Key Points
The rhesus macaque CRISPR RUNX1 gene-editing model recapitulates hematologic abnormalities observed in human RUNX1 deficiency.
RUNX1-mutant HSPCs have a competitive advantage over wildtype or AAVS1-control edited cells, potentially hindering corrective gene therapy.
Abstract
Germ line loss-of-function heterozygous mutations in the RUNX1 gene cause familial platelet disorder with associated myeloid malignancies (FPDMM) characterized by thrombocytopenia and a life-long risk of hematological malignancies. Although gene therapies are being considered as promising therapeutic options, current preclinical models do not recapitulate the human phenotype and are unable to elucidate the relative fitness of mutation-corrected and RUNX1-heterozygous mutant hematopoietic stem and progenitor cells (HSPCs) in vivo long term. We generated a rhesus macaque with an FPDMM competitive repopulation model using CRISPR/Cas9 nonhomologous end joining editing in the RUNX1 gene and the AAVS1 safe-harbor control locus. We transplanted mixed populations of edited autologous HSPCs and tracked mutated allele frequencies in blood cells. In both animals, RUNX1-edited cells expanded over time compared with AAVS1-edited cells. Platelet counts remained below the normal range in the long term. Bone marrows developed megakaryocytic dysplasia similar to human FPDMM, and CD34+ HSPCs showed impaired in vitro megakaryocytic differentiation, with a striking defect in polyploidization. In conclusion, the lack of a competitive advantage for wildtype or control-edited HSPCs over RUNX1 heterozygous–mutated HSPCs long term in our preclinical model suggests that gene correction approaches for FPDMM will be challenging, particularly to reverse myelodysplastic syndrome/ acute myeloid leukemia predisposition and thrombopoietic defects.
Introduction
Germ line monoallelic RUNX1 mutations cause familial platelet disorder with predisposition to myeloid malignancies (RUNX1-FPDMM) characterized by thrombocytopenia, defective platelet function, and an increased risk (up to 40%) of developing hematologic (primarily myeloid) malignancies.1-5 The successes of autologous hematopoietic stem cell (HSC) gene therapies over the past decade6 to ameliorate germ line hematologic disorders have stimulated interest in exploring efficacy in patients with RUNX1-FPDMM.7,8 However, given that current corrective gene-editing does not achieve close to 100% efficiency, it is important to determine whether corrected HSCs will have a competitive advantage over the remaining RUNX1-mutant HSCs, likely required to prevent progression to leukemia.
Zebrafish and mouse models have defined critical roles of RUNX1 in embryonic and adult hematopoiesis.9-11 In addition, induced pluripotent stem cells model derived from patients with RUNX1-FPDMM revealed potentially relevant features, such as decreased generation of HSPCs and impaired megakaryocytic (MK) differentiation.12-14 Nonetheless, these models have neither recapitulated spontaneous leukemic transformation, nor in vivo platelet phenotypes, nor directly measured competitive HSC fitness.
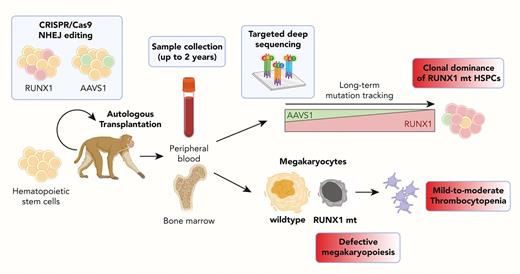
We developed a rhesus macaque RUNX1-FPDMM model utilizing gene editing and showed that RUNX1 heterozygous mutant HSPCs outcompeted AAVS1-control edited and wildtype HSPCs in animals following autologous transplantation. In addition, the animals developed the hallmark clinical characteristics of RUNX1-FPFMM, thrombocytopenia and platelet dysfunction.
Study design
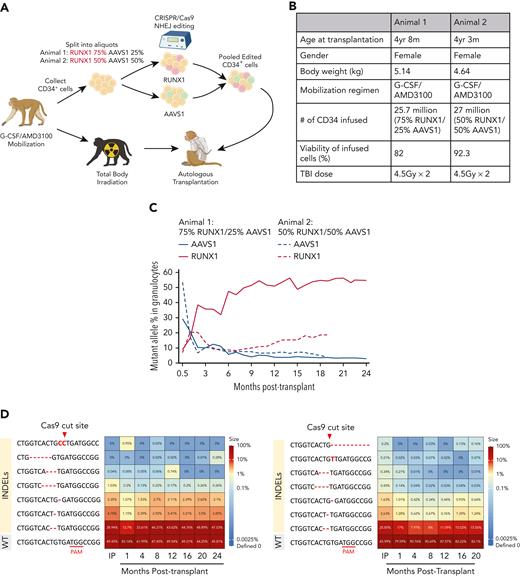
All animal experiments were approved by the NHLBI Animal Care and Use Committee. CD34+ HSPCs were electroporated with RUNX1 or AAVS1 chemically-modified guide RNAs (gRNAs) and Cas9 ribonucleoprotein (Figure 1A-B and supplemental Table 1, available on the Blood website) to create loss-of-function (LOF) indels and were reinfused intravenously. Indel variant allele fraction (VAF) in blood cells across lineages were tracked via targeted deep sequencing using a custom R application (https://github.com/shint3/CH_crispr_analysis.git/).15 Secondary myeloid malignancy mutations were examined using targeted panel sequencing.15 Platelet aggregation was measured using impedance platelet aggregometry. Marrow sampling was conducted for tissue immunohistochemistry and progenitor isolation. Bone marrow CD34+ cells were differentiated into MK cells in the presence of the stem cell factor, thrombopoietin (TPO), interleukin-6, and interleukin-3 for 14 days. MK lineage cells were identified and sorted by flow cytometry based on CD41a expression, and MK ploidy was assessed by staining for DNA content (supplemental Table 2). Additional details about the methods in this study are found in supplemental Methods.
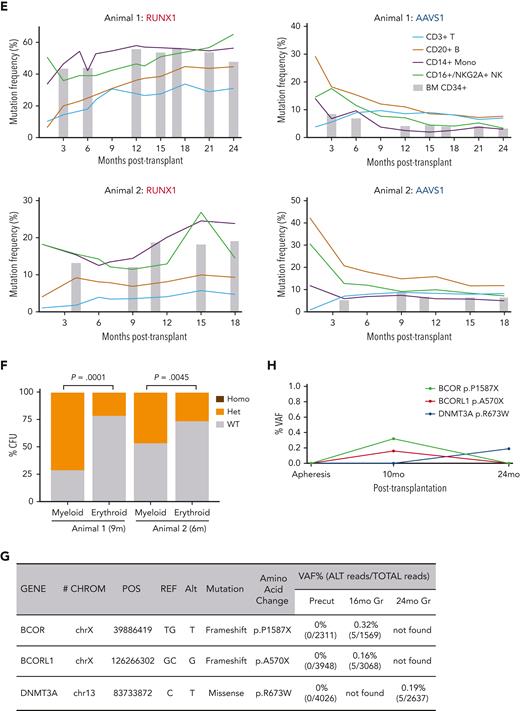
Mutation frequenciesin rhesusmacaque RUNX1/AAVS1 competitive autologous transplantation model across hematopoietic lineages. (A) Schematic outline of RM HSPC gene editing at the RUNX1 and AAVS1 loci and subsequent competitive autologous transplantation. Mobilized CD34+ HSPCs were separated by the ratio of RUNX1 and AAVS1 edited cells (3:1) in the first animal and (1:1) in the second animal. (B) Summary of relevant animal and transplantation parameters. (C) Mutation (indel) frequencies by targeted deep sequencing of PB granulocytes over time post transplantation. (D) Heatmaps show the frequency of each retrieved mutated (indel) read as a percentage of total sequencing reads in granulocyte samples over time. Animal 1 (left); Animal 2 (right). The most abundant indel types for the RUNX1 target site in each animal are listed on the left, with the WT sequence at the bottom. Indels are highlighted in red font, and the predicted Cas9 cut site is indicated by the red arrow. (E) RUNX1 and AAVS1 target site indel frequencies by deep sequencing shown over time in T cells, B cells, monocytes, and mature CD16+ NK cells were analyzed in animal 1 (upper) and animal 2 (lower). (F) BM CD34+ HSPCs obtained at 9 months (animal 1) and 6 months (animal 2) were plated in CFU assays and genotyped for RUNX1 mutations. The percentage of Homo and Het CFUs are shown in myeloid and erythroid colonies for each animal. (G-H) Error-corrected deep sequencing of the exons of the 56 genes most commonly associated with human myeloid malignancies. Detected somatic mutations are summarized for animal 1 (G) annotated mutations (H) VAF over time. BM, bone marrow; CFU, colony-forming unit; Homo, homozygous-mutated; Het, heterozygous; NK, natural killer; PB, peripheral blood.
Mutation frequenciesin rhesusmacaque RUNX1/AAVS1 competitive autologous transplantation model across hematopoietic lineages. (A) Schematic outline of RM HSPC gene editing at the RUNX1 and AAVS1 loci and subsequent competitive autologous transplantation. Mobilized CD34+ HSPCs were separated by the ratio of RUNX1 and AAVS1 edited cells (3:1) in the first animal and (1:1) in the second animal. (B) Summary of relevant animal and transplantation parameters. (C) Mutation (indel) frequencies by targeted deep sequencing of PB granulocytes over time post transplantation. (D) Heatmaps show the frequency of each retrieved mutated (indel) read as a percentage of total sequencing reads in granulocyte samples over time. Animal 1 (left); Animal 2 (right). The most abundant indel types for the RUNX1 target site in each animal are listed on the left, with the WT sequence at the bottom. Indels are highlighted in red font, and the predicted Cas9 cut site is indicated by the red arrow. (E) RUNX1 and AAVS1 target site indel frequencies by deep sequencing shown over time in T cells, B cells, monocytes, and mature CD16+ NK cells were analyzed in animal 1 (upper) and animal 2 (lower). (F) BM CD34+ HSPCs obtained at 9 months (animal 1) and 6 months (animal 2) were plated in CFU assays and genotyped for RUNX1 mutations. The percentage of Homo and Het CFUs are shown in myeloid and erythroid colonies for each animal. (G-H) Error-corrected deep sequencing of the exons of the 56 genes most commonly associated with human myeloid malignancies. Detected somatic mutations are summarized for animal 1 (G) annotated mutations (H) VAF over time. BM, bone marrow; CFU, colony-forming unit; Homo, homozygous-mutated; Het, heterozygous; NK, natural killer; PB, peripheral blood.
Results and discussion
We generated a preclinical rhesus macaque RUNX1-FPDMM model utilizing CRISPR/Cas9 engineering, leading to nonhomologous end joining and indel formation to create LOF mutations in the RUNX1 transcription factor or at the control AAVS1 locus in HSPCs, followed by an intravenous infusion of pooled RUNX1 and AAVS1 edited cells into animals conditioned with myeloablative total body irradiation (4.5 grays daily for 2 days). The ratio of RUNX1 and AAVS1 edited cells (3:1) in the first animal was chosen to conservatively mimic potential gene correction levels in a human clinical trial. Indel frequency was assessed in these mixed infusion products and reflected the ratios of cell fractions edited with each gRNA in the 2 animals with editing efficiencies of 70% to 100% for RUNX1 and 40% to 90% for AAVS1 (Figure 1A, supplemental Figure 1).
Blood count recovery after the transplantation of edited cells was similar to prior animals with transplanted cells edited at other loci (supplemental Figure 2). To address the competitive repopulation fitness of RUNX1-mutant vs AAVS1-control and the unedited WT HSPC, we followed the VAF of indels in circulating granulocytes. Despite the initial dominance of AAVS1-edited cells after the engraftment in both animals, RUNX1-edited cells increased over time, plateauing at a 50% VAF in animal 1 by 10 months after transplantation, suggesting a virtually all-out competition of AAVS1-edited or residual unedited HSPCs if each edited cell is heterozygous. Having observed complete clonal dominance of the RUNX1-mutant HSPCs in animal 1 starting with a 3:1 ratio, we transplanted animal 2 with a lower RUNX1/AAVS1 edited cell ratio (1:1) and observed a similar clonal expansion of RUNX1 edited cells over the AAVS1 edited cells, continuing to increase up to a VAF of 19% by 20 months in animal 2 (Figure 1C), with several predominant predicted LOF indels in RUNX1 (Figure 1D, supplemental Figure 3).
In both animals, the fraction of RUNX1-mutated cells increased over time in all lineages, with higher frequencies in granulocytes, monocytes, and NK cells vs B and T cells (Figure 1E). In contrast, the frequency of AAVS1-mutated cells across lineages did not show myeloid bias and decreased in all lineages in the long term. The RUNX1 VAF increased in BM CD34+ cells over time, whereas the AAVS1 VAF decreased (Figure 1E). RUNX1 mutation frequencies were slightly higher in both purified long-term– and short-term–HSCs than in total CD34+ cells, whereas AAVS1 VAFs were lower in long-term–HSCs than in bulk CD34+ cells and short-term–HSCs (supplemental Figure 4). Despite homozygous editing detected in colonies grown from the infusion product (supplemental Figure 1E), no homozygous RUNX1-mutated CFUs were detected after transplantation, suggesting that all long-term engrafted cells with RUNX1 indels were heterozygous, mimicking the human disorder. The frequencies of RUNX1-mutated CFUs were significantly higher in myeloid vs in erythroid colonies in both animals (Figure 1F), confirming a potential myeloid lineage bias for RUNX1 mutated cells.
Because the most serious outcome for individuals with RUNX1-FPDMM, after the acquisition of additional mutations, is progression to leukemia,16 we screened for common myeloid malignancy driver mutations (supplemental Table 3).17,18 Extremely low VAF mutations (all <1%) were found in BCOR, BCORL1, and DNMT3A; however, there was no expansion of clones with these mutations over time (Figure 1G-H). We also detected no off-target editing (supplemental Figure 5).19
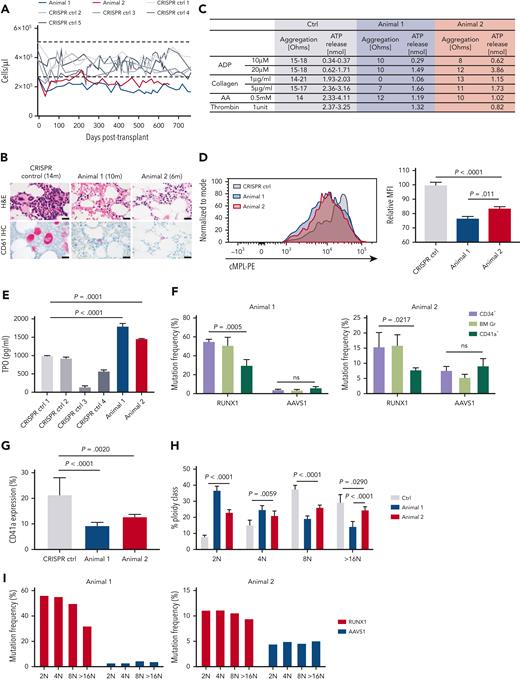
We performed a detailed analysis of peripheral blood counts over time after the engraftment. The total white blood cell counts, hemoglobin concentrations, and hematocrits were normal after transplantation recovery (supplemental Figure 6), whereas both animals showed mild to moderately reduced platelet counts compared with CRISPR-edited transplanted control animals (Figure 2A). Despite the reduced platelet counts, the animals have not shown abnormal bleeding. Both animals showed frequent small and dysplastic MKs in the marrow by 6 months after transplantation, with over 10% to 30% of CD61+ cells and reduced numbers of nuclei, similar to the morphology of MKs observed in patients with RUNX1 deficiencies (Figure 2B).20 No small dysplastic MKs were detected in the control animals with transplants. The overall marrow cellularity and the frequency of MKs were comparable with those of control animals with transplants. There were no clear abnormalities in the level of inflammatory mediators compared with that in control animals with transplants (supplemental Figure 6F).
Analysis of platelet parameters and abnormal megakaryopoiesis in RUNX1/AAVS1 competitive repopulation animals. (A) Platelet counts for animal 1 and animal 2 plotted in comparison with CRISPR control animals. The dotted lines indicate the upper and lower range of normal RM platelet counts (2 SDs from mean). (B) Sections of BM core biopsies obtained from animal 1 and 2 and a CRISPR control animal after transplantation were stained with H&E (upper panel) or via immunohistochemistry for the megakaryocyte marker CD61 (lower panel). Representative microscopic images are shown. CD61+ megakaryocytes stain bright pink. (C) Platelet aggregation results for the RUNX1/AAVS1 and CRISPR control animals. (Animal 1: 23 months–after transplantation, Animal 2: 17 months after transplantation). (D) Mpl (TPO receptor) expression on platelets for animals 1 and 2 and for CRISPR control animals by flow cytometry. Right panel shows relative MFI. (Animal 1: 20 months after transplantation, Animal 2: 14 months after transplantation). (E) Circulating TPO concentrations in PB plasma of RUNX1/AAVS1 animals 1 and 2 and CRISPR control animals by ELISA. CRISPR control animal 1 was used for statistical analyses. (Animal 1: 22 months after transplantation, Animal 2: 16 months after transplantation). (F) Targeted deep sequencing on CD34+ HSPCs, Gr, and CD41a+ megakaryocytes isolated from BM (Animal 1: 15 and 17 months after transplantation, Animal 2: 9 and 11 months after transplantation). The percentages of reads containing indels at the RUNX1 or AAVS1 target sites are shown. (G-I) BM-derived purified CD34+ cells from animals 1 and 2 and controls were differentiated into MKs in vitro for 14 days and analyzed by flow cytometry. (G) CD41a expression. (H) The percentage of each ploidy class within the in vitro differentiated CD41a+ MKs is shown. (I) Each ploidy subpopulation was sorted for targeted deep sequencing. The percentage of reads containing indels at the RUNX1 or AAVS1 target sites are shown. Bar represents 20 μm (B). ELISA, enzyme-linked immunosorbent assay; Gr, granulocyte; H&E, hematoxylin and eosin stain; MFI, mean fluorescence intensity; SD, standard deviation.
Analysis of platelet parameters and abnormal megakaryopoiesis in RUNX1/AAVS1 competitive repopulation animals. (A) Platelet counts for animal 1 and animal 2 plotted in comparison with CRISPR control animals. The dotted lines indicate the upper and lower range of normal RM platelet counts (2 SDs from mean). (B) Sections of BM core biopsies obtained from animal 1 and 2 and a CRISPR control animal after transplantation were stained with H&E (upper panel) or via immunohistochemistry for the megakaryocyte marker CD61 (lower panel). Representative microscopic images are shown. CD61+ megakaryocytes stain bright pink. (C) Platelet aggregation results for the RUNX1/AAVS1 and CRISPR control animals. (Animal 1: 23 months–after transplantation, Animal 2: 17 months after transplantation). (D) Mpl (TPO receptor) expression on platelets for animals 1 and 2 and for CRISPR control animals by flow cytometry. Right panel shows relative MFI. (Animal 1: 20 months after transplantation, Animal 2: 14 months after transplantation). (E) Circulating TPO concentrations in PB plasma of RUNX1/AAVS1 animals 1 and 2 and CRISPR control animals by ELISA. CRISPR control animal 1 was used for statistical analyses. (Animal 1: 22 months after transplantation, Animal 2: 16 months after transplantation). (F) Targeted deep sequencing on CD34+ HSPCs, Gr, and CD41a+ megakaryocytes isolated from BM (Animal 1: 15 and 17 months after transplantation, Animal 2: 9 and 11 months after transplantation). The percentages of reads containing indels at the RUNX1 or AAVS1 target sites are shown. (G-I) BM-derived purified CD34+ cells from animals 1 and 2 and controls were differentiated into MKs in vitro for 14 days and analyzed by flow cytometry. (G) CD41a expression. (H) The percentage of each ploidy class within the in vitro differentiated CD41a+ MKs is shown. (I) Each ploidy subpopulation was sorted for targeted deep sequencing. The percentage of reads containing indels at the RUNX1 or AAVS1 target sites are shown. Bar represents 20 μm (B). ELISA, enzyme-linked immunosorbent assay; Gr, granulocyte; H&E, hematoxylin and eosin stain; MFI, mean fluorescence intensity; SD, standard deviation.
To explore whether the macaque model reproduces RUNX1-FPDMM platelet function defects,21,22 platelet aggregation assays were performed (Figure 2C, supplemental Figure 7). Both animals showed decreased adenosine diphosphate–induced aggregation. Platelets from animal 1 had absent aggregation for low-dose collagen and decreased aggregation for high-dose collagen. Animal 2, with a lower mutant VAF, showed normal aggregation and ATP release to collagen. Similar defects have been found in patients with RUNX1 deficiency.23 Both animals had normal aggregation and adenosine triphosphate release to arachidonic acid. The expression level of the TPO receptor c-Mpl, previously shown to be positively regulated by RUNX1, significantly decreased in RUNX1-mutated animals (Figure 2D).24,25 Notably, TPO levels in the blood increased (Figure 2E), as would be expected in the setting of thrombocytopenia with decreased platelet Mpl cell surface expression.24,26 Platelet ultrastructural morphology examined via electron microscopy revealed some findings similar to those reported in patients with FPDMM,27 including increased heterogeneity in platelet size as well as larger and more frequent membrane structures comprising the open canalicular system, referred to as vacuoles in some prior studies (supplemental Figure 8). Dense granules were rare or difficult to identify in both control and RUNX1-mutated platelets and did not significantly change in number.
Defective megakaryocytopoiesis and the lack of full maturation has been suggested to result in platelet dysfunction and thrombocytopenia in patients with RUNX1-FPDMM.12,27 BM-derived CD41a+ MKs showed decreased RUNX1 VAF compared with that in CD34+ HSPCs and granulocytes, as expected if the RUNX1-mutated HSPCs could not efficiently complete maturation to MKs; whereas AAVS1 CD41a+ MKs had equivalent VAFs to the other cell types (Figure 2F). We further dissected in vitro MK differentiation from CD34+ HSPCs obtained from the animals. After differentiation, the frequency of cells expressing CD41a was lower in the RUNX1-mutated animals than in controls (Figure 2G). The nuclear ploidy of in vitro differentiated CD41a+ MKs was significantly lower in the RUNX1-mutated than in control cultures (Figure 2H). When CD41a+ cells were sorted based on ploidy, RUNX1 VAF inversely correlated with ploidy (Figure 2I), further confirming the defect in maturation to normal polyploid MKs. In contrast, the AAVS1 VAF did not show significant differences across ploidy fractions (Figure 2I). Notably, the severity of platelet and MK defects correlated with the relative RUNX1 VAFs in animal 1 vs animal 2 (Figure 2).
In summary, this novel competitive mosaic nonhuman primate model provided insights into aspects of RUNX1-FPDMM pathophysiology and potential treatments not previously achieved in other preclinical models. We demonstrated that RUNX1 heterozygous mutant HSPCs outcompete AAVS1-control edited cells and WT cells over time after transplantation, reflecting ongoing clonal competition. This suggests that preventing leukemia in patients with RUNX1-FPDMM via current gene correction approaches will be very challenging, given that residual RUNX1 mutant cells would be predicted to expand over time and continue to be at risk for transformation. Thus far, we have not observed progression to leukemia, but this could be attributed to the relatively young age of the animals and the short follow up, compared with the median onset age for acute myeloid leukemia in these patients (33 years).22 Continued follow ups may reveal malignant transformation and allow an investigation of underlying mechanisms. Introduction of additional typical FPDMM secondary leukemia driver mutations into macaque HSPCs can be studied in the same model. Both animals had chronic mild-to-moderate thrombocytopenia and platelet function abnormalities. In addition, we documented markedly impaired megakaryopoiesis both in vitro and in vivo, specifically reduced polyploidization, a sign of MK immaturity, in RUNX1-mutated cells.27 The degree of thrombocytopenia and defective megakaryopoiesis appeared to correlate with the RUNX1 mutation frequency. It suggests that RUNX1 mosaicism can still result in platelet disorders and that gene therapies may not correct the hemostatic abnormalities unless highly efficient.
Acknowledgments
The authors thank Theresa Engels, Justin Golumb, Allen Krouse, and Seth Linde for excellent animal care, Aylin Bonifacino for technical assistance, Eddie Martin for skilled histopathology assistance, and the National Heart, Lung, and Blood Institute Genomics and Flow Cytometry Cores. Some figures were created with BioRender.com.
This work was supported by the intramural research programs of the National Institutes of Health (NIH), National Heart, Lung, and Blood Institute and the NIH, National Genome Research Center, and a gift from the Runx1 Research Program.
Authorship
Contribution: P.L. and C.E.D. conceived and supervised the study; B.-C.L., Y.Z., P.L., and C.E.D. designed the experiments; B.-C.L., Y.Z., E.B., N.O., A.D.-F., B.C., T.-H.S., V.B., Z.A.S., S.G.H., T.Z., and K.R.C. collected and analyzed the data; and B-C.L., Y.Z., P.L. and C.E.D. wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul Liu, Oncogenesis and Development Section, National Human Genome Research Institute, National Institutes of Health, Room 5222C, Building 50, 50 South Dr, Bethesda, MD 20892; e-mail: pliu@nhgri.nih.gov; and Cynthia E. Dunbar, Translational Stem Cell Biology Branch, National Heart, Lung and Blood Institute, National Institutes of Health, Room 5E-3332, Building 10-CRC, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: dunbarc@nhlbi.nih.gov.
References
Author notes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
B.-C.L. and Y.Z. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal