Key Points
PF4/H-binding antibodies were cloned from patients with HIT, a subset of which were platelet activating and presumptively pathogenic.
Specific structural motifs appear to be responsible for platelet-activating properties of “pathogenic” clones.
Abstract
Heparin-induced thrombocytopenia (HIT) is a serious adverse drug reaction characterized by antibodies that recognize platelet factor 4/heparin complexes (PF4/H) and activate platelets to create a prothrombotic state. Although a high percentage of heparin-treated patients produce antibodies to PF4/H, only a subset also makes antibodies that are platelet activating (PA). A close correlation between PA antibodies and the likelihood of experiencing HIT has been demonstrated in clinical studies, but how PA (presumptively pathogenic) and nonactivating (NA) (presumptively benign) antibodies differ from each other at the molecular level is unknown. To address this issue, we cloned 7 PA and 47 NA PF4/H-binding antibodies from 6 patients with HIT and characterized their structural and functional properties. Findings showed that PA clones differed significantly from NA clones in possessing 1 of 2 heavy chain complementarity-determining region 3 (HCDR3) motifs, RX1-2R/KX1-2R/H (RKH) and YYYYY (Y5), in an unusually long complementarity-determining region 3 (≥20 residues). Mutagenic studies showed that modification of either motif in PA clones reduced or abolished their PA activity and that appropriate amino acid substitutions in HCDR3 of NA clones can cause them to become PA. Repertoire sequencing showed that the frequency of peripheral blood IgG+ B cells possessing RKH or Y5 was significantly higher in patients with HIT than in patients without HIT given heparin, indicating expansion of B cells possessing RKH or Y5 in HIT. These findings imply that antibodies possessing RKH or Y5 are relevant to HIT pathogenesis and suggest new approaches to diagnosis and treatment of this condition.
Introduction
Despite being recognized >60 years ago, heparin-induced thrombocytopenia (HIT) remains a major cause of morbidity and mortality in patients treated with heparin, affecting >20 000 patients annually in the United States.1 Although much progress has been made, understanding of HIT pathogenesis is incomplete.2-5 A hallmark of the disorder is the presence of immunoglobulin G (IgG) antibodies specific for conformational changes induced in the platelet-specific chemokine platelet factor 4 (PF4) when it binds to heparin or certain other anionic substances.4,6 However, only a subset of patients who test positive in solid phase assays designed to detect such antibodies experience clinical manifestations of HIT,7-9 the most important of which is a strong predilection for thrombosis. Alternative assays that detect antibodies capable of inducing platelet activation under specialized test conditions produce results correlating much more closely with the clinical condition, HIT.6,8,10 For reasons of convenience, platelets are routinely used to detect such “pathogenic” antibodies, but there is evidence that, in a patient, they also act on other target cells, such as endothelium, monocytes, and leukocytes, to produce the prothrombotic state typical of HIT.2,11-13
Characterization of the molecular properties that distinguish “pathogenic” heparin-induced antibodies from more common but seemingly benign ones that recognize PF4/heparin (PF4/H) complexes but fail to activate platelets would advance understanding of HIT pathogenesis, but achievement of this goal has been frustrated by limitations of conventional serologic methods. Recent advances have enabled cloning of human B cells and production of recombinant human antibodies from patients with immunologic disorders.14,15 In this report, we describe cloning of PF4/H-binding antibodies from patients with HIT, generation of recombinant IgG antibodies from these clones, and characterization of their functional and molecular properties. Findings made provide evidence that certain sequence motifs in heavy chain complementarity-determining region 3 (HCDR3) distinguish “pathogenic” (platelet-activating) antibodies from nonactivating, presumptively nonpathogenic ones and suggest possible new approaches to diagnosis and treatment of HIT.
Materials and methods
Study subjects
Patients with HIT were identified at local hospitals from clinical findings (“4Ts” score ≥ 4)16 and positive test results in both a solid phase PF4/polyvinyl sulfonate enzyme-linked immunosorbent assay (ELISA) (Lifecodes PF4 IgG; Immunocor) and the serotonin release assay (SRA).17 Additional details concerning 8 patients, designated HIT1 through HIT8, whose peripheral blood B cells were used in cloning studies are provided in supplemental Table 1, available on the Blood website. Patients HIT118 and HIT7 had “autoimmune HIT” unrelated to heparin exposure.19 Buffy coat preparations and platelets from deidentified healthy blood donors were obtained through the Versiti Biospecimen Service. The studies were approved by the Institutional Review Boards of the Medical College of Wisconsin and the Froedtert Hospital. All participating patients gave written informed consent.
Isolation and storage of peripheral blood mononuclear cells
Peripheral blood mononuclear cells were isolated from citrated whole blood using Fico-Light-LymphoH (Atlanta Biologicals; I40150). Collected peripheral blood mononuclear cells were washed, centrifuged, and resuspended in freezing medium (90% fetal bovine serum + 10% dimethyl sulfoxide), frozen at −80 °C in a cell freezing container overnight, and transferred to liquid nitrogen the next day. The frozen cells were stored in liquid nitrogen from 1 month to 4 years before being thawed for cloning or studies of B-cell repertoires.
Cloning of peripheral blood B cells
Methods are described in supplemental Materials and methods.
PF4/H ELISA
Antibodies reactive with PF4/H complexes were detected using an ELISA, as previously described.20-22 PF4/H complexes were produced by incubating 10 μg/mL recombinant PF4 (Protein Foundry Inc, Milwaukee, WI) with 0.4 U/mL unfractionated heparin (Sigma-Aldrich; H3393) and applying 50 μL of this mixture to each well of the microtiter plates.
P-selectin expression assay
The assay has been described previously.20,21 Briefly, 106 washed platelets pooled from 3 to 5 normal group O donors were incubated at room temperature (RT) for 20 minutes with 150 μg/mL recombinant PF4 in a total volume of 10 μL. Patient serum (10 μL) + 30 μL phosphate-buffered saline (or 40 μL cultured B-cell supernatant) were then added, and the mixture was incubated for 1 hour at RT without agitation. Phycoerythrin–anti–P-selectin (Invitrogen; 12-0626-82) and Alexa647–anti-glycoprotein (GP) IIb (clone MBC290.5; Versiti Blood Research Institute) were then added, and the mixture was incubated for an additional 20 minutes at RT. Fluorescence-activated cell sorting analysis was then performed, in which P-selectin expression was measured on GPIIb+ cells. Controls included wells containing (1) no PF4, (2) 10 μg/mL IV.3 (FcγRIIa-blocking monoclonal antibody),20 or (3) 100 U/mL unfractionated heparin. Maximum P-selectin expression (100%) was determined by treating platelets with thrombin receptor-activating peptide, 5 μg/mL. Mean fluorescence intensity (MFI) obtained with test samples was subtracted from the MFI obtained with bovine serum albumin controls, and the net signal was expressed as a percentage of the MFI obtained with thrombin receptor-activating peptide.
Antibody binding to PF4-treated platelets
Conditions were the same as for the P-selectin expression assay (PEA), except that following treatment of washed platelets with PF4 and then with recombinant antibody, Alexa647–anti-GPIIb (clone MBC290.5; Blood Research Institute) and fluorescein isothiocyanate–Fab anti-human IgG (Jackson ImmunoResearch; 109-097-003) were added. The mixture was then incubated for an additional 20 minutes and subjected to fluorescence-activated cell sorting analysis, in which fluorescein isothiocyanate–Fab binding to cells gated on GPIIb was measured.
Results
Clonal antibodies from patients with HIT recognize PF4/H complexes, and a subset is platelet activating
In initial studies, a single-cell polymerase chain reaction (PCR) and expression cloning method15 was used to screen 376 single PF4/H-binding, IgG1+ B cells from patient HIT118 (supplemental Figure 1A). Only 1 PF4/H-binding clone (HIT1P3D4) was identified. To improve clonal yield, all 8 patients (HIT1-HIT8; supplemental Table 1) were then studied using a single B-cell culture approach14 (supplemental Figure 1B). Of 5655 B cells subjected to culture, a total of 1944, representing 12% to 48% of cells from individual patients, were expandable (supplemental Table 2). Screening of culture supernatants from these wells led to identification of 53 PF4/H-binding clones from 5 patients with “classic” HIT (HIT2 to HIT6) and 1 patient with “spontaneous” HIT (HIT1)18 (supplemental Figure 2) that constituted 2.7% of all IgG1+ B cells successfully expanded (supplemental Table 2). When the 53 culture supernatants were screened for their ability to induce P-selectin expression in platelets pretreated with PF4 (PEA test), 6 were platelet activating: 5 from patients with “classic” HIT (HIT2 to HIT5) and 1 from a patient with “spontaneous” HIT (HIT1) (supplemental Figure 3). The single PF4/H-binding antibody obtained from patient HIT1 using the single-cell PCR method was also platelet activating (data not shown), making a total of 7 such antibodies. We will subsequently refer to antibodies that bind to PF4/H and activate PF4-treated platelets as “PA,” those that bind to PF4/H but do not activate as “NA,” and those that are nonbinding as “NB.”
Recombinant antibodies produced from PA, NA, and NB clones mirror the behavior of antibodies in patient serum and culture supernatants
Varible regions of the heavy (VH) and light (VL) chains of cultured B cells producing PA, NA, and NB antibodies were amplified by reverse transcription–PCR and sequenced. Unexpectedly, it was found that 100% of 54 PF4/H-binding clones possessed κ light chains, in contrast to 104 NB clones, among which the κ/λ ratio was 61/43. Each of the PF4/H-binding clones and 40 randomly selected κ-chain–bearing NB clones were cloned into expression vectors (IgG1 isotype) and resequenced, and some were expressed in HEK293T cells. Recombinant antibodies derived from B cells producing PA and NA antibodies reacted with PF4/H complexes in ELISA and were variably inhibited by high-dose heparin (Figure 1A), mimicking the behavior of antibodies in serum from the same patients. However, only antibodies cloned from PA cultures activated PF4-treated platelets in reactions that were inhibited by high-dose heparin and the FcγRIIa-specific monoclonal IV.3 (Figure 1B). It has been shown previously that only HIT antibodies capable of activating platelets in the PEA and SRA bind physically to platelets pretreated with PF4.20Figure 1C shows that PA, but not NA or NB, antibodies replicated this behavior, although reactions of PA and NA antibodies with PF4/H did not differ significantly (Figure 1D).
Reactions of the recombinant PA, NA, and NB clonal antibodies. (A) PA and NA but not NB clonal antibodies recognize PF4/H and are inhibited by high-dose heparin (HD Hep). Quantity of antibodies used was 100 μg/mL, except for clones HIT1P4G8, HIT3P1A10, and HIT3P7D2, for which 200 μg/mL was used. Positive controls were plasma from patients HIT2 through HIT5. For convenience, only reactions of one NB clone are shown, but all other NB clones behaved comparably. (B) PA, but not NA or NB, clones (80 μg/mL) activate PF4-treated platelets (P-selectin expression) in reactions that were inhibited by the FcγRIIa-specific monoclonal antibody IV.3 and by HD Hep. Positive controls were HIT plasma from patients HIT2 and HIT3 and thrombin receptor-activating peptide (TRAP). Negative reactions shown for 3 NA and 2 NB clones are typical of all other clones in these categories. (C) PA, but not NA or NB, clones (80 μg/mL) bind physically to PF4-coated, but not uncoated, platelets in reactions that are inhibited by HD Hep. Negative reactions shown for 3 NA and 2 NB clones are typical of all other clones in these categories. (D) Seven PA and 22 NA clones did not differ significantly in their reactions against immobilized PF4/H. Antibodies were diluted threefold serially to produce final concentrations ranging from 300 to 0.14 μg/mL and were tested in PF4/H ELISA. Antibody binding strength (ordinate) was plotted as the area under the curve (AUC) in arbitrary units. Data shown in A through C are representative of at least 4 independent studies. Error bars represent mean ± SD of 2 replicates within each experiment. BSA, bovine serum albumin; N plasma 1, normal plasma 1; ns, not significant.
Reactions of the recombinant PA, NA, and NB clonal antibodies. (A) PA and NA but not NB clonal antibodies recognize PF4/H and are inhibited by high-dose heparin (HD Hep). Quantity of antibodies used was 100 μg/mL, except for clones HIT1P4G8, HIT3P1A10, and HIT3P7D2, for which 200 μg/mL was used. Positive controls were plasma from patients HIT2 through HIT5. For convenience, only reactions of one NB clone are shown, but all other NB clones behaved comparably. (B) PA, but not NA or NB, clones (80 μg/mL) activate PF4-treated platelets (P-selectin expression) in reactions that were inhibited by the FcγRIIa-specific monoclonal antibody IV.3 and by HD Hep. Positive controls were HIT plasma from patients HIT2 and HIT3 and thrombin receptor-activating peptide (TRAP). Negative reactions shown for 3 NA and 2 NB clones are typical of all other clones in these categories. (C) PA, but not NA or NB, clones (80 μg/mL) bind physically to PF4-coated, but not uncoated, platelets in reactions that are inhibited by HD Hep. Negative reactions shown for 3 NA and 2 NB clones are typical of all other clones in these categories. (D) Seven PA and 22 NA clones did not differ significantly in their reactions against immobilized PF4/H. Antibodies were diluted threefold serially to produce final concentrations ranging from 300 to 0.14 μg/mL and were tested in PF4/H ELISA. Antibody binding strength (ordinate) was plotted as the area under the curve (AUC) in arbitrary units. Data shown in A through C are representative of at least 4 independent studies. Error bars represent mean ± SD of 2 replicates within each experiment. BSA, bovine serum albumin; N plasma 1, normal plasma 1; ns, not significant.
Structural differences were identified among PA, NA, and NB clones
As noted, PA and NA clones differed from NB in possessing only κ light chains (Figure 2A). A comparison of V(D)J use by VH and Vκ of 7 PA, 47 NA, and 40 NB clones possessing κ-chains showed that (1) PA and NA clones used Vκ1 more often and Vκ3 less often than NB clones (P < .05) (Figure 2B); (2) VH1 use was greater in NA clones than in NB clones (P < .05) (Figure 2C); and (3) JH6 use differed markedly in the 3 types of clones, in the order of PA > NA >> NB (P < .05 and P < .0001, respectively) (Figure 2C). Increased JH6 use by PA and NA clones led to significantly increased prevalence of VH1-JH6, VH3-JH6, DH3-JH6, DH6-JH6, and VH3-DH3-JH6 combinations relative to the NB clones (Figure 2D). Exclusive use of κ-chain and differential use of certain Vκ genes by the PF4/H-binding clones relative to NB clones suggests that κ-chain is important for binding to PF4/H. Hierarchical use of JH6 suggests this is also true of structural features embedded within JH6.
κ chain and variable gene use by PA, NA, and NB clones. (A) PA and NA clones used κ chains only. (B) Vκ1 was used preferentially by PA clones. (C) There was a striking preference for use of JH6 by PA and NA clones relative to NB clones. (D) Percentage of clones using various heavy chain VDJ gene combinations. Data shown in B to D are based on sequences of 7 PA, 47 NA, and 40 NB clones. P values were calculated by Fisher exact test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. mAb, monoclonal antibody.
κ chain and variable gene use by PA, NA, and NB clones. (A) PA and NA clones used κ chains only. (B) Vκ1 was used preferentially by PA clones. (C) There was a striking preference for use of JH6 by PA and NA clones relative to NB clones. (D) Percentage of clones using various heavy chain VDJ gene combinations. Data shown in B to D are based on sequences of 7 PA, 47 NA, and 40 NB clones. P values were calculated by Fisher exact test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. mAb, monoclonal antibody.
Certain HCDR3 motifs appear to distinguish PA from NA and NB clones
Analysis of HCDR3 in various clones showed that average HCDR3 length of the PA clones was ≥20 amino acids (aa), significantly greater than that expected in randomly selected human IgG clones (average, 15.1 ± 3.5)23 (P < .0001) and that HCDR3 length differed significantly in the 3 types of clones in the order PA > NA > NB (P < .001 and P < .05, respectively) (Figure 3A). Numbers of basic amino acids (Figure 3B) and tyrosine (Tyr) residues (Figure 3C) in the HCDR3 were also different, both in the order of PA > NA > NB. Further analysis showed that HCDR3 of 5 PA clones (from HIT1-HIT3) contained N-terminal RX1-2R/KX1-2R/H sequences, and the other 2 PA clones (from HIT1 and HIT5) had a C-terminal YYYYY (Figure 3D). The N-terminal RX1-2R/KX1-2R/H motif in a long HCDR3 (≥20 aa) are herein designated “RKH,” and the C-terminal YYYYY in a long HCDR3 (≥20 aa) are designated “Y5.” The RKH motif was found in only 2 of 47 NA clones (supplemental Table 3) and was completely absent from 40 NB clones (P < .001). No examples of the Y5 motif were found among either NA (supplemental Table 3) or NB clones (P < .05). Prevalence of RKH and Y5 motifs in PA, NA, and NB clones is depicted in Figure 3E.
Certain structural features differentiate PA, NA, and NB antibodies. Number of total amino acid residues (A), basic residues (B), and Tyr residues (C) in HCDR3 of 7 PA, 47 NA, and 40 NB clones. (D) HCDR3 sequences of PA clones. RX1-2R/KX1-2R/H (RKH) and YYYYY (Y5) motifs are highlighted in red. (E) Prevalence of clones possessing the RKH and Y5 motifs among PA, NA, and NB antibodies. P values were calculated by the unpaired 2-tailed Student t-test (A-C) and by Fisher exact test (E). Error bars indicate mean ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
Certain structural features differentiate PA, NA, and NB antibodies. Number of total amino acid residues (A), basic residues (B), and Tyr residues (C) in HCDR3 of 7 PA, 47 NA, and 40 NB clones. (D) HCDR3 sequences of PA clones. RX1-2R/KX1-2R/H (RKH) and YYYYY (Y5) motifs are highlighted in red. (E) Prevalence of clones possessing the RKH and Y5 motifs among PA, NA, and NB antibodies. P values were calculated by the unpaired 2-tailed Student t-test (A-C) and by Fisher exact test (E). Error bars indicate mean ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
Functions of PA and NA clones could be modified by HCDR3 mutations
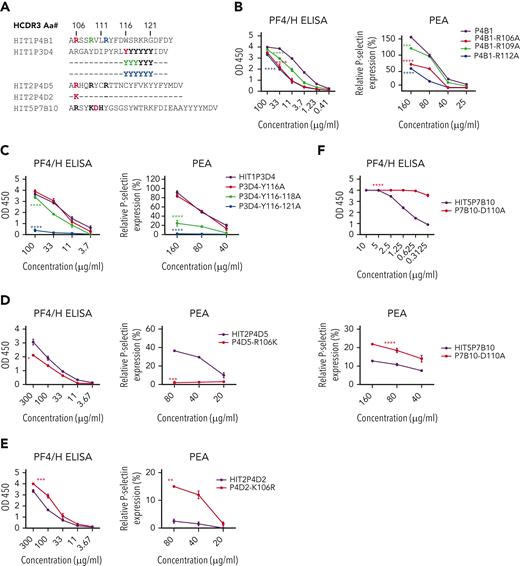
HCDR3 sequences of clones selected for mutation are shown in Figure 4A and supplemental Figure 4A. When alanine (Ala) was substituted for basic amino acid residues at positions 106, 109, and 112 in the RKH motif of PA clone HIT1P4B1, both PF4/H-binding and platelet-activating ability were significantly reduced (P < .001 or P < .0001), particularly when aa 112 was modified (Figure 4B). Comparable effects were seen when basic (RKH) residues of PA clones HIT3P7F6 and HIT3P7H2 (supplemental Figure 4C-D) were similarly modified. In contrast, with PA clone HIT3P1A11, the greatest loss of function occurred when the first basic residue (106) was changed (supplemental Figure 4B). The PA clone HIT1P3D4 contains 6 tyrosine residues in its HCDR3 at positions 116 to 121 (Figure 4A). An Ala substitution at position 116 was without effect. However, substitution at positions 116 to 118 significantly reduced PF4/H binding and platelet activation (P < .0001), and substitutions at all 6 positions (116-121) totally abolished both antibody functions (Figure 4C).
Mutagenesis studies imply a role for RKH and Y5motifs in platelet activation by PA clones. (A) HCDR3 sequences of clones studied. Critical amino acids in RKH and Y5 motifs are bolded. (B) Ala substitution for basic amino acids at positions 106, 109, and 112 of the RKH motif in PA clone HIT1P4B1 impaired its ability to bind PF4/H and to activate PF4-treated platelets. (C) Ala substitutions at positions 116 to 118 and 116 to 121 of the Y5 motif of PA clone HIT1P3D4 impaired its ability to bind to PF4/H and to activate PF4-treated platelets. (D) An R106K substitution in PA clone HIT2P4D5 abolished its ability to activate PF4-treated platelets but had only minimal effect on PF4/H binding. (E) A K106R substitution in NA clone HIT2P4D2 conferred platelet-activating function on the clone, while preserving PF4/H binding. (F) A D110A substitution in NA clone HIT5P7B10 significantly enhanced its platelet-activating ability and markedly increased its avidity for PF4/H. Concentrations of clonal IgG used in each reaction mixture are shown beneath B through F. Data shown are representative of 3 independent experiments for each clone and its mutant. Error bars represent mean ± SD of 2 replicates within each experiment. P values were calculated by 2-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
Mutagenesis studies imply a role for RKH and Y5motifs in platelet activation by PA clones. (A) HCDR3 sequences of clones studied. Critical amino acids in RKH and Y5 motifs are bolded. (B) Ala substitution for basic amino acids at positions 106, 109, and 112 of the RKH motif in PA clone HIT1P4B1 impaired its ability to bind PF4/H and to activate PF4-treated platelets. (C) Ala substitutions at positions 116 to 118 and 116 to 121 of the Y5 motif of PA clone HIT1P3D4 impaired its ability to bind to PF4/H and to activate PF4-treated platelets. (D) An R106K substitution in PA clone HIT2P4D5 abolished its ability to activate PF4-treated platelets but had only minimal effect on PF4/H binding. (E) A K106R substitution in NA clone HIT2P4D2 conferred platelet-activating function on the clone, while preserving PF4/H binding. (F) A D110A substitution in NA clone HIT5P7B10 significantly enhanced its platelet-activating ability and markedly increased its avidity for PF4/H. Concentrations of clonal IgG used in each reaction mixture are shown beneath B through F. Data shown are representative of 3 independent experiments for each clone and its mutant. Error bars represent mean ± SD of 2 replicates within each experiment. P values were calculated by 2-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
We next tested whether PA and NA clones could be interconverted by selectively mutating their HCDR3 domains. For this purpose, we used PA clone HIT2P4D5 and NA clone HIT2P4D2, which differed by 1 aa in the HCDR3, where they possess R and K, respectively, at position 106 and 3 additional amino acids outside HCDR3 (Figure 4A). As shown in Figure 4D, an R106K mutation in PA clone HIT2P4D5 abolished its platelet-activating ability but affected binding to PF4/H only slightly, creating, in effect, an NA antibody. In contrast, the reciprocal mutation in NA clone HIT2P4D2 caused it to become platelet activating and slightly enhanced its avidity for PF4/H, thus converting it to PA (Figure 4E). We then examined 2 other NA antibodies, HIT5P7B10 and HIT6P8C7, which possess the RKH motif (supplemental Table 3) but were unique in failing to activate platelets. Each of these clones had an aspartic acid (D) embedded in the RKH motif of HCDR3 (shown in Figure 4A for HIT5P7B10). We considered whether the acidic D residue might disrupt antibody function because of its proximity to the 3 basic residues of RKH motif and introduced D to alanine (A) mutations in both clones. Only clone HIT5P7B10 was successfully expressed, but the mutation caused it to acquire platelet-activating ability and, interestingly, enhanced its avidity for PF4/H by at least an order of magnitude (Figure 4F).
Further work is required, but these preliminary findings strongly suggest that the RKH and Y5 motifs found in the 7 PA clones identified thus far are critical for PF4-dependent activation of platelets by these antibodies—a property that appears to correlate with antibody pathogenicity in patients with HIT.24
B cells possessing a heavy chain RKH or Y5 motif are present in healthy individuals but are more common in patients experiencing HIT
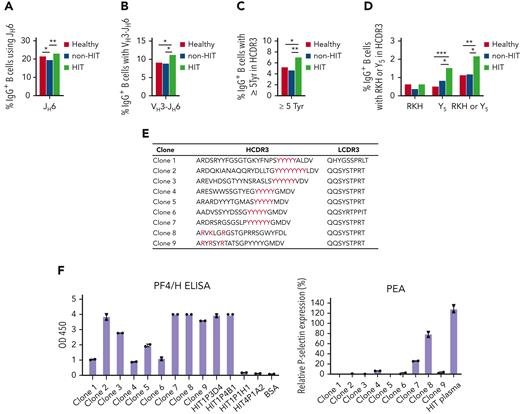
If the RKH or Y5 HCDR3 motifs and other structural properties identified in PA clones contribute to HIT pathogenicity, B cells expressing antibodies with such features might be expanded in blood of patients experiencing HIT. To investigate this possibility, we performed VH repertoire sequencing of peripheral blood IgG+ B cells from (1) 12 patients with HIT (HIT1-HIT7 [from whom B cells were cloned], HIT9-HIT11, and HIT13-HIT14) (supplemental Table 1), (2) 7 patients suspected of having HIT who tested positive in the PF4/H ELISA but were found not to have this condition from clinical findings and negative SRA and/or PEA test results (non–HIT1-HIT7; supplemental Table 4), and (3) 9 healthy subjects using a unique molecular identifier-based full-length VH sequencing protocol.25 Because of limited volume of blood available from each patient with HIT and patients without HIT, the number of IgG+ B cells in individual samples was low. We, therefore, pooled repertoire sequencing data within each of the 3 groups for the purpose of achieving statistical significance in the analysis (supplemental Material). Findings made showed that the frequency of IgG+ B-cell clones that (1) used the JH6 gene or a VH3-JH6 gene combination (Figure 5A-B), (2) possessed ≥5 Tyr residues in the HCDR3 (Figure 5C), or (3) possessed an HCDR3 containing an RKH or Y5 motif (Figure 5D) was significantly greater in confirmed patients with HIT than in healthy subjects or in heparin-treated patients found not to have HIT.
Analysis of peripheral blood IgG+B-cell repertoires in patients with HIT (HIT), patients given heparin without experiencing HIT (non-HIT), and healthy persons. (A) Percentage of IgG+ B cells using the JH6 gene in the 3 populations. (B) Percentage of IgG+ B cells using a VH3-JH6 combination. (C) Percentage of IgG+ B cells possessing at least 5 Tyr residues in HCDR3. (D) Percentage of IgG+ B cells possessing an RKH or Y5 HCDR3 motif. Sequences identified in the 3 groups were pooled for analysis. (E) HCDR3 sequences containing RKH or Y5 identified in repertoire sequencing of peripheral blood IgG+ B cells from patients with HIT that were expressed as recombinant IgG antibodies in a pairing with κ chains containing the indicated κ chain CDR3s selected, as described in text. (F) Functional behavior of 9 clones containing HCDR3 and LCDR3 sequences, shown in Figure 5E. Left: Each of 9 clones reacted strongly with PF4/H in ELISA. Right: Two clones, 1 with RKH and 1 with the Y5 motif, also activated PF4-treated platelets. P values were calculated by Fisher exact test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
Analysis of peripheral blood IgG+B-cell repertoires in patients with HIT (HIT), patients given heparin without experiencing HIT (non-HIT), and healthy persons. (A) Percentage of IgG+ B cells using the JH6 gene in the 3 populations. (B) Percentage of IgG+ B cells using a VH3-JH6 combination. (C) Percentage of IgG+ B cells possessing at least 5 Tyr residues in HCDR3. (D) Percentage of IgG+ B cells possessing an RKH or Y5 HCDR3 motif. Sequences identified in the 3 groups were pooled for analysis. (E) HCDR3 sequences containing RKH or Y5 identified in repertoire sequencing of peripheral blood IgG+ B cells from patients with HIT that were expressed as recombinant IgG antibodies in a pairing with κ chains containing the indicated κ chain CDR3s selected, as described in text. (F) Functional behavior of 9 clones containing HCDR3 and LCDR3 sequences, shown in Figure 5E. Left: Each of 9 clones reacted strongly with PF4/H in ELISA. Right: Two clones, 1 with RKH and 1 with the Y5 motif, also activated PF4-treated platelets. P values were calculated by Fisher exact test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
We then examined whether VH chains found in the repertoire sequencing studies to contain an RKH or Y5 motif in complementarity-determining region (CDR) 3 would behave functionally like a PA clone if paired with a suitable κ chain. For this purpose, 9 VH sequences containing an RKH or Y5 motif were arbitrarily selected from the HIT repertoire and were paired with a Vκ from a PA or NA clone known (supplemental Table 3) to have used the same H-chain VDJ gene combination as these VH sequences. The paired VH and Vκ (HCDR3 and light-chain CDR3 (LCDR3) sequences shown in Figure 5E) were expressed as recombinant IgG1 antibodies and tested for PF4/H binding and PEA activity. Each of 9 antibodies reacted strongly in the PF4/H ELISA, and 2 (1 with RKH and 1 with the Y5 HCDR3 motif) were also positive in the PEA (Figure 5F). The findings provide evidence that B cells found in repertoire sequencing studies to possess an RKH or Y5 motif in HCDR3 are PF4/H specific and that at least a subset of these can produce antibodies capable of PF4-dependent platelet activation.
Discussion
In these studies, we successfully cloned 1986 IgG1+ B cells from 6 patients with HIT, 54 of which produced antibodies that recognized PF4/H complexes. Seven of these (from patients HIT1-HIT3 and HIT5) also induced P-selectin expression in PF4-treated platelets using the PEA. Total yield was 7 antibodies that recognized PF4/H and activated platelets (PA), 47 that recognized PF4/H but did not activate platelets (NA), and 1932 that were nonreactive with PF4/H (NB). When VH and VL regions of PA, NA, and randomly selected NB clones were amplified and sequenced, it was unexpectedly found that PA and NA clones used only κ light chains, in contrast to 104 NB clones in which the κ/λ ratio was about 1.4, close to the expected ratio. Recombinant IgG1 monoclonal antibodies derived from the 7 PA and randomly selected NA and κ-chain–bearing NB clones faithfully replicated the behavior of antibodies in supernatants of B-cell cultures from which they originated.
Studies to identify structural features that differentiate PA and NA antibodies from each other and from NB antibodies showed that, relative to NB clones, PA and NA clones preferentially used Vκ1, VH1 (PA > NA > NB), and JH6 (PA > NA >> NB). Other significant differences were greater HCDR3 lengths and larger numbers of basic amino acid and Tyr residues in HCDR3 in the order PA > NA > NB. In this analysis, 2 HCDR3 motifs designated “RKH” (5 clones) and “Y5” (2 clones) were identified among the 7 PA clones that were of particular interest because RKH was found in only 2 of 47 NA clones and none of 40 NB clones, and Y5 was completely absent from both NA and NB clones.
To define whether the RKH and Y5 motifs were necessary for the PA phenotype, the effects of selected mutations on antibody function were examined. Initially, we found that substituting Ala for R residues at positions 106, 109, and 112 in the PA clone HIT1P4B1 significantly reduced reactivity in PF4/H ELISA and the PEA in the order 112 > 106 > 109. Similar mutations had comparable effects on 3 other RKH-containing PA clones with minor differences. In the Y5-containing clone HIT1P3D4, replacement of all Ys with Ala completely abolished PF4/H binding and PEA activity, whereas replacement of only Y116-118 affected mainly the PEA reaction. The importance of the RKH motif was further examined by determining whether certain mutations could convert an NA clone to PA and vice versa. NA clone HIT2P4D2 and PA clone HIT2P4D5 differed only by possessing K and R, respectively, at position 106 in HCDR3 and 3 other residues outside HCDR3. Introducing a K-to-R mutation at position 106 caused NA clone HIT2P4D2 to acquire a PA phenotype, and a reciprocal mutation in PA clone HIT2P4D5 caused it to revert to NA, demonstrating interconversion of the PA and NA phenotypes. Two of 47 NA clones (HIT5P7B10 and HIT6P8C7) that possessed the RKH HCDR3 motif (supplemental Table 3) were the only examples of RKH-bearing clones that failed to activate PF4-treated platelets. Both clones had an acidic D residue within the basic RKH motif that could have interfered with their function. When Ala was substituted for D in clone HIT5P7B10, it acquired PA function and, remarkably, its avidity for PF4/H was increased about 10-fold. These preliminary mutagenic studies provide evidence that RKH and Y5 motifs identified in 7 PA clones are essential for them to recognize ≥1 epitopes on platelet-associated PF4 and activate platelets—a function that correlates with antibody pathogenicity in patients with HIT.21,26
VH repertoire sequencing of peripheral blood IgG+ B cells showed that structural properties of VH regions found to correlate with the PA phenotype, especially the RKH and Y5 motifs, were more prevalent in a group of 12 patients with HIT than in patients suspected of having HIT who tested negative in SRA/PEA and in healthy individuals. The findings indicate that B cells capable of producing antibodies structurally similar to the PA clones identified in our studies are expanded in peripheral blood of patients with HIT. Identification in healthy people of smaller, but significant numbers of IgG+ B cells possessing the same immunoglobulin structural features found in patients with HIT is consistent with evidence that B cells capable of producing HIT antibodies are prevalent in healthy persons but are normally held in check by immune tolerance mechanisms.27,28 Previous studies suggest that this B-cell reservoir may be a product of the immune response to PF4/bacterial complexes encountered earlier in life.29,30 Our repertoire studies support the view that in some patients given heparin, this preexisting B-cell pool expands to produce pathogenic antibodies capable of causing HIT.
Our finding that both clonal PA and NA antibodies bind to PF4/H and that PA but not NA antibodies bind to PF4-coated platelets indicates that the epitope(s) recognized by PA antibodies are present on both PF4/H and platelet-associated PF4, whereas those recognized by NA antibodies exist only on PF4/H. This provides direct support for the viewpoint that pathogenic (platelet-activating) and nonpathogenic HIT antibodies recognize different epitopes on PF4/H, as suggested by previous studies using antibodies from patients with HIT.8,31,32 It is generally thought that PF4 binds to platelet surface glycosaminoglycans, mainly chondroitin sulfate (CS) and heparan sulfate.33,34 CS is much less heavily sulfated than heparin, and CD-spectroscopic studies by Brandt et al have shown that it reconfigures PF4 differently than heparin,35 creating the possibility that PF4/CS and PF4/H differ in immunogenicity, in which case some patients might produce antibodies specific only for platelet-associated PF4. An example of such a case could be a patient with severe HIT, recently described by Warkentin et al, who had a strong platelet-activating antibody (SRA test) that was nonreactive with PF4/H.36 Studies with PA clones HIT3P1A11 and HIT3P7H2, showing that K109A and R108A mutations, respectively, create antibodies that activate platelets but fail to recognize PF4/H over a wide range of antibody concentrations (supplemental Figure 4B-C), demonstrate that such antibodies are possible. Our preliminary screening of B-cell clones was designed to detect antibodies reactive with PF4/H, which were then screened for PA activity. Accordingly, a population of platelet-activating antibodies that fail to react with PF4/H could have been overlooked. Further studies to examine this possibility are planned.
Our studies have several potential shortcomings. One is that we currently have no explanation for the surprising observation that PA and NA clones identified thus far possess κ light chains only. The fact that NB clones produced in parallel had the expected prevalence of λ chains argues against a technical explanation, however, and preferential use of κ or λ chains has been demonstrated in certain humoral immune responses in humans37-39 and mice.40 For practical reasons, we used only the PEA to evaluate clones for platelet-activating ability, rather than the more widely accepted SRA. However, equivalency of the 2 assays was recently demonstrated in a large clinical trial.26 One of the patients whose B cells were successfully cloned (HIT1) had “autoimmune HIT”19 unrelated to heparin exposure. However, the 2 conditions are almost indistinguishable clinically and serologically. As noted above, our primary screening technique (PF4/H binding) would have excluded hypothetical B cells producing antibodies that are platelet activating but nonreactive against PF4/H. Finally, B-cell cloning studies in larger numbers of patients with HIT are needed to define the extent to which HCDR3 motifs other than the ones identified in this report may affect antibody pathogenicity in HIT. It is known that, in addition to platelets, endothelial cells, monocytes, and neutrophils appear to be involved in HIT pathogenesis.2,11-13 In future studies, it will be of interest to investigate whether the clonal antibodies that activate these types of cells bear the same structural features identified in the PA clones.
In summary, our findings define, for the first time, structural features of a group of HIT antibodies that correlate with their ability to recognize PF4/H complexes and, when 1 of 2 HDCR3 motifs (RKH or Y5) is present, with their ability to bind to and activate PF4-treated platelets. These observations have implications for understanding HIT pathogenesis and for diagnosis and treatment of this condition that deserve further investigation. For example, human monoclonals that could, in the form of F(ab’)2 fragments, compete with pathogenic HIT antibodies for binding to heparin- or cell-associated PF4 could prove useful for treatment of severely affected patients with HIT, and screening of the IgG+ B-cell repertoire could conceivably provide an estimate of HIT risk in patients who are candidates for heparin therapy. Studies to examine these and other implications of current findings are planned.
Acknowledgments
The authors thank Garnett Kelsoe (Duke University, Durham, NC), for providing the CD40Llow stromal cell line and a detailed protocol on single B-cell culture; Patrick Wilson (University of Chicago, Chicago, IL), for help with establishing the single B-cell polymerase chain reaction and expression cloning system; Peter Newman (Versiti, Milwaukee, WI), for providing the purified anti-FcγRIIA antibody IV.3; Brian Curtis (Versiti, Milwaukee, WI) for providing platelet-rich plasma; and Spenser Huang (University of Wisconsin, Milwaukee, WI) for advice on statistical analysis.
The study was supported by National Institutes of Health (NIH), National Heart, Lung, and Blood Institute grant numbers HL148120 (R.W.), HL161127 (R.W.), HL130724 (D.W.), HL158932 (A.P.), and HL13629 (R.A.); NIH, Clinical and Translational Science Institute grant UL1TR001436 (R.W.); NIH, National Institute of Allergy and Infectious Diseases grant AI079087 (D.W.); American Heart Association grant 20PRE35210461 (W.Z.); and Fujian Normal University stipends (Y.W. and J.W.).
Authorship
Contribution: The study was conceptualized and supervised by R.W. and D.W.; experiments were conducted and data were analyzed by R.W., W.Z., Y.Z., M.Y., L.Z., G.F., J.W., Y.W., N.S., and C.J.; clinical consultation was obtained from A.P. and M.I.; and the manuscript was written by R.W. and R.A. and edited by D.W.
Conflict-of-interest disclosure: C.J. reports pending/issued patents (Versiti Blood Center of Wisconsin and Retham Technologies) and reports equity ownership and employment in Retham Technologies. A.P. reports pending/issued patents (Mayo Clinic, Retham Technologies, and Versiti Blood Center of Wisconsin), reports equity ownership in Retham Technologies, and serves on the advisory board of Veralox Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Renren Wen, Versiti Blood Research Institute, 8727 Watertown Plank Rd, Milwaukee, WI 53226; e-mail: rwen@versiti.org; and Demin Wang, Versiti Blood Research Institute, 8727 Watertown Plank Rd, Milwaukee, WI 53226; e-mail: dwang@versiti.org; and Richard Aster, Versiti Blood Research Institute, 8727 Watertown Plank Rd, Milwaukee, WI 53226; e-mail: rhaster@versiti.org.
References
Author notes
For original data and detailed protocols, please contact Renren Wen (rwen@versiti.org).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
W.Z. and Y.Z. contributed equally to the study.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal