Key Points
The structure of fV short reveals the architecture of the entire A1-A2-B-A3-C1-C2 assembly for the first time.
The structure offers molecular context for the functional interactions of the B domain.
Abstract
Coagulation factor V (fV) is the precursor of activated fV (fVa), an essential component of the prothrombinase complex required for the rapid activation of prothrombin in the penultimate step of the coagulation cascade. In addition, fV regulates the tissue factor pathway inhibitor α (TFPIα) and protein C pathways that inhibit the coagulation response. A recent cryogenic electron microscopy (cryo-EM) structure of fV has revealed the architecture of its A1-A2-B-A3-C1-C2 assembly but left the mechanism that keeps fV in its inactive state unresolved because of an intrinsic disorder in the B domain. A splice variant of fV, fV short, carries a large deletion of the B domain that produces constitutive fVa-like activity and unmasks epitopes for the binding of TFPIα. The cryo-EM structure of fV short was solved at 3.2 Å resolution and revealed the arrangement of the entire A1-A2-B-A3-C1-C2 assembly. The shorter B domain stretches across the entire width of the protein, making contacts with the A1, A2, and A3 domains but suspended over the C1 and C2 domains. In the portion distal to the splice site, several hydrophobic clusters and acidic residues provide a potential binding site for the basic C-terminal end of TFPIα. In fV, these epitopes may bind intramolecularly to the basic region of the B domain. The cryo-EM structure reported in this study advances our understanding of the mechanism that keeps fV in its inactive state, provides new targets for mutagenesis and facilitates future structural analysis of fV short in complex with TFPIα, protein S, and fXa.
Introduction
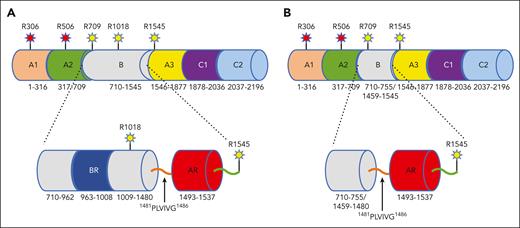
Coagulation factor V (fV) is a large (2224 residues, MW 330 kDa) precursor of the cofactor activated fV (fVa) that together with the enzyme fXa as well as Ca2+ and phospholipids defines the prothrombinase complex and rapidly converts prothrombin to thrombin in the penultimate step of the coagulation cascade.1,2 In addition, fV is directly involved in the regulation of the tissue factor pathway inhibitor α (TFPIα) and protein C pathways that inhibit the coagulation response.2-5 After the removal of a signal peptide of 28 residues, fV secreted to the plasma features the domain structure A1-A2-B-A3-C1-C2 (Figure 1A). The A1 domain (residues 1-316) is connected to the A2 domain (residues 317-709) by a short segment composed mainly of basic amino acids (residues 304-316). An acidic segment (residues 657-709) transitions the A2 domain to the large and poorly conserved11 B domain (residues 710-1545) that continues to the A3 (residues 1546-1877), C1 (residues 1878-2036), and C2 (residues 2037-2196) domains. The structural organization of fV has been elucidated recently using cryogenic electron microscopy (cryo-EM),12 a technique capable of solving the structure of high-molecular weight proteins and their complexes under conditions not biased by crystal contacts10,13-15 and particularly suited for the analysis of coagulation factors.10,12,16-24 The A1-A3-C1-C2 domains are arranged as originally revealed by the bovine inhibited fVa X-ray crystal structure,25 with the C1 and C2 domains forming a membrane-binding module supporting the A1 and A3 domains. New information garnered from the cryo-EM structure of fV includes identification of the sites of activation by thrombin at R709 and R1545, the sites of inactivation by activated protein C (APC) at R306 and R506, and the architecture of the A2 domain housing determinants critical for prothrombin activation, such as the lid (residues 672-691) and the gate (residues 696-702).10,16 The structure has also served as a starting point for the subsequent elucidation of fVa, prothrombinase-free and bound to prothrombin.12,16
Schematic representation of fV and fV short. (A) A1-A2-B-A3-C1-C2 domain organization of fV (2196 residues total) and details of the B domain containing the BR (963KPGKQSGHPKFPRVRHKSLQVRQDGGKSRLKKSQFLIKTRKKKKEK1008) and AR (1493DYIEIIPKEEVQSSEDDYAEIDYVPYDDPYKTDVRTNINSSRDPD1537) that interact to keep fV in its inactive state.1,5-9 The hydrophobic patch 1481PLVIVG1486 is highlighted preceding the AR. The sites of thrombin activation (R709, R1018, R1545) and APC inactivation (R306, R506) are indicated. (B) A1-A2-B-A3-C1-C2 domain organization of the splice variant fV short carrying a deletion of 703 residues, from 756 through 1458, in the B domain (1493 residues total). The deletion removes the site of thrombin activation at R1018 and the entire BR, which unmasks the hydrophobic patch 1481PLVIVG1486 and the AR region to promote TFPIα binding. The cryo-EM structure of fV short (Figure 2; supplemental Figure 3) reveals all residues of the A1-A2-A3-C1-C2 assembly. Reproduced with modifications,10 copyright Elsevier (2022).
Schematic representation of fV and fV short. (A) A1-A2-B-A3-C1-C2 domain organization of fV (2196 residues total) and details of the B domain containing the BR (963KPGKQSGHPKFPRVRHKSLQVRQDGGKSRLKKSQFLIKTRKKKKEK1008) and AR (1493DYIEIIPKEEVQSSEDDYAEIDYVPYDDPYKTDVRTNINSSRDPD1537) that interact to keep fV in its inactive state.1,5-9 The hydrophobic patch 1481PLVIVG1486 is highlighted preceding the AR. The sites of thrombin activation (R709, R1018, R1545) and APC inactivation (R306, R506) are indicated. (B) A1-A2-B-A3-C1-C2 domain organization of the splice variant fV short carrying a deletion of 703 residues, from 756 through 1458, in the B domain (1493 residues total). The deletion removes the site of thrombin activation at R1018 and the entire BR, which unmasks the hydrophobic patch 1481PLVIVG1486 and the AR region to promote TFPIα binding. The cryo-EM structure of fV short (Figure 2; supplemental Figure 3) reveals all residues of the A1-A2-A3-C1-C2 assembly. Reproduced with modifications,10 copyright Elsevier (2022).
An unexpected finding from the cryo-EM structure of fV has been the high degree of disorder in the large B domain, which limited assignments to 4 residues (710SFRN713) at the N-terminal end connecting to R709 and 10 residues (1536PDNIAAWYLR1545) at the C-terminal end preceding R1545.10,12 These small segments do not include the basic region (BR; residues 963-1008) and the acidic region (AR; residues 1493-1537) (Figure 1A) that are assumed to interact intramolecularly to keep fV in its inactive state for the control of both procoagulant and anticoagulant activities.1,5-9 Hence, a key property of the B domain of fV remains unresolved. Elucidation of the architecture of the B domain also bears significant clinical relevance.26-32 The East Texas bleeding disorder is associated with a splice fV isoform, called fV short, resulting in the deletion of residues from 756 to 1458 that include the BR and 31 tandemly arranged repeats of the B domain4,31,32 (Figure 1B). Loss of the BR unmasks a high-affinity binding site in the AR for TFPIα, a key regulator of the assembly of prothrombinase29,30 and blood coagulation.28 Carriers of the East Texas mutation have high plasma concentrations of fV short bound to TFPIα that act in concert with protein S (PS) to inhibit fXa, which is required for the assembly of prothrombinase and procoagulant activity.5-7,26,29,33 A structural understanding of fV short free and in complex with TFPIα, PS, and fXa would significantly advance the basic knowledge on fV and its multiple roles in hemostasis.
In this study, we present a 3.2 Å resolution cryo-EM structure of fV short that reveals the architecture of the entire A1-A2-B-A3-C1-C2 assembly. The shorter B domain is fully resolved and houses several hydrophobic clusters and acidic residues in the AR that likely provide a binding site for the basic C-terminal end of TFPIα. In fV, these epitopes may bind intramolecularly to the BR. The results offer new insights in our understanding of the mechanism that keeps fV in its inactive state and represent an important first step toward the structural elucidation of the anticoagulant properties of fV short in complex with TFPIα, PS, and fXa.
Methods
Materials
Materials obtained commercially were human plasma fV (#HCV-0100; Prolytix, Essex Junction, VT), human fV full-length complementary DNA (cDNA; #40515; ATCC, Manassas, VA), pDest40 expression vector (#12274015; Invitrogen, Waltham, MA), silver stain plus kit (#1610449; Bio-Rad, Hercules, CA), Gibco Expi293 expression kit (#A14635; Thermo Fisher Scientific, Frederick, MD), Quantifoil R 1.2/1.3, Cu 300 mesh grids (Q350CR1.3; Electron Microscopy Sciences, Hatfield, PA), PfuUltra high fidelity DNA polymerase (#600380; Agilent, Santa Clara, CA), NEBuilder HiFi DNA Assembly master mix (#E2621S; NEB, Ipswich, MA), simply Blue safe stain (LC6065; Invitrogen, Waltham, MA), and horseradish peroxidase-conjugated sheep anti-human fV immunoglobulin G (Enzyme Research Laboratory, South Bend, IN). fV short cDNA fragments were cloned via polymerase chain reaction, using full-length fV cDNA (#40515; Manassas, VA) as a template. The resulting fV short cDNA fragments were cloned into the pDest40 expression vector using Gibson assembly. To facilitate purification of recombinant fV short, a 12–amino acid HPC4 tag (EDQVDPRLIDGK) was fused to the C-terminus of the protein, as previously described.34 fV short was then expressed transiently in Expi293 suspension cells per the manufacturer’s instruction. Eighteen or 20 hours after transfection, media was supplemented with the broad serine protease inhibitor PPACK (10 nM, final concentration) and soybean trypsin inhibitor (10 μg/mL) to protect against enzymatic degradation during production. Harvested conditioned media was spun down, further supplemented with benzamidine (10 mM), and purified initially on a Q Sepharose column (HiTrap Q Sepharose, Cytiva) with a salt gradient. Eluent from the Q-column was diluted and supplemented with CaCl2 (5 mM) and purified via immune affinity using the Ca2+-dependent monoclonal antibody against HPC434 (supplemental Figure 1A-B, which is available on the Blood website). The tag fragment attached to the C-terminal of the C2 domain, used in the expression system, is clearly visible in the cryo-EM structure (supplemental Figure 1C). The recombinant construct of fV short corresponds to that of FV755-1459 in the Dahlbäck nomenclature29,33 and carries a deletion of 703 residues compared with plasma fV, from S756 through D1458, including residue D756 (or N756) generated by a new codon across the splice junction between I755 and L1459 in the fV short transcripts from patients with East Texas bleeding disorder or healthy individuals.32 The deletion removes the BR (residues 963-1008), the site of thrombin activation at R1018, and yields a shorter B domain of 133 residues (Figure 1B). Our recombinant fV short cleaves prothrombin in a continuous assay,35 with kcat/Km = 150 ± 3 μM-1s-1 vs 159 ± 1 μM-1s-1 for plasma-derived fVa (data not shown). This is consistent with the properties of fV short variants with residue 756 included7,9 or carrying significantly longer deletions of the B domain, inclusive of the splice site, which retain fVa-like activity9 or the synergism between TFPIα and PS in the inhibition of fXa.29
Cryo-EM grid preparation and data collection
For cryo-EM studies, recombinant fV short was further purified and buffer exchanged at pH 7.4 into 20 mM HEPES, 150 mM NaCl, 5 mM CaCl2 via size-exclusion chromatography, using a Superdex 200 increase 10/300 GL column. Peak fractions were collected and concentrated. At the time of freezing, grids were plasma cleaned on a Gatan Solarus 950 for 60 seconds; 3 μL of fV short (0.1 mg/mL) were applied onto each grid, blotted for 2 seconds, and then immediately plunge frozen in liquid ethane on a FEI Vitrobot Mark IV. Grids were loaded onto a Titan Krios G3 cryo-TEM operating at 300 kV equipped with a FEI Falcon IV (4k × 4k) direct electron detector. Screening and data collection were performed at a pixel size of 0.9 Å, using a dose of 55.09 e⁻/Å2 across 46 frames, using a set defocus range of −1 to −2.4 μm. Exposures were collected from 2 grids under similar conditions. A total of 3316 micrographs were collected.
Image processing, map calculation, and model building
Patch-based motion correction, dose weighting, and patch-based contrast transfer function estimation were performed sequentially in cryoSPARC.36 Particles were initially picked using Blob Picker job. Multiple rounds of 2D classification were performed to clean up the particle stack. Particles from the previous step were used to create an ab initio 3D construction volume. Initial models similar to fV protein data bank (PDB) 7KVE were combined into a single volume to create 2D templates for the template picker. Particles selected by the template picker were cleaned via multiple rounds of 2D classification and used to train Topaz neural network-based picker.37 Particles from different types of picker jobs were combined, duplicate particles removed (using 0.75 particle diameter as threshold), and then cleaned with multiple rounds of 2D classification to produce the final stack. Particles were re-extracted by recentering using a 336 pixel box, and new ab initio 3D construction with multiple classes implementing a high similarity score was carried out to capture potential heterogeneity. 3D volumes were compared in UCSF Chimera.38 The resultant 3D volumes deemed similar were pooled into a single nonuniform refinement, giving a consensus overall resolution of 3.2 Å (EMDB-29010). Masked local refinement of the A2 (EMDB-29008) and C1/C2 (EMDB-29009) with and without parts of the A1 and A3 domains of fV short was carried out to improve the density of mobile components and the disordered loops. To aid with model building, a composite map of the consensus refinement and local refined maps was generated by aligning the 3 maps in UCSF Chimera and taking the maximum value at each voxel (EMDB-29011). Maps were sharpened in Phenix,39 and resolution was estimated using a gold-standard Fourier shell correlation cutoff of 0.143. An initial model for fV short was generated in COOT,40 using PDB 7KVE and AlphaFold41 AF-P12259 for human fV. Initial models were docked and fit as a rigid body using UCSF Chimera. All atoms were refined with restraints, and model building was completed manually in COOT, using the composite map. Manual placement of residues in the B domain was partially guided by models generated using the CR-I-Tasser web server.42 The final model was refined against the composite map, with AlphaFold AF-P12259 and 7KVE as reference models for secondary structure restraints, using Phenix. Relevant parameters of the cryo-EM structure of fV short are summarized in Table 1. Representative 2D class averages and gold-standard Fourier shell correlation of the masked refinement of fV short maps are reported in supplemental Figure 2A-D.
Structural parameters for the cryo-EM structure of fV short
| . | fV short PDB ID 8FDG EMDB-29011 . |
|---|---|
| Data collection and processing | |
| Nominal magnification | 75 000× |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 55.09 |
| Defocus range (μm) | −1 to −2.4 |
| Pixel size (Å) | 0.9 |
| Number of images | 3316 |
| Final particle images | 147 000 |
| Symmetry imposed | C1 |
| Resolution (unmasked, FSC threshold 0.143) | 4.1 |
| Resolution (masked, FSC threshold 0.143) | 3.2 |
| Model composition | |
| Refinement program | Phenix (real space) |
| Number of protein atoms (non-H) | 12 164 |
| Protein residues | 1505 |
| Ligand molecules | 0 |
| Rmsd deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.694 |
| Validation | |
| All-atom clash score | 10 |
| Poor rotamers (%) | 0 |
| Ramachandran disallowed (%) | 0.0 |
| Ramachandran allowed (%) | 11.9 |
| Ramachandran favored (%) | 88.1 |
| MolProbity score | 2.13 |
| . | fV short PDB ID 8FDG EMDB-29011 . |
|---|---|
| Data collection and processing | |
| Nominal magnification | 75 000× |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 55.09 |
| Defocus range (μm) | −1 to −2.4 |
| Pixel size (Å) | 0.9 |
| Number of images | 3316 |
| Final particle images | 147 000 |
| Symmetry imposed | C1 |
| Resolution (unmasked, FSC threshold 0.143) | 4.1 |
| Resolution (masked, FSC threshold 0.143) | 3.2 |
| Model composition | |
| Refinement program | Phenix (real space) |
| Number of protein atoms (non-H) | 12 164 |
| Protein residues | 1505 |
| Ligand molecules | 0 |
| Rmsd deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.694 |
| Validation | |
| All-atom clash score | 10 |
| Poor rotamers (%) | 0 |
| Ramachandran disallowed (%) | 0.0 |
| Ramachandran allowed (%) | 11.9 |
| Ramachandran favored (%) | 88.1 |
| MolProbity score | 2.13 |
Results
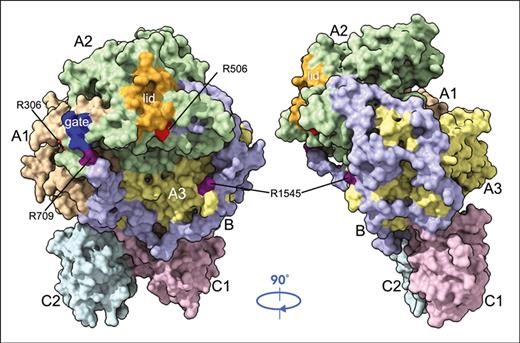
The cryo-EM structure of fV short was solved at atomic (3.2 Å) resolution and reveals the A1-A2-B-A3-C1-C2 assembly (1493 residues) in its entirety for the first time (Figure 2; supplemental Figure 3). Overall, the structure is similar (root-mean-square-deviation (rmsd) = 2.22 Å over 1134 Cα atoms) to that of fV solved recently.12 Unlike the highly disordered B domain of fV, the shorter B domain of fV short fits well in the electron density map, with a backbone and bulky side chains (Figure 3), and reveals new information of functional relevance. The sites of thrombin activation at R709 and R1545 are clearly visible in the A2 and B domains and are >80% exposed to solvent for proteolytic attack (Figure 2A). The sites of APC cleavage at R306 and R506 associated with fVLeiden4,43-47 are 46% and 35% exposed to solvent, respectively, as observed in fV.12 The C domains are similar to each other (rmsd = 1.19 Å over 120 Cα atoms) and shaped as distorted jelly-roll β-barrels, as reported previously in the cryo-EM structure of fV,12 the X-ray crystal structures of bovine inhibited fVa,25 and the recombinant C2 domain of human fVa and fVIIIa.48,49 Together, they define a membrane-binding module and support the A1-A2-B-A3 domains (Figure 2A). The new I755-L1459 junction at the splice site of our fV short construct has a well-defined density (Figure 3B) and sits 10 Å away from R1545 and 57 Å from R709. The C domains are almost identical to those of fV (rmsd = 0.88 Å over 115 Cα atoms for the C1 domain and rmsd = 1.19 Å over 130 Cα atoms for the C2 domain), and so are the A1 (rmsd = 1.02 Å over 291 Cα atoms) and A3 (rmsd = 1.07 Å over 262 Cα atoms) domains. Most of the differences between fV short and fV reside in the A2 domain (rmsd = 2.05 Å over 274 Cα atoms) that, in addition to the site of thrombin activation at R709 and the site of APC cleavage at R506, houses the gate (696YDYQNRL702) and the lid (672ESTVMATRKMHDRLEPEDEE691) that play an important role in the prothrombinase complex.16 In fV short, the gate shifts 18 Å toward R709 and changes its conformation, with the side chains of Y696 and R701 pointing to opposite directions, as observed in fVa in the prothrombinase complex.16 More substantial rearrangements cause the lid to flip almost 90° counterclockwise and to expose the acidic segment 658PDDDEDSYEIFEPP671 in close proximity to the AR (Figures 2A and 4A). The conformational change defines a locale for new interactions with positively charged moieties (see further) and the protease domain of fXa,51,52 consistent with the role of the segment 659DDDED663 in controlling the rate of prothrombin activation.50
Cryo-EM structure of fV short. The cryo-EM structure of fV short was solved at atomic (3.2 Å) resolution and reveals all residues (1493 total), including the short (133 residues total) B domain, of the A1-A2-B-A3-C1-C2 assembly. The protein is rendered in surface representation with the constitutive domains colored in wheat (A1), pale green (A2), light blue (B), pale yellow (A3), light pink (C1), and pale cyan (C2) and features an overall organization similar (rmsd = 2.22 Å over 1134 Cα atoms) to that of fV reported recently.12 (A) The C domains align edge-to-edge to define a membrane-binding platform that supports the A1 and A3 domains side by side, with the A2 domain on top of them. The A2 domain houses the gate (blue; 696YDYQNRL702) and the lid (orange; 672ESTVMATRKMHDRLEPEDEE691) that play an important role in the prothrombinase complex.16 The sites of thrombin activation at R709 and R1545 are clearly visible in the A2 and B domains, as are the sites of APC cleavage at R306 and R506. The B domain (133 residues total) is resolved in its entirety and stretches across the entire width of the protein, from R709 to R1545, making contacts with the A1, A2, and A3 domains (Table 2) and being suspended over the C1 and C2 domains. (B and C) After reaching the opposite side of fV, near the site of thrombin cleavage at R1545, the B domain loops over the A3 domain in the back of the protein, pointing upward toward the lid in the A2 domain. Residues of the HPC4 tag (supplemental Figures 1C and 3) were removed from this rendering for clarity.
Cryo-EM structure of fV short. The cryo-EM structure of fV short was solved at atomic (3.2 Å) resolution and reveals all residues (1493 total), including the short (133 residues total) B domain, of the A1-A2-B-A3-C1-C2 assembly. The protein is rendered in surface representation with the constitutive domains colored in wheat (A1), pale green (A2), light blue (B), pale yellow (A3), light pink (C1), and pale cyan (C2) and features an overall organization similar (rmsd = 2.22 Å over 1134 Cα atoms) to that of fV reported recently.12 (A) The C domains align edge-to-edge to define a membrane-binding platform that supports the A1 and A3 domains side by side, with the A2 domain on top of them. The A2 domain houses the gate (blue; 696YDYQNRL702) and the lid (orange; 672ESTVMATRKMHDRLEPEDEE691) that play an important role in the prothrombinase complex.16 The sites of thrombin activation at R709 and R1545 are clearly visible in the A2 and B domains, as are the sites of APC cleavage at R306 and R506. The B domain (133 residues total) is resolved in its entirety and stretches across the entire width of the protein, from R709 to R1545, making contacts with the A1, A2, and A3 domains (Table 2) and being suspended over the C1 and C2 domains. (B and C) After reaching the opposite side of fV, near the site of thrombin cleavage at R1545, the B domain loops over the A3 domain in the back of the protein, pointing upward toward the lid in the A2 domain. Residues of the HPC4 tag (supplemental Figures 1C and 3) were removed from this rendering for clarity.
Density map of the B domain of fV short. Our recombinant fV short variant is 1 residue shorter than that from the transcripts of patients with East Texas bleeding disorder or healthy individuals32 because it lacks residue D756 or N756 in the B domain generated by a new codon across the I755-L1459 junction. Overall, the B domain fits the electron density map with the backbone and bulky side chains well, but features a higher degree of disorder in the portion proximal to the I755-L1459 junction, especially around the 726TALALENGT734 segment (A). The I755-L1459 junction is well-defined (B) and sits 10 Å away from R1545 and 57 Å from R709 (not shown), with I755 coming in contact with V1544. The C-terminal segment 1534RDPDNIAAWYLR1545 is the best-defined portion of the B domain (E). The fit of the electron density map progressively improves in the portion distal to the I755-L1459 junction (BCDE) because of tighter interactions with the A2 and A3 domains (Table 2; Figures 4 and 6). The preAR, from the splice site to T1492, comprises the hydrophobic patch 1481PLVIVG1486 29 and is well-defined (B), and so are the ascending (C) and descending (D) segments of the AR, from D1493 to D1537. The 1507EDDY1510 wedge (D) penetrates a crevice in the A2 domain and defines the turn where the B domain changes direction (Figures 2B and 4B).
Density map of the B domain of fV short. Our recombinant fV short variant is 1 residue shorter than that from the transcripts of patients with East Texas bleeding disorder or healthy individuals32 because it lacks residue D756 or N756 in the B domain generated by a new codon across the I755-L1459 junction. Overall, the B domain fits the electron density map with the backbone and bulky side chains well, but features a higher degree of disorder in the portion proximal to the I755-L1459 junction, especially around the 726TALALENGT734 segment (A). The I755-L1459 junction is well-defined (B) and sits 10 Å away from R1545 and 57 Å from R709 (not shown), with I755 coming in contact with V1544. The C-terminal segment 1534RDPDNIAAWYLR1545 is the best-defined portion of the B domain (E). The fit of the electron density map progressively improves in the portion distal to the I755-L1459 junction (BCDE) because of tighter interactions with the A2 and A3 domains (Table 2; Figures 4 and 6). The preAR, from the splice site to T1492, comprises the hydrophobic patch 1481PLVIVG1486 29 and is well-defined (B), and so are the ascending (C) and descending (D) segments of the AR, from D1493 to D1537. The 1507EDDY1510 wedge (D) penetrates a crevice in the A2 domain and defines the turn where the B domain changes direction (Figures 2B and 4B).
Shift of the lid in the A2 domain. (A) Most of the differences between fV short and fV reside in the A2 domain (rmsd = 2.05 Å over 274 Cα atoms), which, in addition to the site of thrombin activation at R709 and the site of APC cleavage at R506, houses the gate (blue; 696YDYQNRL702) and the lid (orange; 672ESTVMATRKMHDRLEPEDEE691)that play an important role in the prothrombinase complex.16 In fV short, the gate shifts 18 Å at Y698 toward R709, and the lid flips almost 90° counterclockwise, with M676 and R679 moving 24 Å and 29 Å, respectively. The conformation of the gate becomes similar to that observed in fVa in the prothrombinase complex,16 with the side chains of Y696 and R701 pointing in opposite directions. The rearrangement of the lid is more drastic and exposes the acidic segment 658PDDDEDSYEIFEPP671 in a position where it could provide a locale for binding the protease domain of fXa in the conformation observed in the prothrombinase complex16 (Figure 5). This structural feature of fV short may account for its documented fVa-like activity1,5,8,9,14 and is consistent with the role of the segment 659DDDED663 in controlling the rate of cleavage of prothrombin by prothrombinase.50 The electrostatics support this conclusion because this area is in close proximity to the AR and very acidic (supplemental Figure 3). (B) The segment from 1503 to 1511 (cyan sticks) of the AR in the B domain occupies the top of the loop where the B domain changes direction (Figure 2B). The segment penetrates a crevice in the A2 domain, with the 1507EDDY1510 wedge sandwiched between residues from 505 to 516 and from 665 to 672 (green sticks). Shown are the H-bonding interactions between E1507 and R396, D1508, and R505 next to the site of APC cleavage at R506. Y1510 is in hydrophobic interaction with F668. The O atoms of S1505, S1506, and E1507 are in electrostatic clash with the side chain of E669 and may cause the segment 668FEPP671 to relocate the lid relative to its position in fV.
Shift of the lid in the A2 domain. (A) Most of the differences between fV short and fV reside in the A2 domain (rmsd = 2.05 Å over 274 Cα atoms), which, in addition to the site of thrombin activation at R709 and the site of APC cleavage at R506, houses the gate (blue; 696YDYQNRL702) and the lid (orange; 672ESTVMATRKMHDRLEPEDEE691)that play an important role in the prothrombinase complex.16 In fV short, the gate shifts 18 Å at Y698 toward R709, and the lid flips almost 90° counterclockwise, with M676 and R679 moving 24 Å and 29 Å, respectively. The conformation of the gate becomes similar to that observed in fVa in the prothrombinase complex,16 with the side chains of Y696 and R701 pointing in opposite directions. The rearrangement of the lid is more drastic and exposes the acidic segment 658PDDDEDSYEIFEPP671 in a position where it could provide a locale for binding the protease domain of fXa in the conformation observed in the prothrombinase complex16 (Figure 5). This structural feature of fV short may account for its documented fVa-like activity1,5,8,9,14 and is consistent with the role of the segment 659DDDED663 in controlling the rate of cleavage of prothrombin by prothrombinase.50 The electrostatics support this conclusion because this area is in close proximity to the AR and very acidic (supplemental Figure 3). (B) The segment from 1503 to 1511 (cyan sticks) of the AR in the B domain occupies the top of the loop where the B domain changes direction (Figure 2B). The segment penetrates a crevice in the A2 domain, with the 1507EDDY1510 wedge sandwiched between residues from 505 to 516 and from 665 to 672 (green sticks). Shown are the H-bonding interactions between E1507 and R396, D1508, and R505 next to the site of APC cleavage at R506. Y1510 is in hydrophobic interaction with F668. The O atoms of S1505, S1506, and E1507 are in electrostatic clash with the side chain of E669 and may cause the segment 668FEPP671 to relocate the lid relative to its position in fV.
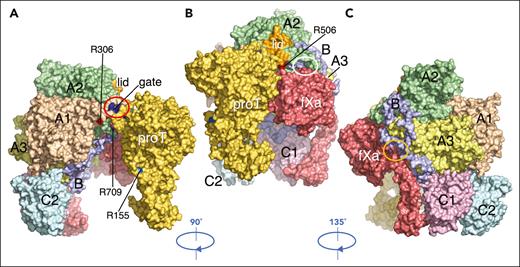
Indeed, the structure of fV short and its electrostatic properties (supplemental Figure 4) define a suitable scaffold for the interaction with fXa and prothrombin that could explain the fVa-like activity of this variant.1,5,8,9,14 A possible complex (Figure 5) built by replacing fVa with fV short in the recent structure of the prothrombin-prothrombinase complex16 would require only small conformational adjustments for productive interactions. Interestingly, the B domain provides no significant hindrance to the binding of prothrombin or fXa aligned against fV short, as observed in the prothrombin-prothrombinase complex.16 Repositioning of the lid in fV short pushes the gate against the backbone of the segment 260LDEDSDRAIE269 of the A chain of prothrombin.10 The clash removes the important interaction between the site of cleavage of prothrombin at R271 with D697 of the gate that directs the A chain to the active site of fXa to promote cleavage at R320 but would be corrected by small adjustments of the gate and the A chain. Repositioning of the lid also removes numerous interactions with fXa observed in the prothrombinase complex16 but exposes the acidic segment 658PDDDEDSYEIFEPP671 (Figures 2A and 4A) for possible new contacts with the enzyme. A backbone clash between the AR segment 1530INSSRDPDNIAAVYLR1545 with the segment 86RKLCSLDN93 of the EGF2 domain of fXa may be removed by a modest (<3 Å) relative rearrangement of the 2 segments. No significant clashes involve fXa and other regions of the B domain, including the I755-L1479 junction at the splice site (Figure 3B).
Putative complex of fV short bound to prothrombin and fXa. Complex of prothrombin (yellow) and fXa (red) bound to fV short (colored as in Figure 2) generated by replacing fVa in the prothrombin-prothrombinase complex16 with the cryo-EM structure of fV short, without further optimization. Overall, the structure of fV short accounts for its fVa-like activity1,5,8,9,14 and points to modest conformational changes needed to optimize interaction with prothrombin and fXa. (A,B) Prothrombin aligns along the C2, A1, and A2 domains and makes no contacts with the B domain. The site of cleavage at R320 (not visible) penetrates the active site of fXa to promote activation along the meizothrombin pathway,16 whereas R271 (not visible) and R155 (light blue) remain widely separated from the enzyme. A backbone clash (red oval) involves the segment 260LDEDSDRAIE269 preceding the site of cleavage of prothrombin at R271 and the segment 693DADYDYQNRL702 of the A2 domain that includes the gate (blue). The clash is triggered by the repositioning of the lid (orange) and removes the important interaction between R271 and D697 of the gate (shifted 12 Å) that directs R320 to the active site of fXa.10,16 Removal of the clash would require slight rearrangement of the gate and/or the A chain of prothrombin. (B,C) fXa aligns along the A2, A3, and C1 domains of fV short. Repositioning of the lid removes numerous interactions with fXa observed in the prothrombinase complex16 and exposes the acidic segment 658PDDDEDSYEIFEPP671 (Figure 2A and 4A) for possible new contacts with the enzyme (white oval). The proximity of this segment to the AR generates an extended surface of negative electrostatic potential (supplemental Figure 4) for the engagement of the protease domain of fXa and explains how intramolecular interaction of this region with the BR in fV may keep the cofactor in its inactive state.1,5-7,9,53 In fV short, this region likely engages the basic C-terminal end of TFPIα, leading to compromised fVa-like activity.7 The proximity to R506 also explains the TFPIα difficult interaction with fVLeiden (R506Q)54,55 and its ability to protect R506 from APC cleavage.27 A backbone clash (orange oval) involves the AR segment 1530INSSRDPDNIAAVYLR1545 of fV short with the segment 86RKLCSLDN93 of the EGF2 domain of fXa. Removal of this clash would require a modest (<3 Å) relative rearrangement of the 2 segments. No significant clashes involve fXa and other regions of the B domain, including the I755-L1459 junction at the splice site of our construct (Figure 3A).
Putative complex of fV short bound to prothrombin and fXa. Complex of prothrombin (yellow) and fXa (red) bound to fV short (colored as in Figure 2) generated by replacing fVa in the prothrombin-prothrombinase complex16 with the cryo-EM structure of fV short, without further optimization. Overall, the structure of fV short accounts for its fVa-like activity1,5,8,9,14 and points to modest conformational changes needed to optimize interaction with prothrombin and fXa. (A,B) Prothrombin aligns along the C2, A1, and A2 domains and makes no contacts with the B domain. The site of cleavage at R320 (not visible) penetrates the active site of fXa to promote activation along the meizothrombin pathway,16 whereas R271 (not visible) and R155 (light blue) remain widely separated from the enzyme. A backbone clash (red oval) involves the segment 260LDEDSDRAIE269 preceding the site of cleavage of prothrombin at R271 and the segment 693DADYDYQNRL702 of the A2 domain that includes the gate (blue). The clash is triggered by the repositioning of the lid (orange) and removes the important interaction between R271 and D697 of the gate (shifted 12 Å) that directs R320 to the active site of fXa.10,16 Removal of the clash would require slight rearrangement of the gate and/or the A chain of prothrombin. (B,C) fXa aligns along the A2, A3, and C1 domains of fV short. Repositioning of the lid removes numerous interactions with fXa observed in the prothrombinase complex16 and exposes the acidic segment 658PDDDEDSYEIFEPP671 (Figure 2A and 4A) for possible new contacts with the enzyme (white oval). The proximity of this segment to the AR generates an extended surface of negative electrostatic potential (supplemental Figure 4) for the engagement of the protease domain of fXa and explains how intramolecular interaction of this region with the BR in fV may keep the cofactor in its inactive state.1,5-7,9,53 In fV short, this region likely engages the basic C-terminal end of TFPIα, leading to compromised fVa-like activity.7 The proximity to R506 also explains the TFPIα difficult interaction with fVLeiden (R506Q)54,55 and its ability to protect R506 from APC cleavage.27 A backbone clash (orange oval) involves the AR segment 1530INSSRDPDNIAAVYLR1545 of fV short with the segment 86RKLCSLDN93 of the EGF2 domain of fXa. Removal of this clash would require a modest (<3 Å) relative rearrangement of the 2 segments. No significant clashes involve fXa and other regions of the B domain, including the I755-L1459 junction at the splice site of our construct (Figure 3A).
Although much shorter than that of fV, the B domain of fV short reveals the architecture of the entire AR (residues 1493-1537) and its preceding segment preAR (residues 1458-1492) housing the hydrophobic patch 1481PLVIVG1486 important for the recognition of TFPIα.26,29,33 Overall, the overlap with the 4 residues at the N-terminal end connecting to R709 and 10 residues at the C-terminal end preceding R1545 detected in the structure of fV12 is very poor (rmsd = 25.2 Å over 14 Cα atoms) because of a very distinct orientation of these segments in the 2 proteins. The B domain of fV short starts after R709 at the C-terminal end of the A2 domain and stretches across the entire width of the protein, making contacts with the A1, A2, and A3 domains (Table 2) but being suspended over and not in contact with the C1 and C2 domains (Figure 2A). After reaching the opposite side, near the site of cleavage at R1545, the B domain loops over the A3 domain in the backbone of the protein, changing direction and pointing upward toward the lid in the A2 domain (Figure 2B-C). The segment from 1503 to 1511 of the AR occupies the top of the loop where the B domain reverses course (Figure 4B) downward, toward R1545 (Figure 2B). The segment penetrates a crevice in the A2 domain with the 1507EDDY1510 wedge, sandwiched between residues from 505 to 516 and from 665 to 672, and pushes the segment 668FEPP671 to trigger relocation of the lid from its position in fV. The shift unmasks the acidic segment 658PDDDEDSYEIFEPP671 (Figures 2A and 4A) next to the AR and generates an extended surface of negative electrostatic potential (supplemental Figure 4) for the engagement of fXa (Figure 5) or positively charged moieties, such as the BR or the C-terminal end of TFPIα. This provides a structural explanation of how TFPIα suppresses fVa-like activity in fV short7 and how intramolecular binding of the BR to this region in fV may keep the cofactor in its inactive state.1,5-7,9,53
Interdomain contacts <5 Å with residues of the B domain
| B domain residue . | Residue . | Domain . |
|---|---|---|
| Q718 | D135 | A1 |
| E720 | H134 | A1 |
| F723 | K167 | A1 |
| E731 | H134 | A1 |
| F736 | W1797 | A3 |
| S738 | K1847 | A3 |
| S739 | R1877 | A3 |
| D1472 | R1603 | A3 |
| F1474 | Y1595 | A3 |
| L1487 | F1666 | A3 |
| I1495 | Y1658 | A3 |
| D1509 | R506 | A2 |
| D1509 | R510 | A2 |
| Y1515 | Y665 | A2 |
| Y1518 | R652 | A2 |
| R1534 | E1768 | A3 |
| B domain residue . | Residue . | Domain . |
|---|---|---|
| Q718 | D135 | A1 |
| E720 | H134 | A1 |
| F723 | K167 | A1 |
| E731 | H134 | A1 |
| F736 | W1797 | A3 |
| S738 | K1847 | A3 |
| S739 | R1877 | A3 |
| D1472 | R1603 | A3 |
| F1474 | Y1595 | A3 |
| L1487 | F1666 | A3 |
| I1495 | Y1658 | A3 |
| D1509 | R506 | A2 |
| D1509 | R510 | A2 |
| Y1515 | Y665 | A2 |
| Y1518 | R652 | A2 |
| R1534 | E1768 | A3 |
The cryo-EM structure of fV short documents a total of 15 residues of the B domain in contact with residues of the A1 (4 total), A2 (4 total), and A3 (8 total) domains. The B domain makes no contacts with the C1 and C2 domains.
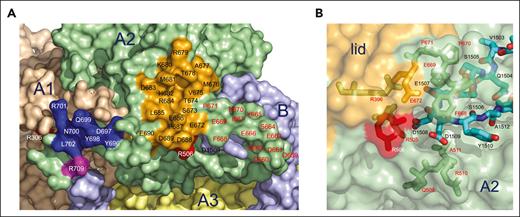
The B domain starts a 180° turn at the 1507EDDY1510 wedge to descend along the A3 domain toward R1545, thereby positioning the entire preAR and AR in the back of fV short (Figure 2C). Intramolecular interactions within the B domain include L1544 with I755, near R1545, and R1534 with N1480 (Figure 6). A strong H-bond connects R1534 to E1668 in the A3 domain and anchors the R1534-N1480 interaction to the surface of the A3 domain. The entire portion from I755 to L1544 defines 4 hydrophobic clusters (HCs) and a patch of acidic residues. The long HC1, from P753 to P1468, occupies the bottom of the B domain and contains 7 residues in a linear arrangement. HC1 continues with the hydrophobic patch 1481PLVIVG1486, identified recently as a locale for the synergistic action of TFPIα and PS in mediating fXa inhibition.29 The patch is part of a more extended cluster (HC2) that also includes F1474, L1475, F1479, and L1487. Residues P1663, W1665, and F1666 are nearby on the surface of the A3 domain (Figure 6), with F1666 interacting with V1487 in HC2. The B domain then continues into HC3, comprising I1497, I1498, V1516, P1517, and Y1518, sealing the ascending and descending portions of the B domain through an intramolecular interaction between I1498 and Y1518. Finally, the B domain reaches R1545 with HC4 defined by V1526, I1530, I1539, A1541, W1542, and L1544, which comes in contact with I755 in HC1. The HCs may contribute to the synergistic recognition of TFPIα and PS. Extensive deletions of fV short support the conclusion that the hydrophobic patch 1481PLVIVG1486 in HC2, but not the segments preceding the patch, are required for this synergism.29 The observation rules out involvement of HC1, but not HC3 and HC4, in TFPIα and PS recognition. The HCs encircle a group of negatively charged residues in the AR that define an acidic cluster (AC) with D1493 and E1496 in the ascending portion, D1519 and D1520 in the descending portion, and D1660, D1661, E1664, and E1668 on the surface of the A3 domain (Figure 5). The AC may work in conjunction with HC2 and the neighboring HC3 and HC4 to provide a locale for interaction with the C-terminal basic region of TFPIα in fV short and the BR in fV.
Functional epitopes of the B domain of fV short. Surface representation of the portion of the B domain bound to the A3 and A2 domains of fV short (Figure 2B) that contains the entire AR (residues 1493-1537) and its preceding segment preAR (residues 1458-1492) housing the hydrophobic patch 1481PLVIVG1486 important for recognition of TFPIα.26,29,33 Residues of the B domain are colored in white (A, F, I, L, M, P, V, W, and Y), cyan (D and E), and light blue (all others) to emphasize the contribution of HCs and acidic residues in the preAR and AR. Intramolecular interactions within the B domain (red ∗) include a hydrophobic coupling between L1544 and I755 near R1545, a H-bond between the side chains of R1534 and N1480 and a hydrophobic contact between I1498 and Y1518. A stronger H-bond connects R1534 to E1668 (cyan) in the A3 domain and anchors the R1534-N1480 interaction to the surface of the A3 domain. The segment I755-L1544 defines 4 HCs and a patch of acidic residues (AC). HC1 occupies the bottom of the B domain and contains 7 residues in a linear arrangement. HC1 continues with the hydrophobic patch 1481PLVIVG148629 as part of HC2 that also includes F1474, L1475, F1479, and L1487. Residues P1663, W1665, and F1666 nearby are colored in white on the surface of the A3 domain, with F1666 interacting with V1487 in HC2. The B domain continues with HC3 that ties the ascending and descending portions of the B domain through an interaction between I1498 and Y1518. The B domain terminates at R1545 with HC4, where L1544 contacts I755 in HC1. The HCs and AC of the B domain of fV short may orchestrate recognition of the C-terminal basic region of TFPIα and its synergistic inhibition of fXa with PS in the fV short/TFPIα/PS/fXa complex. In fV, the HCs and AC of the B domain likely provide the locale for intramolecular engagement of the BR in the B domain that keeps the cofactor in its inactive state. A recent model of the AR has proposed epitopes for binding the BR and the basic C-terminal end of TFPIα.56 Interestingly, the model identifies a hydrophobic core of 9 residues (I1495, I1497, F1498, V1503, I1513, V1516, Y1518, Y1522, and I1530) that may assist the acidic residues E1507, D1509, D1514, D1519, and D1520 in the binding interactions. The general organization is consistent with the features emerged from the cryo-EM structure of fV short, but several residues in the proposed epitopes of the AR56 are unlikely to contribute to binding. Specifically, (i) the 9 hydrophobic residues do not arrange in a well-defined core, with I1495, V1503, and I1530 being widely separated (up to 24 Å); (ii) the side chains of Y1518 and Y1522 are not exposed to solvent; and (iii) E1507 and D1509 are part of the 1507EDDY1510 wedge and completely buried under the A2 domain (Figure 4B).
Functional epitopes of the B domain of fV short. Surface representation of the portion of the B domain bound to the A3 and A2 domains of fV short (Figure 2B) that contains the entire AR (residues 1493-1537) and its preceding segment preAR (residues 1458-1492) housing the hydrophobic patch 1481PLVIVG1486 important for recognition of TFPIα.26,29,33 Residues of the B domain are colored in white (A, F, I, L, M, P, V, W, and Y), cyan (D and E), and light blue (all others) to emphasize the contribution of HCs and acidic residues in the preAR and AR. Intramolecular interactions within the B domain (red ∗) include a hydrophobic coupling between L1544 and I755 near R1545, a H-bond between the side chains of R1534 and N1480 and a hydrophobic contact between I1498 and Y1518. A stronger H-bond connects R1534 to E1668 (cyan) in the A3 domain and anchors the R1534-N1480 interaction to the surface of the A3 domain. The segment I755-L1544 defines 4 HCs and a patch of acidic residues (AC). HC1 occupies the bottom of the B domain and contains 7 residues in a linear arrangement. HC1 continues with the hydrophobic patch 1481PLVIVG148629 as part of HC2 that also includes F1474, L1475, F1479, and L1487. Residues P1663, W1665, and F1666 nearby are colored in white on the surface of the A3 domain, with F1666 interacting with V1487 in HC2. The B domain continues with HC3 that ties the ascending and descending portions of the B domain through an interaction between I1498 and Y1518. The B domain terminates at R1545 with HC4, where L1544 contacts I755 in HC1. The HCs and AC of the B domain of fV short may orchestrate recognition of the C-terminal basic region of TFPIα and its synergistic inhibition of fXa with PS in the fV short/TFPIα/PS/fXa complex. In fV, the HCs and AC of the B domain likely provide the locale for intramolecular engagement of the BR in the B domain that keeps the cofactor in its inactive state. A recent model of the AR has proposed epitopes for binding the BR and the basic C-terminal end of TFPIα.56 Interestingly, the model identifies a hydrophobic core of 9 residues (I1495, I1497, F1498, V1503, I1513, V1516, Y1518, Y1522, and I1530) that may assist the acidic residues E1507, D1509, D1514, D1519, and D1520 in the binding interactions. The general organization is consistent with the features emerged from the cryo-EM structure of fV short, but several residues in the proposed epitopes of the AR56 are unlikely to contribute to binding. Specifically, (i) the 9 hydrophobic residues do not arrange in a well-defined core, with I1495, V1503, and I1530 being widely separated (up to 24 Å); (ii) the side chains of Y1518 and Y1522 are not exposed to solvent; and (iii) E1507 and D1509 are part of the 1507EDDY1510 wedge and completely buried under the A2 domain (Figure 4B).
Discussion
The cryo-EM structure of fV short advances the knowledge recently acquired on fV12 by revealing new features of the B domain, especially the AR that, in fV, likely interacts with the BR to keep the cofactor in its inactive state.1,5-9 The shape of the preAR and AR segments (Figure 6) and the electrostatics in this region (supplemental Figure 4) suggest that the BR may couple with negatively charged residues but also engage several HCs through hydrophobic interactions. These possible BR-AR intramolecular interactions have eluded the recent cryo-EM structure of fV because of the intrinsic disorder of the entire B domain.12 The residues of the preAR and AR regions responsible for these interactions represent new targets for mutagenesis studies that will validate the current structure of fV short and future, high-resolution structures of fV with the B domain resolved in their entirety.
The cryo-EM structure of fV short offers a plausible mechanism for the interaction with TFPIα and PS. The AR has been implicated in the synergistic interaction of TFPIα and PS, leading to sequestration of fXa from the circulation and the mild bleeding phenotype associated with the East Texas bleeding disorder.29,31-33 The preAR and AR of the B domain are organized in 4 HCs of various size that encircle several acidic residues in the AC. Interestingly, a computational model of the AR also identified some hydrophobic and acidic patches,56 but most of their predicted architecture is not confirmed with the cryo-EM structure (Figure 6). HC2 includes the hydrophobic patch 1481PLVIVG1486 responsible for the high-affinity binding of TFPIα and its synergistic cross talk with PS in the inhibition of fXa.29 The effect is retained after deletion of the entire sequence from 713 to 756 or from 1458 to 1480 (67 residues) in the FV712-1481 construct,29 in which only 3 residues separate the site of thrombin cleavage at R709 from the hydrophobic patch. The Cα atom of R709 is >75 Å away from residues of the hydrophobic patch (Figure 2A,B), but that separation may become <10 Å in the FV712-1481 construct. That should bring the hydrophobic patch much closer to R709 and significantly affect the architecture of the B domain. This scenario merits future structural investigation.
The HCs may work in conjunction with the AC in the AR to provide a locale for interaction with TFPIα and account for the much higher (100-fold) affinity compared with fV.6,7,57 TFPIα binds several coagulation factors through its Kunitz domains. The first Kunitz domain inhibits fVIIa bound to tissue factor, and the second inhibits fXa.6,29 Inhibition of fXa is enhanced by PS that binds to the third Kunitz domain58 and functions synergistically with TFPIα in the inhibition of fXa on the surface of negatively charged phospholipids and platelets.59 The third Kunitz domain is also required for optimal cell binding, together with the positively charged C-terminal domain.60 The C-terminal BR of TFPIα has a sequence nearly identical to that of the B domain of fV that maintains the inactive state of the cofactor by binding to the AR.1,5-7,53 Consequently, the C-terminal end of TFPIα binds to the AR of fV and protects it from catalytic attack at R1545.61 The cofactor activity of TFPIα requires the integrity of the R1545 site because cleavage at this site by thrombin abolishes TFPIα binding.6 Peptides corresponding to the C-terminal end of TFPIα competitively inhibit thrombin cleavage at R1545.57,61 Interestingly, binding of TFPIα also takes place in variants of fVa that retain the AR, such as those released from activated platelets.62 The interaction inhibits prothrombinase assembled with these variants on the activated platelet surface by engaging fXa through the second Kunitz domain.57 The tandem engagement of fXa and the AR by TFPIα may be critical to stabilization of the fV short/TFPIα/PS/fXa complex as well, with the third Kunitz domain binding to PS. Indeed, the complex must ensure tight interactions among its components on the surface of negatively charged phospholipids to compensate for the subnanomolar concentrations of fXa, TFPIα, and fV short in the blood.6 The cryo-EM structure of fV short suggests that TFPIα binds to the AR and the nearby acidic segment 658PDDDEDSYEIFEPP671 through its basic C-terminal end and hinders the accessibility of R1545 for thrombin cleavage. In this position, TFPIα would also encroach on R506 (Figure 2AB), thereby explaining the difficulty of interaction with fVLeiden (R506Q)54,55 and the ability to protect R506 from APC cleavage.27 Similar structural interactions are expected of the BR in fV and account for how intramolecular binding to the AR may keep the cofactor in its inactive state.1,5-7,9,53 The third Kunitz domain engages PS likely bound to HC1, HC2, and HC4 of the AR (Figure 6) and in contact with fXa, bound along the A2, A3, C1 domains as observed in the prothrombinase complex16 (see Figure 5) and inhibited by the second Kunitz domain. In this scenario, TFPIα would strongly stabilize the fV short/TFPIα/PS/fXa complex by wrapping around fV short, fXa, and PS, with fXa bound as in the prothrombinase complex and next to PS bound to the back of fV short. A cryo-EM structure of the fV short/TFPIα/PS/fXa complex will test the validity of this hypothesis.
Acknowledgments
The authors gratefully acknowledge Tracey Baird for her assistance with illustrations.
This study was supported in part by the National Institutes of Health National Heart, Lung, and Blood Institute grants HL049413, HL139554, and HL147821 (E.D.C). M.J.R. is supported by the Washington University Center for Cellular Imaging, which is partly funded by Washington University School of Medicine, the Children’s Discovery Institute of Washington University, and St. Louis Children’s Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813), the Foundation for Barnes-Jewish Hospital (3770), the Washington University Diabetes Research Center (DK020579), and The Alvin J. Siteman Cancer Center at Barnes-Jewish Hospital and Washington University School of Medicine (CA091842).
Authorship
Contribution: B.M.M. and L.A.P. performed biochemical preparations, structure determination, and analysis; M.J.R. performed cryo-EM data acquisition; and B.M.M., L.A.P., M.J.R., and E.D.C. analyzed the results and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Enrico Di Cera, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, 1100 South Grand Blvd, St. Louis, MO 63104; e-mail: enrico@slu.edu.
References
Author notes
The structure and maps were deposited in the Protein Data Bank (accession codes 8FDG and EMDB-29011 [composite map], EMDB-29010, EMDB29008, and EMDB-29009). Residues in the PDB file 8FDG are numbered sequentially, and 703 should be added from L756 onward to obtain the corresponding fV number.
Data are available on request from the corresponding author, Enrico Di Cera (enrico@slu.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal