
To explore the mechanisms contributing to the moderate and incomplete responses to IRAK4 inhibitors in MDS/AML, we examined gene expression changes upon targeting IRAK4. RNA sequencing was performed using THP1 cells treated with the IRAK4 inhibitors or a PROTAC (IRAK4 degrader-1) and isogenic WT and IRAK4KO THP1 cells. To identify compensatory pathways following either IRAK4 kinase inhibition or degradation, we focused on upregulated genes (Figure 1D; supplemental Tables 2-5). There were 91 commonly upregulated genes after the treatment with CA-4948 and PF-06650833 relative to vehicle-treated cells (Figure 1E; supplemental Figure 1D). After the deletion (IRAK4KO) or pharmacologic degradation (IRAK4 degrader-1) of IRAK4, 68 commonly upregulated genes were identified (Figure 1E; supplemental Figure 1E). Unexpectedly, pathway analysis of the overlapping upregulated genes corresponded with the enrichment of TLR signaling upon IRAK4 kinase inhibition (Figure 1F) and with that of inflammatory response and IL-6/JAK/STAT signaling upon IRAK4 deletion (Figure 1G). As such, we posited that compensatory signaling via proximal effectors of MyD88-IRAK may contribute to the limited efficacy of IRAK4 inhibitors (Figure 1H). Immunoblotting revealed an increased protein expression of IRAK1 and TRAF6 but not MyD88 or IRAK2 in IRAK4KO MDS/AML cells relative to control cells (Figure 1I). Importantly, IRAK1 phosphorylation, an indication of its activated state, remained elevated upon the deletion of IRAK4 (Figure 1J). Treatment with IRAK4 degrader-1 (Figure 1K) or CA-4948 also resulted in increased IRAK1 phosphorylation (Figure 1L). Thus, IRAK4 deficiency or kinase inhibition corresponds with IRAK1 activation in MDS/AML cells.
Cotargeting IRAK1 and IRAK4 is required to suppress LSPCs
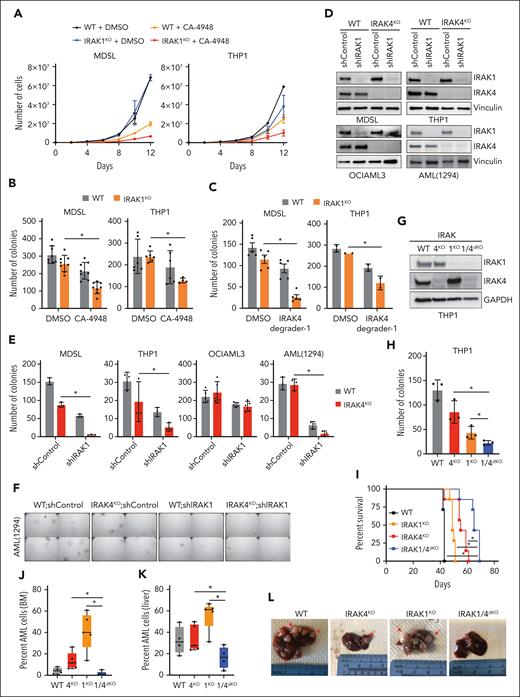
Because IRAK1 is activated upon the inhibition or deletion of IRAK4, we speculated that targeting IRAK1 would augment IRAK4 inhibitors in MDS/AML. Next, we generated IRAK1-deficient (IRAK1KO) THP1 and MDSL cell lines (supplemental Figure 2A). IRAK1KO MDS/AML cells exhibited similar growth kinetics and colony formation as that of WT cells (Figure 2A-B). However, IRAK1KO MDS/AML cells were more sensitive to treatment with IRAK4 inhibitors or PROTAC (Figure 2B-C; supplemental Figure 2B). To confirm these observations, we knocked down IRAK1 in the isogenic WT and IRAK4KO MDS/AML cells (Figure 2D). The combined deficiency of IRAK1 and IRAK4 in THP1, MDSL, and AML (1294) resulted in a significant reduction in LSPC colonies as compared with the deficiency of either IRAK4 or IRAK1 alone (Figure 2E-F). Next, we generated isogenic double IRAK1 and IRAK4-deficient (IRAK1/4dKO) THP1 cells (Figure 2G). IRAK1/4dKO THP1 cells expanded at a slower rate as compared with IRAK1KO or IRAK4KO cells (supplemental Figure 2C). Moreover, IRAK1/4dKO cells formed significantly fewer LSPC colonies as compared with IRAK1KO or IRAK4KO cells after the initial (Figure 2H) and secondary replating (supplemental Figure 2D). To assess the effect of combined IRAK1/4 deficiency on leukemia development, we xenografted the isogenic THP1 cells into immunocompromised mice. Mice that received IRAK1/4dKO THP1 cell engraftment survived longer than mice that received WT, IRAK1KO, or IRAK4KO cell engraftment (Figure 2I; supplemental Figure 2E). Histology samples collected at the time of death revealed the reduced infiltration of IRAK1/4dKO AML cells in the bone marrow (Figure 2J) and liver (Figure 2K) and the absence of liver nodules as compared with mice that received engraftment with WT, IRAK1KO, or IRAK4KO cells (Figure 2L). Thus, concomitant targeting of IRAK1 elicits an exaggerated LSPC defect in IRAK4-deficient MDS/AML models.
The inhibition of IRAK1 confers an exaggerated leukemic defect to IRAK4-deficient AML. (A) Growth curves of WT and IRAK1KO MDSL and THP1 cells treated with CA-4948 (10 μM) or vehicle (2 independent experiments). (B) Colony formation of WT and IRAK1KO MDSL and THP1 cells treated with CA-4948 (30 μM) or vehicle (3 independent experiments). (C) Colony formation of WT and IRAK1KO MDSL and THP1 cells treated with IRAK4 degrader-1 (MDSL, 5 μM; THP1, 10 μM) or vehicle. (D) Immunoblots for IRAK1 and IRAK4 in WT and IRAK4KO cell lines transduced with nontargeting control short hairpin RNA (shRNA; shControl) or shIRAK1. (E) Colony formation of WT and IRAK4KO AML cell lines transduced with nontargeting control shRNA (shControl) or shIRAK1. (F) Representative colony images of WT and IRAK4KO AML (1294) cells transduced with nontarget control shRNA (shControl) or shIRAK1 (original magnification ×40). (G) Immunoblots for IRAK1 and IRAK4 in WT, IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells. (H) Colony formation of isogenic THP1 cells. (I) Kaplan-Meier survival analysis of NSGS mice (n = 7 mice per group) that received engraftment with WT, IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells (Data represent 1 of 2 independent experiments with similar trends). (J) Bone marrow engraftment of WT (n = 4), IRAK4KO (n = 5), IRAK1KO (n = 5), and IRAK1/4dKO (n = 5) THP1 cells in NSGS mice that underwent xenograftment at the time of death. Leukemic engraftment was determined as the percentage of huCD45+huCD33+ cells. (K) Liver engraftment of WT (n = 4), IRAK4KO (n = 5), IRAK1KO (n = 5), and IRAK1/4dKO (n = 5) THP1 cells in NSGS mice that underwent xenograftment at the time of death. Leukemic engraftment was determined as the percentage of huCD45+huCD33+ cells normalized to the number of days. (L) Representative images of livers collected from NSGS mice that underwent xenograftment with WT, IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells. Arrows indicate examples of AML cell infiltration. Significance was determined with a Student t test (∗P < .05). Error bars represent the standard deviation.
The inhibition of IRAK1 confers an exaggerated leukemic defect to IRAK4-deficient AML. (A) Growth curves of WT and IRAK1KO MDSL and THP1 cells treated with CA-4948 (10 μM) or vehicle (2 independent experiments). (B) Colony formation of WT and IRAK1KO MDSL and THP1 cells treated with CA-4948 (30 μM) or vehicle (3 independent experiments). (C) Colony formation of WT and IRAK1KO MDSL and THP1 cells treated with IRAK4 degrader-1 (MDSL, 5 μM; THP1, 10 μM) or vehicle. (D) Immunoblots for IRAK1 and IRAK4 in WT and IRAK4KO cell lines transduced with nontargeting control short hairpin RNA (shRNA; shControl) or shIRAK1. (E) Colony formation of WT and IRAK4KO AML cell lines transduced with nontargeting control shRNA (shControl) or shIRAK1. (F) Representative colony images of WT and IRAK4KO AML (1294) cells transduced with nontarget control shRNA (shControl) or shIRAK1 (original magnification ×40). (G) Immunoblots for IRAK1 and IRAK4 in WT, IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells. (H) Colony formation of isogenic THP1 cells. (I) Kaplan-Meier survival analysis of NSGS mice (n = 7 mice per group) that received engraftment with WT, IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells (Data represent 1 of 2 independent experiments with similar trends). (J) Bone marrow engraftment of WT (n = 4), IRAK4KO (n = 5), IRAK1KO (n = 5), and IRAK1/4dKO (n = 5) THP1 cells in NSGS mice that underwent xenograftment at the time of death. Leukemic engraftment was determined as the percentage of huCD45+huCD33+ cells. (K) Liver engraftment of WT (n = 4), IRAK4KO (n = 5), IRAK1KO (n = 5), and IRAK1/4dKO (n = 5) THP1 cells in NSGS mice that underwent xenograftment at the time of death. Leukemic engraftment was determined as the percentage of huCD45+huCD33+ cells normalized to the number of days. (L) Representative images of livers collected from NSGS mice that underwent xenograftment with WT, IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells. Arrows indicate examples of AML cell infiltration. Significance was determined with a Student t test (∗P < .05). Error bars represent the standard deviation.
IRAK1 and IRAK4 dependency in MDS/AML is independent of MyD88
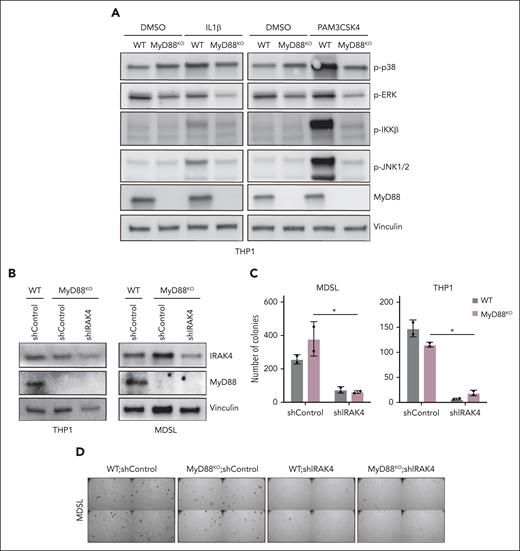
IRAK1 and IRAK4 transduce canonical MyD88 signaling in a mutually dependent manner (Figure 1H). However, this paradigm is inconsistent with the exaggerated defect of leukemic cells upon the concomitant targeting of IRAK1 and IRAK4. Therefore, we hypothesized that IRAK1 and IRAK4 exhibit functional redundancy downstream of MyD88 in leukemic cells. Thus, we first examined the consequences of IRAK1 and IRAK4 deletion on IL1R and TLR2 activation in THP1 cells. The deletion of IRAK4 or IRAK1 was sufficient to prevent the activation of NF-κB upon the stimulation of IL-1R (supplemental Figure 2F). As an exception, phosphorylation of IKKβ upon TLR2 stimulation was partially reduced in IRAK4KO and IRAK1KO THP1 cells but completely ablated in the IRAK1/4dKO cells (supplemental Figure 2G). IRAK4 deletion was also sufficient to block JNK and p38 upon TLR2 stimulation. Similar signaling dependencies were observed in MDSL cells (supplemental Figure 2H). As expected, the deletion of MyD88 resulted in a complete termination of NF-κB and MAPK activation upon IL1R or TLR2 stimulation in AML cells (Figure 3A). Surprisingly, MyD88KO THP1 or MDSL cells formed an equivalent number of colonies as compared with WT cells, indicating that MyD88 is dispensable for LSPCs (Figure 3B-C).
MyD88 is dispensable for MDS/AML LSPCs. (A) Immunoblots for MyD88 and the activation of downstream pathways (phospho-p38, phospho-JNK, phospho-IKK, and phospho- extracellular signal-regulated kinase) in WT and MYD88KO THP1 cells upon a 30-minute treatment with IL-1β (10 ng/µL) or the TLR1/2 ligand PAM3CSK4 (1 µg/mL) as compared with DMSO. (B) Immunoblots for IRAK4 and MyD88 in WT and MYD88KO THP1 and MDSL cells transduced with nontargeting shControl or shIRAK4. (C) Colony formation of WT and MYD88KO THP1 and MDSL cells transduced with nontargeting shControl or shIRAK4. (D) Representative colony images of WT and MyD88KO MDSL cells transduced with nontargeting shControl or shIRAK4 (original magnification ×40). Significance was determined with a Student t test (∗P < .05). Error bars represent the standard deviation. DMSO, dimethyl sulfoxide.
MyD88 is dispensable for MDS/AML LSPCs. (A) Immunoblots for MyD88 and the activation of downstream pathways (phospho-p38, phospho-JNK, phospho-IKK, and phospho- extracellular signal-regulated kinase) in WT and MYD88KO THP1 cells upon a 30-minute treatment with IL-1β (10 ng/µL) or the TLR1/2 ligand PAM3CSK4 (1 µg/mL) as compared with DMSO. (B) Immunoblots for IRAK4 and MyD88 in WT and MYD88KO THP1 and MDSL cells transduced with nontargeting shControl or shIRAK4. (C) Colony formation of WT and MYD88KO THP1 and MDSL cells transduced with nontargeting shControl or shIRAK4. (D) Representative colony images of WT and MyD88KO MDSL cells transduced with nontargeting shControl or shIRAK4 (original magnification ×40). Significance was determined with a Student t test (∗P < .05). Error bars represent the standard deviation. DMSO, dimethyl sulfoxide.
Next, we assessed the dependency of MyD88KO AML cells on the loss of IRAK4. Knockdown of IRAK4 in WT and MyD88KO THP1 and MDSL cell lines resulted in a significant reduction of LSPC function (Figure 3B-D). Thus, MDS/AML cells are dependent on IRAK1/4 signaling but independent of MyD88. As the proximal downstream effector of IRAK1/4, we wanted to discern whether TRAF6 remains relevant to MyD88-independent IRAK1/4 signaling. The deletion of TRAF6 resulted in a significant reduction of LSPC colonies (supplemental Figure 3A-C), suggesting that the MDS/AML cells are dependent on TRAF6-mediated signaling despite the lack of a requirement for MyD88. These findings demonstrate that IRAK1 and IRAK4 mediate MyD88-independent functions that are critical for MDS/AML LSPCs.
Noncanonical IRAK1/4 signaling is essential for maintaining undifferentiated LSPCs
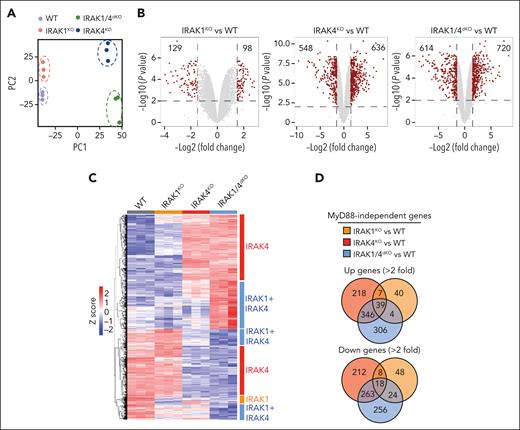
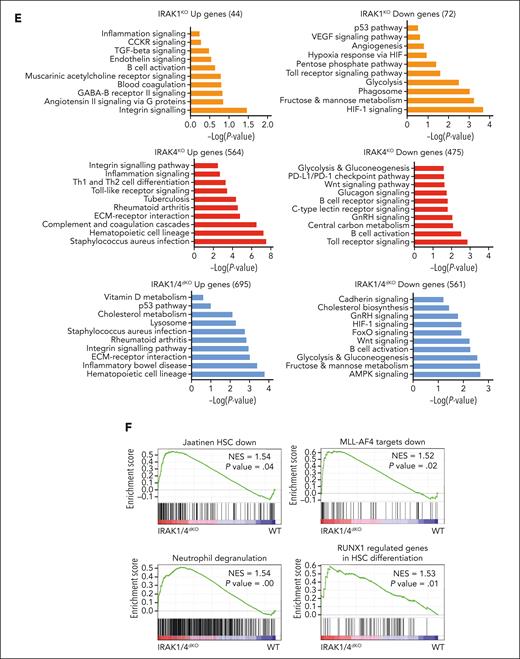
To delineate MyD88-independent IRAK1/4 signaling in MDS/AML, we performed a gene expression analysis in WT, MYD88KO, IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 cells. Principle component analysis of the isogenic cell lines affirmed distinct expression programs (Figure 4A). To focus on MyD88-independent gene programs, we excluded all differentially expressed genes that overlapped with MyD88KO cells (supplemental Figure 4A-B; supplemental Table 6). The deletion of IRAK1 or IRAK4 resulted in 129 and 548 repressed genes, respectively and 98 and 636 overexpressed genes, respectively in comparison with WT cells (Figure 4B). The combined deletion of IRAK1 and IRAK4 resulted in 614 downregulated and 720 upregulated genes. The examination of differentially expressed genes yielded a subset of genes unique to IRAK1KO, IRAK4KO, and IRAK1/4dKO cells (Figure 4C-D; supplemental Table 7). KEGG and PANTHER analysis revealed that the isogenic cell lines corresponded with the enrichment of distinct pathways (Figure 4E). For example, downregulated genes in IRAK1KO cells exhibit the enrichment of pathways related to HIF1α, metabolism, and inflammatory signaling; IRAK4KO cells exhibit enrichment of pathways related to TLR and Wnt, and glycolysis; and IRAK1/4dKO cells exhibit enrichment of pathways related to adenosine monophosphate–activated protein kinase, inflammation, and HSCs (Figure 4E). MyD88KO cells did not show enrichment of these pathways, supporting the observations that IRAK1/4 signaling functions independent of MyD88 (supplemental Figure 4C). Gene set enrichment analysis revealed that the upregulated genes in IRAK1/4dKO cells are associated with the loss of HSC states, suppression of mixed lineage leukemia-rearranged leukemia, and myeloid differentiation programs (Figure 4F).
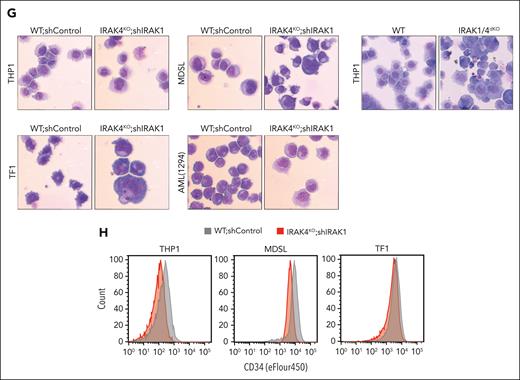
Noncanonical IRAK1/4 signaling is essential for maintaining MDS/AML LSPCs. (A) Principal component analysis of gene expression profiles of WT, IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells. (B) Volcano plots of differentially expressed genes in IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells relative to WT THP1 cells (>2-fold change; ∗P < .05). (C) Heatmap of differentially expressed genes in IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 relative to WT. Bars on the right denote differentially expressed genes attributed to the deficiency of IRAK4 (red) or IRAK1 (orange), or are unique to IRAK1/4dKO (blue). (D) Venn diagrams of overlapping upregulated or downregulated genes in IRAK4KO, IRAK1KO, and IRAK1/4dKO relative to WT THP1. (E) Pathway enrichment of KEGG data sets of upregulated and downregulated genes in IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells. (F) Gene set enrichment analysis of genes dysregulated in IRAK1/4dKO vs WT THP1 cells. Absolute normalized enrichment score and the corresponding P value is shown for each pathway. (G) Representative Wright-Giemsa stains of WT and IRAK4KO cells expressing nontargeting shControl and shIRAK1, respectively (original magnification ×40). (H) Immunophenotyping of the indicated cells for CD34 expression.
Noncanonical IRAK1/4 signaling is essential for maintaining MDS/AML LSPCs. (A) Principal component analysis of gene expression profiles of WT, IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells. (B) Volcano plots of differentially expressed genes in IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells relative to WT THP1 cells (>2-fold change; ∗P < .05). (C) Heatmap of differentially expressed genes in IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 relative to WT. Bars on the right denote differentially expressed genes attributed to the deficiency of IRAK4 (red) or IRAK1 (orange), or are unique to IRAK1/4dKO (blue). (D) Venn diagrams of overlapping upregulated or downregulated genes in IRAK4KO, IRAK1KO, and IRAK1/4dKO relative to WT THP1. (E) Pathway enrichment of KEGG data sets of upregulated and downregulated genes in IRAK4KO, IRAK1KO, and IRAK1/4dKO THP1 cells. (F) Gene set enrichment analysis of genes dysregulated in IRAK1/4dKO vs WT THP1 cells. Absolute normalized enrichment score and the corresponding P value is shown for each pathway. (G) Representative Wright-Giemsa stains of WT and IRAK4KO cells expressing nontargeting shControl and shIRAK1, respectively (original magnification ×40). (H) Immunophenotyping of the indicated cells for CD34 expression.
To determine whether IRAK1 and IRAK4 signaling in LSPCs is required for preserving an undifferentiated LSPC state, we examined morphological changes of MDS/AML cells upon the deletion of IRAK1 and IRAK4. In contrast to WT cells, IRAK1/4dKO cells acquired heterogeneous morphologies and exhibited increased cytoplasmic to nuclear ratios (Figure 4G) and decreased CD34 expression (Figure 4H), which is congruent with myeloid differentiation. Moreover, aberrant myeloid cell differentiation was not observed in MyD88KO cells (supplemental Figure 4D). These findings suggest that IRAK1 and IRAK4 mediate functions that are critical for LSPCs by preserving an immature cell state.
IRAK1 and IRAK4 interactomes reveal noncanonical signaling networks in MDS/AML
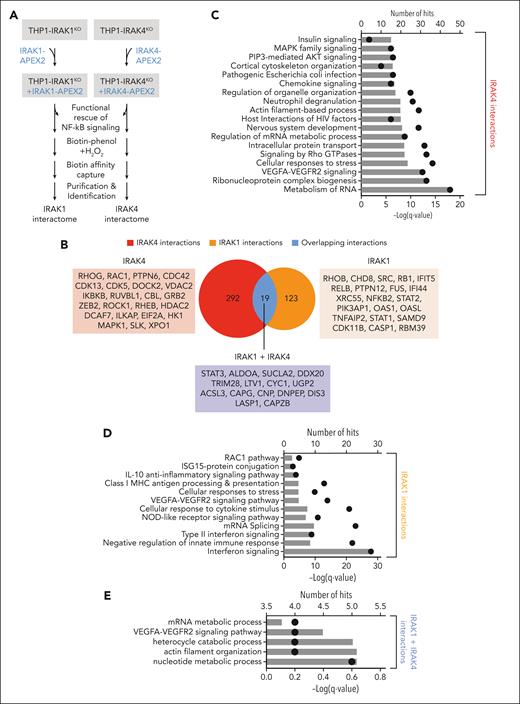
We found that IRAK1 and IRAK4 coordinate complementary gene expression programs implicated in maintaining LSPCs; however, the noncanonical signaling mechanisms by which IRAK1/4 regulates these programs remain unknown. To map the IRAK1 and IRAK4 interactome in AML, we performed proximity labeling of proteins with biotin using APEX2 followed by mass spectrometry25 (Figure 5A; supplemental Figure 5A-B). We identified 311 proteins proximal to IRAK4 and 142 proteins proximal to IRAK1 (Figure 5B; supplemental Table 8-9). In total, 292 proteins were unique to IRAK4 and 123 were unique to IRAK1 (Figure 5B), whereas 19 proteins were common to IRAK1 and IRAK4 (Figure 5B). Several proteins associated with canonical IRAK1/4 signaling were identified, such as IKKβ/IKBKB, RELB, p100/NFKB2, and ERK/MAPK1. To uncover novel IRAK1/4 signaling networks in AML, we performed an ontology pathway analysis26 using the list of IRAK1 and IRAK4 proximal proteins. Proximal IRAK4 proteins included effectors of messenger RNA metabolism, PI3K/AKT, and MAPK (Figure 5C), whereas proximal IRAK1 proteins were enriched for interferon and antiviral signaling (Figure 5D). There were also proteins proximal to both IRAK1 and IRAK4, which include effectors of STAT/interferon and vascular endothelial growth factor receptor signaling and messenger RNA metabolism (Figure 5B,E). The IRAK1 and IRAK4 proximal proteins are predicted to form physical interactions as demonstrated by a protein-protein interaction analysis27 (supplemental Figure 5C-D).
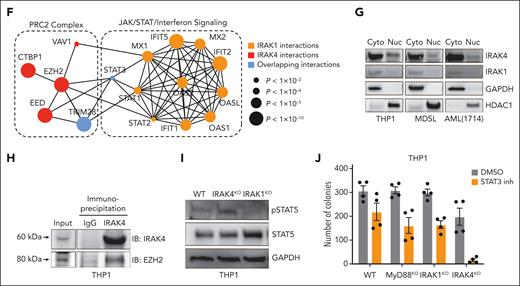
IRAK1 and IRAK4 interactomes reveal noncanonical signaling in AML. (A) Experimental overview of IRAK1 and IRAK4 proximity labeling in THP1 cells: Doxycycline-inducible IRAK4- and IRAK1-APEX2 fusion constructs were transduced into IRAK4KO and IRAK1KO THP1 cells, respectively. Functional rescue of canonical signaling was confirmed in NF-κB assays. Biotin phenol was added to induce proximity labeling with biotin. Biotinylated proteins were isolated and identified via mass spectrometry. (B) Venn diagram of unique and overlapping proteins in the IRAK4 and IRAK1 proximal proteins. (C) Pathway enrichment using IRAK4-specific proximal proteins. Bars represent the number of IRAK4 interacting proteins that appear in the designated pathway. Dots represent the −log(q value) of the pathway enrichment. (D) Pathway enrichment using IRAK1-specific proximal proteins. Bars represent the number of IRAK1 interacting proteins that appear in the designated pathway. Dots represent the −log(q value) of the pathway enrichment. (E) Pathway enrichment using proximal proteins common to IRAK1 and IRAK4. Bars represent the number of interacting proteins that appear in the designated pathway. Dots represent the −log(q value) of the pathway enrichment. (F) Interaction map highlighting IRAK4 interactors in the PRC2 complex and IRAK1 interactors in JAK/STAT/interferon signaling. Circle sizes indicate the adjusted P value for the identified interaction with IRAK1 or IRAK4. (G) Immunoblots for IRAK1 and IRAK4 in the nuclear (Nuc) and cytoplasmic (Cyto) fractions isolated from the indicated cells. (H) Immunoprecipitation of IRAK4 (or immunoglobulin G control) followed by immunoblotting of IRAK4 and EZH2 from THP1 cells. (I) Immunoblots for phospho-STAT5 and STAT5 in WT, IRAK4KO, and IRAK1KO THP1 cells. (J) Colony formation of MyD88KO, IRAK1KO, and IRAK4KO THP1 cells treated with DMSO or BBI608 (STAT3 inhibitor) (500 nM). Error bars represent the standard error of the mean. DMSO, dimethyl sulfoxide.
IRAK1 and IRAK4 interactomes reveal noncanonical signaling in AML. (A) Experimental overview of IRAK1 and IRAK4 proximity labeling in THP1 cells: Doxycycline-inducible IRAK4- and IRAK1-APEX2 fusion constructs were transduced into IRAK4KO and IRAK1KO THP1 cells, respectively. Functional rescue of canonical signaling was confirmed in NF-κB assays. Biotin phenol was added to induce proximity labeling with biotin. Biotinylated proteins were isolated and identified via mass spectrometry. (B) Venn diagram of unique and overlapping proteins in the IRAK4 and IRAK1 proximal proteins. (C) Pathway enrichment using IRAK4-specific proximal proteins. Bars represent the number of IRAK4 interacting proteins that appear in the designated pathway. Dots represent the −log(q value) of the pathway enrichment. (D) Pathway enrichment using IRAK1-specific proximal proteins. Bars represent the number of IRAK1 interacting proteins that appear in the designated pathway. Dots represent the −log(q value) of the pathway enrichment. (E) Pathway enrichment using proximal proteins common to IRAK1 and IRAK4. Bars represent the number of interacting proteins that appear in the designated pathway. Dots represent the −log(q value) of the pathway enrichment. (F) Interaction map highlighting IRAK4 interactors in the PRC2 complex and IRAK1 interactors in JAK/STAT/interferon signaling. Circle sizes indicate the adjusted P value for the identified interaction with IRAK1 or IRAK4. (G) Immunoblots for IRAK1 and IRAK4 in the nuclear (Nuc) and cytoplasmic (Cyto) fractions isolated from the indicated cells. (H) Immunoprecipitation of IRAK4 (or immunoglobulin G control) followed by immunoblotting of IRAK4 and EZH2 from THP1 cells. (I) Immunoblots for phospho-STAT5 and STAT5 in WT, IRAK4KO, and IRAK1KO THP1 cells. (J) Colony formation of MyD88KO, IRAK1KO, and IRAK4KO THP1 cells treated with DMSO or BBI608 (STAT3 inhibitor) (500 nM). Error bars represent the standard error of the mean. DMSO, dimethyl sulfoxide.
Next, we focused on a mechanistic explanation for the aberrant expression of genes implicated in LSPC function after IRAK1/4 inhibition. The integration of enriched pathways derived from the gene expression and interactome analysis highlighted the convergence of IRAK1 on JAK/STAT/interferon-related signaling and IRAK4 on the polycomb repressive complex 2 (PRC2) (Figure 5F). As indicated, several IRAK1 proximal proteins are implicated in JAK-STAT signaling (Figure 5F). We also identified members of PRC2 among the top-ranking IRAK4 proximal proteins (Figure 5F). SUZ12, EED, and EZH2 are part of the core heteromeric PRC2 complex.28 JAK-STAT and PRC2 functions, including activating EZH2 mutations, are well-defined in LSPCs by regulating genes that promote cellular differentiation.29 Because the predicted IRAK4 interactions include nuclear proteins, we examined the localization of IRAK1 and IRAK4 in leukemic cells. IRAK4 localized to the cytoplasm and nucleus of MDS/AML cells (Figure 5G). In contrast, IRAK1 was primarily localized to the cytoplasm. Moreover, we found that EZH2 coimmunoprecipitated with IRAK4 (Figure 5H), suggesting that nuclear IRAK4 can directly associate with the PRC2 complex in AML. To further interrogate the IRAK1-IRAK4 network, we determined whether the deletion of IRAK4 results in a corresponding increase in IRAK1-dependent JAK-STAT signaling. IRAK4KO cells exhibited increased phosphorylated STAT5 as compared with WT cells, whereas IRAK1KO cells expressed phosphorylated STAT5 below the levels observed in WT cells (Figure 5I). Moreover, IRAK4KO THP1 cells were sensitive to a STAT inhibitor, as compared with IRAK1KO, MyD88KO, or WT cells (Figure 5J). Collectively, these findings reveal extensive IRAK1 and IRAK4 signaling networks implicated in maintaining LSPCs.
IRAK1/4 maintains undifferentiated leukemic cell states through chromatin and transcriptional networks
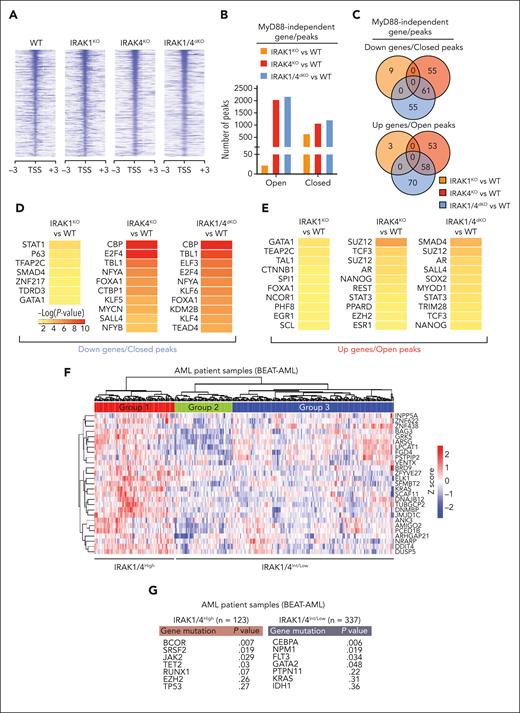
Next, we examined the regulatory effectors downstream of IRAK1/4 signaling that are required for maintaining LSPCs. Although our proteomic and transcriptomic analyses implicated several relevant pathways in MDS/AML, we focused on how IRAK1/4 signaling may coordinate chromatin and transcriptional factor networks. The PRC2 complex, which coordinates transcriptional gene regulation via H3K27 trimethylation, was nominated as one of the top hits among IRAK4-proximal proteins. PRC2-mediated H3K27 trimethylation can induce the compaction of chromatin and loss of transcription factor accessibility at select loci30; as such, we performed an assay for transposase-accessible chromatin sequencing and mapped global changes in chromatin accessibility in isogenic THP1 cells (Figure 6A). To prioritize MyD88-independent pathways, we excluded differential chromatin peaks identified in MYD88KO cells (supplemental Table 10). Although the number of closed peaks was similar in the isogenic cells (Figure 6B; supplemental Table 11), IRAK1KO THP1 cells exhibited fewer open peaks than IRAK4KO and IRAK1/4dKO cells (Figure 6B). IRAK4-regulated chromatin accessibility peaks are preserved in IRAK1/4dKO THP1 cells, whereas the effects of IRAK1 on chromatin accessibility are minimal (Figure 6C). To identify IRAK4-regulated factors that drive the IRAK1/4dKO AML cell phenotype, we assessed genes that are both downregulated and associated with the loss of chromatin accessibility in IRAK4KO and IRAK1/4dKO cells (Figure 6C). We identified a significant overlap of genes that are repressed in IRAK4KO and IRAK1/4dKO cells, which are enriched as targets of CREB-binding protein (CBP) and E2F4 (Figure 6D) and implicated in LSPCs.31,32 Next, we examined the genes associated with open chromatin peaks and upregulated expression that are conserved in IRAK4KO and IRAK1/4dKO cells (Figure 6C). This set of genes were enriched for regulatory targets of PRC2 components (SUZ12 and EZH2) and STAT3 (Figure 6E). Of the genes associated with the loss of chromatin peaks in IRAK1KO cells, only 9 genes were downregulated (Figure 6C), which were the predicted targets of STAT (Figure 6D). Thus, the data suggest that the deletion of IRAK1 and IRAK4 leads to the loss of JAK-STAT signaling and PRC2 function and expression of genes that mediate myeloid leukemia cell differentiation.
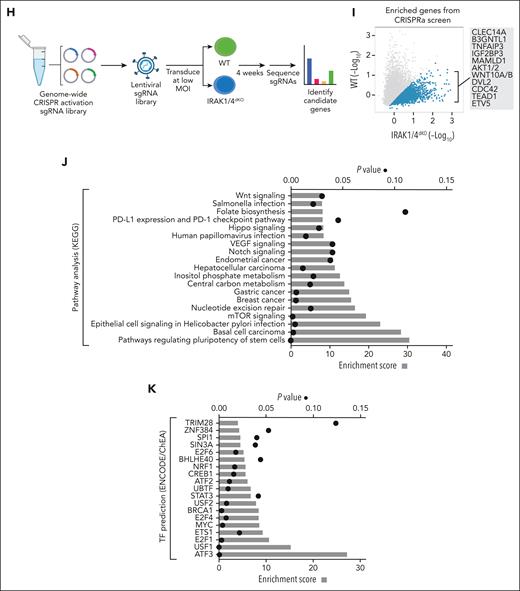
IRAK1/4 maintains undifferentiated leukemic cell states through chromatin and transcription factor networks. (A) Heatmap of chromatin accessibility (assay for transposase-accessible chromatin sequencing) peaks within a 3 kb distance of transcription start sites of genes in WT, IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 cells. (B) The total number of accessibility peaks lost and acquired in IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 relative to WT cells. (C) Venn diagrams of overlap genes that are associated with both differential expression (RNA sequencing) and concordant changes in chromatin accessibility (assay for transposase-accessible chromatin sequencing) in IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 cells. (D-E) Heatmaps of transcription factor enrichment among genes associated with the downregulation and loss of chromatin peaks (D) or the upregulation and acquisition of open chromatin peaks (E) in IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 cells relative to WT cells. Enrichment of transcription factor signatures was determined with the Chromatin Immunoprecipitation (ChIP) Enrichment Analysis (ChEA) 2022 library. Color intensity reflects the Log (P value) of the enrichment score. (F) Heatmap of differential gene expression in patients with AML (relative to healthy controls) using gene expression data curated from the Beat AML data set. The heatmap represents a subset of genes that are downregulated and associated with the loss of chromatin accessibility in IRAK1/4dKO THP1 (IRAK1/4 gene signature). Unsupervised hierarchical clustering analysis resolved distinct cohorts of IRAK1/4-high signature (Group 1) and IRAK1/4-low/intermediate signature (Groups 2 and 3) patients with AML. (G) Enrichment of AML-associated mutations in IRAK1/4-high signature (Group 1) and IRAK1/4-low/intermediate signature (Groups 2 and 3) Patients with AML (from panel F) based on hypergeometric testing. (H) Schematic diagram of the CRISPR activation screen. WT and IRAK1/4dKO THP1 cells were transduced with the pooled sgRNA library targeting more than 18 000 coding isoforms. After 3 weeks, deep sequencing was performed to identify candidate genes. (I) Average Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MAGeCK) score for candidate genes from WT and IRAK1/4dKO THP1 replicate samples. Blue circles represent genes selectively enriched in IRAK1/4dKO THP1 cells. (J) Most significant pathways (KEGG analysis) selectively enriched in IRAK1/4dKO THP1 cells among the top 438 candidate genes (based on fold change and P value). (K) Most significant transcription factors (ENCODE/ChEA analysis) selectively enriched in IRAK1/4dKO THP1 cells among the top 438 candidate genes.
IRAK1/4 maintains undifferentiated leukemic cell states through chromatin and transcription factor networks. (A) Heatmap of chromatin accessibility (assay for transposase-accessible chromatin sequencing) peaks within a 3 kb distance of transcription start sites of genes in WT, IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 cells. (B) The total number of accessibility peaks lost and acquired in IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 relative to WT cells. (C) Venn diagrams of overlap genes that are associated with both differential expression (RNA sequencing) and concordant changes in chromatin accessibility (assay for transposase-accessible chromatin sequencing) in IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 cells. (D-E) Heatmaps of transcription factor enrichment among genes associated with the downregulation and loss of chromatin peaks (D) or the upregulation and acquisition of open chromatin peaks (E) in IRAK1KO, IRAK4KO, and IRAK1/4dKO THP1 cells relative to WT cells. Enrichment of transcription factor signatures was determined with the Chromatin Immunoprecipitation (ChIP) Enrichment Analysis (ChEA) 2022 library. Color intensity reflects the Log (P value) of the enrichment score. (F) Heatmap of differential gene expression in patients with AML (relative to healthy controls) using gene expression data curated from the Beat AML data set. The heatmap represents a subset of genes that are downregulated and associated with the loss of chromatin accessibility in IRAK1/4dKO THP1 (IRAK1/4 gene signature). Unsupervised hierarchical clustering analysis resolved distinct cohorts of IRAK1/4-high signature (Group 1) and IRAK1/4-low/intermediate signature (Groups 2 and 3) patients with AML. (G) Enrichment of AML-associated mutations in IRAK1/4-high signature (Group 1) and IRAK1/4-low/intermediate signature (Groups 2 and 3) Patients with AML (from panel F) based on hypergeometric testing. (H) Schematic diagram of the CRISPR activation screen. WT and IRAK1/4dKO THP1 cells were transduced with the pooled sgRNA library targeting more than 18 000 coding isoforms. After 3 weeks, deep sequencing was performed to identify candidate genes. (I) Average Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MAGeCK) score for candidate genes from WT and IRAK1/4dKO THP1 replicate samples. Blue circles represent genes selectively enriched in IRAK1/4dKO THP1 cells. (J) Most significant pathways (KEGG analysis) selectively enriched in IRAK1/4dKO THP1 cells among the top 438 candidate genes (based on fold change and P value). (K) Most significant transcription factors (ENCODE/ChEA analysis) selectively enriched in IRAK1/4dKO THP1 cells among the top 438 candidate genes.
IRAK1/4 signaling programs define a subset of patients with MDS/AML
To establish whether IRAK1/4 signaling is operational in patients with AML, we performed an unsupervised hierarchical clustering analysis of RNA sequencing data from the BEAT AML trial.33 We used the set of genes that were downregulated and associated with the loss of chromatin accessibility in IRAK1/4dKO cells (supplemental Table 12). Patients with AML were clustered into 3 groups characterized by the expression of the IRAK1/4-associated genes (Figure 6F). Group 1 consisted of patients with elevated expression of the IRAK1/4-associated genes (IRAK1/4High), whereas Group 2 and 3 consisted of patients with decreased and/or variable expression of these genes (IRAK1/4Intermediate/Low) (Figure 6F). IRAK1/4High AML were enriched for mutations in BCOR, SRSF2, JAK2, TET2, RUNX1, and EZH2 (Figure 6G). These mutation profiles are consistent with altered chromatin assembly via the PRC1/2 complex and JAK-STAT signaling and are enriched in AML with myelodysplasia-related changes.34 Independent analysis of the IRAK1/4-associated genes in additional adult and pediatric AML and MDS cohorts also categorized the patients based on the enrichment of IRAK1/4-associated genes (supplemental Figure 6A-C). The IRAK1/4 signature correlated with myelomonocytic subtypes (M4 FAB) of adult (P = 3.28 × 10-5) and pediatric patients with AML (P = .013) and with an antecedent MDS (supplemental Figure 6D). These findings reveal that IRAK1/4 signaling programs are operational in MDS/AML and could define patients with a greater dependency on IRAK1/4.
Low IRAK1/4 levels can be restored by the reactivation of stem cell and leukemia-associated transcriptional programs
To gain an insight into the cellular processes that contribute to the attrition of LSPCs after IRAK1/4 inhibition, we performed a genome-scale CRISPR activation screen to identify genes that restore the growth ability of IRAK1/4-deficient AML cells. WT and IRAK1/4dKO THP1 cells were transduced with a CRISPR activation library consisting of single-guide RNA activating 18 000 coding isoforms (Figure 6H). Transduced cells were grown for 3 weeks and the single-guide RNA libraries were deep-sequenced. MAGeCK was then performed to identify candidate genes that were enriched in IRAK1/4dKO relative to WT cells (Figure 6I; supplemental Table 13). Several candidate genes enriched in IRAK1/4dKO cells have been shown to mediate leukemic activity, such as ETV5, TEAD1, CDC42, AKT1/2, and WNT10A.35-38 The pathway analysis on the candidate genes identified in IRAK1/4dKO cells revealed the enrichment of pathways implicated in stem cell activity and cancer-related pathways (Figure 6J). These overexpressed genes are predicted transcription targets of ATF3, USF1, E2F1, and MYC (Figure 6K), which are implicated in LSPCs. Moreover, we identified STAT3 target genes enriched in IRAK1/4dKO cells supporting our earlier observations that the loss of STAT signaling contributes to the impairment of IRAK1/4-deficient AML cells (Figure 6K). This analysis highlighted that IRAK1/4 inhibition can be undone by the reactivation of stem cell and leukemia-associated transcriptional programs.
A dual IRAK1/4 inhibitor suppresses MDS/AML LSPCs by promoting differentiation
The compensatory nature of IRAK1 and IRAK4 activation in MDS/AML necessitates concomitant inhibition to maximize therapeutic efficacy. Therefore, we sought an inhibitor that simultaneously targets IRAK1 and IRAK4. We used a 3-(pyridine-2-yl)imidazole[1,2-a]pyridine backbone with inhibitory activity against IRAK1 and IRAK4 as a chemical starting point for optimization.39,40 Structure-activity relationship exploration of this core scaffold yielded molecules that potently targeted IRAK1 and IRAK4. KME-2780 exhibited high affinity binding to and inhibition of both IRAK1 and IRAK4 (Figure 7A). We also identified a structurally similar derivative (KME-3859) with selectivity for IRAK4 but not IRAK1. The dual IRAK1/4 inhibitor was significantly more effective at suppressing TLR2-mediated activation of NF-κB and autophosphorylation of IRAK1 in AML cells as compared with the IRAK4 inhibitor (supplemental Figure 7A-B). Next, we compared the gene expression changes in AML cells treated with the dual IRAK1/4 inhibitor (KME-2780) and IRAK4 inhibitor (KME-3859). As expected, there were a common set of downregulated genes (IRAK4-dependent genes) after the treatment with both inhibitors (Figure 7B). These genes were enriched in MAPK/AP1, ATF4, IGFBP, and EGFR signaling (Figure 7C). However, the dual IRAK1/4 inhibitor also downregulated genes that remained expressed with KME-3859 (IRAK1-dependent genes). Consistent with our observation that IRAK1 mediates JAK-STAT signaling, the genes selectively suppressed by the dual IRAK1/4 inhibitor, but not the IRAK4 inhibitor, are enriched in interferon-related pathways (Figure 7C). We also examined whether the inhibitors suppressed the IRAK1/4 signature. KME-2780 suppressed most genes (∼65%) associated with the IRAK1/4 signature in the AML cells, whereas KME-3859 had an unremarkable effect on these genes (Figure 7B). As such, the inhibition of IRAK1 and IRAK4 results in the suppression of gene expression programs that are insufficiently inhibited upon targeting IRAK4 alone.
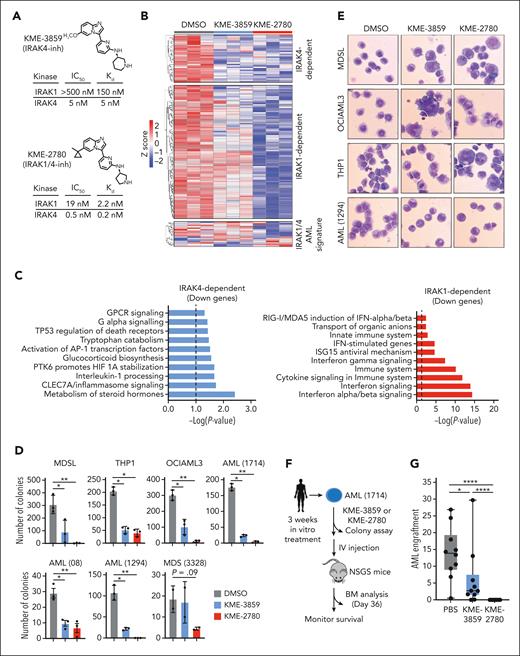
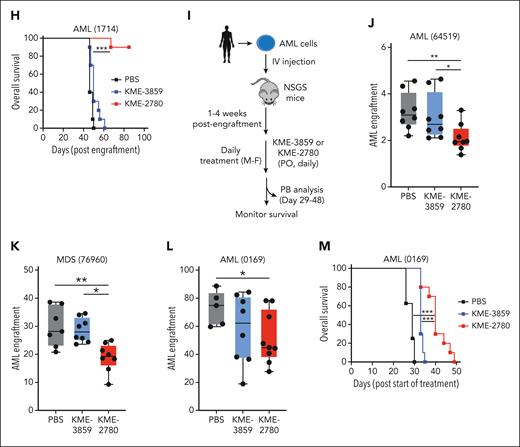
A dual IRAK1/4 inhibitor is more effective at suppressing MDS/AML as compared with a selective IRAK4 inhibitor. (A) Chemical structures of KME-3859 (IRAK4-inh) and KME-2780 (dual IRAK1/4-inh) with IC50 and Kd values for IRAK1 and IRAK4. (B) Heatmap of differentially expressed genes downregulated by both KME-3859 and KME-2780 (IRAK4-dependent), genes downregulated by KME-2780 (“IRAK1-dependent”), and genes representing the IRAK1/4 AML signature (from Figure 6F). (C) Pathway enrichment using KEGG of downregulated genes upon treatment with both KME-3859 and KME-2780 (IRAK4-dependent genes) as compared with vehicle control (top). Pathway enrichment using KEGG of downregulated genes upon the treatment with KME-2780 (IRAK1-dependent genes) as compared with KME-3859 and vehicle control (bottom). (D) Colony formation of MDSL (250 nM), THP1 (1 μM), OCIAML3 (1 μM), AML (1714) (1 μM), AML (1294) (1 μM), AML (08) (250 nM), and MDS (3328) (250 nM) cells treated with DMSO, KME-3859, or KME-2780. (E) Representative Wright-Giemsa stains of cells treated with vehicle (DMSO), KME-3859 (500 nM), or KME-2780 (500 nM) for 12 days (original magnification ×40). (F) Experimental overview: AML (1714) cells derived from patients were treated in vitro with vehicle (DMSO), KME-3859 (500 nM), or KME-2780 (500 nM) for 21 days. After the treatment, live cells were evaluated for colony formation in mice that received xenografts. (G) Bone marrow engraftment of AML (1714) cells in NSGS mice that received xenografts on day 36. Leukemic engraftment was determined as the percentage of huCD45+huCD33+ cells. (H) Kaplan-Meier survival analysis of NSGS mice (n = 10 mice per group) engrafted with AML (1714) cells pretreated with the indicated inhibitors. (I) Experimental overview: AML (64519), AML (0169), and MDS (76960) cells derived from patients were engrafted into NSGS mice. Two weeks after the engraftment, mice were randomly assigned to groups and treated orally daily with vehicle (PBS), KME-3859 (30 mg/kg), or KME-2780 (100 mg/kg). These concentrations were selected to equilibrate the free drug concentrations (supplemental Table 14). (J-L) Peripheral blood engraftment of AML (64519) (J), MDS (76960) (K), and AML (0169) (L) cells in NSGS mice that received xenografts on day 40, 48, and 29 after the treatment. Leukemic engraftment was determined as the percentage of huCD45+huCD33+ cells. (M) Kaplan-Meier survival analysis of NSGS mice (n = 8 mice/group) engrafted with AML (0169) cells and treated with the indicated inhibitors. Significance was determined with a Student t test (∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001). Error bars represent the standard error of the mean or standard deviation.
A dual IRAK1/4 inhibitor is more effective at suppressing MDS/AML as compared with a selective IRAK4 inhibitor. (A) Chemical structures of KME-3859 (IRAK4-inh) and KME-2780 (dual IRAK1/4-inh) with IC50 and Kd values for IRAK1 and IRAK4. (B) Heatmap of differentially expressed genes downregulated by both KME-3859 and KME-2780 (IRAK4-dependent), genes downregulated by KME-2780 (“IRAK1-dependent”), and genes representing the IRAK1/4 AML signature (from Figure 6F). (C) Pathway enrichment using KEGG of downregulated genes upon treatment with both KME-3859 and KME-2780 (IRAK4-dependent genes) as compared with vehicle control (top). Pathway enrichment using KEGG of downregulated genes upon the treatment with KME-2780 (IRAK1-dependent genes) as compared with KME-3859 and vehicle control (bottom). (D) Colony formation of MDSL (250 nM), THP1 (1 μM), OCIAML3 (1 μM), AML (1714) (1 μM), AML (1294) (1 μM), AML (08) (250 nM), and MDS (3328) (250 nM) cells treated with DMSO, KME-3859, or KME-2780. (E) Representative Wright-Giemsa stains of cells treated with vehicle (DMSO), KME-3859 (500 nM), or KME-2780 (500 nM) for 12 days (original magnification ×40). (F) Experimental overview: AML (1714) cells derived from patients were treated in vitro with vehicle (DMSO), KME-3859 (500 nM), or KME-2780 (500 nM) for 21 days. After the treatment, live cells were evaluated for colony formation in mice that received xenografts. (G) Bone marrow engraftment of AML (1714) cells in NSGS mice that received xenografts on day 36. Leukemic engraftment was determined as the percentage of huCD45+huCD33+ cells. (H) Kaplan-Meier survival analysis of NSGS mice (n = 10 mice per group) engrafted with AML (1714) cells pretreated with the indicated inhibitors. (I) Experimental overview: AML (64519), AML (0169), and MDS (76960) cells derived from patients were engrafted into NSGS mice. Two weeks after the engraftment, mice were randomly assigned to groups and treated orally daily with vehicle (PBS), KME-3859 (30 mg/kg), or KME-2780 (100 mg/kg). These concentrations were selected to equilibrate the free drug concentrations (supplemental Table 14). (J-L) Peripheral blood engraftment of AML (64519) (J), MDS (76960) (K), and AML (0169) (L) cells in NSGS mice that received xenografts on day 40, 48, and 29 after the treatment. Leukemic engraftment was determined as the percentage of huCD45+huCD33+ cells. (M) Kaplan-Meier survival analysis of NSGS mice (n = 8 mice/group) engrafted with AML (0169) cells and treated with the indicated inhibitors. Significance was determined with a Student t test (∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001). Error bars represent the standard error of the mean or standard deviation.
Next, we compared the IRAK4 inhibitors’ ability to suppress LSPCs. KME-3859 resulted in a ∼75% reduction in colony formation of MDS/AML cell lines and patient-derived samples (Figure 7D). However, KME-2780 resulted in complete suppression of LSPC colonies in nearly all samples tested (Figure 7D). At these concentrations, KME-2780 treatment of healthy CD34+ cells only modestly inhibited myeloid cell colonies yet increased erythroid colony formation (supplemental Figure 7C). Notably, KME-2780 suppressed OCI-AML3 cells, which were insensitive to the genetic inactivation of IRAK1 and IRAK4. This raises the possibility of off-target growth-inhibitory activity of these compounds in AML cells. KME-2780 was also more effective than KME-3859 at mediating the apoptosis of AML cells (supplemental Figure 7D). Sustained treatment of MDS/AML and patient-derived cell lines with KME-2780 and KME-3859 coincided with heterogeneous morphologies and increased cytoplasmic to nuclear ratios, suggestive of aberrant differentiation (Figure 7E). These morphological changes were more prominent in cells treated with KME-2780 relative to KME-3859. KME-2780 treatment also resulted in a greater expression of CD38, a glycoprotein expressed on mature immune cells, as compared with KME-3859 or vehicle (supplemental Figure 7E).
A dual IRAK1/4 inhibitor suppresses MDS/AML in xenografted mice
Impaired cellular differentiation is a hallmark of MDS/AML, and therapies that promote differentiation are curative in certain subtypes.41 To confirm that the differentiated state of the leukemic cells upon IRAK1/4 inhibition correlates with LSPC suppression in vivo, we first evaluated the leukemic potential of patient-derived AML cells that were exposed to the dual IRAK1/4 or selective IRAK4 inhibitors for 21 days in vitro (Figure 7F). The exposure of patient-derived AML cells to KME-2780 corresponded with diminished colony formation (supplemental Figure 7F) and loss of leukemic engraftment and AML development in vivo (Figure 7G-H). However, KME-3859 only partially suppressed LSPCs. These findings suggest that IRAK1/4 is important for preserving leukemia-propagating cells and that targeting IRAK1/4 can induce the differentiation of LSPCs and suppress AML.
To evaluate the therapeutic potential of targeting IRAK1/4, we also performed xenograft studies using patient-derived MDS/AML samples. Mice that were immunocompromised receivent engraftment with leukemic cells from patients with AML or MDS for 2 weeks and then treated with vehicle, KME-2780, or KME-3859 (Figure 7I). Leukemic cell engraftment in the peripheral blood was significantly reduced with KME-2780 treatment for AML(64519), MDS(76960), and AML(0169) (Figure 7J-K) as compared with vehicle-treated mice. In contrast, KME-3859 did not significantly suppress leukemic cell engraftment during the same time (Figure 7J-L). To determine whether the reduced leukemic cell engraftment affects the overall survival, we monitored mice that received engraftment with AML (0169). Mice treated with the vehicle or KME-3859 achieved a median survival of 29 and 33 days, respectively (Figure 7M). In contrast, mice treated with KME-2780 survived significantly longer (median of 40 days) (Figure 7M). These findings suggest that dual IRAK1 and IRAK4 inhibition more effectively suppresses AML as compared with targeting IRAK4 alone.
Discussion
MDS/AML LSPCs exhibit the dysregulation of immune and inflammatory pathways, many of which converge on IRAK1/4 signaling.6 IRAK1 and IRAK4 have been independently studied in MDS/AML17,18,39,42-46; however, their cooperative functions have not been investigated. Initial data from clinical trials evaluating IRAK4 inhibitors in HR-MDS/AML revealed encouraging responses in patients with activated IRAK4.23 These studies propose that IRAK4 is a relevant target in MDS/AML; however, they also suggest that IRAK4 inhibitors as monotherapy will likely be insufficient to yield durable responses. We confirmed that targeting IRAK4 can impair LSPCs but also elicits compensatory IRAK1 activation. Our study presents a model in which IRAK4 regulates epigenetic machinery and signaling pathways that mediate a differentiation block in leukemic cells, and the targeting of IRAK4 upregulates IRAK1 to activate complimentary pathways. Future studies are needed to determine whether the MyD88-independent functions of IRAK1 and IRAK4 can be extended to other pathologies and normal immunobiology. Functional complementation by proximal effectors and paralogs are reported mechanisms underlying therapy resistance in cancer.47 For example, TP53 can be inactivated by MDM2 or its paralog MDMX; accordingly, dual targeting of MDM2/MDMX is required for superior therapeutic efficacy in AML.48 Therapeutic efforts to target IRAKs have adopted paradigms formed by studies based on the models of acute receptor stimulation, wherein MyD88 recruits IRAK4 to activate IRAK1 and initiate signaling.49 In contrast, we showed that MyD88 is dispensable in MDS/AML and uncovered MyD88-independent IRAK1/4 functions necessary to preserve LSPCs. Proteomic studies identified interactions between IRAK4 and the PRC2 complex, an epigenetic regulator that is essential for LSPCs.28 These findings are further corroborated by the enrichment of BCOR and SRSF2 mutations in patients with AML with the IRAK1/4 signature. BCOR is part of the PRC1 complex, whereas SRSF2 mutations mis-splice EZH2.50,51 We found that IRAK1 interactions mapped to the mediators of JAK-STAT signaling, which could account for the induction of STAT signaling upon targeting IRAK4. A large cohort of adult and pediatric patients with AML and MDS exhibit an IRAK1/4 signature, which suggests that IRAK1/4 signaling is operational and could represent patients with a greater sensitivity to IRAK1/4 inhibitors. Because our findings formed a basis for dual IRAK1/4 targeted therapy, we revealed a therapeutic advantage for dual IRAK1/4 therapy in myeloid malignancies using a small molecule inhibitor. Although the IRAK1/4 inhibitor–induced differentiation of LSPCs and suppressed IRAK1/4-depended signaling more effectively than an IRAK4-selective inhibitor, we anticipate that combination therapies will be necessary to achieve maximal efficacy in the clinic.
Acknowledgments
The authors thank J. Bailey and V. Summey for their assistance with transplantations (Comprehensive Mouse and Cancer Core at Cincinnati Children’s Hospital Medical Center). The authors also thank the Viral Vector Core and DNA sequencing and Genotyping Core at Cincinnati Children’s Hospital Medical Center, and the University of Cincinnati Proteomics laboratory for their assistance; and H. L. Grimes, S. Pasare, L. Lee, and members of the Starczynowski laboratory for their helpful discussion and suggestions.
This work was supported, in part, by the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (U54DK126108), National Heart, Lung, and Blood Institute (NHLBI; R35HL135787), and National Cancer Institute (NCI; R01CA275007); Cincinnati Children’s Hospital Research Foundation; and CancerFree KIDS grants (D.T.S.). J.B. was supported by NIH/NCI T32 training grant (T32CA117846). J.Y. was supported by NIH/NHLBI F32 and American Cancer Society grants. M.W. was supported by NIH/NCI R50 grant (CA211404). E.V. was supported by an NIH/NCI Institutional Research Training Grant (T32CA236764). The mass spectrometry analysis was supported by an NIH instrumentation grant (S10OD026717) (K.D.G.).
Authorship
Contribution: J.B., C.I., J.Y., A.S., E.U., E.V., L.C.B., K.H., M.W., A.K., K.C., S.B.H., P.A., and K.D.G. performed research and analyzed data; J.B., S.B.H., A.V., K.D.G., J.R., C.J.T., and D.T.S. designed the research; and J.B. and D.T.S. wrote the paper.
Conflict-of-interest disclosure: D.T.S. serves on the scientific advisory board at Kurome Therapeutics; is a consultant for and/or received funding from Kurome Therapeutics, Captor Therapeutics, Treeline Biosciences, and Tolero Therapeutics; and has equity in Kurome Therapeutics. L.C.B. consulted for Kurome Therapeutics. J.R. is employed by, and holds equity in, Kurome Therapeutics; holds equity in Airway Therapeutics; and is a consultant for Radius Health and MoglingBio. A.K. is employed by, and holds equity in, Kurome Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Daniel T. Starczynowski, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; e-mail: daniel.starczynowski@cchmc.org.
References
Author notes
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession numbers GSE114922 and GSE58831).
Data that support the findings of this study are available on request from the corresponding author, Daniel T. Starczynowski (daniel.starczynowski@cchmc.org).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal