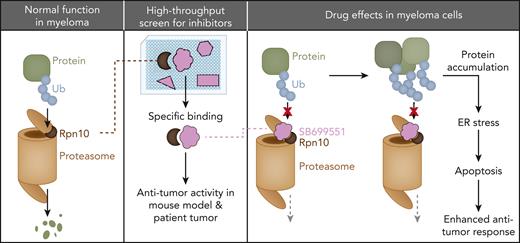
Key Points
Rpn10 plays a pivotal role in MM.
A novel Rpn10 inhibitor, SB, was identified via AlphaScreen.
Abstract
PSMD4/Rpn10 is a subunit of the 19S proteasome unit that is involved with feeding target proteins into the catalytic machinery of the 26S proteasome. Because proteasome inhibition is a common therapeutic strategy in multiple myeloma (MM), we investigated Rpn10 and found that it is highly expressed in MM cells compared with normal plasma cells. Rpn10 levels inversely correlated with overall survival in patients with MM. Inducible knockout or knockdown of Rpn10 decreased MM cell viability both in vitro and in vivo by triggering the accumulation of polyubiquitinated proteins, cell cycle arrest, and apoptosis associated with the activation of caspases and unfolded protein response-related pathways. Proteomic analysis revealed that inhibiting Rpn10 increased autophagy, antigen presentation, and the activation of CD4+ T and natural killer cells. We developed an in vitro AlphaScreen binding assay for high-throughput screening and identified a novel Rpn10 inhibitor, SB699551 (SB). Treating MM cell lines, leukemic cell lines, and primary cells from patients with MM with SB decreased cell viability without affecting the viability of normal peripheral blood mononuclear cells. SB inhibited the proliferation of MM cells even in the presence of the tumor-promoting bone marrow milieu and overcame proteasome inhibitor (PI) resistance without blocking the 20S proteasome catalytic function or the 19S deubiquitinating activity. Rpn10 blockade by SB triggered MM cell death via similar pathways as the genetic strategy. In MM xenograft models, SB was well tolerated, inhibited tumor growth, and prolonged survival. Our data suggest that inhibiting Rpn10 will enhance cytotoxicity and overcome PI resistance in MM, providing the basis for further optimization studies of Rpn10 inhibitors for clinical application.
Introduction
Multiple myeloma (MM), the second most prevalent blood cancer, is incurable and develops from an accumulation of terminally differentiated monoclonal plasma cells (PCs) in the bone marrow (BM). These cells produce high levels of antibodies and other proteins, making them sensitive to disturbances in the proteolytic pathway.1,2 Proteasome inhibitors (PIs) that directly target the 20S core particle (CP), such as the firstline drugs bortezomib and carfilzomib (CFZ), have improved patient survival.3-5 However, they are prone to off-target toxicity and the emergence of drug resistance.6,7 We hypothesized that targeting other parts of the proteosome could enhance PI efficacy or overcome PI resistance, with potentially fewer off-target activities and less toxicity.
The ubiquitin-proteasome system (UPS) mediates protein degradation, largely through8-10 the 26S proteasome, which degrades ubiquitinated proteins.8-10 The proteasome is composed of the 20S CP, which is barrel-shaped, and 1 or 2 19S regulatory particles (RPs), which form a lid that recognizes, binds, and unfolds the ubiquitinated proteins before degradation in the barrel. Within the 19S RP, Rpn10, Rpn13, and Rpn1 capture ubiquitinated proteins through their ubiquitin chain receptors.11-14 Rpn10 is located closest to the substrate translocation channel14 and binds different types of ubiquitin chains but prefers K11 and K48 linkages.15 Targeting Rpn13 triggers cytotoxicity and overcomes bortezomib resistance in MM,16-19 but the role of Rpn10 in MM has not been delineated. We focused on Rpn10 because it is present at the delicate junction of the lid and barrel20; is key to unfolding substrate proteins, such as in yeast21; and has abnormally high expression in different cancers such as breast cancer and hepatocellular carcinoma.22-24 Importantly, hyperexpression of Rpn10 is a high-risk feature in patients with MM treated with a combination of thalidomide, dexamethasone, and bortezomib.25 Here, we validated the clinical and functional significance of Rpn10 in MM, finding that inhibiting it reduces cell viability and improves survival in vivo, thus, suggesting that targeting the 19S RP may improve patient outcomes in MM.
Material and methods
Cell culture and reagents
Normal peripheral blood mononuclear cells (PBMCs) and MM cell lines were cultured in RPMI 1640 complete medium. The other cell lines, including CCRF-CEM, UACC257, SKMEL5, A375, and SU-DHL-4, were cultured in Dulbecco’s modified Eagle medium. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki protocol. MM CD138+ cells, bone marrow stromal cells (BMSCs), and plasmacytoid dendritic cells (pDCs) from patients with MM were isolated and cultured as described previously.26
Drug and reagent sources
Bortezomib was purchased from Selleck Chemicals, LLC. SB699551 (SB) and diUbiquitin (DiUb) were purchased from R&D systems. Bafilomycin A1 (BafA1) and UNC0638 was purchased from Millipore Sigma. NSC697923 and His-tagged Rpn10 recombinant protein were from Enzo Life Sciences, Inc.
Generation of CRISPR/Cas9-KO cell line
For AMO1-CRISPR-Cas9 knockout (KO), AMO1 cells were spinfected with lentivirus expressing inducible Cas9 (GeneCopoeia). Positive cells selected by G418 were then infected with lentivirus expressing single-guide RNA (sgRNA) targeting Rpn10 and selected by puromycin for 7 days. Cas9 protein expression was induced by adding 0.5 μg/mL doxycycline (Dox) in the cultures every other day. KO cells were verified by western blot.
Tandem mass tag–based proteomic analysis
After being cultured with Dox for 4 days, AMO1-sgCT and AMO1 Rpn10–inducible knockout (iKO) cells were collected and lysed, followed by total protein quantification with a Pierce Micro bicinchoninic acid assay. The samples were then reduced, precipitated, and digested using LysC and trypsin. About 50 μg of peptides were then labeled with tandem mass tag and subjected to LC-MS3 proteomic analysis. Peptide spectral matches were filtered to a 1% false discovery rate (FDR) using the target-decoy strategy combined with linear discriminant analysis. P values for the protein differentiation analysis were adjusted for multiple hypothesis testing using the Benjamini-Hochberg method.
Flow cytometric assay
After treatment, cells were washed and resuspended in 2% fetal bovine serum containing phosphate-buffered saline, followed by normal goat serum incubation. Then, the cells were incubated with primary antibody or isotype control (immunoglobulin G). After washing, cells were stained with secondary antibody, washed again, resuspended in staining solution, and subjected to flow cytometry to detect the surface signal.
AlphaScreen high-throughput screening (HTS)
All AlphaScreen assays were performed in a total volume of 10 uL in OptiPlate-384 (PerkinElmer) plates. Briefly, 10 nM of GST-Rpn10 and anti-GST acceptor beads were added to the plate, followed by the addition of compounds. After an hour of incubation, another 4 nM of biotinylated Ub2-7 and streptavidin-donor beads were added to the plate. After incubation in subdued lighting, readings were performed using the EnVision 2102 Multilabel Reader.
In total, 10 000 compounds selected from Institute of Chemistry and Cell Biology (Harvard Medical School, Harvard University) library were used for screening. Results are the percent inhibition relative to the no-compound control from the same assay plate. Assay robustness was confirmed through the calculation of Z-factors.
Field effect biosensor (FEB) assay
Recombinant His-tagged hRpn10 was immobilized on a graphene chip surface (Resistomix LLC). After immobilization and equilibration, the protein surface was exposed to ligands, including DiUb or the compounds SB or UNC0638, separately during the first 5 minutes, followed by washing out. Real-time changes in the electrical current (I-response) and capacitance (C-response) of the chip were measured. The dissociation constant (Kd) is calculated as a median average over the test points. Three independent experiments were performed.
Microscale thermophoresis (MST) analysis
Green fluorescent protein (GFP)–fusion Rpn10 protein was purified from Escherichia coli by immobilized metal affinity chromatography and size exclusion chromatography. The binding affinity between GFP-Rpn10 and SB was measured in a buffer containing Tris-HCl (50 mM) at pH 7.4, NaCl (150 mM), and dithiothreitol (1 mM) using a MONOLITH NT.115 system (NanoTemper Technologies). A total volume of 10 μL of GFP-Rpn10 (1 μM) was mixed with 10 μL of twofold serial-diluted ligand, which ranged from 1 to 25 600 nM. After a 10-minute incubation, samples were transferred into the capillaries and measured. Data were plotted by MO.Affinity Analysis (NanoTemper Technologies).
Human MM xenograft model
Animal model studies were performed as described previously.26 Briefly, CB17 severe combined immunodeficiency mice were subcutaneously inoculated with 5.0 × 106 MM.1S/MM.1S-Rpn10 knockdown (KD) cells or CCRF-CEM cells, or 1.0 × 106 HCT116 or HEK293 cells, or AMO1 Rpn10 short hairpin RNA (shRNA) cells. For the inducible KD experiment, mice were continuously fed with an irradiated 0.0625% Dox diet. For the compound treatment experiment, when tumors were measurable (100 mm3), mice were treated on a 3-times-weekly schedule with vehicle or SB. Mice were euthanized when tumor volume reached the institutional limit (2000 mm3). All animal experimental protocols were approved by and conformed to the relevant regulatory standards of the Institutional Animal Care and Use Committee at the Dana-Farber Cancer Institute.
Statistical analysis
Statistical significance was derived using the 2-tailed Student t test (∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗P ≤ .001, ∗∗∗∗P ≤ .0001). The survival of mice was analyzed by GraphPad Prism software (Systat Software, San Jose, CA).
Results
Expression and clinical significance of Rpn10 in MM
The ubiquitin receptor Rpn13 is a promising therapeutic target that can overcome bortezomib resistance.18 We investigated whether Rpn10 plays a similar role by making CRISPR/Cas9-mediated inducible KOs for Rpn13 and Rpn10, separately. In AMO1 cells, the steady-state depletion of either gene significantly inhibited cell proliferation (Figure 1A; supplemental Figure 1A, available on the Blood website) and caused an accumulation of ubiquitinated proteins (supplemental Figure 1B), with Rpn10 having a larger effect. In addition, we confirmed that Rpn10, but not Rpn13, has a role in the immunoproteasome (iPS)27 by measuring iPS activity (supplemental Figure 1C). These results suggest that Rpn10 is a more attractive target than Rpn13 in MM.
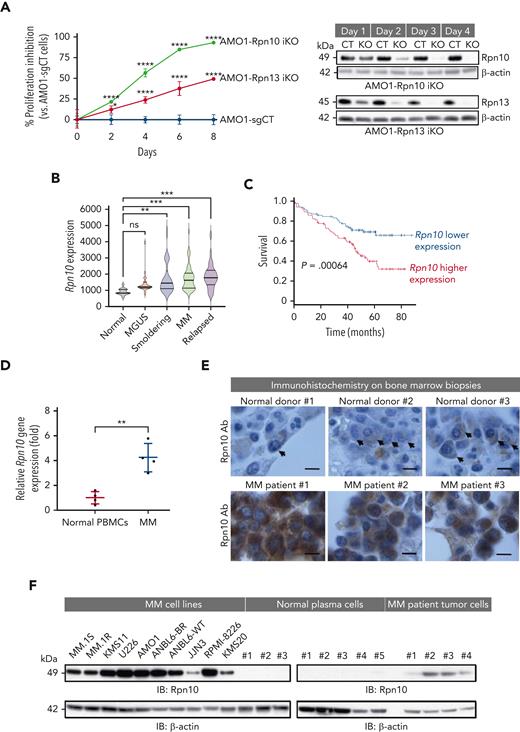
Characterization of ubiquitin receptor Rpn10 in MM cells. (A) Dox-inducible Cas9 AMO1 cells were made by infecting AMO1 cells with lentivirus-packaged inducible Cas9 plasmid followed by G418 selection. These cells then were infected with either control single-guide RNA (sgRNA) or Rpn13- or Rpn10-targeted sgRNA and selected with puromycin to generate stable iKO cells. Cell growth was evaluated by a CellTiter-Glo (CTG) assay. Immunoblot showing Rpn13 and Rpn10 expression, respectively, in corresponding KO cells to test the KO efficiency. Data are shown as the mean ± standard deviation (SD) of triplicates. (B) Gene expression data were collected using Affymetrix Human Genome U133A array platform. Rpn10 expression in the different stages of MM development, including normal CD138+ cells (n = 15), monoclonal gammopathy of undetermined significance (MGUS) (n = 22), smoldering (n = 24), newly diagnosed MM (n = 69), and relapsed MM (n = 28) (data accession number GSE6477). (C) Kaplan-Meier plots of Rpn10 expression vs overall survival of patients with MM. Analysis was performed using samples from 170 newly diagnosed patients with MM (data accession number GSE39754; P = .0064). (D) Total cellular RNA from purified CD138+ cells from BM of patients with MM or normal healthy donor PBMCs was subjected to reverse transcription polymerase chain reaction analysis, followed by real-time polymerase chain reaction using primers designed to recognize sequences internal to Rpn10 (mean ± SD, n = 4). (E) Immunohistochemistry analysis of BM biopsies from normal donors and MM patients showing Rpn10 expression (scale bar, 5 μm). (F) Protein lysates from a panel of MM cell lines, normal PCs, or primary cells from patients with MM were subjected to immunoblotting (IB) using Rpn10 and β-actin antibodies. Ab, antibody; CT, control; ns, not significant.
Characterization of ubiquitin receptor Rpn10 in MM cells. (A) Dox-inducible Cas9 AMO1 cells were made by infecting AMO1 cells with lentivirus-packaged inducible Cas9 plasmid followed by G418 selection. These cells then were infected with either control single-guide RNA (sgRNA) or Rpn13- or Rpn10-targeted sgRNA and selected with puromycin to generate stable iKO cells. Cell growth was evaluated by a CellTiter-Glo (CTG) assay. Immunoblot showing Rpn13 and Rpn10 expression, respectively, in corresponding KO cells to test the KO efficiency. Data are shown as the mean ± standard deviation (SD) of triplicates. (B) Gene expression data were collected using Affymetrix Human Genome U133A array platform. Rpn10 expression in the different stages of MM development, including normal CD138+ cells (n = 15), monoclonal gammopathy of undetermined significance (MGUS) (n = 22), smoldering (n = 24), newly diagnosed MM (n = 69), and relapsed MM (n = 28) (data accession number GSE6477). (C) Kaplan-Meier plots of Rpn10 expression vs overall survival of patients with MM. Analysis was performed using samples from 170 newly diagnosed patients with MM (data accession number GSE39754; P = .0064). (D) Total cellular RNA from purified CD138+ cells from BM of patients with MM or normal healthy donor PBMCs was subjected to reverse transcription polymerase chain reaction analysis, followed by real-time polymerase chain reaction using primers designed to recognize sequences internal to Rpn10 (mean ± SD, n = 4). (E) Immunohistochemistry analysis of BM biopsies from normal donors and MM patients showing Rpn10 expression (scale bar, 5 μm). (F) Protein lysates from a panel of MM cell lines, normal PCs, or primary cells from patients with MM were subjected to immunoblotting (IB) using Rpn10 and β-actin antibodies. Ab, antibody; CT, control; ns, not significant.
We analyzed the gene expression of Rpn10 using a public data set (GSE6477) and found that, except in monoclonal gammopathy of undetermined significance, Rpn10 gene expression was higher in all stages of MM than in normal PCs (Figure 1B). Furthermore, its expression in BM biopsy samples was inversely correlated with overall survival in 170 patients with MM enrolled in a clinical trial (GSE39754) (Figure 1C). We then confirmed by real-time polymerase chain reaction (RT-PCR) that Rpn10 is highly expressed in cells from patients with MM compared with normal PBMCs (Figure 1D). Consistent with these findings, immunohistochemistry studies using BM biopsies from patients with MM and healthy donors showed significantly higher Rpn10 expression in MM cells than in normal cells (Figure 1E). Similar results were observed in both MM cell lines and patient tumor cells compared with normal PCs (Figure 1F). Taken together, these data suggest a role for Rpn10 in the pathogenesis of MM.
Functional significance of Rpn10 in vitro and in vivo
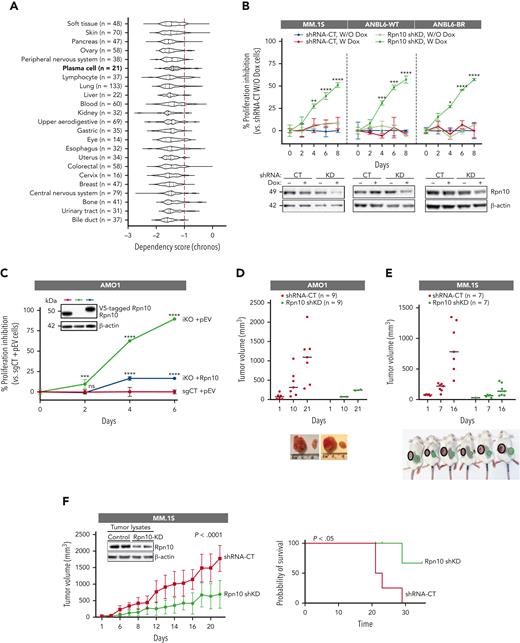
Dependency score analysis by DepMap showed that Rpn10 was essential across many cancer cells, including most of the MM cell lines (Figure 2A). To evaluate the functional significance of Rpn10 in MM, we made an CRISPR/Cas9 KO of Rpn10 (AMO1-iKO cells) in the AMO1 MM cell line, which had one of the highest Rpn10 expression values among cell lines for both messenger RNA (supplemental Figure 2A) and protein (Figure 1F). We also used the HEK293 cell line to model the complete deletion of Rpn10. Steady-state depletion of Rpn10 induced by Dox completely prevented cell proliferation and viability in the AMO1 MM cell line (Figure 1A). KO of Rpn10 in the HEK293 cell line slowed down cell growth (supplemental Figure 2B). Furthermore, we confirmed the growth inhibitory effects by knocking down Rpn10 using 3 different shRNAs in the MM.1S, AMO1, and HCT116 cell lines, as well as in the bortezomib-sensitive and -resistant ANBL6 cell lines (Figure 2B; supplemental Figure 2C-E). When we added Rpn10 back to the AMO1-iKO cells, the inhibited cell growth was restored, indicating that the repression was not because of off-target effects (Figure 2C). Consistent with this, the withdrawal of Dox after a few days of incubation also restored the suppressed cell growth along with the re-expression of Rpn10 (supplemental Figure 2F).
Functional significance of Rpn10. (A) Dependency scores of Rpn10 across cancers based on CRISPR data sets in the DepMap web portal. A lower chronos score indicates that the gene of interest is essential in a given cell line. Score 0 means the gene is not essential, whereas score –1 is comparable with the median of all panessential genes (red line). (B) The proliferation of the MM.1S and ANBL6 inducible Rpn10-shRNA KD cell lines were analyzed using a CTG assay (mean ± SD, n = 3) when cultured with or without Dox. Inducible KD of Rpn10 was achieved using pTRIPz-mCherry vector containing Rpn10-shRNA or scramble control. (C) The stable adding back cell line was achieved when AMO1 Rpn10-iKO cells were transfected with lentivirus-packaged V5-tagged Rpn10 plasmid or empty plasmid (pEV), followed by blasticidin selection. Cell proliferation was measured by a CTG assay (mean ± SD, n = 3). Immunoblot shows the expression levels of Rpn10. Human AMO1 Rpn10-shRNA KD cells (D) or MM.1S Rpn10-shRNA KD cells (E-F) were subcutaneously inoculated into CB17 severe combined immunodeficiency mice. In the early prevention model (D-E), a cohort of mice was treated with an irradiated 0.0625% Dox diet (1-6 mg of Dox per mouse per day) continuously starting 5 days after injection. In the late prevention model (n = 10) (F), mice were treated with an irradiated 0.0625% Dox diet after the tumor became visible and the volume was ∼100 mm3. The average and standard deviation of tumor volume (mm3) are shown vs the time when the tumor was measured (mean tumor volume ± SD). Kaplan-Meier plots show survival in mice (right). Internal blot shows Rpn10 expression of the tumor lysates.
Functional significance of Rpn10. (A) Dependency scores of Rpn10 across cancers based on CRISPR data sets in the DepMap web portal. A lower chronos score indicates that the gene of interest is essential in a given cell line. Score 0 means the gene is not essential, whereas score –1 is comparable with the median of all panessential genes (red line). (B) The proliferation of the MM.1S and ANBL6 inducible Rpn10-shRNA KD cell lines were analyzed using a CTG assay (mean ± SD, n = 3) when cultured with or without Dox. Inducible KD of Rpn10 was achieved using pTRIPz-mCherry vector containing Rpn10-shRNA or scramble control. (C) The stable adding back cell line was achieved when AMO1 Rpn10-iKO cells were transfected with lentivirus-packaged V5-tagged Rpn10 plasmid or empty plasmid (pEV), followed by blasticidin selection. Cell proliferation was measured by a CTG assay (mean ± SD, n = 3). Immunoblot shows the expression levels of Rpn10. Human AMO1 Rpn10-shRNA KD cells (D) or MM.1S Rpn10-shRNA KD cells (E-F) were subcutaneously inoculated into CB17 severe combined immunodeficiency mice. In the early prevention model (D-E), a cohort of mice was treated with an irradiated 0.0625% Dox diet (1-6 mg of Dox per mouse per day) continuously starting 5 days after injection. In the late prevention model (n = 10) (F), mice were treated with an irradiated 0.0625% Dox diet after the tumor became visible and the volume was ∼100 mm3. The average and standard deviation of tumor volume (mm3) are shown vs the time when the tumor was measured (mean tumor volume ± SD). Kaplan-Meier plots show survival in mice (right). Internal blot shows Rpn10 expression of the tumor lysates.
To investigate the in vivo effects of Rpn10 depletion, we used the early prevention model, in which Rpn10 KD was induced by 3 weeks of Dox treatment starting 5 days after the cells were subcutaneously inoculated. For the AMO1 cells, 7 of 9 mice with scrambled shRNA developed tumors, whereas only 2 of 9 mice with Rpn10 shRNA did (Figure 2D). For MM.1S cells, shKD of Rpn10 significantly delayed tumorigenesis and produced smaller tumors than the control mice (Figure 2E). In the late treatment model, in which the mice were treated with Dox when the tumors from MM.1S cells had grown to ∼100 mm3, Rpn10 shKD inhibited tumor growth and prolonged survival (Figure 2F). This was confirmed in HEK293 cells in the 3D bioprinting model and the late treatment model (supplemental Figure 2G-H). These data nominate Rpn10 as a potential therapeutic target in MM cells.
Proteomic analysis of AMO1 Rpn10 iKO
To characterize the overall effects of Rpn10 in MM, we performed proteomic analysis on AMO1-iKO and control cells under Dox induction (Figure 3A) and found that 209 proteins were significantly upregulated and 149 proteins were downregulated (FDR < 0.05) upon Rpn10 KO (Figure 3B). Using gene set enrichment analysis (GSEA), we found an enrichment of downregulated proteins mainly in cell cycle–related pathways, including the G2-M phase transition, DNA replication, nuclear division, and chromosome segregation (Figure 3C). Our data show that Rpn10 KD triggered similar, albeit not identical, downstream pathways in MM.1S vs AMO1 cells. (supplemental Figure 3A). We found an enrichment of upregulated proteins in the mitochondrial and antigen presentation–related pathways, as well as the lysosomal transport pathway, which is related to autophagy, in Rpn10 iKO cells (Figure 3C).
Proteomic analysis of Rpn10 iKO. After being cultured with Dox for 4 days, AMO1 Rpn10-iKO cells were subjected to proteomic analysis by multiplexed proteomics with tandem mass spectrometry. (A) Volcano plot of differentially expressed genes in AMO1 Rpn10-iKO cells compared with those in control cells. Blue and red dots represent the genes (P < .05) that were downregulated or upregulated, respectively. (B) Heatmap showing significantly (false discovery rate [FDR] < .05) altered genes in Rpn10 iKO cells. (C) Gene set enrichment analysis (GSEA) and gene ontology biological process (GOBP) significantly enriched after Rpn10 iKO. For all pathways shown, FDR < 10%. (D) GSEA-derived enrichment plots for the lysosomal transport pathway (left). AMO1 Rpn10-iKO cells were cultured with Dox as described for the indicated time points (middle) and when cultured with Dox, 25 nM BafA1 was added to the culture of Rpn10 iKO cells for the last 16 hours of day 4 (right). Cells were then lysed and subjected to immunoblot using antibodies against p62, LC3, LAMP2, or β-actin. (E) GSEA-derived enrichment plots for antigen processing and presentation of peptide antigen (left). AMO1 Rpn10-iKO cells were cultured with Dox for 4 days. Cells were subjected to flow cytometry after staining with anti–HLA-DR or anti–HLA-DQ Abs and 7AAD (middle). 7AAD– cells were gated out, and the surface expression of HLA-DR or DQ expression was quantified. Rpn10-iKO cells, from which Dox was washed out, were added to total BM-derived mononuclear cells of patients with MM (n = 4) (right). Degranulation marker CD107a was measured on CD3+CD4+ T cells or CD3–/CD56+ natural killer (NK) cells. FC, fold change; MFI, mean fluorescence intensity.
Proteomic analysis of Rpn10 iKO. After being cultured with Dox for 4 days, AMO1 Rpn10-iKO cells were subjected to proteomic analysis by multiplexed proteomics with tandem mass spectrometry. (A) Volcano plot of differentially expressed genes in AMO1 Rpn10-iKO cells compared with those in control cells. Blue and red dots represent the genes (P < .05) that were downregulated or upregulated, respectively. (B) Heatmap showing significantly (false discovery rate [FDR] < .05) altered genes in Rpn10 iKO cells. (C) Gene set enrichment analysis (GSEA) and gene ontology biological process (GOBP) significantly enriched after Rpn10 iKO. For all pathways shown, FDR < 10%. (D) GSEA-derived enrichment plots for the lysosomal transport pathway (left). AMO1 Rpn10-iKO cells were cultured with Dox as described for the indicated time points (middle) and when cultured with Dox, 25 nM BafA1 was added to the culture of Rpn10 iKO cells for the last 16 hours of day 4 (right). Cells were then lysed and subjected to immunoblot using antibodies against p62, LC3, LAMP2, or β-actin. (E) GSEA-derived enrichment plots for antigen processing and presentation of peptide antigen (left). AMO1 Rpn10-iKO cells were cultured with Dox for 4 days. Cells were subjected to flow cytometry after staining with anti–HLA-DR or anti–HLA-DQ Abs and 7AAD (middle). 7AAD– cells were gated out, and the surface expression of HLA-DR or DQ expression was quantified. Rpn10-iKO cells, from which Dox was washed out, were added to total BM-derived mononuclear cells of patients with MM (n = 4) (right). Degranulation marker CD107a was measured on CD3+CD4+ T cells or CD3–/CD56+ natural killer (NK) cells. FC, fold change; MFI, mean fluorescence intensity.
The UPS and autophagy are the 2 main protein degradation pathways, and they can compensate for deficits in the other.28,29 Therefore, with proteasome function diminished by Rpn10 KO, we investigated whether there was an increase in autophagy. We found elevated expression of p62,30 LC3,30 and LAMP231 in AMO1-iKO cells and MM.1S KD cells and confirmed this increase was owing to autophagy by applying BafA132 (Figure 3D; supplemental Figure 3B-C).
Autophagy can deliver peptide antigens from their intracellular source proteins to major histocompatibility complex (MHC) class II molecules for presentation to CD4+ T cells.33,34 Interestingly, our proteomic data showed that antigen processing and presentation via the MHC class II pathway were enriched in AMO1-iKO cells. We then evaluated the level of MHC class II cell-surface receptors HLA-DR and HLA-DQ and found that both were increased on the cell surface (Figure 3E). This was confirmed in MM.1S Rpn10 KD cells (supplemental Figure 3D). Moreover, we found activation of CD3+/CD4+ T cells and CD3−/CD56+ natural killer (NK) cells after coculture with Dox-induced AMO1-iKO cells and BM-derived mononuclear cells from patients with MM. We also found that pathways related to T-cell activation and immune response were enriched in MM.1S KD cells (supplemental Figure 3A). Together, these data indicate an enhanced immune response, but unlike CD4+ cells, we did not find changes in CD8+ T cells in Rpn10-targeted cells (data not shown), which can be because of different cytogenetic characteristics and/or MM surfaceomes of MM.1S and AMO1 cells.
Rpn10 blockade induces cell death by cell cycle arrest and apoptosis
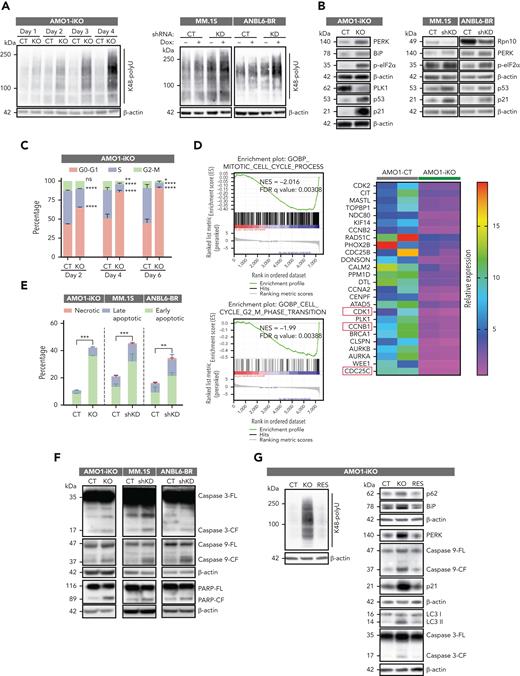
We observed a robust accumulation of K48 polyubiquitinated proteins during steady-state–inducible depletion of Rpn10 in AMO1-iKO cells, MM.1S KD cells, and ANBL6-BR KD cells (Figure 4A). A similar accumulation was noted in Rpn10–small interfering RNA (siRNA)–transfected MM.1S or AMO1 cells and in Rpn10 KO HEK293 cells, as well as in a reporter cell line expressing ubiquitin-tagged GFPu-1, which is constitutively targeted for proteasome degradation (supplemental Figure 4A-B). These data validate Rpn10 as a ubiquitin receptor in the proteasome and that Rpn10 blockade, like PIs, inhibits proteasome-mediated protein degradation.
Rpn10 blockade induces cell death. (A) Polyubiquitylated proteins were detected in AMO1 Rpn10-iKO cells, MM.1S Rpn10-shKD cells, and ANBL6 Rpn10-shKD cells by western blot analysis. (B) Protein lysates from indicated cell lines were subjected to immunoblotting using antibodies against Rpn10, p-eIF2α, PERK, BiP, p53, p21, PLK1, or β-actin. (C) AMO1 Rpn10-iKO cells were cultured with Dox for indicated time points and fixed in 70% ethanol. After washing with phosphate-buffered saline, cells were stained with propidium iodide, and the DNA content of cells was then analyzed using fluorescence-activated cell sorter. Bar graph shows percentage of cell populations in the G1, G2/M, or S phases of the cell cycle. (D) GSEA-derived enrichment plots for mitotic cell cycle pathway and cell cycle G2-M phase transition pathway (left) and heatmap showing proteins related to G2-M phase transition enriched in AMO1-CT cells (right) have been shown. MM.1S shRNA KD cells, ANBL6-BR shRNA KD cells, and AMO1 Rpn10-iKO cells were cultured with Dox for indicated time points and then analyzed for apoptosis with annexin V/4′,6-diamidino-2-phenylindole double staining assay (mean ± SD, n = 3) (E) or protein lysates were then subjected to immunoblotting using antibodies against caspase-3, caspase-9, PARP, or β-actin (F). (G) AMO1 sgRNA-CT cells transfected with lentivirus-packaged pEV, referred to as CT; AMO1 Rpn10-iKO cells transfected with lentivirus-packaged pEV vector, referred to as KO; and AMO1 Rpn10-iKO transfected with lentivirus-packaged V5-tagged Rpn10 plasmid, referred to as RES. Protein lysates from CT, KO, or RES cells were then subjected to immunoblotting using antibodies against Rpn10 or K48 polyubiquitin, p62, LC3, BiP, PERK, caspase-3, caspase-9, p21, or β-actin.
Rpn10 blockade induces cell death. (A) Polyubiquitylated proteins were detected in AMO1 Rpn10-iKO cells, MM.1S Rpn10-shKD cells, and ANBL6 Rpn10-shKD cells by western blot analysis. (B) Protein lysates from indicated cell lines were subjected to immunoblotting using antibodies against Rpn10, p-eIF2α, PERK, BiP, p53, p21, PLK1, or β-actin. (C) AMO1 Rpn10-iKO cells were cultured with Dox for indicated time points and fixed in 70% ethanol. After washing with phosphate-buffered saline, cells were stained with propidium iodide, and the DNA content of cells was then analyzed using fluorescence-activated cell sorter. Bar graph shows percentage of cell populations in the G1, G2/M, or S phases of the cell cycle. (D) GSEA-derived enrichment plots for mitotic cell cycle pathway and cell cycle G2-M phase transition pathway (left) and heatmap showing proteins related to G2-M phase transition enriched in AMO1-CT cells (right) have been shown. MM.1S shRNA KD cells, ANBL6-BR shRNA KD cells, and AMO1 Rpn10-iKO cells were cultured with Dox for indicated time points and then analyzed for apoptosis with annexin V/4′,6-diamidino-2-phenylindole double staining assay (mean ± SD, n = 3) (E) or protein lysates were then subjected to immunoblotting using antibodies against caspase-3, caspase-9, PARP, or β-actin (F). (G) AMO1 sgRNA-CT cells transfected with lentivirus-packaged pEV, referred to as CT; AMO1 Rpn10-iKO cells transfected with lentivirus-packaged pEV vector, referred to as KO; and AMO1 Rpn10-iKO transfected with lentivirus-packaged V5-tagged Rpn10 plasmid, referred to as RES. Protein lysates from CT, KO, or RES cells were then subjected to immunoblotting using antibodies against Rpn10 or K48 polyubiquitin, p62, LC3, BiP, PERK, caspase-3, caspase-9, p21, or β-actin.
Accumulation of ubiquitinated (misfolded/unfolded) proteins triggers the unfolded protein response and apoptosis35 through stress on the endoplasmic reticulum (ER).35 Importantly, Rpn10 depletion in AMO1, MM.1S, and ANBL6-BR upregulates ER stress proteins (p-eIF2α, PERK, and BiP) and p53/p21 apoptotic signaling (Figure 4B), which is known to inhibit cyclin-dependent kinase.
Dox-treated AMO1-iKO cells showed increase in G0-G1 cell cycle arrest phase (Figure 4C) and decrease in proteins related to mitosis and the G2-M phase (Figure 4D). In particular, genes in the E2F pathway were downregulated, indicating cell cycle arrest via the p53-p21-RB-E2F axis (supplemental Figure 4C). These results were confirmed using MM.1S cells (supplemental Figure 4D). The apoptosis analysis in Rpn10 AMO1-iKO, Rpn10-shRNA MM.1S, and ANBL6.BR cells showed an increase in early and late apoptotic cell populations (Figure 4E; supplemental Figure 4E) associated with PARP cleavage and activation of caspase-3 and caspase-9 (Figure 4F). Importantly, the depletion of Rpn10 caused the accumulation of K48 polyubiquitinated proteins, increased autophagy, triggered the ER stress response, and cleaved caspases, and these signaling events were restored by adding back wild-type Rpn10 in AMO1 Rpn10 iKO cells (Figure 4G).
Biochemical characterization of novel Rpn10 inhibitor SB
To find small molecules that target Rpn10, we developed a HTS assay with AlphaScreen technology, as described previously.16 Briefly, GST-tagged Rpn10 was immobilized on anti-GST–coated acceptor beads, and biotinylated Ub2-7 was immobilized on streptavidin-tagged donor beads. Upon laser excitation at 680 nm, the donor bead releases a singlet-state oxygen molecule. If the donor and acceptor beads are in proximity because of Rpn10 binding to Ub2-7, then the acceptor bead emits light at 520 to 620 nm. The binding of small molecules interrupts this recognition event and eliminates or reduces the signal. The assay was miniaturized and optimized for HTS in a 384-well format (Figure 5A). Two rounds of screening a library of 10 000 small molecules were performed independently; 787 hits were identified that showed over 30% inhibition with 30 μM treatment. False-positive hits were excluded by counter-screen using biotin-GST fusion protein. The top hits that were commercially available were further confirmed by a dose-response AlphaScreen assay (data not shown) and polyubiquitin accumulation assay. We then did a MM cell viability assay (data not shown) and identified the compound with the largest effect to be SB, as evidenced by a dose-response curve (Figure 5B-C).
Biochemical characterization of a novel inhibitor of Rpn10, SB. (A) AlphaAssay screening for Rpn10 inhibitor (binder). Human recombinant Rpn10-GST and biotinylated Ub2-7 were used. (B) Percentage of inhibition of Rpn10-Ub2-7 binding by the 10 000 compounds screened. Internal figure: chemical structure of Rpn10 inhibitor (binder) candidate SB. (C) Dose response AlphaScreen assay of SB binding to Rpn10 or GST protein. (D) Recombinant human Rpn10 protein was incubated with control, SB (10μM), or NSC697923 (10μM) for 30 minutes at real time, then Ub2-7 was added and incubated for 1 hour at real time. The mixture was then immunoprecipitated with Rpn10 antibody and subjected to immunoblotting using antibodies against Rpn10 and poly-Ub. (E) Measurement of Kd of hRpn10 with SB by FEB assay. Ten micrometers of compound or DiUb was applied on a recombinant His-hRpn10–immobilized graphene chip separately. DiUb and compound UNC0638 were used as positive and negative controls, respectively. The Kd is calculated as a median average over the test points. Three independent experiments were performed. Real-time changes of the I-response are shown in circles and correspond to the 1:1 binding model shown as solid lines. (F) Measurement of Kd of hRpn10 with SB by MST assay. Kd was derived from the binding response as a function of the GFP-hRpn10 concentration. Error in Kd represents fitting errors. (G) After being cultured with Dox for 3 days, AMO1 sgRNA-CT cells, AMO1 Rpn10-iKO cells, and iKO with Rpn10 adding back cells were treated with SB at different concentrations for 24 hours, followed by the cell viability being measured by a CTG assay (mean ± SD, n = 3).
Biochemical characterization of a novel inhibitor of Rpn10, SB. (A) AlphaAssay screening for Rpn10 inhibitor (binder). Human recombinant Rpn10-GST and biotinylated Ub2-7 were used. (B) Percentage of inhibition of Rpn10-Ub2-7 binding by the 10 000 compounds screened. Internal figure: chemical structure of Rpn10 inhibitor (binder) candidate SB. (C) Dose response AlphaScreen assay of SB binding to Rpn10 or GST protein. (D) Recombinant human Rpn10 protein was incubated with control, SB (10μM), or NSC697923 (10μM) for 30 minutes at real time, then Ub2-7 was added and incubated for 1 hour at real time. The mixture was then immunoprecipitated with Rpn10 antibody and subjected to immunoblotting using antibodies against Rpn10 and poly-Ub. (E) Measurement of Kd of hRpn10 with SB by FEB assay. Ten micrometers of compound or DiUb was applied on a recombinant His-hRpn10–immobilized graphene chip separately. DiUb and compound UNC0638 were used as positive and negative controls, respectively. The Kd is calculated as a median average over the test points. Three independent experiments were performed. Real-time changes of the I-response are shown in circles and correspond to the 1:1 binding model shown as solid lines. (F) Measurement of Kd of hRpn10 with SB by MST assay. Kd was derived from the binding response as a function of the GFP-hRpn10 concentration. Error in Kd represents fitting errors. (G) After being cultured with Dox for 3 days, AMO1 sgRNA-CT cells, AMO1 Rpn10-iKO cells, and iKO with Rpn10 adding back cells were treated with SB at different concentrations for 24 hours, followed by the cell viability being measured by a CTG assay (mean ± SD, n = 3).
SB is an antagonist of the 5-hydroxytryptamine (serotonin) receptor HTR5A, which is coupled to Gαi/o protein.36,37 To rule out the effects of HTR5A, we found that (1) gene dependency score analysis using DepMap showed that HTR5A is not essential for MM cells (supplemental Figure 5A); (2) unlike Rpn10, HTR5A is very lowly expressed in MM cell lines, including AMO1 cells (supplemental Figure 5B-C); (3) genetic blockade (siRNA) of HTR5A in MM cell lines (AMO1, MM.1S, ANBL6-WT, and ANBL6-BR) showed no alteration in their viability (supplemental Figure 5D); and (4) no significant correlation was noted between HTR5A expression and MM patient overall survival (GSE2658) (supplemental Figure 5E).
We confirmed the binding between SB and Rpn10 in vitro. We used a competition assay between SB, Ub2-7, and Rpn10 via immunoprecipitation to confirm that SB disturbs the binding of Ub2-7 to Rpn10 (Figure 5D). To further determine the specificity and characterize the Kd of Rpn10 with SB, we used the FEB and MST assays. In the FEB assay, the real-time changes of I-response and capacitance (as C-response) of the chip were measured and returned robust curves with Kd = 116 nM (R2 = 0.9961), confirming the interaction of SB with Rpn10 (Figure 5E). In the MST assay, GFP-tagged Rpn10 (supplemental Figure 5F) was incubated with varying concentrations of SB, and we found a notably high affinity (Kd = 51.16 nM) (Figure 5F; supplemental Figure 5G-H).
Finally, in vivo binding was validated using AMO1-iKO and HEK293 KO cells. When SB was applied to AMO1-iKO, there was a marked reduction in cytotoxicity, whereas adding back Rpn10 restored the sensitivity to SB (Figure 5G). This was confirmed in HEK293 Rpn10-KO cells, and transient transfection of Rpn10 into HEK293 Rpn10-KO cells partially restored the sensitivity to SB (supplemental Figure 5I). Together, we identified and validated the compound SB as an inhibitor of Rpn10, demonstrating the specificity of SB against Rpn10 and, importantly, excluding a role for HTR5A in SB–mediated MM cell death.
Antitumor activity of SB
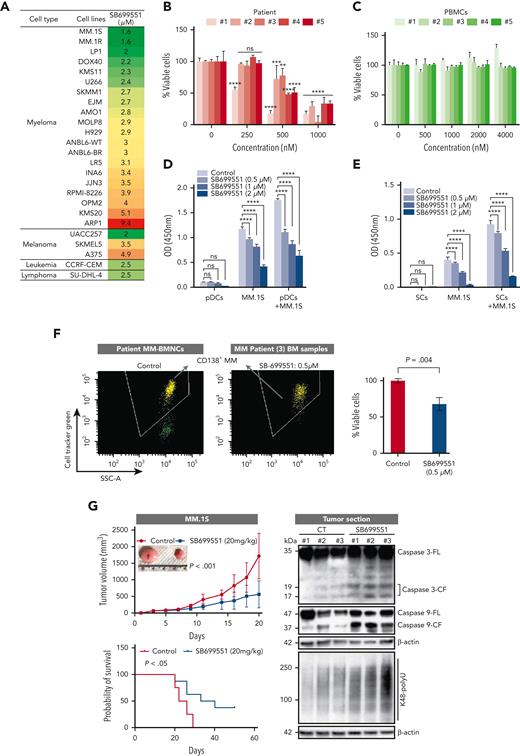
SB induced a dose-dependent decrease in viability in various MM cell lines (50% inhibitory concentration [IC50] range, 1.6-9.4 μM) (Figure 6A; supplemental Table 1). To determine whether SB similarly affects cells from patients with MM, tumor cells isolated from patients relapsing after multiple previous therapies, including bortezomib, lenalidomide, and dexamethasone, were treated with SB. MM was considered refractory to specific therapy when the disease progressed during therapy or relapsed within 2 months of discontinuing therapy. We found a dose-dependent decrease in the viability of all cells from patient with CD138+ MM after SB treatment (Figure 6B) without affecting the viability of normal PBMCs (Figure 6C). These data suggest selective anti-MM activity and a favorable therapeutic index for SB.
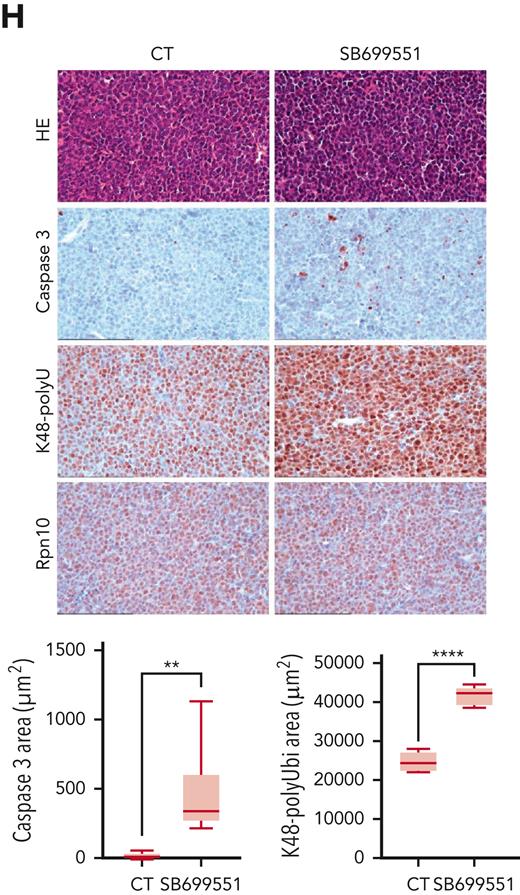
Anti-MM activity of SB. (A) MM, melanoma, leukemia, and lymphoma cell lines were treated with dimethyl sulfoxide (DMSO) control or SB at different concentrations for 48 hours, followed by assessment for cell viability using a WST assay (P < .05 for all cell lines, n = 3). IC50 was calculated and presented in the table. (B) Purified CD138+ cells from a patient with MM were treated with DMSO or SB for 48 hours, followed by assessment for cell viability using a CTG assay (mean ± SD of triplicate cultures). (C) Normal PBMCs from healthy donors were treated with DMSO or the indicated concentrations of SB for 48 hours, and then analyzed for cell viability using a CTG assay (mean ± SD, n = 3, P value is ns). (D-E) MM.1S cells were cultured with or without pDCs or BMSCs in the presence or absence of indicated concentrations of SB for 48 hours, and then cell viability was measured by a WST assay (mean ± SD, n = 3). (F) Total BM-derived mononuclear cells from patients with MM (n = 3) were treated with Rpn10 inhibitor SB (nontoxic concentration of 0.5 μM) or DMSO control for 2 days, and multicolor flow analysis was used to assess MM cell lysis. CD138+ MM cells were quantified by staining with CD138-FITC Ab. Representative fluorescence-active cell sorter scatter plot showing a decrease in the number of viable fluorescein isothiocyanate–positive MM cells after treatment with SB (left) and bar graph shows quantification of CD138+ MM cells in the left panel (right) are shown. The fold change was obtained after normalization with control data and presented as percentage of viable cells in the presence vs absence of SB (mean ± SD, P < .05). (G) Mice bearing human MM.1S MM tumors were treated with either vehicle control or SB (20 mg/kg, intraperitoneally) 3 times weekly for 14 days. Average and SD of tumor volume (mm3) is shown vs time when tumor was measured (mean tumor volume ±SD, 10 mice per group) (left top) and Kaplan-Meier plots show survival in mice (left bottom) are shown. SB-treated mice showed increased survival vs control vehicle-treated mice. Tumor lysates from control vehicle- and SB-treated mice were subjected to immunoblot analysis using anti–caspase-3, caspase-9, K48 polyubiquitin, and β-actin (right). (H) Representative hematoxylin and eosin (HE) and immunohistochemistry stains of caspase-3, K48 polyubiquitin and Rpn10 in tumor tissue from control- and SB-treated mice. Scale bars, 100 μM.
Anti-MM activity of SB. (A) MM, melanoma, leukemia, and lymphoma cell lines were treated with dimethyl sulfoxide (DMSO) control or SB at different concentrations for 48 hours, followed by assessment for cell viability using a WST assay (P < .05 for all cell lines, n = 3). IC50 was calculated and presented in the table. (B) Purified CD138+ cells from a patient with MM were treated with DMSO or SB for 48 hours, followed by assessment for cell viability using a CTG assay (mean ± SD of triplicate cultures). (C) Normal PBMCs from healthy donors were treated with DMSO or the indicated concentrations of SB for 48 hours, and then analyzed for cell viability using a CTG assay (mean ± SD, n = 3, P value is ns). (D-E) MM.1S cells were cultured with or without pDCs or BMSCs in the presence or absence of indicated concentrations of SB for 48 hours, and then cell viability was measured by a WST assay (mean ± SD, n = 3). (F) Total BM-derived mononuclear cells from patients with MM (n = 3) were treated with Rpn10 inhibitor SB (nontoxic concentration of 0.5 μM) or DMSO control for 2 days, and multicolor flow analysis was used to assess MM cell lysis. CD138+ MM cells were quantified by staining with CD138-FITC Ab. Representative fluorescence-active cell sorter scatter plot showing a decrease in the number of viable fluorescein isothiocyanate–positive MM cells after treatment with SB (left) and bar graph shows quantification of CD138+ MM cells in the left panel (right) are shown. The fold change was obtained after normalization with control data and presented as percentage of viable cells in the presence vs absence of SB (mean ± SD, P < .05). (G) Mice bearing human MM.1S MM tumors were treated with either vehicle control or SB (20 mg/kg, intraperitoneally) 3 times weekly for 14 days. Average and SD of tumor volume (mm3) is shown vs time when tumor was measured (mean tumor volume ±SD, 10 mice per group) (left top) and Kaplan-Meier plots show survival in mice (left bottom) are shown. SB-treated mice showed increased survival vs control vehicle-treated mice. Tumor lysates from control vehicle- and SB-treated mice were subjected to immunoblot analysis using anti–caspase-3, caspase-9, K48 polyubiquitin, and β-actin (right). (H) Representative hematoxylin and eosin (HE) and immunohistochemistry stains of caspase-3, K48 polyubiquitin and Rpn10 in tumor tissue from control- and SB-treated mice. Scale bars, 100 μM.
The interaction of MM cells with BMSCs triggers cytokine secretion, which mediates the paracrine growth of MM cells and protects against drug-induced apoptosis.38 In particular, pDCs are relatively resistant to therapies, protect MM cells from therapy-induced cytotoxicity, suppress immune responses, and promote tumor growth and survival.26,39 SB inhibited BMSC- or pDC-induced MM.1S cell proliferation (Figure 6D-E). Treatment of total BM-derived mononuclear cells from patients with MM, including CD138+ MM cells, with low concentrations of SB (0.5 μM for 48 hours) decreased their viability (Figure 6F). These data suggest that SB not only targets MM cells but also overcomes the cytoprotective effects of the BM microenvironment.
We next examined the in vivo efficacy of SB using the human plasmacytoma MM.1S xenograft mouse model.40-42 Treatment of MM.1S tumor-bearing mice with an intraperitoneal injection of SB (20 mg/kg) inhibited MM tumor growth and prolonged survival (Figure 6G). SB was well tolerated without significant weight loss. Western blot analysis and immunohistochemistry of tumors from SB-treated mice showed an increase in ubiquitinated proteins, activation of caspases, and polyubiquitinated proteins in SB-treated mice compared with those in control vehicle-treated mice (Figure 6G-H). Similar effects of SB were observed in xenograft models of HEK293, the colorectal cell line HCT116, and the leukemia cell line CCRF-CEM (supplemental Figure 6A).
Mechanisms of SB-induced MM cell death
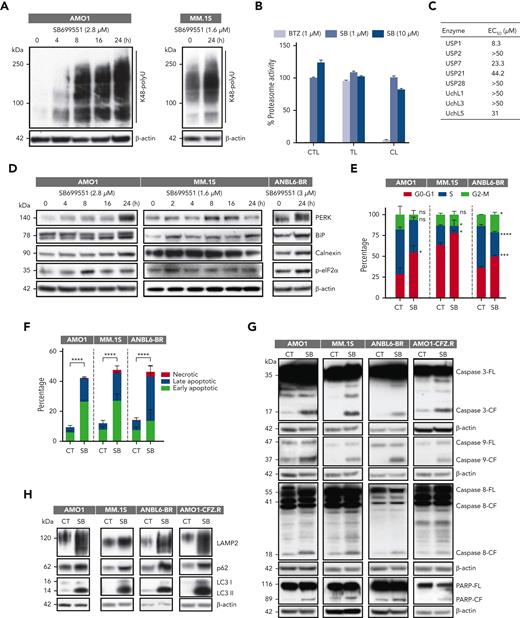
We first confirmed that SB treatment did not change the expression of Rpn10 in either tumor sections or MM cell lines (Figure 6H; supplemental Figure 7A). We next examined the effect of SB on proteasome function. Similar to Rpn10 depletion, SB induced polyubiquitylated protein accumulation in AMO1 and MM.1S cells (Figure 7A). To determine whether SB blocks the activity of the 20S CP, MM.1S cells were treated with SB (1 and 10 μM) for 3 hours and analyzed for chymotrypsin-, trypsin-, or caspase-like proteasome activities. We found that SB did not affect 20S activity (Figure 7B). Inhibiting deubiquitinating enzymes (DUBs) results in the accumulation of polyubiquitylated proteins.41,43 Therefore, we tested DUB activity using the recombinant human proteins USP1, USP2, USP7, USP21, USP28, UchL1, UchL3, and UchL5. We observed no reduction in DUB enzymatic activity (Figure 7C). Similar to Rpn10 genetic depletion, treatment with SB produced similar effects on ER stress, the cell cycle, apoptosis, and activation of caspases in MM (Figure 7D-G). Consistent with these findings, treatment of MM cells with SB induced the expression of autophagy-associated proteins, such as LAMP2, p62, and LC3 (Figure 7H). Interestingly, on SB-treated cells, we found elevated “eat-me” signals in the form of calreticulin, indicating that targeting Rpn10 may induce immunogenic cell death (supplemental Figure 7B).
Mechanisms of SB-induced MM cell death. (A) AMO1 and MM.1S cells were treated with DMSO control or SB at the IC50 concentration for 24 hours; protein lysates were subjected to immunoblot analysis using anti-K48 polyubiquitin or anti–β-actin Abs. (B) MM.1S cells were treated with DMSO, SB, or bortezomib (BTZ) for 3 hours; protein lysates were analyzed for proteasome activities. The percentage of proteasome activity was normalized to a DMSO control (mean ± SD, n = 3). (C) Recombinant human proteins USP1, USP2, USP7, USP21, USP28, UchL1, UchL3, and UchL5 were incubated with SB for 30 minutes at 37°C and then analyzed for DUB activity (mean ± SD, n = 3). (D-H) Indicated cells were treated with DMSO or SB; protein lysates were then subjected to immunoblotting using ER stress-related antibodies against p-eIF2α, PERK, BiP, and calnexin (D); caspase-related antibodies against caspase-3, caspase-8, caspase-9, and PARP (G) (caspase-3/-9 (AMO1, MM.1S, ANBL6-BR) have same actin reprobe; PARP/caspase-9 (AMO1-CFZ.R) have same actin reprobe); and autophagy-related antibodies against LAMP2, p62, LC3, or β-actin (H); treated cells were subjected to cell cycle analysis (E) or apoptosis analysis (F). CL, caspase-like proteasome activity; CTL, chymotrypsin-like proteasome activity; TL, trypsin-like proteasome activity.
Mechanisms of SB-induced MM cell death. (A) AMO1 and MM.1S cells were treated with DMSO control or SB at the IC50 concentration for 24 hours; protein lysates were subjected to immunoblot analysis using anti-K48 polyubiquitin or anti–β-actin Abs. (B) MM.1S cells were treated with DMSO, SB, or bortezomib (BTZ) for 3 hours; protein lysates were analyzed for proteasome activities. The percentage of proteasome activity was normalized to a DMSO control (mean ± SD, n = 3). (C) Recombinant human proteins USP1, USP2, USP7, USP21, USP28, UchL1, UchL3, and UchL5 were incubated with SB for 30 minutes at 37°C and then analyzed for DUB activity (mean ± SD, n = 3). (D-H) Indicated cells were treated with DMSO or SB; protein lysates were then subjected to immunoblotting using ER stress-related antibodies against p-eIF2α, PERK, BiP, and calnexin (D); caspase-related antibodies against caspase-3, caspase-8, caspase-9, and PARP (G) (caspase-3/-9 (AMO1, MM.1S, ANBL6-BR) have same actin reprobe; PARP/caspase-9 (AMO1-CFZ.R) have same actin reprobe); and autophagy-related antibodies against LAMP2, p62, LC3, or β-actin (H); treated cells were subjected to cell cycle analysis (E) or apoptosis analysis (F). CL, caspase-like proteasome activity; CTL, chymotrypsin-like proteasome activity; TL, trypsin-like proteasome activity.
We next looked at the effectiveness of SB in PI-resistant cell lines. The IC50 of SB for bortezomib-sensitive (ANBL6-WT) and bortezomib-resistant (ANBL6-BR) cells were almost the same, at ∼3 μM, indicating that SB overcomes bortezomib resistance. In addition, we found that knocking down Rpn10 in ANBL6-BR cells resensitizes the cells to bortezomib (supplemental Figure 7C). Furthermore, the IC50 ratio of SB for AMO1 cells resistant to CFZ and wild-type AMO1 cells was 1.17, although the ratio of CFZ is 185.8 and that of bortezomib is 14.244 (supplemental Figure 7D). SB did not change the expression of Rpn10 in any of the resistant cell lines but did induce the accumulation of K48 polyubiquitinated protein, ER stress responses, caspase cleavage, and autophagy (Figure 7G-H; supplemental Figure 7E). Together, these data demonstrate that targeting Rpn10 can overcome bortezomib and CFZ resistance in MM.
Discussion
Overall, depleting Rpn10 more efficiently inhibited MM cell growth and caused greater accumulation of polyubiquitylated protein compared with the depletion of Rpn13, nominating it as a better target in MM. We found that inhibiting it caused cell cycle arrest and ER stress, which agrees with a study in esophageal cancer cells.45 Interestingly, we found that the expression of polo-like kinase 1 (PLK1), which is involved in multiple stages of cell cycle progression and early mitosis in mammals, was significantly suppressed in KO cells. On one hand, PLK1 expression can be promoted by myc or E2F transcription factor and repressed by the binding of p53 or p21 at its promoter.46,47 In contrast, PLK1 can modulate p53 function in a negative feedback loop by interacting directly with p53 or indirectly through different factors such as MDM2, TOPORS, or GTSE1.46 These factors can either modulate the phosphorylation or ubiquitylation of p53, thus, changing its stability. How Rpn10 depletion suppresses PLK1, and whether its suppression contributes to the upregulation of p53, needs further investigation.
The UPS maintains normal cellular proteostasis and indirectly affects redox homeostasis. In fact, PIs targeting 20S or 19S can increase the reactive oxygen species/reactive nitrogen species ratio and oxidative stress.48,49 The disruption of redox homeostasis damages the mitochondria, resulting in their degradation. Consistent with this, our proteomic analysis showed a robust enrichment in mitochondria-related pathways in iKO cells, reflecting an attempt to produce functional mitochondria to rebuild the electron-respiratory chain. Furthermore, we found that the observed apoptosis is associated with activation of caspases and PARP cleavage, suggesting that Rpn10 depletion induced both mitochondria-dependent and independent signaling pathways.
We found that the autophagy pathway was induced in iKO cells, which is consistent with reports that the proteasome and autophagy pathways can compensate for each other.29,50 Autophagy can function as a source of peptide antigens for presentation to CD4+ T cells,33,34 and concurrently, we found that several pathways related to antigen processing and presentation were enriched in Rpn10 iKO cells. Combined with the fact that Rpn10 plays a role in the iPS,27 whose defects can enhance the NK cell killing in MM51 and the ability of dendritic cells to stimulate antitumor immunity in melanoma,52 depleting or inhibiting Rpn10 may enhance the immune response to tumors, thus, overcoming the immune suppression in MM.
In conclusion, we propose a model (supplemental Figure 8) by which SB binds Rpn10 and prevents it from feeding K48 polyubiquitinated proteins into the proteasome. These proteins accumulate, thus, triggering autophagy, ER stress, and eventually cell death by apoptosis. This cell death is likely immunogenic53 and can promote an antitumor immune response. Furthermore, as SB shuts down proteasome function in a manner distinct from other PIs, we believe that it can overcome PI resistance.
Acknowledgments
The authors thank Christina Usher for manuscript edits, the Y. Shrike Zhang laboratory for the 3D bioprinting model studies, Dmytro Kovalskyy for the FEB binding assays, Jennifer Smith for HTS assays, and Sirano Dhe-Paganon and Hyuk-Soo Seo for the His tagged-Rpn10 recombinant protein.
This work was supported by grants from the National Cancer Institute, National Institutes of Health (P01: CA155258-10, R01: CA207237-05, and R01: CA050947) and SPORE grant (P50-CA100707-18) (N.C.M. and K.C.A.); a Paula and Rodger Riney Foundation grant; and the Adelson Program in Multiple Myeloma Research grant (K.C.A.). K.C.A. is an American Cancer Society Clinical Research Professor.
Authorship
Contribution: D.C. conceptualized the project, designed research, and analyzed data; T.D. and Y.S. performed all experiments and analyzed the data; A.R. helped in flow cytometry assays; Y.Y., M.F., and N.C.M. helped in CRISPR knockout studies; X.W. and M.K.S. helped in proteomic data analysis; C.S. and H.W. helped in microscale thermophoresis analysis; J.P., T.S., and R.D.C. helped in immunohistochemistry analysis; Y.-T.T. provided clinical samples; and T.D., Y.S., D.C., and K.C.A. wrote the manuscript with input from all authors.
Conflict-of-interest disclosure: K.C.A. is a consultant to Pfizer, Amgen, AstraZeneca, Janssen, and Precision Biosciences; is a board member of C4 Therapeutics, Dynamic Cell Therapies, Window, and Mana; and an equity owner in C4 Therapeutics, Oncopep, NextRNA, and Dynamic Cell Therapies. D.C. is a consultant to Stemline Therapeutics, Inc and equity owner in C4 Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Dharminder Chauhan, Dana-Farber Cancer Institute, SM-601A, 450 Brookline Ave, Boston, MA 02215; e-mail: Dharminder_Chauhan@dfci.harvard.edu; and Kenneth C. Anderson, Dana-Farber Cancer Institute, SM-601D, 450 Brookline Ave, Boston, MA 02215; e-mail: Kenneth_Anderson@dfci.harvard.edu.
References
Author notes
∗T.D. and Y.S. contributed equally to this study.
†D.C. and K.C.A. are joint senior authors.
The proteomic data are available at Mendeley Data (https://doi.org/10.17632/t2pmyzzt6n.1 and https://doi.org/10.17632/vdtjzmdgkk.1).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Proteomic analysis of Rpn10 iKO. After being cultured with Dox for 4 days, AMO1 Rpn10-iKO cells were subjected to proteomic analysis by multiplexed proteomics with tandem mass spectrometry. (A) Volcano plot of differentially expressed genes in AMO1 Rpn10-iKO cells compared with those in control cells. Blue and red dots represent the genes (P < .05) that were downregulated or upregulated, respectively. (B) Heatmap showing significantly (false discovery rate [FDR] < .05) altered genes in Rpn10 iKO cells. (C) Gene set enrichment analysis (GSEA) and gene ontology biological process (GOBP) significantly enriched after Rpn10 iKO. For all pathways shown, FDR < 10%. (D) GSEA-derived enrichment plots for the lysosomal transport pathway (left). AMO1 Rpn10-iKO cells were cultured with Dox as described for the indicated time points (middle) and when cultured with Dox, 25 nM BafA1 was added to the culture of Rpn10 iKO cells for the last 16 hours of day 4 (right). Cells were then lysed and subjected to immunoblot using antibodies against p62, LC3, LAMP2, or β-actin. (E) GSEA-derived enrichment plots for antigen processing and presentation of peptide antigen (left). AMO1 Rpn10-iKO cells were cultured with Dox for 4 days. Cells were subjected to flow cytometry after staining with anti–HLA-DR or anti–HLA-DQ Abs and 7AAD (middle). 7AAD– cells were gated out, and the surface expression of HLA-DR or DQ expression was quantified. Rpn10-iKO cells, from which Dox was washed out, were added to total BM-derived mononuclear cells of patients with MM (n = 4) (right). Degranulation marker CD107a was measured on CD3+CD4+ T cells or CD3–/CD56+ natural killer (NK) cells. FC, fold change; MFI, mean fluorescence intensity.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/21/10.1182_blood.2022017897/2/m_blood_bld-2022-017897-gr3ad.jpeg?Expires=1765961729&Signature=xptC-bladhM2eOSA5AIWXGAsJ~tjbb9u-gJaVXkgXjjOvYKsHSlzixLTDvdl~krqR2lp95ojR~bRjE-T5vtrpGXRWMAbacR4M0LJ4~juZMR5slbSAPSuyaxMPa6C-9bnUMMwr4RtQIh7WBJkMF0Itjg-4OeWvlqSPNCDa5pPrBcHmRzIDKIBRn~prasfcPSU4uwXjiWokMZ27GOWHw5ZL8h9BSCcYIG5KUbJb-iB9c7Iqd7Se19R2sZhI2-iu04u1w-IT0ob3bLjjWYVeZA~NWgfUu4n5j-CYCODkgnlNS3JuatG0TV759mLkixQrKdNRkeNSd18MLpgoJIIv-Lo2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Proteomic analysis of Rpn10 iKO. After being cultured with Dox for 4 days, AMO1 Rpn10-iKO cells were subjected to proteomic analysis by multiplexed proteomics with tandem mass spectrometry. (A) Volcano plot of differentially expressed genes in AMO1 Rpn10-iKO cells compared with those in control cells. Blue and red dots represent the genes (P < .05) that were downregulated or upregulated, respectively. (B) Heatmap showing significantly (false discovery rate [FDR] < .05) altered genes in Rpn10 iKO cells. (C) Gene set enrichment analysis (GSEA) and gene ontology biological process (GOBP) significantly enriched after Rpn10 iKO. For all pathways shown, FDR < 10%. (D) GSEA-derived enrichment plots for the lysosomal transport pathway (left). AMO1 Rpn10-iKO cells were cultured with Dox as described for the indicated time points (middle) and when cultured with Dox, 25 nM BafA1 was added to the culture of Rpn10 iKO cells for the last 16 hours of day 4 (right). Cells were then lysed and subjected to immunoblot using antibodies against p62, LC3, LAMP2, or β-actin. (E) GSEA-derived enrichment plots for antigen processing and presentation of peptide antigen (left). AMO1 Rpn10-iKO cells were cultured with Dox for 4 days. Cells were subjected to flow cytometry after staining with anti–HLA-DR or anti–HLA-DQ Abs and 7AAD (middle). 7AAD– cells were gated out, and the surface expression of HLA-DR or DQ expression was quantified. Rpn10-iKO cells, from which Dox was washed out, were added to total BM-derived mononuclear cells of patients with MM (n = 4) (right). Degranulation marker CD107a was measured on CD3+CD4+ T cells or CD3–/CD56+ natural killer (NK) cells. FC, fold change; MFI, mean fluorescence intensity.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/21/10.1182_blood.2022017897/2/m_blood_bld-2022-017897-gr3e.jpeg?Expires=1765961729&Signature=AWl-iYLpZjfJtNvDTyl9JhwarUJ0uUKu3xFAGlNx3g01-9ZrMXjOt5EtVR1mWeXVvSopgOmQFM~6NZLYxQQ5lvghHunUXsQQ1pLtQAgQYsrhSkR1qtwL3gEAMnrFkcSy6FIVrpiV6LMpb5sHkSObkz37IYmHr6~B58LyteYR16lMhktElLRofpZk2Xu2NF3pZR0NjS4VfxEZhFMuZF2nY2ScL4X8R~3swgiyO~WS2E2JQW8GKeiMhXa~LbQfS15f~PKR8IwsEbNqGlMSeZTxE8X4GzMLtTyDqUdw4VNevUzPfVa9mcAJ4FUQgbZ19pIXFw2V6Z7ealFQtZi5YlXUeA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal