Key Points
The NFIA-ETO2 fusion impairs erythroid differentiation and cooperates with mutant TP53 to induce PEL in mice.
NFIA-ETO2 binds and represses erythroid regulators, most likely by binding to NFI motifs and/or interaction with ETO2 decorating these loci.
Abstract
The NFIA-ETO2 fusion is the product of a t(1;16)(p31;q24) chromosomal translocation, so far, exclusively found in pediatric patients with pure erythroid leukemia (PEL). To address the role for the pathogenesis of the disease, we facilitated the expression of the NFIA-ETO2 fusion in murine erythroblasts (EBs). We observed that NFIA-ETO2 significantly increased proliferation and impaired erythroid differentiation of murine erythroleukemia cells and of primary fetal liver–derived EBs. However, NFIA-ETO2–expressing EBs acquired neither aberrant in vitro clonogenic activity nor disease-inducing potential upon transplantation into irradiated syngenic mice. In contrast, in the presence of 1 of the most prevalent erythroleukemia-associated mutations, TP53R248Q, expression of NFIA-ETO2 resulted in aberrant clonogenic activity and induced a fully penetrant transplantable PEL-like disease in mice. Molecular studies support that NFIA-ETO2 interferes with erythroid differentiation by preferentially binding and repressing erythroid genes that contain NFI binding sites and/or are decorated by ETO2, resulting in a activity shift from GATA- to ETS-motif-containing target genes. In contrast, TP53R248Q does not affect erythroid differentiation but provides self-renewal and survival potential, mostly via downregulation of known TP53 targets. Collectively, our work indicates that NFIA-ETO2 initiates PEL by suppressing gene expression programs of terminal erythroid differentiation and cooperates with TP53 mutation to induce erythroleukemia.
Introduction
Acute erythroleukemia (AEL) is a rare but aggressive human cancer characterized by uncontrolled accumulation of erythroid progenitor cells. Different subtypes, including pure acute erythroleukemia (PEL; also known as acute myeloid leukemia-M6a [AML-M6a] or Di Giuglielmo disease), have been described, with blasts committed exclusively to the erythroid lineage and AML-M6a characterized by the presence of erythroid precursors together with myeloid blasts, which often makes the diagnosis of AEL challenging.1,2 Because of the low reproducibility of erythroid blast counts and the close biological relationship, the World Health Organization reclassified AML-M6a into the category of myelodysplastic syndrome.3
Sequencing studies revealed a heterogeneous, genomic AEL landscape in which mutations of the tumor suppressor TP53 gene were the most prevalent lesions with some recurrent mutational hotspots, including TP53R248.4-9 Notably, tumor cells of almost all analyzed patients with PEL were found to carry mono or biallelic TP53 mutations mostly affecting its DNA binding domain.10,11 Earlier mouse models of progression of myeloproliferative neoplasms driven by constitutive active tyrosine kinases also showed that loss of TP53 resulted in erythroleukemia-like phenotypes.12-14
Cytogenetic analysis and RNA sequencing of leukemic cells from pediatric patients with PEL revealed a chromosomal translocation t(1;16)(p13;q24) leading to the expression of a fusion between the nuclear factor 1-A (NFIA) and ETO2 (also known as, core binding factor runt domain αsubunit 2 translocated to 3).15-18 The transcription factor NFIA has previously been shown to control erythroid fate of hematopoietic progenitors, whereas ETO2 was characterized as transcriptional coregulator controlling the differentiation of hematopoietic stem cells and erythroid progenitor cells.19,20 More recently, a novel t(1;8)(p13;q21) translocation fusing NFIA to ETO (ie, RUNXT1) was identified in pediatric PEL, providing additional evidence for the significant association between a fusion of NFIA to ETO/ETO2 domains and the malignant transformation of cells of the erythroid lineage.21 Here, we report that retroviral NFIA-ETO2 expression impairs in vitro terminal differentiation of murine erythroleukemia (MEL) cells and primary murine erythroblasts (EBs) but is not sufficient to induce a disease in vivo upon transplantation into lethally-irradiated mice. In the presence of a leukemia-associated TP53R248Q mutation, however, NFIA-ETO2–expressing EBs gain serial replating activity and induce a fully penetrant PEL-like disease upon transplantation into irradiated mice. Molecular studies suggest that NFIA-ETO2 interferes with differentiation by binding and repressing erythroid regulatory genes that contain NF-binding sites and/or are decorated by ETO2. This results in an activity shift from GATA- to ETS-motif–containing genes. On the other hand, TP53R248Q supports cell self-renewal and survival. Collectively, our work functionally characterizes the NFIA-ETO2 fusion as an initiation lesion toward PEL.
Methods
Cloning of the NFIA-ETO2 fusion ORF into the pMSCV retroviral expression vector
Full-length complementary DNA for murine Nfia was acquired from Addgene (#112698) and full-length complementary DNA for human ETO2 was obtained from Thomas Mercher (Paris, France). NFIA-ETO2 has an open reading frame (ORF) leading to a chimeric protein containing 203 amino acid residues from Nfia (NP_001128145.1) and 603 residues from ETO2 (NP_005178.4). The fusion gene as well as Nfia or ETO2 ORFs were cloned into pMSCV and pcDNA3.1 expression vectors containing selection markers such as green fluorescent protein (GFP) and neomycin, respectively. To clone NFIA-ETO2 domain mutants, Q5 Site-Direct Mutagenesis Kit (New England Biolabs) was used per the manufacturer’s protocol. Primers are listed in supplemental Table 1, available on the Blood website.
To knock down putative NFIA-ETO2 targets, we facilitated the expression of short hairpin RNAs from the pLT3GEPIR backbone (kind gift from Johannes Zuber, Vienna, Austria). Target-selective oligonucleotides were amplified via polymerase chain reaction (PCR) using the primers miRE-XhoI-fw (5′-TGAACTCGAGAAGGTATATTGC TGTTGACAGTGAGCG-3′) and miRE-EcoRI-rev (5′-TCTCGAATTCTAGCCCCTT GAAGTCCGAGGCAGTAGGC-3′) and cloned into pLT3GEPIR. Emerging vectors were sequence-validated. The sequences of 97-mer oligonucletides targeting Myb and Spi1 are available in supplemental Table 1.
BM transplantation
E14.5 fetal liver (FL)–derived, adult (6-10 weeks) bone marrow [BM]-derived NFIA-ETO2, or vector-control transduced EBs (1.5 × 106) were transplanted together with normal total BM (4 × 106) from ∼6 to 9-week-old mice into lethally- or sublethally-irradiated recipient mice (BC57BL/6NCrl) (2 × 600 cGy). For secondary transplantation, 2 × 106 total BM cells of symptomatic mice were transplanted into lethally- or sublethally-irradiated recipients. The irradiation dose did not affect the emerging phenotype.
Cytospin preparation
Cytospins were prepared on noncoated cytoslides (Catalog 5991051; Thermo Fisher Scientific, Reinach, Switzerland) via centrifugation of 1 × 105 cells for 3 minutes at 200 revolutions per minute using a Shandon cytospin 3 centrifuge, followed by Wright-Giemsa staining.
Additional information is provided in the supplemental Materials.
Results
NFIA-ETO2 blocks in vitro differentiation of MEL and primary murine EBs
To address the activity in erythroid cells, we first facilitated the expression of the NFIA-ETO2 fusion in MEL cells. These cells undergo partial erythroid maturation after exposure to polar compounds, such as dimethyl sulfoxide (DMSO), reflected by reduced CD71 and increased Ter119 surface expression associated with augmented hemoglobin production. This leads to reddish cell pellets and benzidine-staining positive cells.22 Of note, although NFIA-ETO2 expression slightly increased MEL cell proliferation, it impaired DMSO-induced erythroid differentiation, as shown by reduced benzidine staining, Ter119 expression, and hemoglobin messenger RNA (mRNA) levels (Figure 1A-D).
NFIA-ETO2 blocks in vitro terminal differentiation of MEL and primary murine EBs. (A-C) NFIA-ETO2 expression (compared with vector-transduced cells) slightly increased proliferation (A) and the number of benzidine-positive cells (B) and reduced Ter119 surface expression on MEL cells grown for 4 days in 2% DMSO (C). (D) NFIA-ETO2–expressing MEL cells expressed significantly lower Hbα1 and Hbβ1 mRNA level than vector control-transduced cells. (E-F) NFIA-ETO2–expressing BM-derived mouse EBs proliferated significantly faster than vector-transduced control cells (CTRL) over 6 days in MM (E) or in DM (F) (n = 4). (G) Representative images of Wright-Giemsa-stained cytospin preparations from NFIA-ETO2–expressing primary mouse BM-derived EBs (top) compared with vector-transduced control cells (bottom). The left panels show EBs in MM (day 0), the middle panel EBs after 2 days in DM, and the right panel EBs after 4 days in DM. The small insert shows a reddish cell pellet in CTRL cells, whereas the pellet of NFIA-ETO2–expressing cells appeared white. Images were recorded with a 60× objective using a Nikon-TI. Scale bar, 50 μm. (H) NFIA-ETO2–expressing, FL-derived (E14.5) EBs continued proliferating, whereas cells overexpressing ETO2 started to decrease after 5 weeks of culture in MM. NFIA- and CTRL vector (pMSCV-GFP)–expressing cells could not be expanded. Data represent 1 out of 2 independent experiments. (I-J) NFIA-ETO2–expressing BM-derived EBs expressed lower levels of Hbα1 mRNA (I) and Gypa mRNA (J) than CTRL cells grown in MM (n = 3) and 24 hours in DM (n = 5), as assessed via qRT-PCR. (K) CD71 and Ter119 surface expression (in % of cells) on NFIA-ETO2– and CTRL, BM-derived EBs after 6 days in DM (n = 4). Values are presented as individual points; bar graphs represent the mean value of independent biological replicates (n); error bars are standard error of the mean. Statistical significances in was tested with paired two-tailed t tests.
NFIA-ETO2 blocks in vitro terminal differentiation of MEL and primary murine EBs. (A-C) NFIA-ETO2 expression (compared with vector-transduced cells) slightly increased proliferation (A) and the number of benzidine-positive cells (B) and reduced Ter119 surface expression on MEL cells grown for 4 days in 2% DMSO (C). (D) NFIA-ETO2–expressing MEL cells expressed significantly lower Hbα1 and Hbβ1 mRNA level than vector control-transduced cells. (E-F) NFIA-ETO2–expressing BM-derived mouse EBs proliferated significantly faster than vector-transduced control cells (CTRL) over 6 days in MM (E) or in DM (F) (n = 4). (G) Representative images of Wright-Giemsa-stained cytospin preparations from NFIA-ETO2–expressing primary mouse BM-derived EBs (top) compared with vector-transduced control cells (bottom). The left panels show EBs in MM (day 0), the middle panel EBs after 2 days in DM, and the right panel EBs after 4 days in DM. The small insert shows a reddish cell pellet in CTRL cells, whereas the pellet of NFIA-ETO2–expressing cells appeared white. Images were recorded with a 60× objective using a Nikon-TI. Scale bar, 50 μm. (H) NFIA-ETO2–expressing, FL-derived (E14.5) EBs continued proliferating, whereas cells overexpressing ETO2 started to decrease after 5 weeks of culture in MM. NFIA- and CTRL vector (pMSCV-GFP)–expressing cells could not be expanded. Data represent 1 out of 2 independent experiments. (I-J) NFIA-ETO2–expressing BM-derived EBs expressed lower levels of Hbα1 mRNA (I) and Gypa mRNA (J) than CTRL cells grown in MM (n = 3) and 24 hours in DM (n = 5), as assessed via qRT-PCR. (K) CD71 and Ter119 surface expression (in % of cells) on NFIA-ETO2– and CTRL, BM-derived EBs after 6 days in DM (n = 4). Values are presented as individual points; bar graphs represent the mean value of independent biological replicates (n); error bars are standard error of the mean. Statistical significances in was tested with paired two-tailed t tests.
To corroborate these findings in primary cells, we established EBs from E14.5 FLs or from lineage marker–depleted BM from mice aged between 6 and 10 weeks. Before viral transduction, the cells were expanded for ∼8 to 10 days in maintenance medium (MM), containing stem cell factor, dexamethasone, insulin-like growth factor 1, cholesterol, and erythropoietin (EPO; supplemental Figure 1A).23 NFIA-ETO2 expression (confirmed at the mRNA and protein level; supplemental Figure 1B) significantly increased EB proliferation in MM (Figure 1E). Although control vector-transduced cells almost completely differentiated into mature erythrocytes within 4 to 6 days in differentiation medium (DM; containing EPO and stem cell factor), NFIA-ETO2 expressing cells continued proliferating (Figure 1E-F) and did not exhibit any major morphologic signs of differentiation, nor did they form any red cell pellets (Figure 1G; data not shown). Although the retroviral overexpression of NFIA-ETO2 resulted in stable immortalization of FL-derived EBs, ETO2–overexpressing cells started decreasing after 5 weeks, and NFIA-overexpressing cells could not be successfully expanded (Figure 1H; supplemental Figure 1C). Independent of the cellular origin (FL or adult BM) impaired erythroid differentiation of NFIA-ETO2–expressing EBs was associated with increased Kit, decreased Ter119 surface expression, and significantly reduced mRNA expression of several erythroid marker genes, including Hbα1, Hbβ1, EpoR, Gypa, and Gata1 (Figure 1I-K; supplemental Figure 1D-I). Notably, NFIA-ETO2–expressing EBs proliferated equally in the presence and absence of exogenous EPO (supplemental Figure 1J). Taken together, retroviral NFIA-ETO2 expression increased EPO-independent proliferation and impaired induced terminal erythroid differentiation of primary FL- and adult BM-derived mouse EBs. Importantly, however, NFIA-ETO2–expressing EBs could not be serially propagated in methylcellulose cultures (MCs), and transplantation of the cells into lethally-irradiated syngeneic mice did not result in any disease (supplemental Figure 1K-Q).
Expression of NFIA-ETO2 in MEL cells and primary EBs reduces expression of erythroid master regulators
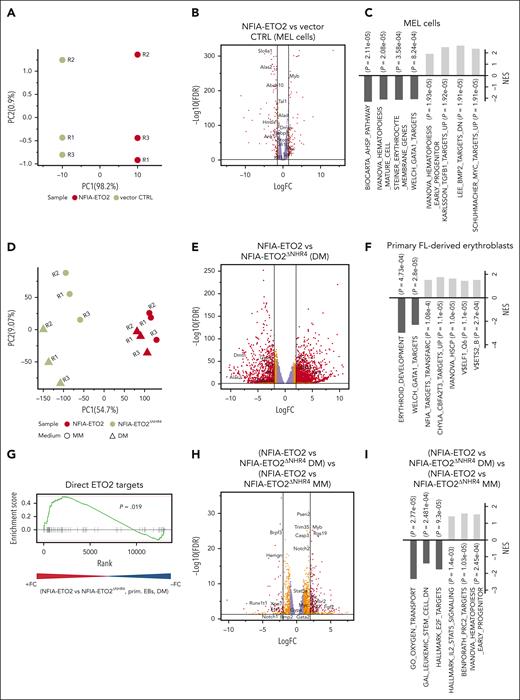
To better understand how NFIA-ETO2 impairs terminal erythroid differentiation, we first determined the gene expression profiles of MEL cells expressing NFIA-ETO2 exposed to DMSO (2%) for 48 hours. Performing principle component analysis (PCA) helped clearly separate the gene expression profiles of NFIA-ETO2 and vector-transduced controls (Figure 2A). DMSO-induced differentiation of NFIA-ETO2–expressing MEL cells was associated with significant up and downregulation of 866 and 742 genes, respectively (false discovery rate [FDR] ≤0.05, fold change [FC] >1.5). In particular, several known targets of the erythroid master transcription factor GATA1, such as 5'-aminolevulinate synthase 2), solute carrier family 4 member 1, and hemoglobins were among the most significantly less expressed genes in the presence of NFIA-ETO2 (Figure 2B; supplemental Table 2). In addition, several master regulators of erythroid differentiation including Tal1 (TAL BHLH transcription factor 1, erythroid differentiation factor), Gfi1B (growth factor independent 1B transcriptional repressor), or Klf1 (Kruppel-like factor 1) were also significantly less expressed. In contrast, hematopoietic proto-oncogenes, including Myb, Myc, and some Ets family members were among the most significantly highly expressed genes. Gene set enrichment analysis (GSEA) revealed a negative enrichment of gene signatures with erythroid differentiation and positive enrichment of gene signatures regulated by the MYC proto-oncogene and TGF-β signaling (Figure 2C; supplemental Table 3). Notably, short hairpin RNA–mediated knockdown of Myb or Spi1 helped overcome the NFIA-ETO2–mediated differentiation block in MEL cells, as measured by Ter119 and CD71 expression (supplemental Figure 2A).
Reduced expression of master regulators of erythroid differentiation in NFIA-ETO2+ MEL cells and primary EBs. (A) PCA of the gene expression signatures of NFIA-ETO2–expressing MEL cells compared with CTRL cells grown for 2 days in 2% DMSO. Each point represents 1 sample, colored for NFIA-ETO2 (red) or CTRLs (olive). (B) Volcano plot showing DEGs from the NFIA-ETO2 and CTRL MEL cells grown for 2 days in 2% DMSO (FDR <0.05; logFC >±1.5 black lines). Red and orange points represent statistically significant dysregulated genes (FDR < 0.05; logFC >±1.25 or 1.5 respectively), whereas purple points represent nonsignificantly regulated genes. (C) Selected positive and negative GSEA enrichment scores of DEG between NFIA-ETO2–expressing MEL and CTRL cells grown for 2 days in 2% DMSO (padj < 0.05). (D) PCA of the gene expression signatures of FL-derived EBs expressing NFIA-ETO2 or NFIA-ETO2ΔNHR4 grown in MM or for 24 hours in DM. Each point represents 1 sample, colored and shaped based on NFIA-ETO2 (red), or NFIA-ETO2ΔNHR4 (olive) expression based on the medium in which cells were cultured in (MM, dots; DM, triangles). (E) Volcano plot showing DEGs from the NFIA-ETO2- vs NFIA-ETO2ΔNHR4–expressing, FL-derived EBs after 24 hours in DM (FDR <0.05; logFC >±1.5 black lines). Orange and red points represent statistically significant dysregulated genes (FDR <0.05; logFC >±1.25; or FDR <0.01; logFC >±1.5 respectively), whereas purple points represent nonsignificantly regulated genes. (F) Selected positive and negative GSEA enrichment scores of DEGs between NFIA-ETO2- or NFIA-ETO2ΔNHR4–expressing, FL-derived EBs grown 24 hours in DM (P < .05). (G) GSEA shows significant enrichment for previously proposed24 direct ETO2 targets among upregulated DEGs in NFIA-ETO2–expressing FL-derived EBs) compared with NFIA-ETO2ΔNHR4–expressing cells grown for 24 hours in DM (P < .05). (H) Volcano plot showing DEGs from the NFIA-ETO2 vs NFIA-ETO2ΔNHR4-expressing EBs grown in MM vs NFIA-ETO2 vs NFIA-ETO2ΔNHR4–expressing, FL-derived EBs grown in DM for 24 hours (FDR >0.05; logFC >±1.5 black lines). Orange and red points represent statistically significant dysregulated genes (FDR <0.05; logFC >±1.25; or FDR <0.01; logFC >±1.5 respectively), whereas purple points represent nonsignificantly regulated genes. (I) Selected GSEA correlations of DEG from NFIA-ETO2 vs NFIA-ETO2ΔNHR4–expressing, FL-derived EBs grown in MM vs NFIA-ETO2 vs NFIA-ETO2ΔNHR4–expressing EBs grown in DM for 24 hours.
Reduced expression of master regulators of erythroid differentiation in NFIA-ETO2+ MEL cells and primary EBs. (A) PCA of the gene expression signatures of NFIA-ETO2–expressing MEL cells compared with CTRL cells grown for 2 days in 2% DMSO. Each point represents 1 sample, colored for NFIA-ETO2 (red) or CTRLs (olive). (B) Volcano plot showing DEGs from the NFIA-ETO2 and CTRL MEL cells grown for 2 days in 2% DMSO (FDR <0.05; logFC >±1.5 black lines). Red and orange points represent statistically significant dysregulated genes (FDR < 0.05; logFC >±1.25 or 1.5 respectively), whereas purple points represent nonsignificantly regulated genes. (C) Selected positive and negative GSEA enrichment scores of DEG between NFIA-ETO2–expressing MEL and CTRL cells grown for 2 days in 2% DMSO (padj < 0.05). (D) PCA of the gene expression signatures of FL-derived EBs expressing NFIA-ETO2 or NFIA-ETO2ΔNHR4 grown in MM or for 24 hours in DM. Each point represents 1 sample, colored and shaped based on NFIA-ETO2 (red), or NFIA-ETO2ΔNHR4 (olive) expression based on the medium in which cells were cultured in (MM, dots; DM, triangles). (E) Volcano plot showing DEGs from the NFIA-ETO2- vs NFIA-ETO2ΔNHR4–expressing, FL-derived EBs after 24 hours in DM (FDR <0.05; logFC >±1.5 black lines). Orange and red points represent statistically significant dysregulated genes (FDR <0.05; logFC >±1.25; or FDR <0.01; logFC >±1.5 respectively), whereas purple points represent nonsignificantly regulated genes. (F) Selected positive and negative GSEA enrichment scores of DEGs between NFIA-ETO2- or NFIA-ETO2ΔNHR4–expressing, FL-derived EBs grown 24 hours in DM (P < .05). (G) GSEA shows significant enrichment for previously proposed24 direct ETO2 targets among upregulated DEGs in NFIA-ETO2–expressing FL-derived EBs) compared with NFIA-ETO2ΔNHR4–expressing cells grown for 24 hours in DM (P < .05). (H) Volcano plot showing DEGs from the NFIA-ETO2 vs NFIA-ETO2ΔNHR4-expressing EBs grown in MM vs NFIA-ETO2 vs NFIA-ETO2ΔNHR4–expressing, FL-derived EBs grown in DM for 24 hours (FDR >0.05; logFC >±1.5 black lines). Orange and red points represent statistically significant dysregulated genes (FDR <0.05; logFC >±1.25; or FDR <0.01; logFC >±1.5 respectively), whereas purple points represent nonsignificantly regulated genes. (I) Selected GSEA correlations of DEG from NFIA-ETO2 vs NFIA-ETO2ΔNHR4–expressing, FL-derived EBs grown in MM vs NFIA-ETO2 vs NFIA-ETO2ΔNHR4–expressing EBs grown in DM for 24 hours.
In chimeric NFIA-ETO2, the NFIA-DNA–binding domain DNA is fused to ETO2 Nervy-homology (NHR) domains previously shown to mediate contacts with transcriptional coregulators.19,20 To explore their individual impact in the biological activity of the fusion, we cloned a series of C-terminal FLAG epitope-tagged NFIA-ETO2 deletion mutants. After exploring comparable expression of proteins of the calculated size in HeLa cells (not shown), we facilitated their expression in MEL cells and primary EBs. Despite expression levels similar to the full-length fusion (measured via quantitative reverse transcription PCR ; not shown), the mutants lacking the NFIA-DNA binding domain (DBD) or the ETO2-NHR2–4 domains were unable to enhance proliferation and impair erythroid differentiation, indicating that both the NFIA-DBD and the ETO2-NHR2–4 domains are essential for the fusion’s oncogenic activity (supplemental Figure 2B-D). Notably, induced differentiation of NFIA-ETO2ΔNHR4 mutant-expressing EBs was associated with a gene expression signature of normal terminal erythroid differentiation (supplemental Figure 2E-F; supplemental Table 4), indicating loss of activity compared with that of the full fusion protein.
Next, we compared the gene expression profiles of FL-derived primary murine EBs expressing either full-length or the inactive NFIA-ETO2ΔNHR4 mutant grown for 24 hours in DM. Performing PCA helped clearly separate the gene expression profiles of NFIA-ETO2 and NFIA-ETO2ΔNHR4 (PC1 = 54%) (Figure 2D). NFIA-ETO2 was associated with 2317 significantly up- and 1443 downregulated genes (FDR ≤0.05; logFC >1.5; Figure 2E; supplemental Table 5). Among the highest upregulated genes were several ETS family transcription factors, including Erg as well as the signaling effectors Stat5a/b , all previously shown to be involved in malignancies of the erythroid lineage.25-27 Several erythroid maturation-associated GATA1 targets, such as Alas2 or dematin actin-binding protein, a structural erythrocyte protein, were among the most significantly downregulated genes.28 Similar to what we observed in MEL cells, NFIA-ETO2–expressing primary EBs also expressed reduced levels of genes coding for transcription factors that functionally interact with GATA1 such as Tal1 or Klf1. GSEA revealed negative enrichment of gene signatures related to the development of the erythroid lineage and with GATA1 target genes. Positive enrichment was seen with NFIA target genes as well as with genes upregulated in Eto2−/− BM cells (Figure 2F; supplemental Table 6). Comparative GSEA results illustrate the significant upregulation of previously proposed direct ETO2 target genes in cells expressing NFIA-ETO2 compared with those expressing the inactive NFIA-ETO2ΔNHR4 mutant (Figure 2G; supplemental Table 7).24
To identify the genes affected by NFIA-ETO2 upon induction of erythroid differentiation, we compared differentially expressed genes (DEGs) in cells expressing NFIA-ETO2 or the inactive NFIA-ETO2ΔNHR4 mutant expanded in MM, with those grown in DM for 24 hours. We found 984 significantly up- and 436 downregulated genes (FDR >0.05; logFC >1.5) in NFIA-ETO2–expressing cells (Figure 2H; supplemental Table 8). Among the significantly upregulated genes was Myb, known to be repressed during normal erythroid differentiation.29 Moreover, GSEA results revealed positive enrichment to gene signatures upregulated in hematopoietic progenitor cells, STAT5 signaling mediators, and PRC2 targets. GSEA negatively-enriched gene signatures were previously reported to be downregulated in leukemic stem cells, involved in cell cycle regulation and oxygen transport (Figure 2I; supplemental Table 9). Comparing DEGs in NFIA-ETO2–expressing MEL and primary EBs revealed more than 4000 commonly dysregulated genes (FDR ≤0.05), including transcriptional regulators, such as Tal1, Klf1, or Gfi1b as well as markers of erythroid differentiaton, including Ache, Gypa, Gypc, Tmcc2, or Alas2 (supplemental Figure 2G; supplemental Table 10). Collectively, these data show that the NFIA-ETO2–mediated impaired erythroid differentiation is associated with aberrant expression of erythroid key regulators and known hematopoietic proto-oncogenes.
NFIA-ETO2 expression alters chromatin accessibility and aberrantly binds to gene loci encoding regulators of erythroid differentiation
To link our findings of NFIA-ETO2–induced impaired terminal differentiation, enhanced EB proliferation, and altered gene expression with specific chromatin states, we next studied differential chromatin accessibility by assay for transposase-accessible chromatin coupled with sequencing (ATAC-seq). In differentiation-induced, vector-transduced, FL-derived control EBs, HOMER-mediated motif analysis revealed enrichment in GATA and KLF family transcription factor binding motifs.30 In contrast, NFIA-ETO2–expressing FL-derived EBs showed a predominant enrichment in ETS-family transcription factors and RUNX motifs (Figure 3A; supplemental Table 11). Comparing the target sequences confirmed the enrichment for the ETS- and RUNX motifs in NFIA-ETO2–expressing cells, whereas GATA- and NFIA-motifs were more enriched in control cells (supplemental Figure 3A). Comparison of ATAC-seq results from NFIA-ETO2+ cells with data from ETO2-expressing primary EBs9 showed significant enrichment of accessibility for ETS-family transcription factors in NFIA-ETO2 EBs indicating that NFIA-ETO2 does not simply phenocopy of increased ETO2 activity (Figure 3B; supplemental Table 12). Collectively, these data suggest that blocked erythroid differentiation by NFIA-ETO2 is associated with dysbalanced activation of ETS/RUNX vs GATA/NFIA motif binding factors and that the NFIA-ETO2 effects on chromatin are different from ETO2 overexpression.
NFIA-ETO2 expression alters chromatin accessibility and binds to gene loci encoding regulators of erythroid differentiation. (A) Comparative motif enrichment in primary EBs expressing NFIA-ETO2 or CTRL of ATAC-seq peaks via HOMER. Motifs were ranked according to the variation of normalized P values calculated for peaks detected in the CTRL (negative values) and the NFIA-ETO2 expressing EBs (positive values). (B) Comparative motif enrichment in primary EBs expressing NFIA-ETO or ETO2 from ATAC-seq peaks via HOMER. Motifs were ranked based on the variation of normalized P values calculated for peaks detected in the ETO2- (negative values) and NFIA-ETO2–expressing primary EBs (positive values). Waterfall plot was made after substraction of the peaks detected in control conditions of both experiments.9 (C) Heatmaps representing ChIPseq read intensities of the NFIA-ETO2-HA (red), H3K4me1 (blue), H3K27ac (green), H3K27me3 (orange), and H3K9me1 (yellow). Each heatmap corresponds to the sum of reads mapped to the region focused on NFIA-ETO2-HA peak centers with ±5 kb. (D) Waterfall plot showing enriched motifs associated with NFIA-ETO2-HA peaks in MEL cells expressing NFIA-ETO2-HA built using HOMER. Motifs are ranked according to their P values. (E) Barplot representing DEGs (obtained in the RNA-seq analysis of NFIA-ETO2 and vector-transduced MEL cells) located in the vicinity of NFIA-ETO2-HA peaks. DEGs related to erythropoiesis were acquired through literature-based search (Erythroid DEGs). (F) Genome browser snapshots of ChIPseq tracks showing the binding of NFIA-ETO2-HA (red), H3K27ac (green), H3K4me1 (blue), H3K9me1 (yellow,) and H3K27me3 (orange) to the Gypc and Alas2 loci, and their RNA-seq tracks (dark green) showing their normalized expression in MEL cells. The known cis-regulatory element in the mm10 mouse genome from ENCODE data are indicated in the yellow track. (G) Bargraph representing NFIA-ETO2-HA peaks containing NFIA motif and/or overlaping with ETO2 peak, which were located in the vicinity of DEGs (obtained via the RNA-seq analysis of NFIA-ETO2 and vector-transduced MEL cells). HA, hemagglutinin.
NFIA-ETO2 expression alters chromatin accessibility and binds to gene loci encoding regulators of erythroid differentiation. (A) Comparative motif enrichment in primary EBs expressing NFIA-ETO2 or CTRL of ATAC-seq peaks via HOMER. Motifs were ranked according to the variation of normalized P values calculated for peaks detected in the CTRL (negative values) and the NFIA-ETO2 expressing EBs (positive values). (B) Comparative motif enrichment in primary EBs expressing NFIA-ETO or ETO2 from ATAC-seq peaks via HOMER. Motifs were ranked based on the variation of normalized P values calculated for peaks detected in the ETO2- (negative values) and NFIA-ETO2–expressing primary EBs (positive values). Waterfall plot was made after substraction of the peaks detected in control conditions of both experiments.9 (C) Heatmaps representing ChIPseq read intensities of the NFIA-ETO2-HA (red), H3K4me1 (blue), H3K27ac (green), H3K27me3 (orange), and H3K9me1 (yellow). Each heatmap corresponds to the sum of reads mapped to the region focused on NFIA-ETO2-HA peak centers with ±5 kb. (D) Waterfall plot showing enriched motifs associated with NFIA-ETO2-HA peaks in MEL cells expressing NFIA-ETO2-HA built using HOMER. Motifs are ranked according to their P values. (E) Barplot representing DEGs (obtained in the RNA-seq analysis of NFIA-ETO2 and vector-transduced MEL cells) located in the vicinity of NFIA-ETO2-HA peaks. DEGs related to erythropoiesis were acquired through literature-based search (Erythroid DEGs). (F) Genome browser snapshots of ChIPseq tracks showing the binding of NFIA-ETO2-HA (red), H3K27ac (green), H3K4me1 (blue), H3K9me1 (yellow,) and H3K27me3 (orange) to the Gypc and Alas2 loci, and their RNA-seq tracks (dark green) showing their normalized expression in MEL cells. The known cis-regulatory element in the mm10 mouse genome from ENCODE data are indicated in the yellow track. (G) Bargraph representing NFIA-ETO2-HA peaks containing NFIA motif and/or overlaping with ETO2 peak, which were located in the vicinity of DEGs (obtained via the RNA-seq analysis of NFIA-ETO2 and vector-transduced MEL cells). HA, hemagglutinin.
To identify putative direct target gene loci, we sequenced NFIA-ETO2–associated immunoprecipitated chromatin (ChIPseq). Based on the robustness of MEL cells as model for erythroid differentiation, we studied NFIA-ETO2 chromatin binding after 48 hours of DMSO-induced maturation. Because the precipitation of chromatin-associated FLAG-tagged NFIA-ETO2 yielded poor and mostly nonspecific enrichment, we subsequently generated an NFIA-ETO2-3XHA–tagged fusion that similarly impaired DMSO-induced erythroid differentiation (supplemental Figure 3B). ChIPseq revealed 1350 putative NFIA-ETO2 binding sites mostly located in the distal intergenic region (47%) and/or exonic regions (28%) and predominantly located in active chromatin regions, as visualized based on significantly higher overlap with H3K27ac and H3K4me1 than with repressive H3K27me3 and H3K9me1 histone marks (Figure 3C; supplemental Figure 3C; supplemental Table 13). Analysis of the NFIA-ETO2 binding regions showed enrichment for motifs of several known erythroid regulators, such as GATA1, SCL/TAL1, and GFI1B but also of NF1 family transcription factors (Figure 3D; supplemental Figure 3D; supplemental Table 14).30,31 Integration of ChIPseq and RNA-seq showed that 62% of DEGs bound by NFIA-ETO2 were downregulated (Figure 3E), indicating a repressive effect of NFIA-ETO2 on transcription (χ2 test; P = 3.664e-06). Importantly, the vast majority of NFIA-ETO2 gene targets associated with erythroid lineage commitment were downregulated, including key transcriptional regulators such as Tal1, and terminal erythroid differentiation markers, including Gypc or Alas2, suggesting that the fusion operates in transcriptionally active regions where it locally fine-tunes transcription (Figure 3F). In contrast, NFIA-ETO2 was bound only to a relatively low number (38%) of upregulated genes, which included the proto-oncogenes Spi1 and Lmo2 (supplemental Figure 3E; supplemental Table 13). To examine potential binding partners of NFIA-ETO2 at up- and downregulated genes, we analyzed TF binding sites in the vicinity of NFIA-ETO2–bound loci via HOMER motif analysis. Interestingly, we found RUNX motifs significantly enriched at upregulated genes, whereas GATA and NF1 motifs were enriched at downregulated genes, in line with the findings obtained via ATAC-seq (supplemental Table 15). A comparison of the ChIPseq and RNA-seq signatures obtained in MEL cells with the expression signatures from NFIA-ETO2–expressing primary EBs revealed that a large fraction of potentially NFIA-ETO2–bound gene loci in MEL cells were repressed (χ2 test; P- = 1.8e-04) in fusion-expressing primary EBs, including multiple erythroid differentiation factors (supplemental Figure 3F-G). We also investigated the relationship between NFIA-ETO2–bound DEGs and NF1 DNA motifs to which the NF1 family of transcription factors bind. We detected NF1 motifs in >30% of NFIA-ETO2 targeted DEGs and in ≈50% DEGs related to erythroid differentiation (literature search–based; supplemental Table 16), which were predominantly downregulated (Figure 3G). Finally, we compared NFIA-ETO2–bound DEGs with previously reported ETO2-bound genes in MEL cells and found that >40% of fusion-bound DEGs were associated with ETO2, of which the majority were downregulated (supplemental Figure 3H).32 Taken together, these observations indicate that NFIA-ETO2 binds to mostly active chromatin and represses several key regulators of terminal erythroid differentiation that contain NF-binding sites and/or are decorated with ETO2.
Functional cooperation of NFIA-ETO2 with TP53R248Q in vitro
Mutations in the TP53 DNA–binding domain is a molecular hallmark of AEL and, in particular, of PEL.10,11 Based on the reported unique PEL phenotypes in patients whose tumor cells carry the NFIA-ETO2 fusion, we hypothesized that the fusion functionally cooperates with altered TP53 activity. To test this, we took advantage of a mouse strain in which mouse Tp53 exons 4 to 9 are replaced with the human TP53 sequence containing the hotspot R248Q missense mutation.33
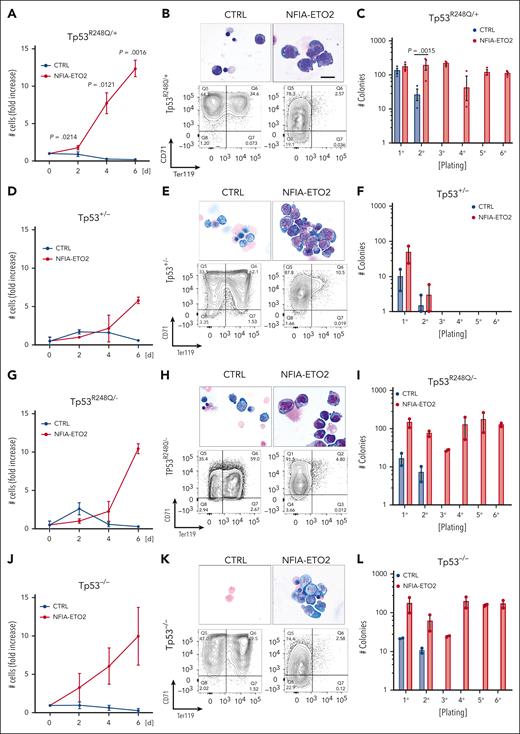
Similarly to NFIA-ETO2 expression in wild type (WT) cells, its expression in heterozygous TP53R248Q/+ primary mouse EBs impaired their terminal differentiation as shown by increased proliferation, immature cellular morphology and reduced TER119 expression while maintaining Kit and CD71 expression (Figure 4A-B; supplemental Figure 4A-B). Importantly, however, in contrast to its expression in WT cells, we observed a significant increase in clonogenic activity of NFIA-ETO2–expressing TP53R248Q/+ cells in serial replating assay MCs (Figure 4C; supplemental Figure 4C). Notably, although the effect on proliferation and differentiation was similar, we did not observe any impact on colony formation upon NFIA-ETO2 expression in Tp53+/− EBs (Figure 4D-F). In contrast, NFIA-ETO2 expression in TP53R248Q/− or Tp53−/− EBs again enabled propagation with the sixth passage (Figure 4G-L).
Functional cooperation of NFIA-ETO2 with TP53R248Q in vitro. (A,D,G,J) Growth curves of TP53R248Q/+ (A), Tp53+/− (D), TP53R248Q+/− (G), and Tp53−/− (J) BM-derived EBs expressing NFIA-ETO2 (red line) compared with CTRL cells (blue line) over 6 days in DM (n = 2-3). (B,E,H,K) Representative images of Wright-Giemsa-stained cytospin preparations (top) and flow cytometry panels showing CD71/Ter119 expression (bottom) of TP53R248Q/+ (B), Tp53+/− (E), TP53R248Q/− (H), and Tp53−/− (K) BM-derived EBs expressing NFIA-ETO2 compared with CTRL grown in MM (left) and in DM (right). Data represent 1 out of 2 to 3 independent experiments. Images were recorded with a 60× objective using a Nikon-TI. Scalebars, 50 μm. (C,F,I,L) Colony formation in MC by TP53R248Q/+ (C), Tp53+/− (F), TP53R248Q/− (I), and Tp53−/− (L) BM-derived EBs expressing NFIA-ETO2 compared with CTRL. Shown are absolute numbers of colonies formed in 6 consecutive platings (n = 2-3). Note that control cells cannot self-renew after 2 rounds of plating. Values are presented as individual points, bar graphs represent the mean value of independent biological replicates (n); and error bars are standard error of the mean. Statistical significance was tested with paired two-tailed t tests.
Functional cooperation of NFIA-ETO2 with TP53R248Q in vitro. (A,D,G,J) Growth curves of TP53R248Q/+ (A), Tp53+/− (D), TP53R248Q+/− (G), and Tp53−/− (J) BM-derived EBs expressing NFIA-ETO2 (red line) compared with CTRL cells (blue line) over 6 days in DM (n = 2-3). (B,E,H,K) Representative images of Wright-Giemsa-stained cytospin preparations (top) and flow cytometry panels showing CD71/Ter119 expression (bottom) of TP53R248Q/+ (B), Tp53+/− (E), TP53R248Q/− (H), and Tp53−/− (K) BM-derived EBs expressing NFIA-ETO2 compared with CTRL grown in MM (left) and in DM (right). Data represent 1 out of 2 to 3 independent experiments. Images were recorded with a 60× objective using a Nikon-TI. Scalebars, 50 μm. (C,F,I,L) Colony formation in MC by TP53R248Q/+ (C), Tp53+/− (F), TP53R248Q/− (I), and Tp53−/− (L) BM-derived EBs expressing NFIA-ETO2 compared with CTRL. Shown are absolute numbers of colonies formed in 6 consecutive platings (n = 2-3). Note that control cells cannot self-renew after 2 rounds of plating. Values are presented as individual points, bar graphs represent the mean value of independent biological replicates (n); and error bars are standard error of the mean. Statistical significance was tested with paired two-tailed t tests.
Independent of the TP53 genotype, serially propagated NFIA-ETO2–expressing cells maintained erythroblastic characteristics, as shown with CD71 and Kit expression (data not shown). Notably, the TP53 genotype did not affect in vitro proliferation or induced terminal differentiation of murine EBs, as tracked by Kit, CD71, and Ter119 surface marker expression (supplemental Figure 4D-I). Collectively, these data show that in the presence of a mutant TP53R248Q allele or in the absence of 2 WT Tp53 alleles, NFIA-ETO2 expression results in aberrant serial replating activity of primary mouse EBs.
NFIA-ETO2–expressing TP53R248Q/+ hematopoietic cells induce erythroleukemia in mice
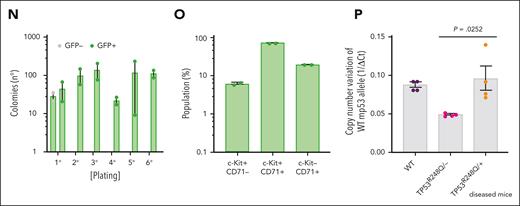
To address the impact of TP53 alterations on the oncogenic potential of the NFIA-ETO2 fusion in vivo, we transplanted FL-derived TP53R248Q/+ or WT (C57/B6) murine EBs retrovirally expressing NFIA-ETO2 into lethally-irradiated WT recipients. Importantly, all mice that had received NFIA-ETO2–transduced TP53R248Q/+ EBs developed signs of leukemia after a median latency of 3 months (2-6.5 months, n = 8), whereas mice receiving vector-transduced cells remained healthy (Figure 5A). Symptomatic mice presented with hepatosplenomegaly, erythroid progenitors in peripheral blood smears, and significant infiltration in the BM, liver, and spleen (Figure 5B-G), which was associated with elevated white blood cell paired with reduced red blood cell and platelet counts (Figure 5H-J). The BM of symptomatic mice was infiltrated to a variable degree by NFIA-ETO2 (GFP+)–expressing cells (mean = 45.4%; range, 8.3% to 82.5%; n = 8) and contained a significantly higher number of Kit+/CD71+ and CD71+/Ter119+ cells than mice that received vector-transduced cells (P = .0018; and P = .0002, respectively) (Figure 5K-M). Enriched (GFP+) NFIA-ETO2–expressing BM-derived EBs from mice that were affected with the disease formed dense and round colonies in MC, composed of Kit+ and CD71+ cells that could be serially propagated (Figure 5N-O; supplemental Figure 5A). Transplantation of 2 × 106 total BM cells from symptomatic secondary mice into lethally-irradiated syngeneic secondary recipients propagated the disease after a short median latency of 25 days (20-29 days; n = 4; Figure 5A). Similar to the primary recipients, symptomatic mice presented with anemia, thrombocytopenia, hepatosplenomegaly, and extensive organ infiltration (supplemental Figure 5B-H). Notably, although a loss of the remaining WT allele was previously proposed to stabilize TP53 mutants,34 quantitative PCR analysis suggested that the NFIA-ETO2–expressing TP53R248Q/+ tumor cells of symptomatic mice did not lose the WT Tp53 allele (Figure 5P). Taken together, these data indicate functional collaboration between the NFIA-ETO2 fusion and the TP53R248Q/+ allele in a murine model of PEL.
NFIA-ETO2–expressing TP53R248Q/+ hematopoietic cells induce erythroleukemia in mice. (A) Kaplan Meier plot of disease-free mice recipients of NFIA-ETO2–expressing (red line = primary recipients, n = 9; purple line = secondary recipients, n = 8) or CTRL TP53R248Q/+ cells (blue dashed line n = 4). (B) Spleens of a control (left) and a symptomatic mouse (right) that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells (right). (C) Wright-Giemsa–stained peripheral blood smear from a diseased mouse showing 2 EBs (original magnification ×100; scale bar, 10 μm). (D-F) Images of H&E-stained sections of BM (D), spleen (E), and liver (F) of diseased mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ EBs (original magnification ×40; scale bar, 30 μm). (G) Spleen and liver weight of symptomatic mice that received transplantation with NFIA-ETO2–expressing (red dots; n = 8) or CTRL cells (blue dots; n = 4). (H-J) Peripheral WBC (×109/L) (H), RBC counts (×1012/L) (I), and PLT counts (×109/L) (J) in symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ (red dots, n = 8) compared with mice that received transplantation with CTRL cells (blue dots; n = 4). (K) Fraction of transduced GFP+ cells (%) before transplantation (T) and at termination of the experiment in mice that received transplantation with NFIA-ETO2- (red dots; n = 8) or CTRL (blue dots; n = 4) EBs. (L) Kit and CD71 surface expression in total BM cells (in percentage) from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells (n = 8) or vector-transduced control (CTRL) cells (n = 4). (M) CD71 and Ter119 surface expression in total BM cells (in %) from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells (n = 8) or CTRL cells (n = 4). (N) Colony formation by GFP+ and GFP− EBs harvested from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells. Shown are absolute numbers of colonies from first to the sixth consecutive plating in MC. (O) Kit and CD71 surface expression in cells (in %) from the second MC plating of GFP+ cells from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells. (P) Quantification of Tp53 alleles in BM cells from WT (purple dots), TP53R248Q/− (pink dots), and from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells (orange dots) assessed via PCR analysis. Values are presented as individual points; bar graphs represent the mean value of independent biological replicates (n); and error bars are standard error of the mean. Statistical significance was tested with unpaired two-tailed t tests. H&E, hematoxylin and eosin.
NFIA-ETO2–expressing TP53R248Q/+ hematopoietic cells induce erythroleukemia in mice. (A) Kaplan Meier plot of disease-free mice recipients of NFIA-ETO2–expressing (red line = primary recipients, n = 9; purple line = secondary recipients, n = 8) or CTRL TP53R248Q/+ cells (blue dashed line n = 4). (B) Spleens of a control (left) and a symptomatic mouse (right) that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells (right). (C) Wright-Giemsa–stained peripheral blood smear from a diseased mouse showing 2 EBs (original magnification ×100; scale bar, 10 μm). (D-F) Images of H&E-stained sections of BM (D), spleen (E), and liver (F) of diseased mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ EBs (original magnification ×40; scale bar, 30 μm). (G) Spleen and liver weight of symptomatic mice that received transplantation with NFIA-ETO2–expressing (red dots; n = 8) or CTRL cells (blue dots; n = 4). (H-J) Peripheral WBC (×109/L) (H), RBC counts (×1012/L) (I), and PLT counts (×109/L) (J) in symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ (red dots, n = 8) compared with mice that received transplantation with CTRL cells (blue dots; n = 4). (K) Fraction of transduced GFP+ cells (%) before transplantation (T) and at termination of the experiment in mice that received transplantation with NFIA-ETO2- (red dots; n = 8) or CTRL (blue dots; n = 4) EBs. (L) Kit and CD71 surface expression in total BM cells (in percentage) from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells (n = 8) or vector-transduced control (CTRL) cells (n = 4). (M) CD71 and Ter119 surface expression in total BM cells (in %) from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells (n = 8) or CTRL cells (n = 4). (N) Colony formation by GFP+ and GFP− EBs harvested from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells. Shown are absolute numbers of colonies from first to the sixth consecutive plating in MC. (O) Kit and CD71 surface expression in cells (in %) from the second MC plating of GFP+ cells from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells. (P) Quantification of Tp53 alleles in BM cells from WT (purple dots), TP53R248Q/− (pink dots), and from symptomatic mice that received transplantation with NFIA-ETO2–expressing TP53R248Q/+ cells (orange dots) assessed via PCR analysis. Values are presented as individual points; bar graphs represent the mean value of independent biological replicates (n); and error bars are standard error of the mean. Statistical significance was tested with unpaired two-tailed t tests. H&E, hematoxylin and eosin.
NFIA-ETO2 expression in TP53R248Q/+ EBs alters TP53 target gene expression
To better understand this functional cooperation, we compared the gene expression signatures of WT and TP53R248Q/+ NFIA-ETO2–expressing EBs expanded in MM and after 24 hours, induced differentiation in DM. PCA results revealed a consistent shift on the second PC (22.3% variation), induced by the presence of TP53R248Q, even if the majority of the variance was explained by differences between replicates and growth conditions (MM and DM) (Figure 6A). We found only 13 significantly up- and 8 significantly (FDR ≤0.05) downregulated genes in MM-growing NFIA-ETO2–expressing cells with or without TP53R248Q (Figure 6B; supplemental Table 17). Under DM conditions, the presence of TP53R248Q was associated with significant upregulation of 7 and downregulation of 5 genes (Figure 6C; supplemental Table 18). The most significantly downregulated gene in both conditions was Eda2r, encoding for the ectodermal dysplasia receptor that belongs to the TP53-controlled tumor necrosis factor receptor family.35 Additional previously annotated TP53 targets include the pleckstrin homology-like domain family A member 3, polo-like kinase 2, anoctamin 3, and the zinc finger protein 365.36-38 GSEA revealed positive enrichment of gene signatures up regulated in hematologic and other malignant disorders (module 46), to signatures downregulated in leukemic stem cells, involved in heme metabolism and other metabolic pathways. Conversely, a negative enrichment was found for chromatin silencing and epigenetic processes that impair gene expression (Figure 6D; supplemental Table 19). Comparison of chromatin accessibility by ATAC-seq revealed a very similar overall pattern between WT and TP53R248Q/+ EBs expressing NFIA-ETO2 cultured 24 hours in DM, without any statistically significant difference in global motif enrichment (Figure 6E). Projection of gene expression signatures into a PCA-reduced transcriptional space of normal hematopoietic differentiation9 indicates varying patterns of a differentiation block for all NFIA-ETO2 samples remaining in proximity to progenitor states, regardless of the TP53 status or the medium in which the cells were grown (supplemental Figure 6A).
Downregulation of known TP53 target genes in NFIA-ETO2–expressing TP53R248Q/+ EBs. (A) PCA of gene expression signatures of WT or TP53R248Q/+ EBs expressing NFIA-ETO2 (WT, red; and TP53R248Q/+, blue) grown in MM (dot) or 24 hours in DM (triangle). Each point represents one sample, colored and shaped based on its TP53 status (WT or TP53R248Q/+) and the medium in which the cells were cultured (MM vs DM), respectively. (B) Volcano plot showing DEGs in NFIA-ETO2–expressing WT- vs TP53R248Q/+ EBs grown in MM. Dysregulated genes are colored in red (FDR <0.05). (C) Volcano plot showing DEGs in NFIA-ETO2–expressing WT- vs TP53R248Q/+ EBs grown in DM for 24h. Dysregulated genes are colored in red (FDR <0.05). (D) Selected positive and negative GSEA enrichment scores of DEGs obtained in NFIA-ETO2–expressing TP53R248Q/+ EBs when compared with NFIA-ETO2–expressing WT EBs cultured in DM (p-adj <0.1). (E) Profile plots representing ATAC-seq signals from WT and TP53R248Q/+ NFIA-ETO2–expressing EBs. Profile-plots were focused on peaks centers with ±5 kb. The heatmap represents the hierarchical clustering of ATAC-seq signals, performed with WT and TP53R248Q/+ NFIA-ETO2–expressing EBs. Heatmaps were also focused on peak centers with ±5 kb. (F) BART-predicted transcription factors that most likely suppress the downregulated genes in TP53R248Q/+ NFIA-ETO2–expressing EBs. (G) Selected positive and negative GSEA enrichment scores of DEGs of patients with AEL with TP53 mutations vs patients with AEL without TP53 mutations (padj <0.05). Data from Iacobucci et al.8 (H) Mean expression (count per million) of DEG-up and DEG-down in patients with AEL with (red dots) or without (green dots) TP53 mutations. DEG-up and DEG-down are the gene signatures comprising significantly upregulated (left) and downregulated (right) found to be differentially expressed in NFIA-ETO2–expressing TP53R248Q/+ EBs. Shown are AEL RNA-seq samples from Iacobucci et al.8 (I) Relative mean expression of DEG-up and DEG-down in patients with AML with (red dots) or without (green dots) TP53 mutations. DEG-up and DEG-down are the gene signatures comprising significantly upregulated (left) and downregulated (right) found to be differentially expressed in NFIA-ETO2–expressing TP53R248Q/+ EBs. Shown are AML microarray samples from the TCGA project. BART, binding analysis for regulation of transcription,
Downregulation of known TP53 target genes in NFIA-ETO2–expressing TP53R248Q/+ EBs. (A) PCA of gene expression signatures of WT or TP53R248Q/+ EBs expressing NFIA-ETO2 (WT, red; and TP53R248Q/+, blue) grown in MM (dot) or 24 hours in DM (triangle). Each point represents one sample, colored and shaped based on its TP53 status (WT or TP53R248Q/+) and the medium in which the cells were cultured (MM vs DM), respectively. (B) Volcano plot showing DEGs in NFIA-ETO2–expressing WT- vs TP53R248Q/+ EBs grown in MM. Dysregulated genes are colored in red (FDR <0.05). (C) Volcano plot showing DEGs in NFIA-ETO2–expressing WT- vs TP53R248Q/+ EBs grown in DM for 24h. Dysregulated genes are colored in red (FDR <0.05). (D) Selected positive and negative GSEA enrichment scores of DEGs obtained in NFIA-ETO2–expressing TP53R248Q/+ EBs when compared with NFIA-ETO2–expressing WT EBs cultured in DM (p-adj <0.1). (E) Profile plots representing ATAC-seq signals from WT and TP53R248Q/+ NFIA-ETO2–expressing EBs. Profile-plots were focused on peaks centers with ±5 kb. The heatmap represents the hierarchical clustering of ATAC-seq signals, performed with WT and TP53R248Q/+ NFIA-ETO2–expressing EBs. Heatmaps were also focused on peak centers with ±5 kb. (F) BART-predicted transcription factors that most likely suppress the downregulated genes in TP53R248Q/+ NFIA-ETO2–expressing EBs. (G) Selected positive and negative GSEA enrichment scores of DEGs of patients with AEL with TP53 mutations vs patients with AEL without TP53 mutations (padj <0.05). Data from Iacobucci et al.8 (H) Mean expression (count per million) of DEG-up and DEG-down in patients with AEL with (red dots) or without (green dots) TP53 mutations. DEG-up and DEG-down are the gene signatures comprising significantly upregulated (left) and downregulated (right) found to be differentially expressed in NFIA-ETO2–expressing TP53R248Q/+ EBs. Shown are AEL RNA-seq samples from Iacobucci et al.8 (I) Relative mean expression of DEG-up and DEG-down in patients with AML with (red dots) or without (green dots) TP53 mutations. DEG-up and DEG-down are the gene signatures comprising significantly upregulated (left) and downregulated (right) found to be differentially expressed in NFIA-ETO2–expressing TP53R248Q/+ EBs. Shown are AML microarray samples from the TCGA project. BART, binding analysis for regulation of transcription,
To better understand the differential regulation of this small number of TP53 targets, we applied binding analysis for regulation of transcription to obtain predictive information about potential upstream regulators.39 Binding analysis for regulation of transcription combines ATAC-seq binding information with model-based analysis of regulation of gene expression in MARGE-predicted genomic cisregulatory regions. Interestingly, in addition to TP53, several polycomb repressive complex 2 (PRC2) proteins, such as JARID2, EZH2, and SUZ12 were predicted to regulate the downregulated genes in NFIA-ETO2–expressing TP53R248Q/+ cells (Figure 6F). Negative enrichment of PRC2 targets was also observed in GSEA-based comparisons of the transcriptomes of a cohort of primary human AEL samples with or without TP53 mutations (Figure 6G; supplemental Table 20).9 Collectively, these observations suggest that TP53R248Q cooperates with NFIA-ETO2 primarily by altering the expression of tumor suppressive TP53 target genes, which eventually augmented PRC2 activity, extending previous findings and suggesting the regulation of self-renewal through interaction of mutant TP53 with EZH2.40
Because TP53 DNA binding mutations are highly prevalent among patients with AEL, we also compared the DEGs (DEG-up and DEG-down) from NFIA-ETO2–expressing TP53R248Q mouse EBs with expression profiles from primary human pediatric erythroleukemia samples.8 Assuringly, there was an excellent mouse/human data overlap. Interestingly, DEG-up genes were significantly enriched (P = 6.2e-06), and DEG-down genes decreased (P = .055; not significant at alfa 0.05) in patients with AEL carrying TP53 mutations (Figure 6H). However, the genes did follow the expression pattern (DEG-up was downregulated, P = .0052; and DEG-down was downregulated, P = 3.7e-05) in adult AML subtypes from TCGA carrying TP53 mutations (Figure 6I),41,42 suggesting that these TP53-related expression profiles are dependent on the erythroid disease phenotype and/or age of the affected patients.
Discussion
The exclusive association of NFIA-ETO2 with PEL suggests a critical role in the erythroid disease phenotype.15-18 Our molecular analytic results indicate that the NFIA-ETO2 fusion binds to and controls some key regulators of erythroid differentiation leading to dysbalanced transcriptional activity favoring ETS-motif factors while repressing GATA and KLF motif factors associated with downregulation of genes of erythroid maturation and upregulation of several proto-oncogenes. Notably, previous studies indicated that the level of ETO2 coordinates differentiation and proliferation of erythroid cells via the coregulation of TAL1/GATA1 complexes and EPO-dependent signaling.43,44
NFIA-ETO2 contains the N-terminal DBD of NFIA fused to almost the entire ETO2 ORF containing 4 NHR domains known to affect transcription and providing the interface for multiple protein-protein interactions.16,20 We found that the NFIA-DBD and the ETO2-NHR domains are necessary for NFIA-ETO2 to interfere with terminal differentiation of primary EBs, suggesting that the fusion controls gene expression by interaction of its NFIA moiety with DNA and/or by interaction of its NHR motifs with other NHRmotif–containing proteins. The observation that >40% of potential NFIA-ETO2 binding sites overlap with previously reported ETO2-bound loci suggest that heterodimerization of NFIA-ETO2 with endogenous ETO2 could be involved.32,44 Although not statistically significant, it is also likely that NFIA-ETO2 finds some of its targets by binding to NFI-motifs located within or very close to differentially expressed gene loci. In the future, it will be interesting to validate these findings in a more physiological setting, such as with primary NFIA-ETO2+ PEL cells.
In stark contrast to other AML forms, alterations of TP53 are the most prevalent lesions in AEL, reported to be present in almost all PEL cases. However, their functional role remains poorly understood.4-11 Recent work in human cells suggested that the activation of TP53 is not only a mediator of EBs cell cycle arrest and apoptosis upon impaired ribosome biogenesis in diseases such as Diamond-Blackfan anemia but also plays a critical role for normal erythroid differentiation.45 Similar to other human cancers, TP53 DBD hotspot mutations are the most prevalent in AEL and PEL.4-11 Whether DBD mutations such as TP53R175H or TP53R248Q act as gain of function or in a dominant-negative manner remains a matter of ongoing debates, but the answer is most likely context-dependent. The murine homolog to TP53R175H has recently been shown to act as a gain-of-function aberration, initiating and promoting myeloid complex karyotype AML.46 Engineering of TP53 mutations, including TP53R248Q in human AML cell lines, suggested a dominant-negative effect driving clonal selection.47 We observed that the TP53 genotype did not affect in vitro proliferation and differentiation of murine EBs. Importantly, however, the expression of NFIA-ETO2 impaired erythroid differentiation independently of the TP53 genotype. In contrast, as in WT cells, NFIA-ETO2 expression in Tp53+/− EBs did not elicit aberrant serial propagation, supporting a dominant-negative activity of the TP53R248Q mutant allele in this particular setting.
Earlier studies showed that a mutated TP53 provided self-renewal advantage to murine HSPC when transplanted in presence or absence of genotoxic stress.48 More recent work suggested that upon oncogenic stress, for example, by a constitutively active JAKV617F mutant, reduced TP53 levels are functionally cooperating events toward secondary AML. Notably, in contrast to NFIA-ETO2–expressing Tp53R248Q/+ EBs, alteration of both Tp53 alleles appears to be necessary for transformation of JAK2V617F+ myeoproliferative neoplasms toward erythroid secondary AML.49,50
Similar to NFIA-ETO2, the presence of TP53R248Q was also permissive to induce an erythroleukemic phenotype because of the overexpression of the ETS-transcription factor ERG.9 Likewise, transplantation of TP53R172H BM cells, which were virally expressing an AEL-associated TRKAH498R mutation, also resulted in an erythroleukemia-like disease in mice.8 These studies collectively suggest that functional collaboration by TP53 mutations (or aberrant TP53 pathway regulation) is critical for the development of PEL. Although we did not obtain any information on the TP53 genotype of the patients with NFIA-ETO2+ whose data were published, TP53 mutations are generally rare in pediatric AML.8,51 The lack of respective material prevented us to investigate whether pediatric PEL cells with NFIA-ETO2/ETO fusion also carry any TP53 alterations. Nevertheless, we obtained information about a 6-week-old patient with PEL in whom the tumor cells carried an NFIA-ETO fusion, a somatic KITp.D816Y but also a MUTYHc.91delG,p.A31fs variant at 50% variant allele fraction in both the germ line and the tumor DNA, whereas no TP53 variants were detected. The MutY DNA glycosylase seems also to control TP53-mediated tumor suppression, suggesting that a germ line MUTYH mutation may cooperate with a NFIA-ETO/ETO2 fusion, though impairing TP53 function.52 Further studies will be necessary to explore the role of any germ line mutations that affect TP53 function in PEL.
Acknowledgments
The authors thank R. Santoro and L. Lerra (Zürich, Switzerland) and R. Slany (Erlangen, Germany) for their precious input into this work. The authors also appreciate the service of the flow- and animal experimentation facilities of the Department of Biomedicine. The authors also thank the Bioinformatics Core Facility and, particularly, Florian Geier for his help in the use of their bioinformatic analysis pipeline.
This work was supported by Swiss Cancer Research (KFS-4258-08-2017, the San Salvatore Foundation, Lugano (201525), and the Wilhelm-Sander Foundation, München (2017-035.1) (J. Schwaller). I.N. was supported by the Alexander S. Onassis Public Benefit Foundation.
Authorship
Contribution: M.-R.P.-B., and Z.J. designed and performed experiments, analyzed data, and wrote the manuscript; J. Seguin, F.O.B., R.I., and A.F. analyzed data; M.F.B., R.S., I.N., A.L., S.J., and O.C. performed experiments; A.T., U.M.M., and R.S.-H. provided critical materials; T.M. designed experiments, analyzed data, and wrote the manuscript; and J. Schwaller designed and performed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Juerg Schwaller, Department of Biomedicine, University Children’s Hospital Basel, University of Basel, ZLF, Lab 202, Hebelstr 20, Basel CH-4031, Switzerland; e-mail: j.schwaller@unibas.ch.
References
Author notes
∗M.-R.P.-B. and Z.J. are joint first authors.
†F.O.B. and J. Seguin contributed equally to this study.
Sequencing data sets for the MEL cells and primary EBs reported in this article have been deposited in the Gene Expression Omnibus database (different SubSeries grouped in the SuperSeries; accession number GSE205437).
Data are available on request from the corresponding author, Juerg Schwaller (j.schwaller@unibas.ch).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement”in accordance with 18 USC section 1734.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal