Key Points
Data provide an association between HDM treatment in myeloma and mutational load increases at relapse.
Increased mutational load also increases clonal complexity but does not affect the outcome later.
Abstract
High-dose melphalan (HDM) improves progression-free survival in multiple myeloma (MM), yet melphalan is a DNA-damaging alkylating agent; therefore, we assessed its mutational effect on surviving myeloma cells by analyzing paired MM samples collected at diagnosis and relapse in the IFM 2009 study. We performed deep whole-genome sequencing on samples from 68 patients, 43 of whom were treated with RVD (lenalidomide, bortezomib, and dexamethasone) and 25 with RVD + HDM. Although the number of mutations was similar at diagnosis in both groups (7137 vs 7230; P = .67), the HDM group had significantly more mutations at relapse (9242 vs 13 383, P = .005). No change in the frequency of copy number alterations or structural variants was observed. The newly acquired mutations were typically associated with DNA damage and double-stranded breaks and were predominantly on the transcribed strand. A machine learning model, using this unique pattern, predicted patients who would receive HDM with high sensitivity, specificity, and positive prediction value. Clonal evolution analysis showed that all patients treated with HDM had clonal selection, whereas a static progression was observed with RVD. A significantly higher percentage of mutations were subclonal in the HDM cohort. Intriguingly, patients treated with HDM who achieved complete remission (CR) had significantly more mutations at relapse yet had similar survival rates as those treated with RVD who achieved CR. This similarity could have been due to HDM relapse samples having significantly more neoantigens. Overall, our study identifies increased genomic changes associated with HDM and provides rationale to further understand clonal complexity.
Introduction
The beneficial role of high-dose melphalan (HDM) with autologous stem cell transplant (ASCT) in newly diagnosed patients with multiple myeloma (MM) has been demonstrated in a number of studies. However, with the adaptation of newer therapeutic options, higher response rates are associated with significantly increased progression-free survival (PFS) and overall survival.1-5 Three-drug regimens containing a combination of lenalidomide, bortezomib, and dexamethasone (RVD) provide nearly a 100% response rate and significantly longer PFS. The role of HDM after such an effective induction regimen had been unclear, which had provided the rationale for our IFM/DFCI 2009 study. The clinical results of this aforementioned study demonstrated a significantly higher complete remission (CR) rate, minimal residual disease negative rate (29.5% vs 20%; P < .001), and longer median PFS for patients undergoing ASC transplantation than for the those who received RVD alone (50 vs 36 months; adjusted hazard ratio, 0.65; P < .001).6 Despite these clinical successes, the majority of patients relapse and receive novel single agent and combination salvage therapies with variable efficacies.
HDM followed by ASCT has been used for patients with MM as a standard of care,7-10 however, the impact of HDM on surviving MM cells is not well characterized. Melphalan, an alkylating agent, inhibits DNA and RNA synthesis by chemically altering the guanine bases in DNA.11 The melphalan-introduced base adducts can compromise genome integrity by blocking essential biological processes and inducing DNA breaks and subsequent apoptotic cell death.12,13 However, myeloma cells that survive the alkylating agent–induced DNA damage may undergo activation of alternative resistance mechanisms or induction of DNA repair via mechanisms such as base excision repair, mismatch repair, or homologous recombination (HR).13 This unintentional repair of chemically induced mutations can eventually affect the mutational load of tumor cells.
The impact of various alkylating agents on normal cells as well as primary tumors has been previously evaluated in vitro. Among 113 treatment conditions tested for an impact on human induced pluripotent stem cells,14 approximately half of them induced additional and/or double substitutions and/or insertions and deletions (indels), and various treatments produced unique mutational patterns clustered in distinct groups.14 Similarly, in a Caenorhabditis elegans model, alkylating agents and cisplatin induced DNA damage and repair-related mutational patterns similar to those observed in human cancer genomes.15 Therapy-related neoplasms predominantly due to DNA damage induced by cytotoxic therapy, such as melphalan have been reported in a number of malignancies.16,17 In MM, systemic seeding of residual surviving MM cells or subclones with accelerated growth at relapse, after HDM and ASCT,18,19 associated with a unique mutational pattern have been reported.18,20 Here, we investigated these possible DNA-damaging effects of alkylating agent on MM cells especially following HDM in a larger cohort of patients who were uniformly treated.21 The phase 3 IFM 2009 study in which newly diagnosed patients were randomized to either RVD treatment alone or RVD + HDM/ASCT treatments provided an ideal setting for such an investigation.
Methods
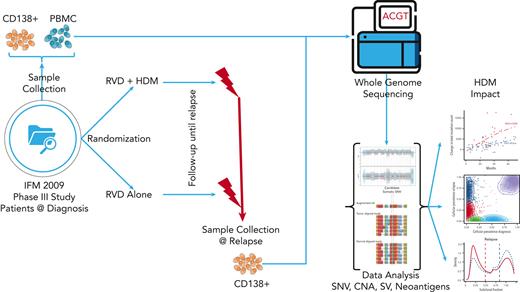
Patient samples
All patient samples were collected from the IFM/DFCI 2009 clinical trial, a phase 3, multicenter, randomized, open-label study designed to evaluate the clinical benefit from the drug combination RVD followed by lenalidomide maintenance (arm A) vs RVD + HDM and ASCT followed by lenalidomide maintenance (arm B). After written informed consent was obtained from each patient, bone marrow samples were collected at the time of diagnosis and at the first relapse after the first line of therapy. All clinical and genomic data were deidentified, in accordance with the Declaration of Helsinki. This study was approved by the Toulouse ethics committee. All bone marrow samples were purified using CD138+ magnetic beads followed by DNA extraction. Purity of the CD138+ fraction was assessed via anti-CD138 immunocytochemistry after sorting it, and only samples that passed quality control were sequenced. Peripheral blood mononuclear cells from the same individuals collected at diagnosis were used as constitutional DNA.
WGS and processing
All tumor and constitutional DNA samples were profiled using whole-genome sequencing (WGS). In brief, short-insert genomic libraries were constructed using a minimum of 200 ng of DNA from each sample. Flow cells were prepared, and sequencing clusters were generated in accordance with Illumina protocols. All WGS libraries were sequenced with 150 bp paired-end sequencing with an average sequence coverage of 81× for tumor samples (supplemental Table 1, available on the Blood website). Paired-end reads from WGS were aligned to GRCh38 using BWA-mem. SAMtools22 were used for file conversions. Duplicated reads were marked, and base quality score recalibration was performed using MarkDuplicates and ApplyBQSR. Single-nucleotide variants (SNVs) and small indels were marked with Mutect2.23 We created a panel of normal (PoN) calls to reduce false positive somatic calls by using all the germ line samples and the CreateSomaticPanelOfNormals function in GATK4 workflow. PoN were used to filter sites, and the population variant resource containing allele-specific frequencies from gnomAD and matched germ line samples were used to filter alleles. Raw mutation calls were filtered using FilterMutectCalls. Only the mutations that passed all the filters applied by FilterMutectCalls and had ≥10× coverage for both tumor and normal samples were extracted with BCFtools for further analysis. We used FACETS24 to identify allele-specific copy number alterations and ploidy and purity estimates. Structural variants (SVs) were analyzed using Manta.25 SNVs and indels were annotated using Variant Effect Predictor (VEP).26
Power analysis for mutation detection
We calculated the probability of correctly rejecting the null hypothesis that an alternative allele is a sequencing error rather than a true mutation for diploid, monosomy, and trisomy regions. As shown in supplemental Table 1, our average sequencing depth was 81× and the median purity was 83%. We calculated power from 5× to 100× coverage for diploid regions, which was decreased to half for monosomies and increased by 50% for trisomies. We used a framework developed by Carter and et al27 to calculate the detection power. This framework allowed us to account for tumor purity, ploidy, local depth of sequencing, random sequencing error rates, and the required false positive rate for mutation vs sequencing errors.27 The sequencing error rate was set to 0.001, and the false positive rate was set to 5e−07. The fraction of cells harboring a mutation was set to within a range of between 20% and 100%, with 10% incremental steps (supplemental Figure 5).
Driver events
The union of the previously described driver events was collected from multiple publications.28-30 We intersected all driver point mutations and translocations in 68 patients (136 samples) and retained mutations based on the frequency. Additional events between diagnosis and relapse time points were detected using the McNemar test in R. The null hypothesis was that there is an equal probability of detecting mutation X at diagnosis and at relapse. Visualization for the driver events was created with ComplexHeatmap.31
Polynomial model
The change in the total mutational load of patients was modeled via a second-degree polynomial regression adjusted for no intercept, PFS time in months, the study arm, and the interaction between time and study arm, and was implemented in R using the lm() function.
Transcriptional strand bias analysis and motifs around SNVs
Mutations overlapping with RNA and intergenic regions were annotated with VEP. Transcriptional strand bias was evaluated using the strand_bias_test in the MutationalPatterns R package, which applies a Poisson test, for strand asymmetry, and a multiple testing correction.32
Motif discovery
Motifs around SNVs were detected with MEME Suite.33 Each SNV locus was extended by 12 bp on either side. Sequences were extracted in FASTA format. All sequences around the mutations in the RVD + HDM group were used as discovery sequences, and those in the RVD-alone group were used as control sequences. Minimum motif length was set to 3 bp and maximum length was set to 10 bp. FASTA files were analyzed with the discriminative regular expression motif elicitation module in the MEME Suite.34 Both forward and reverse strands were processed without limiting the maximum motif count. The E value was set to .05, and all motifs below this threshold have been reported.
SNVs and ID signatures
Mutational signatures were estimated with signeR35 and sigminer.36 SNVs were mapped onto trinucleotide sequences by including the 5′ and 3′ neighboring-base context to construct a 96 × 68 matrix of mutation counts. The optimal number of mutational signatures was estimated via a saddlepoint approximation of the Bayesian information criterion. Sigminer was also run on the data by using 100 nonnegative matrix factorization runs and 20 bootstraps. Contributions of each identified signature in the study arms were extracted from normalized and absolute exposure matrices. Exposures between treatment arms were then compared with a Wilcoxon signed-rank test. Similarities between de novo signatures and known COSMIC mutational signatures37 were estimated using cosine similarity. Insertion/deletion (indel) signatures were analyzed with similar workflows and applied to an 83-letter alphabet of all possible indel groups based on microhomology-mediated end-joining and repeats.
Creating a predictive model of treatment
All 10 SNV signatures identified by the de novo signature extraction process were used in the classification model creation process without assigning any a priori weights. The outcome for the model was “arm” and the input for the model was normalized signature exposures. Boosted classification trees implemented in the R/caret package was used with 150 iterations. The tuning parameter ν was held constant at a value of 0.1, and this accuracy was used to select the optimal model using the largest value. Leave-one-out cross-validation was used to avoid overfitting the model. Once the model was final, a confusion matrix was created using the training set and an independent validation set to show model accuracy. Final confusion matrices are given in supplemental Files.
Clonal evolution analysis
After correcting for purity and ploidy, we estimated the clonality of mutations, starting from variant allele frequencies. We use an expectation-maximization algorithm based on the probability of observing a specific number of reads supporting a mutation, given the number of reads overlapping the position, the contamination, and the cellularity of a clone. Once we calculate the clonality of mutations, we define the number of clones based on minimizing Bayesian information criteria. We used QuantumClone38 to cluster the mutations at diagnosis and relapse. To reduce false negatives, a custom script was used to evaluate the variant allele frequencies of all mutations detected only at a single time point. We probabilistically modeled the clonal evolution trees constraints to account for statistical variability obtained from ClonEvol.39 For each patient, the cumulative fraction of mutations at a given cancer cell fraction (CCF) was calculated using the trapezoidal rule. Mean values were visualized using ggplot. Contributions of SNV signatures in CCF groups and their uncertainties were quantified using 1000 bootstraps with a multiple linear regression model with the restriction that any coefficient must be >0.
Neoantigen detection
We used an HLA typing tool, xHLA,40 with normal DNA samples from each patient for typing both class I and II HLA genes. Detected HLA types were then used for neoantigen detection with pVACtools,41 a cancer immunotherapy suite for neoantigen detection. All VCF files were annotated with VEP using wild-type and frameshift plugins. HLA types detected with xHLA and annotated VCF files were then used to detect neoantigens. We ran MHCflurry, MHCnuggetsI, and MHCnuggetsII epitope prediction algorithms with a length of between 8 to 11 bp for MHC class I subpeptides and between 12 and 18 bp for class II predictions. We removed neoantigen candidates that did not meet the criteria of a 50% concentration binding score of <500 and/or minimum mutant/wild-type fold change of >1. A top-score filter was used to select the most promising peptide candidate for each variant.
Survival and other statistical analysis
All other analyses were completed in R programming language. Survival analysis and Kaplan–Meier plots were generated using survival and survminer packages. Data were prepared using the readr, readxl, bedr, and maftools packages. Color schemas for the figures were selected using RColorBrewer. Genomic Scar Score (GSS) scores with clinical annotations were visualized using ComplexHeatmap, whereas other figures were visualized using ggpubr and ggplot2. Driver mutations were identified using dNdScv and MutSigCV.
Results
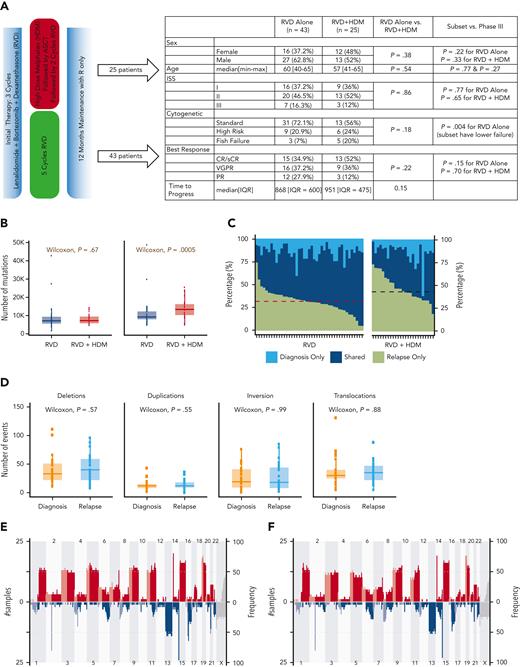
We performed deep WGS on CD138+ MM cells from 136 samples collected from 68 patients at the time of diagnosis and at first relapse. Forty-three patients were enrolled in the RVD-alone arm, and 25 patients in the RVD + HDM followed by ASCT arm (Figure 1A). The median age was 60 years (range, 40-65 years) for RVD and 57 years (range, 41-65 years) for RVD + HDM arm. The disease characteristics including those in the International Staging System (ISS), cytogenetic/fluorescence in situ hybridization–risk groups, and overall response categories were similar between patients studied here and the overall study population6 (Figure 1A).
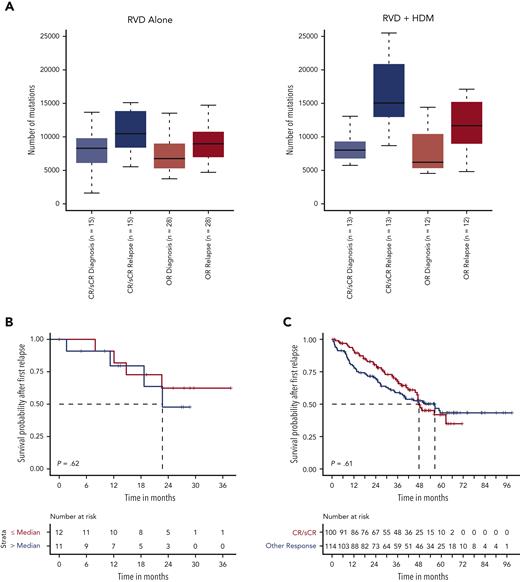
Overall differences between the RVD-only and the RVD + HDM arm. (A) Phase 3 study design and clinical features of patients in the IFM 2009 study. Overall comparisons between the 2 arms in this study, and selected samples analyzed in this study are shown in the table. (B) Number of mutations for samples in RVD-alone arm and RVD + HDM arm at diagnosis (left) and relapse (right). y-axis shows number of mutations in each group and x-axis shows groups. P values are shown (top). (C) For patients, (x-axis) all detected mutations at diagnosis and relapse are taken, and the percentage of contribution from 3 categories (light blue: mutations only detected at diagnosis; dark blue: mutations detected at both time points (shared), 6; and green: mutations only detected at relapse) are shown (y-axis). Patients treated with RVD alone (left) and patients treated with RVD + HDM (right) are shown. (D) Number of SVs (y-axis) for deletions, duplications, inversions, and translocations (left to right) at diagnosis (orange) and relapse (light blue) (x-axis) for patients treated with RVD + HDM. (E-F) Copy number profiles for patients treated with RVD + HDM at diagnosis (E) and relapse (F). Left side of each panel shows the number of patients (% is given on right) with gain (red) and deletions (blue) from chromosome 1 to X (x-axis). Light and dark blue and red colors are separating p and q arms of each chromosome. IQR, interquartile range; max, maximum; min, minimum; PR, partial response; R, lenalidomide; sCR, stringent complete response; VGPR, very good partial response.
Overall differences between the RVD-only and the RVD + HDM arm. (A) Phase 3 study design and clinical features of patients in the IFM 2009 study. Overall comparisons between the 2 arms in this study, and selected samples analyzed in this study are shown in the table. (B) Number of mutations for samples in RVD-alone arm and RVD + HDM arm at diagnosis (left) and relapse (right). y-axis shows number of mutations in each group and x-axis shows groups. P values are shown (top). (C) For patients, (x-axis) all detected mutations at diagnosis and relapse are taken, and the percentage of contribution from 3 categories (light blue: mutations only detected at diagnosis; dark blue: mutations detected at both time points (shared), 6; and green: mutations only detected at relapse) are shown (y-axis). Patients treated with RVD alone (left) and patients treated with RVD + HDM (right) are shown. (D) Number of SVs (y-axis) for deletions, duplications, inversions, and translocations (left to right) at diagnosis (orange) and relapse (light blue) (x-axis) for patients treated with RVD + HDM. (E-F) Copy number profiles for patients treated with RVD + HDM at diagnosis (E) and relapse (F). Left side of each panel shows the number of patients (% is given on right) with gain (red) and deletions (blue) from chromosome 1 to X (x-axis). Light and dark blue and red colors are separating p and q arms of each chromosome. IQR, interquartile range; max, maximum; min, minimum; PR, partial response; R, lenalidomide; sCR, stringent complete response; VGPR, very good partial response.
A significant increase in mutational load at relapse following HDM
In both RVD and RVD + HDM groups, a similar number of mutations was observed at diagnosis (7137 vs 7230, respectively; P = .67) (Figure 1B). At relapse, a significant increase in the number of SNVs was observed (supplemental Figure 1). However, this increase was significantly higher at the time of relapse after treatment with HDM than after treatment with RVD alone (median total SNVs, 13 383 vs 9242, respectively; P = .0005) (Figure 1B), with corresponding increases in the overall mutational load of 73% (mean increase in SNVs, 5952) vs 24% (mean increase in SNVs, 2153; P = .0005). Of all the detected mutations at both sampling time points, 45% were observed only at relapse for HDM group compared with 34% for RVD (Wilcoxon P = 4.69e−05) (Figure 1C). Neither mutational loads only observed at diagnosis nor mutations shared at both time points were different between the 2 arms (1538 vs 2058, Wilcoxon P = .17; 5943 vs 5994, Wilcoxon P = .88) (Figure 1C). Interestingly, the number of SVs including deletions (P = .57), duplications (P = .55), inversions (P = .99), and translocations (P = .88) were similar at diagnosis in both arms and did not change significantly at relapse (Figure 1D; supplemental Figure 1). Similarly, no significant difference in the copy number alterations, including the frequency of indels across the genome, was observed at each time point for either treatments (Figure 1E-F; supplemental Tables 6 and 7), indicating no impact of HDM treatment on large-scale structural changes.
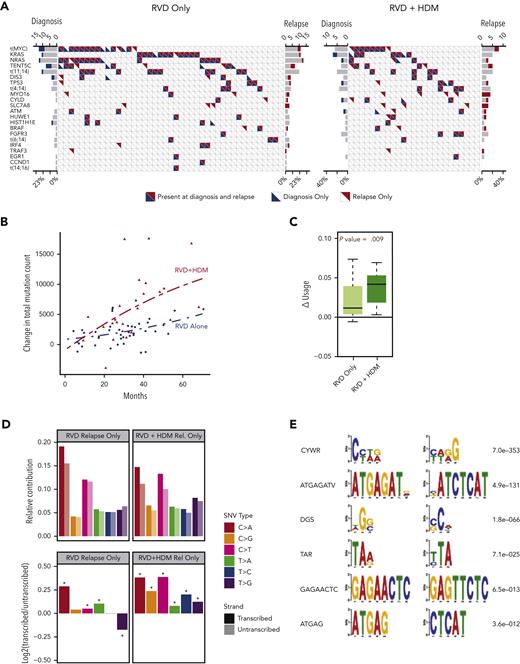
Next, we evaluated the induction or enrichment of driver mutations and translocations after therapy. All translocations involving immunoglobulin heavy chain (t[4;14], t[6;14], t[11;14], t[14;16]) were observed at diagnosis and maintained at relapse. Translocations involving the MYC region (involving immunoglobulin H [IgH], IgL, or IgK, superenhancers proximal to FAM46C) were the most common translocation events in both study arms. Unlike IgH translocations, their presence was not always consistent between diagnosis and relapsed samples (Figure 2A). We also observed a similar frequency of other driver mutations in both arms (Figure 2A; supplemental Tables 3-5), except for a significantly higher frequency of mutations in the SLC7A8 and MYO16 genes than in the other genes at relapse. Overall, 10.3% of all patients had MYO16 mutations at relapse vs 1.5% at diagnosis (McNemar P < .05), and 8.8% had SLC7A8 mutations at relapse vs 0% at diagnosis (McNemar P < .05). Mutational frequency for these 2 genes were not different between the 2 treatment arms, indicating a possible role of RVD treatment rather than HDM.
Driver events and mutational locations. (A) Frequency of driver mutations and translocations (y-axis) are shown for RVD-alone (left) and RVD + HDM (right) arms. Each column shows 1 patient; blue and red triangles show mutations detected at diagnosis and relapse, respectively. (B) Change in total mutational load from diagnosis to relapse (y-axis). Red triangles (patients treated with RVD + HDM) and blue points (patients treated with RVD alone) show the total mutational load difference between diagnosis and relapse for each patient. Polynomial regression curves are shown for the 2 arms over time (x-axis). (C) Change in genomic region use (y-axis) for mutations overlapping with RNAs for RVD-alone and RVD + HDM arms (left to right). (D) Transcription strand bias analysis. Relative contribution (y-axis, top) for 6 possible SNV types (color coded) for relapse-only mutations are shown. Contributions from mutations overlapping with transcribed strand are shown in darker colors, and transcribed strand shown in lighter colors. log2 ratio of transcribed/untranscribed strands are shown (bottom) with statistically significant differences marked with an asterisk (∗). (E) De novo motifs identified 12 bp upstream and downstream of SNVs. Consensus motifs (left) and significance values (right) are shown.
Driver events and mutational locations. (A) Frequency of driver mutations and translocations (y-axis) are shown for RVD-alone (left) and RVD + HDM (right) arms. Each column shows 1 patient; blue and red triangles show mutations detected at diagnosis and relapse, respectively. (B) Change in total mutational load from diagnosis to relapse (y-axis). Red triangles (patients treated with RVD + HDM) and blue points (patients treated with RVD alone) show the total mutational load difference between diagnosis and relapse for each patient. Polynomial regression curves are shown for the 2 arms over time (x-axis). (C) Change in genomic region use (y-axis) for mutations overlapping with RNAs for RVD-alone and RVD + HDM arms (left to right). (D) Transcription strand bias analysis. Relative contribution (y-axis, top) for 6 possible SNV types (color coded) for relapse-only mutations are shown. Contributions from mutations overlapping with transcribed strand are shown in darker colors, and transcribed strand shown in lighter colors. log2 ratio of transcribed/untranscribed strands are shown (bottom) with statistically significant differences marked with an asterisk (∗). (E) De novo motifs identified 12 bp upstream and downstream of SNVs. Consensus motifs (left) and significance values (right) are shown.
HDM-induced SNVs predominantly involve transcribed strand
Acquisition of mutations increased over time in both arms; however, the rate of accumulation of mutations was significantly higher in the HDM group vs the RVD group (Figure 2B). Moreover, patients treated with HDM accumulated significantly more mutations within the regions between transcription start and transcription termination sites than those treated with RVD (P = .009) (Figure 2C). We converted all mutations to 6 possible SNV types and identified mutations overlapping with transcribed or nontranscribed strands to investigate any strand bias in newly acquired mutations. Patients receiving HDM had significantly increased asymmetry favoring the transcribed strand for all SNV types (Figure 2D). The SNVs observed at diagnosis were similarly distributed between the transcribed and nontranscribed strand. Furthermore, we analyzed DNA sequences around mutations by considering 12-bp regions around SNV locations. We identified CYWR (false discovery rate [FDR] = 7e−353) and ATGAGATV (FDR = 4.9e−131) as motifs significantly enriched around mutations observed after HDM treatment (Figure 2E).
Mutational patterns in myeloma cells after HDM treatment
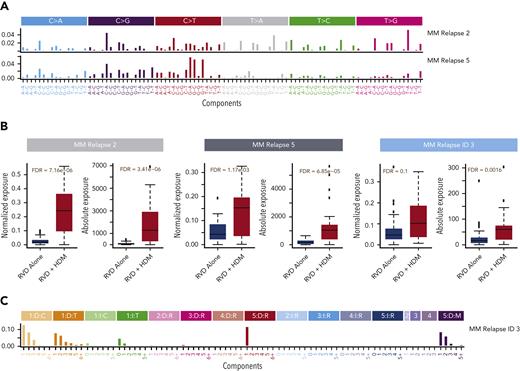
Next, we evaluated the underlying mutational processes affecting the newly acquired mutations by evaluating the clusters of mutations using a Bayesian variant of the nonnegative matrix factorization algorithm. We identified 10 distinct mutational patterns for newly acquired mutations (supplemental Figure 2). Among these signatures, MM relapse signature 2 and MM relapse signature 5 had a significantly higher relative (FDR = 7.16e−06 and FDR = 1.17e−03, respectively) (Figure 3A-B) and absolute contribution (FDR = 3.41e−06 and FDR = and 6.85e−05, respectively) in the HDM group than in the RVD group (Figure 3B). Both signatures showed high cosine similarity with the HR deficiency signature in the COSMIC database (cosine similarity = 0.7 and 0.77, respectively) (supplemental Figure 3) and previously described MM signature 1.18,42 Similar to SNVs, we also evaluated newly acquired small indels and identified 11 ID signatures (supplemental Figure 4). ID signature 3 had significantly higher relative (FDR = 0.1) and absolute (FDR = 0.0016) contributions for patients treated with HDM (Figure 3B-C) than for those treated with RVD. Similar to SNV signatures, MM relapse ID signature 3 is also associated with the repair of DNA double-strand breaks by nonhomologous end-joining mechanisms (cosine distance = 0.7) (supplemental Figure 3). To estimate the significance of the identified signatures and their relationship to therapy, we developed a boosted classification trees model and observed 97% accuracy, with 92% sensitivity and 100% specificity in predicting exposure to HDM. The positive and negative prediction values of the model were 100% and 95%, respectively. Furthermore, we created a validation set using the data of 32 patients at relapse who were also enrolled in the IFM2009 study (18 in the RVD-alone arm and 14 in the RVD + HDM arm) and applied the predictive model to it. On the independent data set, the model accuracy was 90% (95% confidence interval, 74%-98%) (supplemental Table 2). Samples from the validation cohort were sequenced and analyzed using the same sequencing protocol and analytical pipeline as that of the original sample set.
Mutational processes associated with HDM. (A) Two mutational patterns that are significantly higher in patients after HDM treatment were identified by de novo mutational signature analysis. Both mutational patterns are highly similar to that in COSMIC SBS3. Six possible substitutions (C→A, C→G, C→T, T→A, T→C, and T→G) of pyrimidines have been illustrated for the bases from 5′ to 3′, which creates 96 (4 × 6 × 4) possible contexts. Color coding was used to separate 6 possible options and the relative contribution of each context (x-axis) is given on the y-axis. Etiology of each signature is estimated by using their cosine distance to published SBS signatures in COSMIC mutational signatures data files. (B) Absolute (right) and relative (left) contributions (y-axis) of single nucleotide mutational signatures (SBS) and ID signature in the RVD-alone and RVD + HDM groups (x-axis in each panel). (C) De novo extracted ID signature that is significantly higher in the HDM group. Small indels were separated into 84 possible options and color coded by major event classification. Single–base pair deletions (orange) and insertions (green) are further broken down into groups based on the homopolymer (repeating the same base after the indel) length. Insertions (blue color shades) and deletions (red color shades) that are 2, 3, 4, or 5 bp or longer were separated into subgroups based on repeated sequence of event on the 3′ end of each sequence. Deletions that have microhomology (presence of the same short sequence) on either the 5′ or 3′ site of the event (purple shades) were also separated into subgroups based on the lengths of the deleted sequence and microhomology site. Similar to SBS signatures, cosine distance to published ID signatures on the COSMIC website was calculated by downloading the ID signature database using the GRCh38 genome.
Mutational processes associated with HDM. (A) Two mutational patterns that are significantly higher in patients after HDM treatment were identified by de novo mutational signature analysis. Both mutational patterns are highly similar to that in COSMIC SBS3. Six possible substitutions (C→A, C→G, C→T, T→A, T→C, and T→G) of pyrimidines have been illustrated for the bases from 5′ to 3′, which creates 96 (4 × 6 × 4) possible contexts. Color coding was used to separate 6 possible options and the relative contribution of each context (x-axis) is given on the y-axis. Etiology of each signature is estimated by using their cosine distance to published SBS signatures in COSMIC mutational signatures data files. (B) Absolute (right) and relative (left) contributions (y-axis) of single nucleotide mutational signatures (SBS) and ID signature in the RVD-alone and RVD + HDM groups (x-axis in each panel). (C) De novo extracted ID signature that is significantly higher in the HDM group. Small indels were separated into 84 possible options and color coded by major event classification. Single–base pair deletions (orange) and insertions (green) are further broken down into groups based on the homopolymer (repeating the same base after the indel) length. Insertions (blue color shades) and deletions (red color shades) that are 2, 3, 4, or 5 bp or longer were separated into subgroups based on repeated sequence of event on the 3′ end of each sequence. Deletions that have microhomology (presence of the same short sequence) on either the 5′ or 3′ site of the event (purple shades) were also separated into subgroups based on the lengths of the deleted sequence and microhomology site. Similar to SBS signatures, cosine distance to published ID signatures on the COSMIC website was calculated by downloading the ID signature database using the GRCh38 genome.
Clonal evolution and heterogeneity after HDM treatment
To understand the patterns of clonal expansion during MM progression after therapy, we reconstructed the clonal structure using an expectation-maximization algorithm at diagnosis and relapse for all patients. Majority the of patients in both arms showed a branching or differential evolution pattern (Figure 4A-B). However, we observed a static evolution pattern (similar clonal composition between diagnosis and relapsed pairs) in 4 patients (9%), all in the RVD arm (Figure 4C). Although clonal evolution patterns were similar in general between the 2 arms, at the time of relapse, the majority of the mutational load observed after HDM treatment was at the subclonal level (Figure 4D). Next, we quantified heterogeneity as the cumulative fraction of mutations at a sliding CCF and observed significantly higher heterogeneity in the HDM cohort (P = .01) (Figure 4E). We further stratified mutations into 4 groups based on CCFs, and found that the change in clonal mutational load was similar between the 2 arms; however, mutations with <50% CCF were more common in the HDM arm (P = .003 and P = .004, respectively) (Figure 4F). Using the mutational signatures defined previously, we also observed that the 2 significantly high mutational signatures in the HDM group were also high in these subclonal mutations (Figure 4G).
Clonal evolution and complexity after HMD treatment. (A-C) Three representative examples of positive selection of subclones (A,B) and static evolution (C) patterns observed in patients after treatment. For each panel, CCF of mutations are shown at diagnosis (x-axis) and relapse (y-axis). Mutations were clustered using expectation-maximization algorithm, and clusters are color coded with densities around the clusters shown in shaded colors. For each pattern, most likely clonal evolution trees are shown (bottom). Fish plots (bottom left) are showing the clonal composition at a given time point (diagnosis [top] or relapse [bottom]), and a clonal evolution tree is also shown (bottom right). (D) Density (y-axis) of mutations at given a CCF (x-axis) for RVD-only (blue dashed line) and RVD + HDM (red solid line) groups at diagnosis (left) and relapse (right). (E) Mean cumulative percentage of mutations (y-axis) at a given CCF (x-axis) from all patients in the RVD + HDM (red curve) and RVD-alone (blue curve) arms. Boxplots (right) show area under the cumulative percentage of mutations curve for each patient at relapse for RVD + HDM and RVD-only arms. (F) Number of mutations (y-axis) at relapse by clonality levels (x-axis) between 2 treatment arms (color coded). (G) Relative mutational signature contribution (y-axis) from HDM-specific signatures (signatures 2 and 5) at 4 different clonality levels (x-axis). Rel, relative.
Clonal evolution and complexity after HMD treatment. (A-C) Three representative examples of positive selection of subclones (A,B) and static evolution (C) patterns observed in patients after treatment. For each panel, CCF of mutations are shown at diagnosis (x-axis) and relapse (y-axis). Mutations were clustered using expectation-maximization algorithm, and clusters are color coded with densities around the clusters shown in shaded colors. For each pattern, most likely clonal evolution trees are shown (bottom). Fish plots (bottom left) are showing the clonal composition at a given time point (diagnosis [top] or relapse [bottom]), and a clonal evolution tree is also shown (bottom right). (D) Density (y-axis) of mutations at given a CCF (x-axis) for RVD-only (blue dashed line) and RVD + HDM (red solid line) groups at diagnosis (left) and relapse (right). (E) Mean cumulative percentage of mutations (y-axis) at a given CCF (x-axis) from all patients in the RVD + HDM (red curve) and RVD-alone (blue curve) arms. Boxplots (right) show area under the cumulative percentage of mutations curve for each patient at relapse for RVD + HDM and RVD-only arms. (F) Number of mutations (y-axis) at relapse by clonality levels (x-axis) between 2 treatment arms (color coded). (G) Relative mutational signature contribution (y-axis) from HDM-specific signatures (signatures 2 and 5) at 4 different clonality levels (x-axis). Rel, relative.
Association between change in mutational load, clinical response, and long-term outcome
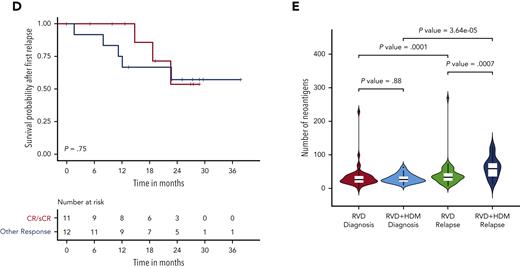
Having identified a high mutational load in subclonal cell populations after HDM treatment, we evaluated whether patient response affected the mutational change. Although the mutational load increased at relapse in all patients, in HDM, the increase was greater in patients achieving CR than that in others (Wilcoxon P = .07). Moreover, the increase in mutational burden was further accentuated in patients relapsing from CR after HDM treatment compared with those who received RVD only (median, 15 053 vs 10 468, respectively; Wilcoxon P = .02) (Figure 5A). As we observed deeper responses in patients with relatively higher mutational burdens, we investigated whether high mutational burden at relapse, especially after HDM treatment, had any impact on the subsequent outcome. Importantly, neither patients with high mutational load after HDM treatment (Figure 5B) nor patients who achieved CR had significantly different survival after PFS1 compared with other patients (Figure 5C-D). In addition, we investigated the presence of neoantigens for each patient at diagnosis and relapse. We observed a significantly higher number of neoantigens in patients relapsing after treatment with HDM than that in patients relapsing after treatment with RVD alone (Figure 5E; supplemental Figure 6). Among the mutational signatures, the single base substitutions 3 (SBS3), SBS-MM1, and AID/somatic hypermutation signatures were predicted to contribute to neoantigen creation (supplemental Figure 6).
Association between mutational load chance and response and clinical outcome. (A) Number of mutations (x-axis) by response groups (sCR/CR) or other responses (≤VGRP) at diagnosis and relapse for the RVD-alone (left) and RVD + HDM group (right). (B) Survival probability after first relapse from RVD + HDM treatment by mutational load. Patients with mutational load more than the median are shown in red curve and patients with low mutational load are shown in blue. (C) Survival probability after first relapse from RVD + HDM treatment by best response to initial RVD + HDM treatment. Patients with CR/sCR are shown in red and patients with other responses are shown in blue. All patients who had relapse after RVD + HDM in IFM 2009 are shown. (D) Same as in panel C but only patients in our study are shown. (E) Number of neoantigens (y-axis) detected in each group (x-axis) at diagnosis (left 2 violins) and relapse (right 2 violins). RVD-only and RVD + HMS groups compared using Wilcoxon test and patients at diagnosis and relapse from the same group compared with paired analysis. P values for each comparison are shown (top). OR, other response.
Association between mutational load chance and response and clinical outcome. (A) Number of mutations (x-axis) by response groups (sCR/CR) or other responses (≤VGRP) at diagnosis and relapse for the RVD-alone (left) and RVD + HDM group (right). (B) Survival probability after first relapse from RVD + HDM treatment by mutational load. Patients with mutational load more than the median are shown in red curve and patients with low mutational load are shown in blue. (C) Survival probability after first relapse from RVD + HDM treatment by best response to initial RVD + HDM treatment. Patients with CR/sCR are shown in red and patients with other responses are shown in blue. All patients who had relapse after RVD + HDM in IFM 2009 are shown. (D) Same as in panel C but only patients in our study are shown. (E) Number of neoantigens (y-axis) detected in each group (x-axis) at diagnosis (left 2 violins) and relapse (right 2 violins). RVD-only and RVD + HMS groups compared using Wilcoxon test and patients at diagnosis and relapse from the same group compared with paired analysis. P values for each comparison are shown (top). OR, other response.
Discussion
HDM with ASCT has demonstrated superior PFS after very effective novel-agent combination therapy (RVD).6,43-45 However, the impact of HDM, with its inherent DNA-damaging effects, on surviving MM cells may affect the genomic makeup of MM cells at relapse, with potential implications for relatively longer term outcomes among these patients.19,46-48 The IFM/DFCI 2009 study provided an ideal setting to evaluate the genomic impact of HDM, because patients were treated with a regimen not containing an alkylating agent in 1 arm and the same regimen with added HDM in the other. Although available samples at diagnosis and relapse from a smaller cohort of patients were sequenced here, they are representative of the data of larger patient population. Importantly, both arms in this subset are also balanced for all risk features and clinical outcomes, including relatively similar time to relapse.
We report a significant increase in single-nucleotide changes in MM cells at relapse with both treatments. However, the mutational burden was significantly higher after HDM treatment than that after RVD treatment. Previous studies have evaluated genomic changes in either relapsed/refractory MM samples or compared unpaired samples from diagnosis and relapse, reporting an increased number of mutations.18,20,49 Here, we observe the increase in paired samples of a randomized study to clearly establish the impact of HDM on the MM genome. Melphalan significantly increases DNA adducts,50-52 forms covalent DNA interstrand crosslinks, and causes DNA breaks that prevent RNA and DNA replication. This explains the predominant transcriptional strand bias for mutations observed with HDM, because transcribed regions are more susceptible than nontranscribed regions to melphalan binding.53 Perhaps unexpectedly, we did not observe frequent, new, large-scale somatic events or the new SVs formation within the same genomic coordinates, despite the fact that DNA breaks are expected, and combined genotoxic exposure and DNA repair deficiency alters mutation rates.15 Several individuals did have a change in copy number alterations or the number of SVs. However, none of these events were frequent enough to be considered an effect of HDM or RVD. Future studies with larger cohorts should look at the enrichment of these infrequent events. The observation of the differential use of HR and DNA damage signatures in newly acquired mutations in HDM cohort also confirms the impact of melphalan and the pattern of DNA lesions generated by an alkylating agent.13 Genomic changes caused by melphalan treatment can be repaired or processed by several enzymes and pathways, which may counteract the induced genomic damage. However, the lack of changes in large-scale events and significant increase in point mutations observed in our study suggests dysfunction of specific DNA repair pathways.
Previous studies in MM have identified a number of driver mutations, albeit mostly at a subclonal level.28,29 Subsequent studies looking at relapse have suggested the enrichment of clones with certain mutations, such as that in TP53; however, the incremental changes were relatively small and not adequate to explain relapse. We did not observe significant changes, either in absolute number, CCF levels or driver mutations at relapse after either of the treatments. The small but significant change in mutations involving MYO16 and SLC7A8 from diagnosis to relapse was observed in both treatment arms and may indicate a relationship with RVD treatment and/or lenalidomide maintenance. The potential functional role of these genes in cell cycle and protein transport requires further investigation to delineate their molecular role associated with relapse.54-57 In addition to driver events, HDM may also cause changes in hematopoietic stem cells or myeloid cells. The DETERMINATION study reported similar secondary primary hematologic cancers between the RVD-alone group (2.5%) and HDM group (3.6%) (5-year cumulative incidence, 1.6% and 3.5%, respectively; P = .32), with acute myeloid leukemia or myelodysplastic syndromes reported in none of the patients in the RVD-alone group as compared with 10 patients in the transplant group (2.7%) (P = .002).58 In another study, Sperling et al found that lenalidomide treatment can also cause clonal selection and promote TP53-mutated therapy-related myeloid neoplasms.58,59 Because of our study design, we were not able to evaluate the impact on myeloid cells or stem cells after HDM exposure. However, the impact of HDM should be evaluated in future studies in addition to MM cells.
An important issue raised here is the clinical importance of significantly increased the mutational burden after HDM. Does this reflect an increase in the clonal content, the number of mutations per clone, or both? Although melphalan is a known mutagen and may be responsible for the increased mutations observed, it is by no means the only reason for this, and other alternative possibilities do coexist. Additional mechanisms, such as causing a clonal selection of cells with high mutational burdens, would be an alternative hypothesis that could be addressed in the future. We do see increased mutation levels in the RVD-alone treatment as well. Additional sequencing methods, such as single-cell DNA sequencing or WGS/whole exome sequencing followed by an error-corrected sequencing approach with the use of unique molecular identifiers and high-coverage depths (>30 000×)60 could be used on longitudinal samples in future studies to better address such questions. Samples not only collected at diagnosis and relapse but also at the time of response, the end of induction, and after transplant can also help understand this phenomenon. The current depth of sequencing and the type of methodology used do not allow a clear delineation of the clonal complexity, but we hypothesize that it results from a combination of both the number of mutations per cell and the number of clones. The increased number of mutations might suggest the possibility of more aggressive disease. However, this study and all other available data sets do not indicate that posttransplant relapses have worse outcome. We acknowledge that our study is not optimal to answer this question, because the majority of the patients in the RVD arm received a delayed transplant as salvage therapy. Intriguingly, patients achieving CR in our cohort had a significantly higher number of mutations. However, despite increased mutational burden, the survival after first relapse was similar in patients achieving initial CR compared with the rest. This may be, at least in part, because of the higher immunogenic nature of MM cells with higher mutational burden and hence neoantigen repertoire. This is supported by our neoantigen analysis (Figure 5E). This may also explain the lack of poor outcome after relapse following HDM. Importantly, immunological interventions may thus be an attractive therapeutic modality in these patients. Our study provides initial data to generate hypotheses regarding possible immunotherapeutic intervention after transplantation; future experimental validation together with retrospective and prospective studies are needed. Analysis using deeper response measurements, such as minimum residual disease, should be considered in the future, because it may highlight the true impact of mutational increase after deep responses as opposed to only looking at complete response.
In conclusion, our study, using unique paired samples, identifies increased genomic changes associated with HDM and provides the rationale to further understand clonal complexity at diagnosis and relapse after both standard-dose and HDM therapy.
Acknowledgments
This work was supported by grants from the National Cancer Institute, National Institutes of Health (grants P01-155258 and 5P50 CA100707) (N.C.M., K.C.A., M.K.S., H.A.-L., M.F., and M.A.S.); a Department of Veterans Affairs merit review award (I01BX001584-01) (N.C.M.); and a Riney Family Foundation (N.C.M., K.C.A., M.K.S.) grant. M.R. received a fellowship grant from the American Italian Cancer Foundation.
Authorship
Contribution: M.K.S., N.C.M., and H.A.-L. designed and conducted the study; M.K.S., M.R., A.A.S., A.H.B., and G.P. analyzed the data; and all authors collected data, discussed the results, and wrote the manuscript.
Conflict-of-interest disclosure: K.C.A. has received consulting fees from Pfizer, Amgen, AstraZeneca, Janssen, Precision Biosciences and serves the board of directors of C4 Therapeutics, OncoPep, Mana, Raqia Therapeutics, NextRNA, Window, and Starton. N.C.M. is a consultant for Bristol Myers Squibb, Janssen, OncoPep, Amgen, Pfizer, Karyopharm, Legend, NextRNA, Raqia, AbbVie, Takeda, and GSK and is on the board of directors of OncoPep. G.P. holds equity in Phaeno Biotechnologies, is on the SAB of Realm, and currently consults for Delphi Diagnostics and for Martingale Labs. A.T. is employed by Bristol Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Mehmet Kemal Samur, Department of Data Science, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: mehmet_samur@dfci.harvard.edu; and Nikhil C. Munshi, Department of Medical Oncology, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: nikhil_munshi@dfci.harvard.edu.
References
Author notes
The somatic call data reported in this article have been deposited in the European Genome-phenome archive (accession number EGAS00001006731).
Data are available on request from the corresponding author Nikhil C. Munshi (nikhil_munshi@dfci.harvard.edu).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Clonal evolution and complexity after HMD treatment. (A-C) Three representative examples of positive selection of subclones (A,B) and static evolution (C) patterns observed in patients after treatment. For each panel, CCF of mutations are shown at diagnosis (x-axis) and relapse (y-axis). Mutations were clustered using expectation-maximization algorithm, and clusters are color coded with densities around the clusters shown in shaded colors. For each pattern, most likely clonal evolution trees are shown (bottom). Fish plots (bottom left) are showing the clonal composition at a given time point (diagnosis [top] or relapse [bottom]), and a clonal evolution tree is also shown (bottom right). (D) Density (y-axis) of mutations at given a CCF (x-axis) for RVD-only (blue dashed line) and RVD + HDM (red solid line) groups at diagnosis (left) and relapse (right). (E) Mean cumulative percentage of mutations (y-axis) at a given CCF (x-axis) from all patients in the RVD + HDM (red curve) and RVD-alone (blue curve) arms. Boxplots (right) show area under the cumulative percentage of mutations curve for each patient at relapse for RVD + HDM and RVD-only arms. (F) Number of mutations (y-axis) at relapse by clonality levels (x-axis) between 2 treatment arms (color coded). (G) Relative mutational signature contribution (y-axis) from HDM-specific signatures (signatures 2 and 5) at 4 different clonality levels (x-axis). Rel, relative.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/14/10.1182_blood.2022017094/3/m_blood_bld-2022-017094-gr4.jpeg?Expires=1769089807&Signature=3lVnF1CrfFfRtnPhhzUklmArRI~o5YCfE8C~SHGqw0plxfH4rXkdRfEsmSG~IRRhyWWuNG1BxOqarDwwRudMt1o~CcWFvST~QZQdUBwFtClFePu-r3BkAV2RNvH2iOad3tDDXpRrVw25CLgnnJteduCvSTGqunQ1SWD2UtOPBkIKR7qaJzxa1leiVrrJqhNzXIwiFAbJc2e1xjBZkI8RBAu3hg3NhvrI2C3YdZcdfkMrlbpxysj~TXZcZyXhMDU82DO1Yy5QEI4Jvupek8pD6iBfqHDCpL6s4nTMdyYlvuaSJ6kmTA8x6V8pyhWapvSwBixk07zB3UOjK4cl5AWDSg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal