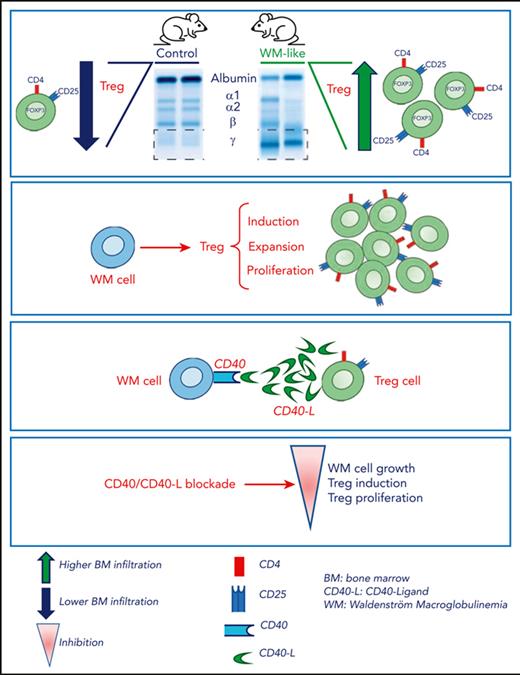
Key Points
A CD40/CD40-ligand interaction mediates WM cell–Tregs cross talk.
Blocking the CD40/CD40-ligand axis inhibits Treg-mediated immunosuppression and reduces WM cell growth.
Abstract
Recent investigations have improved our understanding of the molecular aberrations supporting Waldenström macroglobulinemia (WM) biology; however, whether the immune microenvironment contributes to WM pathogenesis remains unanswered. First, we showed how a transgenic murine model of human-like lymphoplasmacytic lymphoma/WM exhibits an increased number of regulatory T cells (Tregs) relative to control mice. These findings were translated into the WM clinical setting, in which the transcriptomic profiling of Tregs derived from patients with WM unveiled a peculiar WM-devoted messenger RNA signature, with significant enrichment for genes related to nuclear factor κB–mediated tumor necrosis factor α signaling, MAPK, and PI3K/AKT, which was paralleled by a different Treg functional phenotype. We demonstrated significantly higher Treg induction, expansion, and proliferation triggered by WM cells, compared with their normal cellular counterpart; with a more profound effect within the context of CXCR4C1013G-mutated WM cells. By investigating the B-cell–to–T-cell cross talk at single-cell level, we identified the CD40/CD40-ligand as a potentially relevant axis that supports WM cell–Tregs interaction. Our findings demonstrate the existence of a Treg-mediated immunosuppressive phenotype in WM, which can be therapeutically reversed by blocking the CD40L/CD40 axis to inhibit WM cell growth.
Introduction
Waldenström macroglobulinemia (WM) is a clonal B-cell malignancy characterized by the accumulation of lymphoplasmacytic cells within the bone marrow (BM) and a serum monoclonal immunoglobulin M (IgM) component.1-3 The pathogenesis of WM stems from an asymptomatic precursor stage, named IgM monoclonal gammopathy of undetermined significance, defined by a BM infiltration of <10% and no significant symptoms.1,2 IgM monoclonal gammopathy of undetermined significance may evolve into active WM, and patients may present with anemia, low platelet count, adenopathy, organomegaly, peripheral neuropathy, and symptomatic hyperviscosity.2,3
Recent studies have demonstrated the occurrence of somatic mutations of myeloid differentiation primary response 88 (MYD88) and C-X-C chemokine receptor type 4 (CXCR4), showing their crucial role in supporting WM pathogenesis and disease progression.4-10
Despite the significant improvement in the understanding of the molecular mechanisms supporting WM biology and disease progression,4-10 whether immunosuppressive mechanisms could contribute to WM pathogenesis remains unexplored. Recent studies have shown how changes in T cells, with a decline in interferon response and tumor-specific immunity, could support WM biology.11
T regulatory immune cell (Treg) expansion represents 1 of the most crucial immunosuppressive mechanisms. In particular, Tregs are a subset of CD4+CD25+ T lymphocytes that express transcription factor forkhead box P3 (FOXP3) and play a vital role in the self-tolerance maintenance and the control of immune homeostasis, supporting cancer cells in immune suppression and evasion.12 The role of Tregs in facilitating tumor progression has been shown both in solid tumors and hematologic malignancies;12-17 however, their functional relevance in mediating WM pathogenesis has not been investigated.
Firstly, we observed Treg involvement in WM biology, using a murine lymphoplasmacytic/WM transgenic model,18 in which we could demonstrate an increased number of CD4+CD25+Foxp3+ Tregs, relative to control mice. Next, these data were validated within the setting of human WM disease. We subsequently showed that Tregs derived from patients with WM (WM-Tregs) exhibit a higher suppressive activity compared with that of Tregs derived from healthy donors (HD-Tregs). Moreover, WM cells can induce Treg activation and expansion and the Treg-induced phenotype is more significantly enhanced within the context of CXCR4-mutated WM cells. By performing single-cell RNA sequencing (scRNA-seq) studies, we have unveiled that Treg immunosuppressive activity may result from the interaction mediated by the tumor necrosis factor (TNF) family ligands CD40-ligand (CD40L) and CD40, expressed on Tregs and WM cells, respectively.
Our findings provide evidence for the existence of a Treg-mediated immunosuppressive phenotype in WM and suggest how halting the CD40L/CD40 axis may represent a strategy to inhibit the Treg-mediated immunosuppressive scenario in WM.
Methods
Cells and reagents
Approval for these studies was obtained from the local ethics committee. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki protocol. A detailed description is provided within the supplemental File.
Flow cytometric (FACS) and cell proliferation analysis
Anti-human antibodies against CD4 (clone L3T4) Allophycocyanin (APC) (BD Biosciences; #555349), CD25 (clone 2A3) PE-Cy7 (BD Biosciences; #335824), and FOXP3 (clone 259 D/C7) Alexa Fluor 488 (BD Biosciences; #560047) were used for the fluorescence-activated cell sorter (FACS) analyses. Cells were surface stained, fixed/permeabilized, and stained intracellularly using the FOXP3/transcription factor staining buffer set (BD Biosciences, #562574) per the manufacturer’s protocol. To evaluate the proliferation of each cell population, Ki-67 (clone B56) BV421 (BD Biosciences; #562899) detection was performed per the manufacturer’s protocol. All data were acquired using a FACSCanto II flow cytometer (BD Biosciences) and analyzed using Cytobank (Santa Clara, CA).
Tregs induction, expansion, and suppression assays
A detailed description is provided within the supplemental File, available on the Blood website.
Bulk RNA-seq
RNA was extracted from age-matched HD-Tregs and WM-Tregs, as previously reported.19-21 RNA samples were processed using a WT PLUS reagent kit, per the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA). Gene set enrichment analysis (GSEA) was applied on global protein-coding gene expression profiles: significant gene sets were selected based on a nominal P value < .05 and false discovery rate < 0.25.12,20-23
A detailed description is provided within the supplemental File.
Gene expression profiling
Differential gene expression analysis was performed for Affymetrix CEL data using the LIMMA package.24 A detailed description is provided within the supplemental File.
scRNA-seq library preparation, sequencing, and data processing and analysis
scRNA libraries were generated using a Chromium microfluidics instrument (10x Genomics) coupled with a Next GEM Single Cell 3′ kit. Sequences were generated using a NovaSeq 6000 instrument (Illumina) with a target of 25 000 reads per cell. Raw fastq reads were initially processed using Cell Ranger count version 6.1 and aligned against the GRCh38 human genome (GENCODE version 32/Ensembl 98). Subsequently, count matrixes generated with Cell Ranger were processed with Seurat version 4.0.25 A detailed description is provided within the supplemental File.
In vivo studies: M88B2m transgenic mice, flow cytometry immunophenotyping, quantification of serum gamma fraction, IgM isotyping, and ex vivo expansion assays
M88B2m transgenic mice were generated as previously reported.18 A detailed description is provided within the supplemental File.
Statistics
All statistical analyses were performed with GraphPad (La Jolla, CA) Prism software. The data presented represent the mean ± standard deviation (SD). Results from experiments with 2 groups were analyzed by using 2-tailed unpaired Student t test. One-way analysis of variance (ANOVA) was used when ≥3 independent groups were compared. A P value < .05 was considered statistically significant.
Results
A transgenic murine lymphoplasmacytic/WM model points toward a role of Treg in supporting WM biology
We interrogated the potential role of Tregs in supporting WM pathogenesis by using a previously reported lymphoplasmacytic/WM transgenic murine model,18 which recapitulates features of human WM disease, including a progressive development of lymphomas with BM infiltration of lymphoplasmacytic cells, and monoclonal serum IgM (Figure 1A and B). Characterization of the BM immune microenvironment by flow cytometry showed an increased number of infiltrating CD4+ T lymphocytes over CD8+ T cells, most of which exhibited expression of PD-1 and TIGIT as exhaustion markers (Figure 1C). Further characterization of the T-cell compartment revealed a significantly higher level of CD4+CD25+FoxP3+ Tregs in M88B2m mice compared with that in control mice (Figure 1D). Finally, we investigated whether WM cells could exert a different effect on Treg proliferation, compared with normal B cells. By performing a coculturing ex vivo assay, we demonstrated how murine WM cells recruited a significantly higher number of more abundant Ki67+ Tregs compared with B lymphocytes from healthy mice (Figure 1E). Collectively, these results suggest that immunosuppressive Tregs may play in supporting WM disease biology.
A transgenic murine lymphoplasmacytic/WM model points toward a role for Treg in supporting WM biology. (A) Representative flow cytometry analyses showing the percentages of B220+CD138− B cells, B220+CD138+ plasmablasts, and B220−CD138+ plasma cells in the BM of M88B2m transgenic mice developing lymphoplasmacytic lymphoma/WM in comparison with mb1-cre control mice. (B) Representative serum protein electrophoresis images of M88B2m and control mice. One representative image is shown. Quantification of gamma fractions is represented. (right) Enzyme-linked immunosorbent assay in serum of M88B2m mice compared with control mice (n = 6 per cohort) measuring the levels of secreted IgM.(C) Flow cytometry analyses of CD4+ and CD8+ T-cell subsets, including CD62L+CD44– naïve subpopulations. Quantification of the percentage of CD4+ and CD8+ T cells with expression of PD-1 or TIGIT (n = 6-10 mice per cohort) is also shown. (D) Representative flow cytometry analyses measuring the percentage of CD4+CD25+Foxp3+ Tregs in M88B2m mice (n = 10) compared with control mice (n = 6). (E) Ex vivo coculturing assays including Tregs isolated from healthy mice with lymphoma B cells from M88B2m mice or control B lymphocytes. Quantification of Ki67+ vs Ki67− Treg numbers are shown for each coculture (n = 2 independent experiments, each in duplicate). ∗P < .05; ∗∗P < .01; and ∗∗∗P < .001.
A transgenic murine lymphoplasmacytic/WM model points toward a role for Treg in supporting WM biology. (A) Representative flow cytometry analyses showing the percentages of B220+CD138− B cells, B220+CD138+ plasmablasts, and B220−CD138+ plasma cells in the BM of M88B2m transgenic mice developing lymphoplasmacytic lymphoma/WM in comparison with mb1-cre control mice. (B) Representative serum protein electrophoresis images of M88B2m and control mice. One representative image is shown. Quantification of gamma fractions is represented. (right) Enzyme-linked immunosorbent assay in serum of M88B2m mice compared with control mice (n = 6 per cohort) measuring the levels of secreted IgM.(C) Flow cytometry analyses of CD4+ and CD8+ T-cell subsets, including CD62L+CD44– naïve subpopulations. Quantification of the percentage of CD4+ and CD8+ T cells with expression of PD-1 or TIGIT (n = 6-10 mice per cohort) is also shown. (D) Representative flow cytometry analyses measuring the percentage of CD4+CD25+Foxp3+ Tregs in M88B2m mice (n = 10) compared with control mice (n = 6). (E) Ex vivo coculturing assays including Tregs isolated from healthy mice with lymphoma B cells from M88B2m mice or control B lymphocytes. Quantification of Ki67+ vs Ki67− Treg numbers are shown for each coculture (n = 2 independent experiments, each in duplicate). ∗P < .05; ∗∗P < .01; and ∗∗∗P < .001.
Differential transcriptome profiling characterizes WM-Tregs, leading to a different functional phenotype between WM- and HD-Tregs
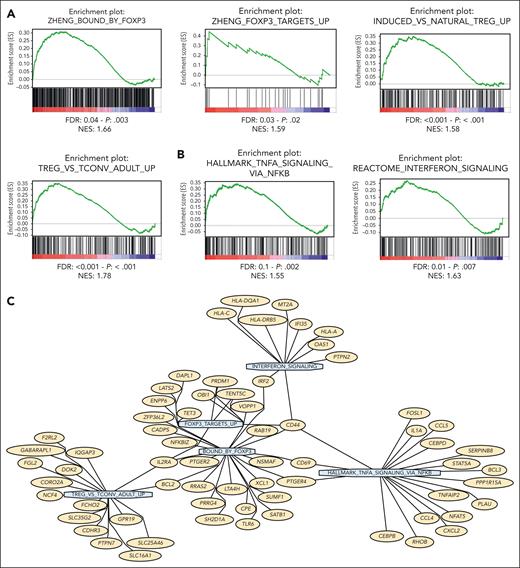
Next, in order to evaluate the role of Tregs in mediating WM pathogenesis, we moved within the human setting and performed bulk RNA-seq of HD- and WM-Tregs, which showed a peculiar transcriptome signature characterizing WM-Tregs (n = 14) compared with their normal cellular counterpart (n = 8). Specifically, GSEA allowed the identification of WM-Tregs presenting with, relative to HD-Tregs, transcriptome profiling enriched for FOXP3-target genes and Treg induction (Figure 2A). Importantly, WM-Tregs showed a significant enrichment for genes related to interferon- and TNF-α–related genes via nuclear factor κB (Figure 2B and C). Overall, these findings provide evidence for the occurrence of a peculiar transcriptome signature, which differentiates WM-Tregs from HD-Tregs.
Tregs obtained from patients with WM present with a peculiar transcriptome signature that differs from HD-Tregs. (A,B) Bulk RNA-seq comparing WM-Tregs (n = 14) and HD-Tregs (n = 8). WM-Tregs present with a significant enrichment of FOXP3, and Treg-, TNF-α–, interferon-related genes, as assessed by GSEA. Normalized enrichment score (NES) was generated by comparing WM-Tregs and HD-Tregs. NES, nominal P value, and false discovery rate q values are shown for each plot. (C) Network plot showing the top leading-edge genes of the GSEA signatures associated with Treg activity. Genes are reported in yellow and GSEA signatures in light blue.
Tregs obtained from patients with WM present with a peculiar transcriptome signature that differs from HD-Tregs. (A,B) Bulk RNA-seq comparing WM-Tregs (n = 14) and HD-Tregs (n = 8). WM-Tregs present with a significant enrichment of FOXP3, and Treg-, TNF-α–, interferon-related genes, as assessed by GSEA. Normalized enrichment score (NES) was generated by comparing WM-Tregs and HD-Tregs. NES, nominal P value, and false discovery rate q values are shown for each plot. (C) Network plot showing the top leading-edge genes of the GSEA signatures associated with Treg activity. Genes are reported in yellow and GSEA signatures in light blue.
To elucidate the functional sequelae secondary to the observed transcriptome differences occurring between WM- and HD-Tregs, we performed Treg induction assay. We first cultured CD4+CD25− non-Tregs either alone or in the presence of WM cells (BCWM.1; MWCL.1) or CD19+ B cells derived from healthy donors, in the presence of a transwell, in order to account for possible soluble factor–mediated Treg induction mechanisms. After 72 hours, we observed a significantly greater ability of WM cells to favor induction of CD4+CD25+FoxP3+ Tregs, compared with CD19+ B cells derived from healthy donors (n = 4), which were used as control. Importantly, no significant differences, in terms of percentage of induced Tregs, were observed between CD4+CD25− non-Tregs cultured either alone or in the presence of CD19+ B cells derived from healthy donors (Figure 3A). In parallel, modulation of Treg proliferation was also interrogated. By using Ki67 flow cytometric analysis, we found a significantly greater increase of Treg growth induced by WM cells compared with CD19+ B cells derived from healthy donors (Figure 3B).
WM cells enhance Treg induction. (A) CD4+CD25− non-Tregs were cultured either alone or in the presence of WM cells (BCWM.1; MWCL.1) or CD19+ B cells derived from healthy donors, for 72 hours, using a transwell. Significant Tregs induction was observed upon exposure to both BCWM.1 and WMCL.1. The percentage indicates the CD25+FOXP3+ cells gated on CD4+ cells. A representative experiment is shown. The experiment was repeated 3 times, using different normal CD19+ B cells (n = 4). ∗∗P < .01 determined using one-way ANOVA. Error bars indicate mean ± SD. (B) The proliferative rate of CD4+CD25+FOXP3+ Tregs was assessed by evaluating the percentage of Ki67+ cells. A significant increase in Ki-67 expression in Tregs was observed in the BCWM.1 and WMCL1 coculture groups compared with in the absence of cells or with CD19+ B-cell groups. A representative experiment is shown. The experiment was repeated 3 times. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, determined using one-way ANOVA. Error bars indicate mean ± SD. (C) CD4+CD25− non-Tregs were cultured either alone or in the presence of CD19+ cells derived from the BM of patients with WM (WM-1; WM-2), or CD19+ B cells derived from healthy donors, for 72 hours, using a transwell. Significantly higher Treg induction was observed upon exposure to WM tumor cells. The percentage indicates the CD25+FOXP3+ cells gated on CD4+ cells. (D) The proliferative rate of CD4+CD25+FOXP3+ Tregs was assessed by evaluating the percentage of Ki67+ cells. WM and HD, as in panel C. P indicates P value; one-way ANOVA; Error bars indicate mean ± SD. (E) Tregs derived from patients with WM present with a significant enrichment of MAPK, PI3K/AKT-related genes, as assessed by GSEA. NESs were generated by comparing WM-Treg and HD-Tregs. NES, nominal P value, and false discovery rate q values are shown for each plot.
WM cells enhance Treg induction. (A) CD4+CD25− non-Tregs were cultured either alone or in the presence of WM cells (BCWM.1; MWCL.1) or CD19+ B cells derived from healthy donors, for 72 hours, using a transwell. Significant Tregs induction was observed upon exposure to both BCWM.1 and WMCL.1. The percentage indicates the CD25+FOXP3+ cells gated on CD4+ cells. A representative experiment is shown. The experiment was repeated 3 times, using different normal CD19+ B cells (n = 4). ∗∗P < .01 determined using one-way ANOVA. Error bars indicate mean ± SD. (B) The proliferative rate of CD4+CD25+FOXP3+ Tregs was assessed by evaluating the percentage of Ki67+ cells. A significant increase in Ki-67 expression in Tregs was observed in the BCWM.1 and WMCL1 coculture groups compared with in the absence of cells or with CD19+ B-cell groups. A representative experiment is shown. The experiment was repeated 3 times. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, determined using one-way ANOVA. Error bars indicate mean ± SD. (C) CD4+CD25− non-Tregs were cultured either alone or in the presence of CD19+ cells derived from the BM of patients with WM (WM-1; WM-2), or CD19+ B cells derived from healthy donors, for 72 hours, using a transwell. Significantly higher Treg induction was observed upon exposure to WM tumor cells. The percentage indicates the CD25+FOXP3+ cells gated on CD4+ cells. (D) The proliferative rate of CD4+CD25+FOXP3+ Tregs was assessed by evaluating the percentage of Ki67+ cells. WM and HD, as in panel C. P indicates P value; one-way ANOVA; Error bars indicate mean ± SD. (E) Tregs derived from patients with WM present with a significant enrichment of MAPK, PI3K/AKT-related genes, as assessed by GSEA. NESs were generated by comparing WM-Treg and HD-Tregs. NES, nominal P value, and false discovery rate q values are shown for each plot.
To further corroborate our findings, we explored Treg induction using CD19+ cells derived from patients with BM WM, and confirmed a significantly greater induction of CD4+CD25+FoxP3+ Tregs when CD4+CD25− non-Tregs were cocultured with primary WM cells, compared with B cells derived from healthy donors (Figure 3C). We also confirmed that the primary WM cell–induced Tregs showed a significant increase of Treg proliferative activity, compared with that observed in Tregs exposed to B cells derived from healthy donors, as shown by performing Ki67 assay (Figure 3D).
Next, we interrogated the transcriptome signature of WM- vs HD-Tregs and found a significant enrichment of several proproliferative and prosurvival pathways in WM-Tregs, including MAPK- and PI3K/AKT-related genes (Figure 3E), thus providing evidence for the underlying molecular mechanisms responsible for the induced WM-Treg growth.
Taken together, the aforementioned data suggest that WM cells enhance Treg induction and proliferation, thus favoring an immunosuppressive milieu that could support WM biology. We subsequently addressed the question whether WM tumor cells could also affect Tregs in terms of modulating their expansion, and, importantly, investigated the occurrence of a WM-to-Treg interaction, which could further define novel insights into WM disease pathogenesis.
WM cells promote the expansion and immunosuppressive activity of Tregs
To further dissect the ability of WM cells to favor the immunosuppressive activity of Tregs, we investigated whether WM cells could also promote the expansion of Tregs. We cocultured CD4+CD25+ Tregs with either WM cells (BCWM.1; MWCL.1) or CD19+ B cells derived from healthy donors, which were used as control. After 72 hours, we observed a significantly greater increase in enriched CD4+CD25+FOXP3+ Tregs in the presence of WM cells, compared with that observed in the presence of CD19+ B cells derived from healthy donors (Figure 4A). Within the same experimental setting, we confirmed that the observed Treg expansion induced by WM cells was also supported by the significantly greater increase in Treg proliferation (Figure 4B). To investigate the conventional T-cell (Tcon)-suppression capacity of WM-Tregs, we performed a carboxyfluorescein succinimidyl ester (CFSE) proliferation assay, showing reduction of Tcon proliferation upon coculture with WM-Tregs. Specifically, we isolated CD4+CD25+ Tregs from patients with WM and from healthy individuals and performed coculture with CD4+CD25− Tcons, pretreated with CFSE, at the indicated ratios. After 5 days, analysis demonstrated a significant reduction of Tcon proliferation when cocultured with WM-Tregs compared with coculture with HD-Tregs (Figure 4C). Overall, these findings demonstrate how primary WM-Tregs are more capable of suppressing Tcon proliferation than HD-Tregs. To further corroborate the occurrence of molecular mechanisms underlying the observed higher suppressive activity of WM-Tregs, we evaluated the expression of Treg effector molecules that reportedly suppress immune effector cells, such as Epstein-Barr virus–induced 3 (EBI3), fibrinogen-like protein 2 (FGL2), granzyme B (GZMB), interleukin 10 (IL-10), perforin 1 (PRF-1), and transforming growth factor–β26-28; and found that EBI3, FGL2, GZMB, and PRF-1 were significantly upregulated in primary WM-Tregs compared with that in their normal cellular counterpart. In parallel, WM-Tregs presented with a significant upregulation of TIM3 and enrichment for cytotoxic T-lymphocyte antigen 4, a well-known player in Treg-mediated suppression (Figure 4D,E).29
WM cells enhance Treg expansion. (A) Treg expansion assay. CD4+CD25+ Tregs cultured with either CD19+ B cells derived from healthy donors or WM cells (BCWM.1; WMCL.1), for 72 hours using a transwell. FOXP3+ cells within the CD4+ cells are shown. Significantly higher Treg expansion was observed upon exposure to WM cells compared with B cells derived from healthy donors. A representative experiment is shown. The experiment was repeated 3 times. ∗∗∗P < .001 determined using one-way ANOVA. Error bars indicate mean ± SD. (B) The proliferative rate of CD4+CD25+FOXP3+ Tregs was assessed by evaluating the percentage of Ki67+ cells. ∗∗∗P < .001. (C) Treg-mediated suppression as measured by CFSE dilution. CD4+CD25− Tcons were isolated from HDs and labeled with 5 μM CFSE. Cells were activated with anti-CD3– and anti-CD28–coated beads and cultured either alone or in the presence of either CD4+CD25+ Tregs obtained from patients with WM or HDs, at the indicated Tcon-to-Treg ratio. After 72 hours, proliferation was determined by CFSE dilution and flow cytometric analysis. ∗∗P < .001 determined using two-way ANOVA. Error bars indicate mean ± SD. (D) Heatmap obtained from RNA-seq comparing WM-Tregs (n = 14) and HD-Tregs (n = 8) (EBI3: P = .03; FGL2: P < .001; GZMB: P < .0001; PRF1: P < .001; TIM3: P = .02). (E) WM-Tregs present with a significant enrichment of CTLA4-related genes, as assessed by GSEA. NESs were generated by comparing WM-Tregs and HD-Tregs. NES, nominal P value, and false discovery rate q values are shown.
WM cells enhance Treg expansion. (A) Treg expansion assay. CD4+CD25+ Tregs cultured with either CD19+ B cells derived from healthy donors or WM cells (BCWM.1; WMCL.1), for 72 hours using a transwell. FOXP3+ cells within the CD4+ cells are shown. Significantly higher Treg expansion was observed upon exposure to WM cells compared with B cells derived from healthy donors. A representative experiment is shown. The experiment was repeated 3 times. ∗∗∗P < .001 determined using one-way ANOVA. Error bars indicate mean ± SD. (B) The proliferative rate of CD4+CD25+FOXP3+ Tregs was assessed by evaluating the percentage of Ki67+ cells. ∗∗∗P < .001. (C) Treg-mediated suppression as measured by CFSE dilution. CD4+CD25− Tcons were isolated from HDs and labeled with 5 μM CFSE. Cells were activated with anti-CD3– and anti-CD28–coated beads and cultured either alone or in the presence of either CD4+CD25+ Tregs obtained from patients with WM or HDs, at the indicated Tcon-to-Treg ratio. After 72 hours, proliferation was determined by CFSE dilution and flow cytometric analysis. ∗∗P < .001 determined using two-way ANOVA. Error bars indicate mean ± SD. (D) Heatmap obtained from RNA-seq comparing WM-Tregs (n = 14) and HD-Tregs (n = 8) (EBI3: P = .03; FGL2: P < .001; GZMB: P < .0001; PRF1: P < .001; TIM3: P = .02). (E) WM-Tregs present with a significant enrichment of CTLA4-related genes, as assessed by GSEA. NESs were generated by comparing WM-Tregs and HD-Tregs. NES, nominal P value, and false discovery rate q values are shown.
These results demonstrate that WM cells not only increase Treg numbers but are also capable of increasing Treg suppressive activity, which may lead to a more suppressed WM immune milieu.
Effects of CXCR4C1013G-mut WM cells on Tregs
It has been shown how the CXCR4C1013G somatic mutation acts as an oncogenic mutation, leading to WM disease progression and drug resistance.4,10,30,31 We therefore hypothesized that CXCR4-mutated (CXCR4-mut) WM cells could possibly modulate the Treg phenotype differently than CXCR4–wild-type (CXCR4-wt) WM cells. WM cell–dependent Treg induction was evaluated by comparing CXCR4-wt and CXCR4-mut WM cells, which showed a significantly higher Treg induction upon exposure of CD4+CD25− non-Tregs to both CXCR4-mut BCWM.1 and MWCL.1 (supplemental Figure 1A-B). Of note, the induced Tregs presented a higher proliferative rate in the CXCR4-mut scenario, compared with CXCR4-wt control cells (supplemental Figure 1C-D). Overall, these findings further confirm the ability of CXCR4-mut WM cells to affect Treg induction and growth.
scRNA-seq analyses
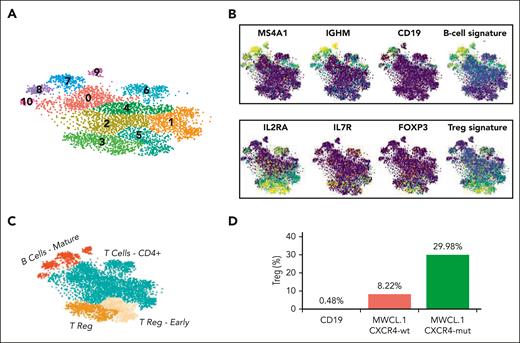
To gain insight into the potential molecular mechanisms responsible for the observed Treg induction in WM, we performed scRNA-seq using the whole cell population harvested at the end of the induction assay. This approach allowed the study of both populations (WM cells/normal B cells and CD4+CD25− non-Treg/induced CD4+CD25+FOXP3+ Treg) at single-cell resolution, thus also investigating the potential cross talk between WM and Tregs, which may support WM pathogenesis. Given the importance of CXCR4 mutation in WM biology,4,10,30,31 the analysis was performed with both CXCR4C1013G-mut and CXCR4-wt cells. T-distributed stochastic neighbor embedding (t-SNE) and cluster identification via the Louvain algorithm,32 allowed us to identify a total of 12 clusters (Figure 5A), with a B-cell compartment corresponding to clusters 7, 8, 9, and 11 and enriched for B-cell markers such as CD19, MS4A1 (CD20), and IgHM (Figure 5B,C). In parallel, Tregs were mapped to cluster 3, as also demonstrated by the enrichment for IL-2RA (CD25), IL-7R, and FOXP3 genes (Figure 5B,C). Milder expression of the same gene set was also shown in cluster 5, marking T cells likely to transition into Treg.
Single-cell transcriptome analysis. (A) t-distributed stochastic neighbor embedding (tSNE) plot of the integrated WMCL1 analysis. The colors highlight the different cell clusters; the numbers reported at the top of each cluster indicate the associated cluster index. (B) tSNE plots highlighting the expression level of B-cell (top) and Tregs (bottom) markers in a yellow (high expression) to blue (low expression) color scale. (C) tSNE plot representing the different cell types at cluster resolution. (D) Percentage of Tregs after exposure to normal CD19 cells, to WMCL1, or to WMCL1-CXCR4C1013G cells.
Single-cell transcriptome analysis. (A) t-distributed stochastic neighbor embedding (tSNE) plot of the integrated WMCL1 analysis. The colors highlight the different cell clusters; the numbers reported at the top of each cluster indicate the associated cluster index. (B) tSNE plots highlighting the expression level of B-cell (top) and Tregs (bottom) markers in a yellow (high expression) to blue (low expression) color scale. (C) tSNE plot representing the different cell types at cluster resolution. (D) Percentage of Tregs after exposure to normal CD19 cells, to WMCL1, or to WMCL1-CXCR4C1013G cells.
In line with previous data, comparison of the Treg fraction under different experimental conditions revealed a significant induction of Tregs when non-Tregs were cocultured in the presence of CXCR4-wt compared with in the presence of CD19+ B cells derived from healthy donors (Figure 5D). Notably, this induction was even more profound when non-Tregs were cocultured with CXCR4C1013G-mut cells (Figure 5D).
Interaction between TNF-family ligands and their receptors supports Treg functions in WM: identification and validation of the WM-to-Treg interaction mediated by CD40/CD40L
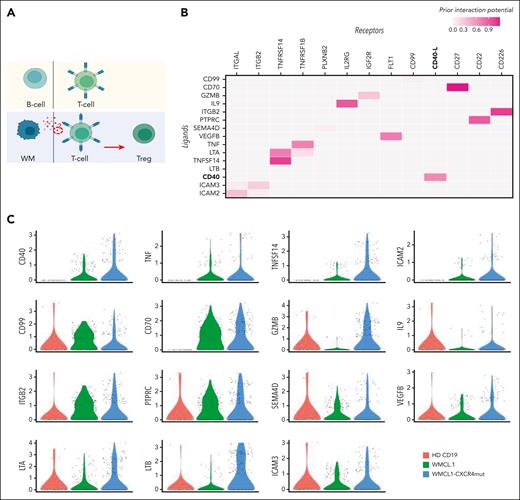
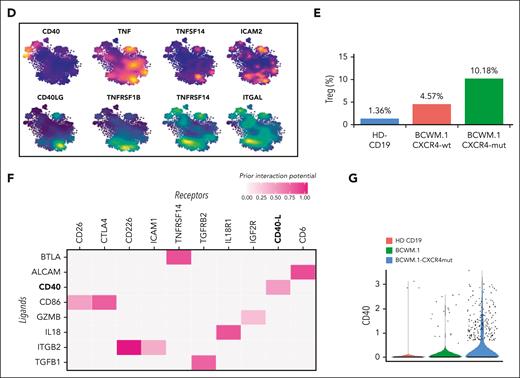
To characterize the specific signaling events responsible for the induction of Treg differentiation (Figure 6A), we evaluated the B-cell–T-cell interactions at single-cell level, adopting a B->T cross talk model (Figure 6A). Briefly, WMCL1 or WMCL1-CXCR4C1013G clusters were identified as potential sender cells, that is, cells responsible for the release of the immunosuppressive signal. Similarly, cluster-3 Tregs were considered as receiver cells, and the Treg transcriptional program was identified by comparing Treg normalized counts with the CD4+ T-cell cluster that is transcriptionally the closest (supplemental Methods). This cross talk model allowed the identification of a set of candidate protein-to-protein interactions common between WMCL1 and WMCL1-CXCR4C1013G (Figure 6B). To prioritize this set, the expression pattern of the candidate sender markers was analyzed in normal CD19, WMCL1, and WMCL1-CXCR4C1013G cells (Figure 6C). Higher priority was given to genes showing a WMCL1-CXCR4C1013G > WMCL1 > CD19 expression pattern, because the effect of this interaction in the induction of the immunosuppressive phenotype was shown to be more pronounced in WMCL1-CXCR4C1013G cells compared with WMCL1 or normal CD19 cells. This approach allowed us to identify a set of 4 high-priority genes, namely CD40, TNF, TNFSF14, and ICAM2, with the first 3 all being part of the TNF gene family and the latter being an intercellular adhesion molecule belonging to the ICAM family. We then analyzed, in detail, the expression pattern of the 4 genes. Although all of these genes were expressed in B cells, as expected, only the expression of CD40 was both maximal in, and virtually restricted to, the B clusters (Figure 6D), with the other 3 genes showing a more diffuse pattern.
B-cell–to–T-cell cross talk model at single-cell level. (A) Schematic representation of the expected B-cell–to–T-cell crosstalk model. In the top panel the normal B/T condition is shown; the bottom panel depicts the model of the proposed B-cell–to–T-cell interaction, with clonal B cells releasing factor(s) responsible for the induction of CD4+ T-cell differentiation to Tregs. (B) Heatmap representing the top candidate protein-protein interactions occurring between B (ligands, y-axis) and T (receptors, x-axis) cells in WMCL1 and WMCL1-CXCR4C1013G cells. CD40/CD40LG interactors are shown in bold. (C) Violin plots showing the normalized expression level of all the candidate receptors expressed by B cells (B) in CD19, WMCL1, and WMCL1-CXCR4C1013G cells. (D) tSNE plots highlighting the expression level of top B-cell (top) and T-cell (bottom) interactors. B-cell markers are represented in an orange (high expression) to blue (low expression) color scale; Treg markers are represented in a yellow (high expression) to blue (low expression) color scale. (E) Percentage of Tregs after exposure to normal CD19, BCMW.1, or BCWM.1-CXCR4C1013G cells. (F) Heatmap representing the top candidate protein-protein interactions occurring between B (ligands, y-axis) and T (receptors, x-axis) cells in BCMW.1 and BCWM.1-CXCR4C1013G cells. CD40/CD40LG interactors are shown in bold. (G) Violin plots showing the normalized expression level of CD40 in normal CD19, BCWM.1, or BCWM.1-CXCR4C1013G cells.
B-cell–to–T-cell cross talk model at single-cell level. (A) Schematic representation of the expected B-cell–to–T-cell crosstalk model. In the top panel the normal B/T condition is shown; the bottom panel depicts the model of the proposed B-cell–to–T-cell interaction, with clonal B cells releasing factor(s) responsible for the induction of CD4+ T-cell differentiation to Tregs. (B) Heatmap representing the top candidate protein-protein interactions occurring between B (ligands, y-axis) and T (receptors, x-axis) cells in WMCL1 and WMCL1-CXCR4C1013G cells. CD40/CD40LG interactors are shown in bold. (C) Violin plots showing the normalized expression level of all the candidate receptors expressed by B cells (B) in CD19, WMCL1, and WMCL1-CXCR4C1013G cells. (D) tSNE plots highlighting the expression level of top B-cell (top) and T-cell (bottom) interactors. B-cell markers are represented in an orange (high expression) to blue (low expression) color scale; Treg markers are represented in a yellow (high expression) to blue (low expression) color scale. (E) Percentage of Tregs after exposure to normal CD19, BCMW.1, or BCWM.1-CXCR4C1013G cells. (F) Heatmap representing the top candidate protein-protein interactions occurring between B (ligands, y-axis) and T (receptors, x-axis) cells in BCMW.1 and BCWM.1-CXCR4C1013G cells. CD40/CD40LG interactors are shown in bold. (G) Violin plots showing the normalized expression level of CD40 in normal CD19, BCWM.1, or BCWM.1-CXCR4C1013G cells.
To further corroborate this finding, we assessed the functional impact of BCWM.1 and BCWM.1-CXCR4C1013G WM cells in the support of Treg induction. In line with the WMCL1 model, BCWM.1 cells and, to a greater extent, BCWM.1-CXCR4C1013G cells fostered a more pronounced increase in the CD4+CD25+FoxP3+ Treg population, relative to their normal CD19+ B-cell counterpart (Figure 6E). Furthermore, Treg proliferation was significantly enhanced within the context of CXCR4C1013G-mut WM cells (Figure 6E). Finally, BCWM.1 B->T cross talk analysis also supported CD40/CD40L as a potentially relevant cell-to-cell interaction (Figure 6F and G).
Taken together, our data indicate the existence of CD40/CD40L-mediated cross talk between WM cells (CD40+) and Tregs (CD40L+), which is responsible for the induction of an immunosuppressive milieu. To further corroborate our findings, we interrogated the transcriptional profile of an independent series of tumor-depleted WM BM milieu (n = 22), compared with the related normal counterpart (n = 10). In line with our results, we could confirm the existence of a significant enrichment for a Treg signature, and for CD40/CD40L signaling–related genes (supplemental Figure 2A-B).
Given the identified CD40/CD40L axis as a potential regulator of the WM cell/Treg cross talk, we performed functional validation assays by using DRI-C21045, a potent inhibitor of the CD40/CD40L interaction,33 with the aim to neutralize the axis and to investigate whether CD40/CD40L abrogation could result in reversion of the WM cell–dependent Treg induction.
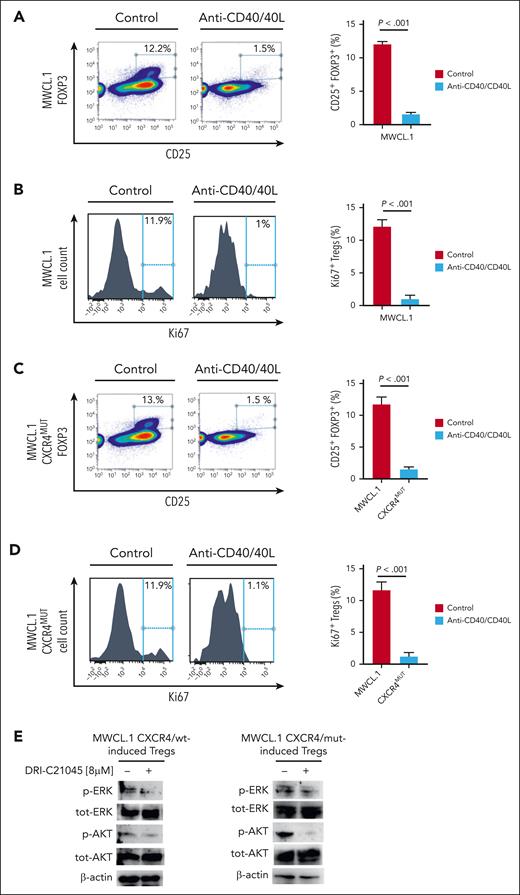
Treg induction was tested in the presence and absence of WM cells (BCWM.1; MWCL.1), exposed to DRI-C21045 and we observed a significant decrease in Treg induction upon CD40/CD40L inhibition (Figure 7A; supplemental Figure 3A). In parallel, by abrogating the CD40/CD40L interaction, a significant reduction of Treg proliferation was observed (Figure 7B; supplemental Figure 3B). Importantly, DRI-C21045 was tested in the absence of WM cells, showing the lack of any significant effect on CD4+/CD25− non-Tregs in terms of Treg induction modulation (supplemental Figure 4A).
CD40/CD40L blockade is functionally relevant in halting WM-cell–dependent Treg induction, even in the context of CXCR4C1013G-mut WM cells. (A) CD4+CD25− non-Tregs were cultured either alone or in the presence of WM cells (MWCL.1) for 72 hours, in presence or absence of CD40/CD40L-inhibitor (DRI-C21045; 8 μM), using a transwell. Inhibition of Treg induction was observed upon exposure to DRI-C21045. The percentage indicates the CD25+FOXP3+ cells gated on CD4+ cells. A representative experiment is shown. The experiment was repeated 3 times. P indicates P value, determined using one-way ANOVA. Error bars indicate mean ± SD. (B) The proliferative rate of CD4+CD25+FOXP3+ Tregs was assessed by evaluating the percentage of Ki67+ cells. Significant inhibition of Ki-67 in Tregs was observed. A representative experiment is shown. The experiment was repeated 4 times. P indicates P value, determined using one-way ANOVA. Error bars indicate mean ± SD. (C) Same experimental approach as in panel A, using MWCL.1-CXCR4C1013G mutated cells. (D) Same experimental approach as in panel B, using MWCL.1-CXCR4C1013G mutated cells. (E) Both induced and noninduced Tregs, prepared as in panel A, were harvested, and cell lysates were subjected to western blot analysis using antiphoshpo(p)-AKT, anti–total (tot)-AKT, anti–p-ERK, anti–tot-ERK, and anti–β actin.
CD40/CD40L blockade is functionally relevant in halting WM-cell–dependent Treg induction, even in the context of CXCR4C1013G-mut WM cells. (A) CD4+CD25− non-Tregs were cultured either alone or in the presence of WM cells (MWCL.1) for 72 hours, in presence or absence of CD40/CD40L-inhibitor (DRI-C21045; 8 μM), using a transwell. Inhibition of Treg induction was observed upon exposure to DRI-C21045. The percentage indicates the CD25+FOXP3+ cells gated on CD4+ cells. A representative experiment is shown. The experiment was repeated 3 times. P indicates P value, determined using one-way ANOVA. Error bars indicate mean ± SD. (B) The proliferative rate of CD4+CD25+FOXP3+ Tregs was assessed by evaluating the percentage of Ki67+ cells. Significant inhibition of Ki-67 in Tregs was observed. A representative experiment is shown. The experiment was repeated 4 times. P indicates P value, determined using one-way ANOVA. Error bars indicate mean ± SD. (C) Same experimental approach as in panel A, using MWCL.1-CXCR4C1013G mutated cells. (D) Same experimental approach as in panel B, using MWCL.1-CXCR4C1013G mutated cells. (E) Both induced and noninduced Tregs, prepared as in panel A, were harvested, and cell lysates were subjected to western blot analysis using antiphoshpo(p)-AKT, anti–total (tot)-AKT, anti–p-ERK, anti–tot-ERK, and anti–β actin.
Considering the relevance of CXCR4C1013G mutation in WM biology,4,10,30,31 we investigated whether CD40/CD40L inhibition could also modulate the Treg phenotype within the context of CXCR4-mut WM cells. Our findings demonstrated that halting CD40/CD40L interaction could also target Treg inhibition and Treg growth within the context of CXCR4-mut WM (Figure 7C,D; supplemental Figure 3C-D). In order to further define modulation of prosurvival signaling pathways, total protein lysates were obtained from Tregs induced by either CXCR4/wt or CXCR4C1013G mut WM cells, in the presence and absence of DRI-C21045. The results indicate that CD40/CD40L blockade led to inhibition of p-AKT and p-ERK in Tregs (Figure 7E).
Having shown that targeting the CD40/CD40L axis resulted in inhibition of Tregs, we then investigated whether this could lead to modulation of WM cell growth. We therefore interrogated the proliferative potential of WM cells harvested at the end of the induction assay. We evaluated whether DRI-C21045–mediated CD40/CD40L blockade could result in modulation of WM cell proliferation, within the context of the halted Treg induction, and we observed a significant inhibition of WM cell growth (supplemental Figure 5A-B). We also tested WM cells alone exposed to DRI-C21045, showing lack of cytotoxicity in DRI-C21045–treated cells (supplemental Figure 5C).
Discussion
The literature has shown that Tregs may play a role in either favoring tumor immune evasion, leading to tumor progression and poor prognosis,12-17 or prediction of both survival and risk of transformation, as shown in patients with follicular lymphoma.34 However, the functional relevance of Tregs in supporting WM pathogenesis has not been elucidated. Our aim was therefore to assess how Tregs are potentially involved in WM pathogenesis by using a transgenic lymphoplasmacytic/WM murine model, and to translate the observations obtained within the in vivo setting to a scenario involving patients with WM. We therefore described the transcriptome profiling of primary WM-Treg, and dissected the cross talk between WM and Tregs, thus identifying the interactions that may be responsible for immunosuppression and WM disease progression.
The transgenic lymphoplasmacytic/WM murine model allowed us to unveil a biological role of Tregs in the support of WM disease progression. These findings were corroborated by showing the existence of a transcriptome signature that clearly differentiates WM-Tregs from HD-Tregs, thus confirming the hypothesis that, indeed, Tregs represent a novel component of WM pathogenesis. Our findings show that WM-Tregs are characterized by a significant enrichment of prosurvival signaling cascades, including MAPK- and PI3K/AKT-related genes. We then confirmed how these changes were functionally mirrored by enhanced proliferation of primary WM-Tregs, relative to their normal cellular counterpart. Of note, Treg induction and expansion were both significantly enhanced within the context of WM, compared with normal B cells. Notably, primary Tregs derived from patients with WM showed a significantly higher suppressive activity, compared with HD-Tregs, which can be explained, at least in part, by the upregulation of EBI3, FGL2, GZMB, and PRF1, reported to play an essential role in Treg-mediated suppression of tumor clearance.35 Our findings show, to the best of our knowledge, for the first time, that patients with WM present with a Treg transcriptome and phenotype that may support WM tumor biology.
Given the occurrence of CXCR4C1013G somatic aberration in ∼30% of patients with WM, and its role in favoring WM disease progression,4,10,30,31 we also investigated whether CXCR4-mut WM cells could present with a different ability to mold the Treg phenotype. We provide evidence that demonstrates a higher Treg induction, expansion, and proliferation, exerted by CXCR4C1013G-mut WM cells.
Recent studies have shown how, despite the presence of low levels of programmed cell death 1 (PD-1) within the BM niche of patients with WM, abundant levels of soluble PD-1 ligands (s-PDL-1/2) were reportedly observed within the peripheral blood, leading to the formation of a hypothesis of how s-PDL-1/2 could contribute to WM biology by conditioning a T-cell function attenuation, thus resulting in WM disease progression.36 It is known that PDL-1 can enhance immunosuppressive activity of Tregs and convert naïve CD4+ T cells to Tregs,37 thus confirming our findings that indicate the occurrence of Treg dysregulation, characterized by a profound immunosuppressive phenotype.
To further define the specific signaling events responsible for the induction of Treg differentiation, we dissected B-cell–T-cell interactions at the single-cell level, adopting a B-to-T cross talk model. This approach allowed us to identify the CD40/CD40-ligand axis as a putative link to support WM-Tregs interaction. We therefore hypothesized that the CD40/CD40-ligand axis could act as a facilitator of WM cell–driven Treg induction and expansion. Indeed, by using a pan-CD40/CD40-ligand inhibitor, a reverted phenotype with reduced Treg induction and expansion was demonstrated, suggesting the therapeutic relevance for a novel approached with the aim of at halting the Treg-dependent immunosuppression in WM disease.
Moreover, previous studies have pointed toward the occurrence of mast-cell infiltration in close association with WM cells; Tournilhac et al showed the potential role of the CD40/CD40-ligand axis in mediating the interaction between WM cells and mast cells, expressing CD40 and CD40-ligand, respectively.38 Therefore, halting the CD40/CD40-ligand axis could have a dual effect, ultimately leading to inhibition of WM tumor growth.
FOXP3+ Tregs reportedly inhibit anticancer immune response, leading to tumor immune evasion and disease progression. These findings have prompted researchers to consider the possibility of achieving an antitumor response as a consequence of Treg depletion. However, this may not be the best approach because Treg ablation could be responsible for the induction of autoimmune disease. Therefore, the alternative and more promising strategy is the possibility of targeting WM tumor cell–specific Tregs by halting the vital part of this interaction represented by the CD40/CD40-ligand axis.
Taken together, our studies demonstrate the occurrence of Treg-mediated immunosuppression in WM, regulated via the CD40/CD40-ligand axis, which creates a functional cross talk between WM and Tregs. Moreover, the results suggest how halting CD40/CD40-ligand interaction may represent a strategy to inhibit the Treg-mediated immunosuppressive scenario in WM, resulting in inhibition of WM cell proliferation.
Acknowledgments
The authors thank Cinzia Caprio for assisting with flow cytometry sample preparation; M. Reth (University of Freiburg) and H.C. Reinhardt (University Hospital Essen) for providing Mb1-cre and Myd88p.L252P mice, respectively; and G. Knittel (University Hospital Essen) for providing the M191 cell line.
This study was supported by the European Hematology Association, Italian Association for Cancer Research, Associazione Italiana contro le Leucemie-linfomi e Mieloma Brescia, Fondazione Spedali Civili Brescia, Pro Loco Mompiano, Brescia (A.M.R.) and by the Italian Association for Cancer Research and the Department of Excellence project IMPACT MEDICINE (IMaging e PAtologia digitale CombinaTi per nuove traiettorie di diagnosi e terapia) (R.P.). J.C. and J.A.M.-C. were supported through the Accelerator Award Program from the Spanish Association Against Cancer, Cancer Research United Kingdom, and Associazione Italiana per la Ricerca sul Cancro and by Spanish Ministry of Health–Instituto de Salud Carlos III (FIS) grants PI19/00818 and CIBERONC #CB16/12/00489.
Authorship
Contribution: A.S. and A.M.R. conceived and designed the study; A.S., V.D., A.G.S., and R.P. contributed to writing of the manuscript; A.M.R. and R.P. performed study supervision; A.S., F.R., and F.D.S. performed in vitro studies; V.G. and M.Ch. performed flow cytometry studies; Y.K. assessed flow cytometry analysis; M.Ce., D.S., A.C., C.A., A.A., A.T., and M.M. collected patient samples; A.S., F.R., and F.D.S. processed patient samples; A.M.R., A.S., Y.K., R.P, M.Ce., A.G.S, A.C., and J.A.M.-C. contributed to manuscript reviewing; D.D, S.S, and D.R. performed RNA sequencing and single-cell RNA sequencing profiling; K.T. and A.N. performed gene expression profiling; R.P. performed bulk RNA sequencing, scRNA sequencing, and GEP analysis; and J.C. and J.A.M.-C. generated the murine WM model and performed in vivo studies.
Conflict-of-interest disclosure: A.M.R. receives research funding from AstraZeneca, European Hematology Association, Transcan2-ERANET, and Italian Association for Cancer Research; and honoraria from Amgen, Celgene, Janssen, and Takeda. J.A.M.-C. receives research funding from Roche/Genentech, Bristol-Myers Squibb, Janssen, Regeneron, Palleon, and Priothera. The remaining authors declare no competing financial interests.
Correspondence: Aldo M. Roccaro, ASST Spedali Civili di Brescia, Clinical Trial Center, Translational Research and Phase I Unit, P.le Spedali Civili di Brescia, n. 1, 25123, Brescia, Italy; e-mail: aldomaria.roccaro@asst-spedalicivili.it.
References
Author notes
∗R.P. and A.M.R. are joint last authors.
Files related to transcriptome profiling of WM- and healthy individual-derived Tregs have been deposited in the Sequence Read Archive (accession number PRJNA863084). Files related to single-cell RNA sequencing data have been deposited in the from Sequence Read Archive (accession number PRJNA861837).
Data are available on request from the corresponding author, Aldo M. Roccaro (aldomaria.roccaro@asst-spedalicivili.it).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal