Key Points
High-risk t(4;14) MM patients can be identified by using the coordinates of the translocation breakpoints in NSD2 gene on chromosome 4.
Large-scale genomic analysis of NDMM patient data identifies mutations and copy number aberrations enriched in t(4;14) patients.
Abstract
Although translocation events between chromosome 4 (NSD2 gene) and chromosome 14 (immunoglobulin heavy chain [IgH] locus) (t(4;14)) is considered high risk in newly diagnosed multiple myeloma (NDMM), only ∼30% to 40% of t(4;14) patients are clinically high risk. We generated and compared a large whole genome sequencing (WGS) and transcriptome (RNA sequencing) from 258 t(4;14) (n = 153 discovery, n = 105 replication) and 183 non-t(4;14) NDMM patients with associated clinical data. A landmark survival analysis indicated only ∼25% of t(4;14) patients had an overall survival (OS) <24 months, and a comparative analysis of the patient subgroups identified biomarkers associated with this poor outcome, including translocation breakpoints located in the NSD2 gene and expression of IgH-NSD2 fusion transcripts. Three breakpoints were identified and are designated as: “no-disruption” (upstream of NSD2), “early-disruption” (in the 5' UTR), and “late-disruption” (within the NSD2 gene). Our results show a significant difference in OS based on the location of DNA breakpoints (median OS 28.6 “late-disruption” vs 59.2 “early disruption” vs 75.1 months “no disruption”). These findings have been replicated in an independent replication dataset. Also, univariate and multivariate analysis suggest high-risk markers such as del17p, 1p independently contribute to poor outcome in t(4;14) MM patients.
Introduction
A translocation event between chromosome 4 (NSD2 gene) and chromosome 14 (immunoglobulin heavy chain [IgH] locus) (t(4;14)) is a primary clonal event in 11% to 15% of newly diagnosed multiple myeloma (NDMM) patients.1-3 t(4;14) translocation results in the generation of 2 derivative chromosomes that place fibroblast growth factor receptor 3 (FGFR3) and nuclear receptor SET domain protein 2 (NSD2, also known as WHSC1 and MMSET) under the control of the IgH super enhancer elements.2 t(4;14) is included in the definition of Revised International Staging System (R-ISS) stage III classification4 and is an accepted feature of high-risk NDMM. However, previous analyses indicated that only 30% to 40% of t(4;14) patients are in fact high risk, with some achieving outcomes similar to intermediate- or standard-risk patients.5,6 Previous analysis of t(4;14) patients showed a positive association with FGFR3 mutations, 1q gain, and deletions in 1p, 4p, 11q, 12p, 13q, and 14q.7 In a large cohort of 1905 NDMM t(4;14) “double-hit” (having 2 adverse genomic lesions) patients, those with 1q gain or deletions in 13q or BIRC2/3 had poor outcome.8 However, these studies did not specifically analyze the high-risk t(4;14) subgroup.
Multiple groups have investigated t(4;14) biology, highlighting the roles of FGFR3 and NSD2 as potential drivers of aggressive disease and poor clinical outcome. Overexpression of these putative oncogenes was associated with the t(4;14) population, and the ectopic expression of FGFR3 was used as a t(4;14) detection method in vitro.1,9 Reverse transcription polymerase chain reaction (RT-PCR) analyses by Keats et al of 31 t(4;14) patients indicated that ∼75% express FGFR3, but adverse prognosis was not correlated with FGFR3 expression.10 Multiple studies have subsequently discounted the causal role of FGFR3 overexpression with poor prognosis.1,2,11,12 In the Keats analysis, as well as in another study (n = 11 t(4;14)),13 both full-length NSD2 (designated as MB4-1) and 2 truncated transcript isoforms (MB4-2 and MB4-3) were detectable by RT-PCR, but they did not investigate association of the NSD2 transcripts with clinical outcome. Due to the limitation of small data sets and lack of robust genomic and clinical data, the molecular characteristics of t(4;14) multiple myeloma (MM) have not been fully described. Previous efforts have attempted to describe the variability across the DNA breakpoint locations on chromosome 4 and the possible impact of the resultant fusion transcripts on survival. One of these efforts included whole genome sequencing (WGS) (n = 14 t(4;14)) and identified t(4;14) breakpoint locations upstream of the NSD2 gene (resulting in a full-length transcript) or within the coding sequence (expressing truncated isoforms MB4-2 and MB4-3).14 Analysis of 256 patient tumors for the fusion transcripts indicated the prognostic role of MB4-2, especially in association with del17p, and implied the potential biological role of the truncated NSD2 protein.15 In another RT-PCR study (n = 53 t(4;14)), the expression of both NSD2 truncated isoforms were associated with poor prognosis, but corresponding DNA breakpoint locations were not analyzed.11
To comprehensively explore the genomic and transcriptomic characteristics of t(4;14) NDMM patients, we collected and generated the largest cohort of data from 258 t(4;14) NDMM patients (153 discovery, 105 independent replication) with WGS, RNA sequencing (RNA-seq), and clinical data and compared it with data from 183 non-t(4;14) MM patients. These data allowed us to characterize the genomic landscape of t(4;14) patients, including mutations, copy number aberrations (CNAs), translocation breakpoints, and their association with clinical outcome. Importantly, this combined WGS/RNA-seq data set allowed more precise identification of the translocation breakpoints than is possible with fluorescent in situ hybridization (FISH), as well as the ability to associate these breakpoints with resultant NSD2 transcripts and assess their impact on outcome.
We first compared t(4;14) vs non-t(4;14) NDMM patients to identify genomic features enriched in the t(4;14) population. Then, we focused on the t(4;14) cohort and performed a supervised analysis of patients with overall survival (OS) <24 months from diagnosis (designated as high-risk [HR]) versus those with OS >24 months (designated non-high-risk [NHR]) to identify features associated with poor clinical outcome. This analysis led us to identify 3 translocation breakpoints and associated fusion transcripts of NSD2 and their association with poor clinical outcome. Notably, expression of neither NSD2 nor FGFR3 nor mutations in FGFR3 correlated with poor outcome in HR t(4;14). A multivariate analysis (including various molecular and clinical features) showed that translocation breakpoints as well as deletions 17p and 1p and amplification (amp) 1q were independently associated with OS. We further replicated these findings in an independent cohort of 105 t(4;14) NDMM patients. Our data provide a potential path to developing a DNA sequencing-based assay to molecularly identify t(4;14) patients, to refine risk stratification criteria, and to potentially develop therapies for HR t(4;14) MM patients.
Methods
Patients and data sets
WGS and transcriptome data were generated from NDMM patients (N = 129) with confirmed t(4;14) by FISH from the Institut Universitaire du Cancer Oncopole in Toulouse (TOU). Genomic data from the Myeloma Genome Project (MGP) were available, which included both t(4;14) and non-t(4;14) patients from the Multiple Myeloma Research Foundation (N = 34 t(4;14)) (MMRF, [CoMMpass, IA20] NCT01454297), and Intergroupe Francophone du Myélome (N = 24 t(4;14)) (IFM2009, NCT01191060). Details on the MGP data set including demographics, data generation, and computational pipelines have been described.7,16-18 Patients with t(4;14) from the TOU and IFM2009 comprised the discovery data set. To harmonize the t(4;14) and non-t(4;14) data sets based on treatment, 34 t(4;14) patients from the MMRF replication data set were included who were treated with lenalidomide in combination with bortezomib and dexamethasone (RVd), which was the induction regimen in the IFM2009 trial and a common regimen in the TOU data set (Table 1). An additional cohort of t(4;14) patients from Toulouse/DFCI (T/D, N = 71) was combined with MMRF (N = 34) and used as a replication cohort (N = 105) for analysis of breakpoint association with outcome in t(4;14) patients (Table 1). The IFM2009 non-t(4;14) population (N = 183) WGS data set was used as a comparison group to the t(4;14) population for comparison of genomic features between populations.
Demographic data of NDMM patients
| . | Non-t(4;14) . | t(4;14) . | |||
|---|---|---|---|---|---|
| Discovery . | Replication . | ||||
| IFM2009 . | IFM2009 . | Toulouse . | MMRF . | T/D . | |
| N | 183 | 24 | 129 | 34 | 71 |
| Median age, y (range) | 60 (35-65) | 56 (41-65) | 63 (43-85) | 58 (36-83) | 59 (39-71) |
| Male, % (no.) | 60.58 (83/137) | 75 (12/16) | 56.59 (73/129) | 67.65 (23/34) | 67.6 (48/71) |
| Female, % (no.) | 39.42 (54/137) | 25 (4/16) | 43.41 (56/129) | 32.35 (11/34) | 32.4 (23/71) |
| ISS1, % (no.) | 35.62 (52/146) | 27.78 (5/18) | 19.23 (20/104) | 23.53 (8/34) | 21.1 (15/71) |
| ISS2, % (no.) | 45.89 (67/146) | 22.22 (4/18) | 46.15 (48/104) | 55.88 (19/34) | 46.5 (33/71) |
| ISS3, % (no.) | 18.49 (27/146) | 50 (9/18) | 34.62 (36/104) | 20.59 (7/34) | 32.4 (23/71) |
| Induction | RVd | RVd | Investigator's choice | RVd | Investigator's choice |
| Stem cell transplant, % (no.) | 46.78 (80/171) | 43.48 (10/23) | 62.02 (80/129) | 61.76 (21/34) | 100 (71/71) |
| Maintenance, % (no.) | 100 (183/183) | 100 (24/24) | 79.65 (90/113) | 44.12 (15/34) | NA |
| . | Non-t(4;14) . | t(4;14) . | |||
|---|---|---|---|---|---|
| Discovery . | Replication . | ||||
| IFM2009 . | IFM2009 . | Toulouse . | MMRF . | T/D . | |
| N | 183 | 24 | 129 | 34 | 71 |
| Median age, y (range) | 60 (35-65) | 56 (41-65) | 63 (43-85) | 58 (36-83) | 59 (39-71) |
| Male, % (no.) | 60.58 (83/137) | 75 (12/16) | 56.59 (73/129) | 67.65 (23/34) | 67.6 (48/71) |
| Female, % (no.) | 39.42 (54/137) | 25 (4/16) | 43.41 (56/129) | 32.35 (11/34) | 32.4 (23/71) |
| ISS1, % (no.) | 35.62 (52/146) | 27.78 (5/18) | 19.23 (20/104) | 23.53 (8/34) | 21.1 (15/71) |
| ISS2, % (no.) | 45.89 (67/146) | 22.22 (4/18) | 46.15 (48/104) | 55.88 (19/34) | 46.5 (33/71) |
| ISS3, % (no.) | 18.49 (27/146) | 50 (9/18) | 34.62 (36/104) | 20.59 (7/34) | 32.4 (23/71) |
| Induction | RVd | RVd | Investigator's choice | RVd | Investigator's choice |
| Stem cell transplant, % (no.) | 46.78 (80/171) | 43.48 (10/23) | 62.02 (80/129) | 61.76 (21/34) | 100 (71/71) |
| Maintenance, % (no.) | 100 (183/183) | 100 (24/24) | 79.65 (90/113) | 44.12 (15/34) | NA |
Cohort demographics for discovery dataset consisting of a t(4;14) specific cohort from TOU, and the t(4;14) subset of IFM2009. The replication cohort was comprised of data from the MMRF CoMMpass trial (IA20) and a Toulouse/Dana-Farber Cancer Institute (T/D) cohort. Patients from the IFM2009 trial without a t(4;14) were used as a comparator set. Per protocol, the induction regimen in IFM2009 was RVd followed by either stem cell transplant or additional cycles of RVd, followed by lenalidomide maintenance. The TOU dataset is composed of t(4;14) patients treated off study in which treatments were by investigator’s choice of standard of care doublet and triplet regimens. Patients in the TOU data set may have received lenalidomide maintenance or continuous therapy at relapse (grouped together in the table). Patients from MMRF included in the replication cohort received RVd, and patients in the T/D dataset received investigator’s choice induction regimens.
Samples and fluorescence in situ hybridization analysis for TOU data set
Cytogenetic analysis for the TOU data set was performed by FISH. Diagnostic bone marrow (BM) samples (pretreatment) were shipped overnight to a central laboratory. Plasma cells (PCs) were isolated from BM using CD138+ MAC-Sorting (Miltenyi Biotec, Paris, France). Postsorting purity was checked by cytologic analysis of a spin from positive fraction. Only samples with ≥70% PCs after sorting were kept for the cytogenetic analysis. FISH analysis was performed with specific probes targeting t(4;14), and percentage of involved PCs was evaluated by counting at least 100 total cells. Patients with 30% positive cells were considered positive for t(4;14).
Generation of genomic data for TOU data set
For the TOU data set, CD138+ PCs and matched germline controls (peripheral blood mononuclear cells) were stored in RLT buffer (Qiagen), and DNA/RNA was extracted using the Qiagen AllPrep DNA/RNA Mini Kit. WGS was performed on tumor/normal pairs at an average of 60×/30× depth. RNA-seq libraries were produced using TruSeq Stranded RNA HT kit. RNA-seq (100 million reads, paired-end) was performed on tumor samples. All sequencing was performed on Illumina HiSeq2500 instruments. Data were processed and analyzed as described (supplemental Methods, available on the Blood website).
Results
Data sets and characteristics
Our discovery data set (N = 153) comprised TOU and IFM2009 and replication (N = 105) from MMRF and T/D. Patient characteristics and OS were similar between the 4 t(4;14) patient cohorts in terms of median age (56, 63 years discovery [IFM, TOU] versus 58, 59 [MMRF, T/D]), ISS3 (50% [9/18], 34.62% [36/104] discovery [P = .15] vs 20.59% [7/34], 32.4% [23/71] replication) (Table 1, supplemental Figure 1A). Induction regimens were either RVd (IFM2009, MMRF) or investigator’s choice doublet/triplet regimens (TOU, T/D). The T/D cohort all received a stem cell transplant; otherwise, approximately one-half of patients in the other cohorts received a stem cell transplant (43.48%, 62.02% discovery [IFM, TOU] vs 61.76% MMRF and 46.78% non-t(4;14)). All patients in the IFM2009 cohort received lenalidomide maintenance. TOU and MMRF cohorts received investigator’s choice for maintenance/continuous therapy (TOU: 79.65% [lenalidomide maintenance 16.28% (n = 21); continuous lenalidomide at relapse 67.26% (n = 76)] and MMRF: 44.12% lenalidomide maintenance). T/D cohort did not receive maintenance therapy (Table 1). Patient characteristics for the non-t(4;14) cohort were similar to the t(4;14) cohorts in terms of age (median, 60 years), but prevalence of ISS3 was lower. Per IFM2009 protocol, treatment regimens were the same for non-t(4;14) and t(4;14) patients (Table 1; supplemental Figure 1B).
Genomic landscape of t(4;14) NDMM
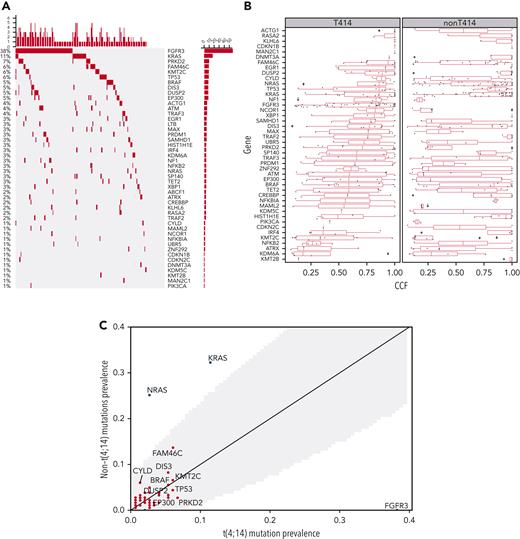
We first explored the landscape of mutations and CNAs in t(4;14) and contrasted that with non-t(4;14) NDMM and our previously published data (Walker et al).7 In the current analysis, we found enrichment of mutations in FGFR3 and PRKD2 as well as amplifications in 1q21 and deletions in 1p, 4q, 11q, 12p, 13q, and 14q in t(4:14) patients compared with the non-t(4:14) NDMM population consistent with previous analysis (Figure 1).7Figure 1A shows the prevalence of the top 30 driver mutations down to the 1% level in our data set. Among these, FGFR3 (38%), KRAS (11%), PRK2D (7%), FAM46C (6%), KMT2C (6%), TP53 (6%), BRAF (5%), DIS3 (5%), DUSP2 (5%), and EP300 (5%) were the most frequently mutated. We confirmed the high prevalence of FGFR3 mutation in our t(4;14) cohort, which appeared to be mutually exclusive with NRAS/KRAS/BRAF mutations (cumulatively, 20% of patients [Figure 1A]). In addition, KRAS and BRAF mutations in t(4;14) patients had a lower median CCF than in non-t(4;14) patients (Figure 1B). In comparison, in non-t(4;14) patients there were no FGFR3 mutations and significantly more NRAS (46/183 vs 4/149, Fisher P = 2e-9) and KRAS (59/183 vs 17/149, Fisher P = 6e-6) mutations (Figure 1C). FAM46C, DIS3, and CYLD mutation frequencies were higher in non-t(4:14), and TP53, EP300, and PRK2D mutations were higher in t(4;14), but they were not statistically significant. A small nonsignificant increase in mutations (∼10% overall) was noted in histone lysine methyltransferases (KTM2C, KTM2B, KDM6A) in t(4;14) patients, indicating their potential biological role along with NSD2 protein, also a histone lysine methyl transferase. In addition, there was a subset of t(4;14) patients without mutations in any of the top 30 driver genes (Figure 1A).
Comparison of mutations and CNAs between t(4;14) and non-t(4;14) patients. (A) Waterfall plot of nonsilent mutations in the discovery cohort ordered by prevalence. Bars on the left and percentages on the left of the plot indicate prevalence of mutations in each gene. Bars at the top of the plot indicate prevalence of mutations in any of the driver genes in each patient. (B) Comparison of cancer clonal fraction (CCF) of mutations shown in panel A, starting from clonal (CCF = 1) and decreasing. t(4;14) population on the left, non-t(4;14) on the right. (C) Mutation prevalence in MM driver genes in t(4;14) patients versus non-t(4;14) patients. Shaded gray region highlights difference in mutation prevalence that are not significant. (D) Comparison of CNA gains (copy number 3+, red lines) and deletions (copy number 0-1, blue lines) between t(4;14) (solid lines) and non-t(4;14) patients (dashed lines). Black dots represent false discovery rate (FDR) P < .05.
Comparison of mutations and CNAs between t(4;14) and non-t(4;14) patients. (A) Waterfall plot of nonsilent mutations in the discovery cohort ordered by prevalence. Bars on the left and percentages on the left of the plot indicate prevalence of mutations in each gene. Bars at the top of the plot indicate prevalence of mutations in any of the driver genes in each patient. (B) Comparison of cancer clonal fraction (CCF) of mutations shown in panel A, starting from clonal (CCF = 1) and decreasing. t(4;14) population on the left, non-t(4;14) on the right. (C) Mutation prevalence in MM driver genes in t(4;14) patients versus non-t(4;14) patients. Shaded gray region highlights difference in mutation prevalence that are not significant. (D) Comparison of CNA gains (copy number 3+, red lines) and deletions (copy number 0-1, blue lines) between t(4;14) (solid lines) and non-t(4;14) patients (dashed lines). Black dots represent false discovery rate (FDR) P < .05.
For CNAs, we confirmed the previous findings of a significant decrease in hyperdiploidy (FDR < 0.05) and an increase (≥3 copies) in 1q (FDR = 2e-09) and 4p (FDR = 8e-09) and a number of deletions (including 1p [FDR = 3e-02], 4p [FDR = 2e-03], 11q [FDR = 3e-06], 12p [FDR = 5e-06], 13q [FDR = 2e-09], and 14q [FDR = 1e-05]) (Figure 1D, supplemental Table 1). In addition, we found significant novel associations (FDR < 0.05) with deletions in 2q, 3p, 4q, 5p, 6p, 7p, 9p, 11q, 18p, chr20, and 22q. Del17p had no significant enrichment or depletion in t(4;14) (Figure 1D, supplemental Table 1). NSD2 and FGFR3 expression was significantly higher in the t(4;14) group (supplemental Figure 2). Within the t(4;14) patients, there was a strong association of del11p with mutations in FGFR3. Together, this analysis confirmed genomic abnormalities previously associated with t(4;14) and also identified significant novel deletions (2q, 3p, 5p, 6p, 7p, 9p, 18p, 20p, 20q, and 22q).
Landmark survival analysis and genomic features of HR t(4;14)
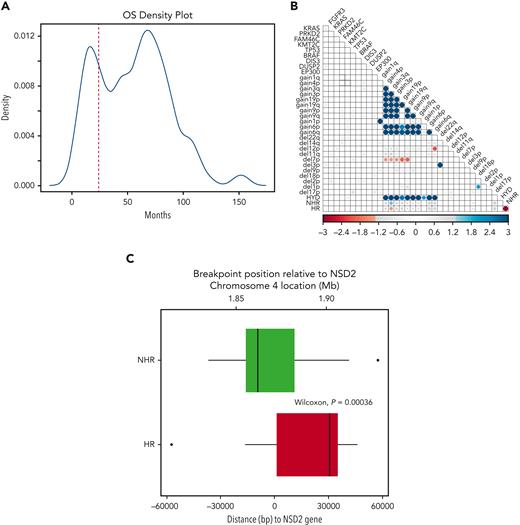
The OS of patients in our discovery cohort presented a bimodal distribution with peaks at ∼15 and ∼68 months, demonstrating a clear difference in survival in 2 subgroups (Figure 2A) consistent with previously described prognostic heterogeneity among t(4;14) MM patients.5 To identify genomic features accounting for these outcome differences, we performed a landmark analysis defining HR patients (those with an OS event before 24 months) and NHR as an event or censoring after 24 months. The cutoff of 24 months was chosen as it is the point at which the 2 OS distributions crossed (Figure 2A). To further dissect the putative HR and NHR subgroups, we looked for specific associations of genomic features in the HR group (Figure 2B). Of the highly prevalent mutations and CNAs, the only significant association was amplification of 3q with the NHR group. This analysis showed expression of NSD2 or FGFR3, mutations in FGFR3, and amp1q were not significantly associated with HR t(4;14) patients (Figure 2B, supplemental Figure 3). Unlike our analysis, previous reports suggested the role of FGFR3 mutation or expression in HR t(4;14)2,19,20 (supplemental Table 2).
HR and NHR t(4;14) based on landmark survival analysis. (A) Density plot of patient overall OS in t(4;14) NDMM patients. (B) Association of genomic and risk features in t(4;14) NDMM patients. Blue indicates significant positive association; red indicates significant negative association. Size and color intensity scaled to −log(fdr_p). FDR P values >.05 are set to gray. (C) Translocation breakpoint of HR and NHR patients relative to the start of NSD2. Zero on the x-axis indicates the position of NSD2 gene, with positive numbers representing breakpoints occurring within the NSD2 gene. Negative numbers indicate locations upstream of NSD2.
HR and NHR t(4;14) based on landmark survival analysis. (A) Density plot of patient overall OS in t(4;14) NDMM patients. (B) Association of genomic and risk features in t(4;14) NDMM patients. Blue indicates significant positive association; red indicates significant negative association. Size and color intensity scaled to −log(fdr_p). FDR P values >.05 are set to gray. (C) Translocation breakpoint of HR and NHR patients relative to the start of NSD2. Zero on the x-axis indicates the position of NSD2 gene, with positive numbers representing breakpoints occurring within the NSD2 gene. Negative numbers indicate locations upstream of NSD2.
WGS data allowed us to examine the exact breakpoint locations and to identify how the reciprocal break and joining event between chromosomes 4 and 14 created the recombinant chromosomes. We compared the size of the translocated segment in the chr4p arm between HR and NHR patients and found that a larger region, ie, ones with a breakpoint further into chr4, was translocated in HR patients (Wilcoxon P = 4e-4). Anchoring the breakpoint location relative to the start of the NSD2 gene, we found the median of the distribution of breakpoint locations in the NHR group was upstream of the NSD2 gene, whereas in the HR group it was within the NSD2 gene itself (Figure 2C). This result led us to investigate the precisely mapped coordinates of the breakpoint location beyond the binary grouping relative to the start of the NSD2 gene.
Stratification of risk based on breakpoint location and other genomic markers
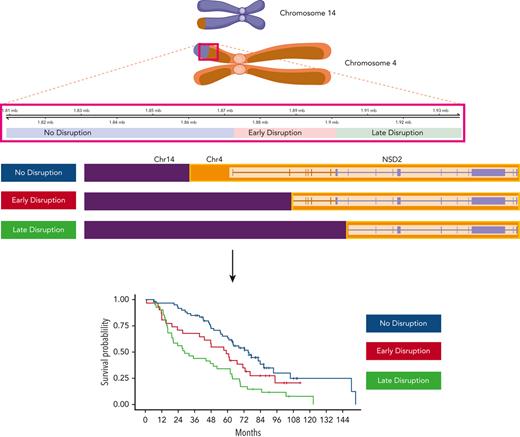
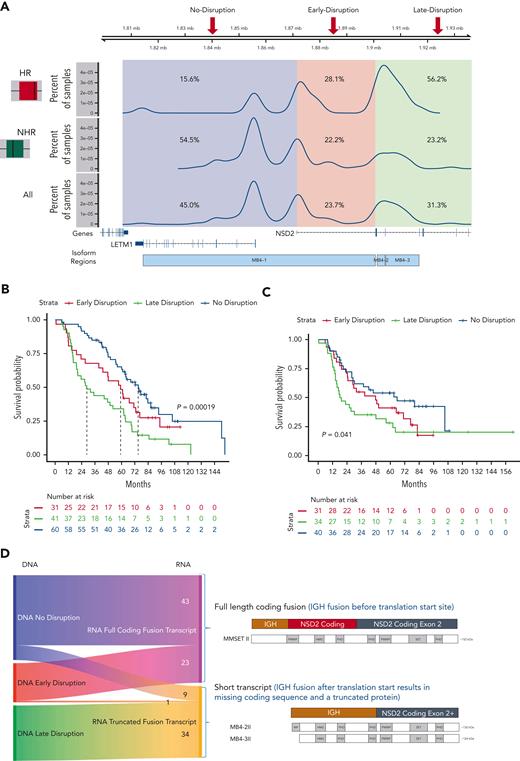
Analysis of the breakpoint locations on chr4 showed 3 main distribution peaks: (1) corresponding to a region upstream of the NSD2 transcription start site (named no-disruption, 45.0%), (2) between the NSD2 transcription start site and the translation start site (early-disruption, 23.7%), and (3) downstream of the translation start site (late-disruption, 31.3%) (Figure 3A).14 Previously, the genomic regions were annotated as the region upstream of the exon, which could be translated to form the different resulting transcriptional fusion of IgH-NSD2 transcripts. Our early- and late-disruption peaks are in positions similar to those in cases previously described in a small cohort,14 in which the region upstream of the first coding exon of NSD2 was designated as MB4-1, the region upstream of the second exon as MB4-2, and the region upstream of the third exon as MB4-3. The breakpoint regions we identify differ from the previous definitions defined by predicted isoform expression. Our no-disruption and early-disruption region both map to the previous MB4-1 region. Our late-disruption region maps to both the previous MB4-2 and MB4-3 regions. The 3 breakpoint groups had little association with other t(4;14) genomic features, including NSD2 overexpression. We did find an association between no-disruption breakpoints and both FGFR3 mutations and increased FGFR3 expression (supplemental Figure 4).
Translocation breakpoint location identifies HR t(4;14) patients. (A) Breakpoint locations form 3 peaks corresponding to regions relative to NSD2. The no-disruption region (blue) occurs upstream of the start of NSD2. The early-disruption region (red) is between the start of NSD2 and the translation start of NSD2. The late-disruption region (green) is the region downstream of the NSD2 translation start site. (B) Kaplan Meir plot showing the OS of patients based on their breakpoint location. (C) Independent replication cohort showing OS of patients based on breakpoint location. (D) Sankey plot showing the relationship between sample breakpoint location and identified fusion transcript.
Translocation breakpoint location identifies HR t(4;14) patients. (A) Breakpoint locations form 3 peaks corresponding to regions relative to NSD2. The no-disruption region (blue) occurs upstream of the start of NSD2. The early-disruption region (red) is between the start of NSD2 and the translation start of NSD2. The late-disruption region (green) is the region downstream of the NSD2 translation start site. (B) Kaplan Meir plot showing the OS of patients based on their breakpoint location. (C) Independent replication cohort showing OS of patients based on breakpoint location. (D) Sankey plot showing the relationship between sample breakpoint location and identified fusion transcript.
The 3 breakpoint locations were then associated with the 2 risk groups, identifying that in the HR t(4;14) group most patients (56.2%) have a late-disruption translocation, whereas in the NHR group most patients (54.5%) have a breakpoint in the no-disruption category (Fisher exact P = 8.5e-4) (Figure 3A). Finally, categorizing patients based on their translocation breakpoint group, we found a significant difference in outcome between the late-disruption breakpoint and the no-disruption breakpoint (median OS [mOS] 28.6 vs 75.1 months, P = 2e-4), whereas the early-disruption group had an intermediate outcome (mOS 59.4 months) (Figure 3B, also see Discussion). This outcome association was replicated in an independent cohort of t(4;14) patients (N = 105), in which patients with late-disruption breakpoint translocations had the shortest mOS of 18.0 months, early disruption group had an mOS of 48.0 months, and patients in the no-disruption group had an mOS of 66.0 months (P = 4e-2) (Figure 3C). We also performed univariate and multivariate analyses to further analyze the association of breakpoint location, age, ISS2, ISS3, del17p, trisomy 5/21, and amp1q with impact on OS (supplemental Table 3). In the univariate analysis, the late-disruption breakpoint (P = 6e-5), 17p, and 1p were significantly associated with poor OS, and trisomy 5 was associated with good OS, whereas in the multivariate analysis, both the late-disruption (P = 3.6e-3) and the early-disruption breakpoint were significantly associated with poor OS (P = .04) as well as deletions 17p and 1p and 1q amplification. Of note, age, ISS2, and ISS3 were not significant after correcting for the molecular features (supplemental Table 3).
Relationship between breakpoint location and fusion NSD2 transcripts and translation products
The DNA breakpoint analysis predicted the precise location and formation of the fusion transcripts. To directly address the effect of the breakpoints on the fusion transcripts and translated NSD2 protein, we analyzed the fusion transcripts from the RNA-seq data generated from the same patients. We identified fusions between the IgH transcript and exons of NSD2 and were able to detect a fusion transcript in 87.3% of the t(4;14) samples, corresponding to either the expected full-length isoform, MB4-1/MMSETII,13 or truncated isoforms (MB4-2/3)13 (Figure 3D). Categorizing patients based on the expressed fusion transcript into 3 groups (MB4-1 vs MB4-2 vs MB4-3) or 2 groups (MB4-1 vs combined MB4-2 and MB4-3) shows a significant difference in OS (median 71.2 months vs 44.3 months, supplemental Figure 5A-C).
Almost all patients expressed the expected fusion transcript isoform based on their breakpoint location (Figure 3D), with patients with a no-disruption or early-disruption breakpoint expressing a full-length fusion transcript (82.7% and 95.8%, respectively), and those with a late-disruption breakpoint expressing a truncated fusion transcript. There was a small subset of patients (n = 10), however, with a no- or early-disruption breakpoint that expressed a truncated fusion transcript (Figure 3D left middle panel blue/pink to orange). We found that 9 out of 10 patients had a variable internal deletion, downstream of the translocation breakpoint, that removed exon 2, the first coding exon of NSD2 (supplemental Figure 6), and the presence of this deletion could explain the observed shorter fusion transcript.
Discussion
In order to identify the genomic features associated with clinical heterogeneity of t(4;14) NDMM, we analyzed and compared a large cohort of 153 t(4;14) tumor samples with 183 non-t(4;14) NDMM. Our analysis substantially confirms the association of mutations and CNAs associated with t(4;14) described before.7,16 In addition, we also identified novel mutations that trended higher in t(4;14) patients, including TP53, EP300, and several histone lysine methyltransferases (KMT2C, KMT2B, KDM6A), although these mutations were not significantly different between t(4;14) and non-t(4;14) patients. Patients with t(4;14) had significantly fewer KRAS/NRAS mutations and trended toward fewer mutations in FAM46C, DIS3, CYLD, and BRAF. For CNAs, our analysis indicated that patients with t(4;14) had more frequent deletions in 2q, 3p, 5p, 6p, 7p, 9p, 20p, 20q, and 22q than non-t(4;14) patients. Next, we focused on those genomic features that could identify within t(4;14) patients those with poor prognosis. We found significant association of the location of the translocation breakpoint within the NSD2 gene as an independent biomarker that separated the high-risk from low-risk t(4;14) patients. This finding was confirmed with an independent data set of 105 t(4;14) NDMM cases. Univariate and multivariate analysis of the genomic and clinical features demonstrated additional biomarkers, such as the presence of deletion 17p or 1p or 1q amplification, can create “double-hit” genotypes adding to poor survival in this group of patients. In contrast, we found no difference in the expression of FGFR3 or NSD2 between the HR and NHR groups, nor did we find any significant difference in the frequency of FGFR3 mutation in the HR (34.1% vs NHR 39.1%, P = .68) group or prevalence of amp1q (22.2% vs 22.7%, P = 1) (supplemental Figure 3). Based on our analysis, we suggest potential clinical utility of the NSD2 breakpoint analysis as an additional tool to identify and risk stratify t(4;14) patients.
We used a landmark survival analysis for the initial discovery of HR and NHR t(4;14) patients and chose a shorter cutoff of 24 months OS to separate the patients into 2 groups. Although we used this cutoff based on the peaks of the bimodal survival distribution of the discovery data set, it is reasonable to use longer cutoffs for OS such as 36 months to define HR vs NHR groups. Indeed, when we compared patients based on a 36-month cutoff, the percentage distribution of patients across the 3 breakpoint groups remained consistent (supplemental Table 4), and the associated genomic features of HR t(4;14) were also similar (data not shown). However, using 48 months as a cutoff did not separate the 3 breakpoint groups due to increased loss of patients from censoring. We should note that multiple factors could impact OS, including differences in treatment regimens. Notwithstanding, the outcome in HR t(4;14) patients remained consistently poor in our analysis.
Importantly, translocation breakpoint analysis identified associated NSD2 fusion transcripts. This analysis indicated that the breakpoint location and expression of NSD2 fusion transcripts are key molecular features distinguishing HR vs NHR patients within the t(4;14) population. Although the expression of various NSD2 fusion transcripts have been previously associated with OS,11 the association of these transcripts and the breakpoint locations was not explored side by side. In our analysis, we identified precise translocation breakpoints for each patient and classified them in 3 groups: no-disruption, early-disruption, and late-disruption, depending on their distance relative to the NSD2 transcription start site. These 3 groups showed a significant association with outcome, even stronger than fusion transcript expression (mOS 28.6 months vs 75.1 months breakpoint groups, mOS 44.3 months vs 71.2 months expression groups). Unlike the initial description of HR based on OS density that suggested a bimodal population of patients, breakpoint analysis identified 3 subsets of patients with differing outcome. The OS of the early-disruption group that was identified by breakpoint analysis was intermediate risk compared with the other 2 groups, but more work would be needed to validate the prognosis of early-disruption patients. How the genomic features, the breakpoints, and the type of fusion transcripts affect the biology and prognosis in these patients would require further analysis.
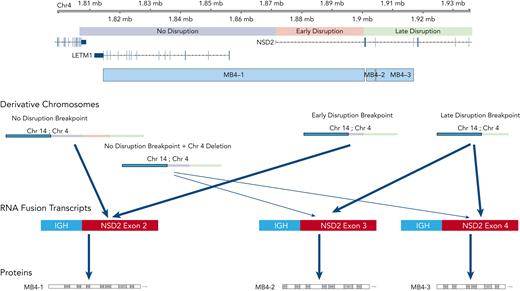
In general, there was a correlation between the breakpoint location and the expression of the type of fusion transcripts. All patients with a late-disruption breakpoint expressed a truncated isoform, and the majority of those with no-disruption expressed the full-length transcript. However, there were some patients with a no-disruption breakpoint who nonetheless expressed a short, truncated transcript. This was due to an internal deletion of a genomic region right after the translocation breakpoint, which explains the missing NSD2 region in the expressed truncated transcript (Figure 4). The expression of the truncated fusion transcripts showed a significant association with poor outcome; however, the separation of OS between these NSD2-disrupted groups was smaller than the one between no-disruption vs late-disruption breakpoint groups. This observation suggests that the breakpoint locations are driving the observed separation in outcome and that the effect of fusion isoforms on outcome is weaker. Diagnostically, FISH probes traditionally used in the identification of 4;14 translocations are not suitable for precisely locating the breakpoints and thus identifying high-risk late-disruption patients. As a practical solution, we suggest incorporating a next generation sequencing–based approach, such as the recently described MGP Myeloma Panel,21 for identifying these patients.
Summary view of t(4;14) patients based on breakpoint location, expression of fusion transcripts, and predicted NSD2 protein. Chromosome 4 shown on top with breakpoint locations (no- vs early- vs late-disruption) shown relative to genes on chromosome 4. Resultant derivative chromosomes from different translocation breakpoints along with RNA fusion transcripts and proteins are shown.
Summary view of t(4;14) patients based on breakpoint location, expression of fusion transcripts, and predicted NSD2 protein. Chromosome 4 shown on top with breakpoint locations (no- vs early- vs late-disruption) shown relative to genes on chromosome 4. Resultant derivative chromosomes from different translocation breakpoints along with RNA fusion transcripts and proteins are shown.
Our breakpoint-based outcome analysis led to reassessment of the NSD2 fusion transcripts from the previous analysis of samples that explored either full length (MB4-1) or truncated (MB4-2/MB4-3) isoforms. The first, as mentioned above, is the small number of cases in which a no-disruption breakpoint expresses a truncated isoform due to a deletion, creating a mismatch between the correspondence between breakpoint fusion transcripts. It is hard to draw conclusions from a small set of patients, but these patients did have the longer survival that matches the no-disruption breakpoint group and not the poor survival that matches the truncated isoform expression. The second difference is the separation of OS in those cases expressing a full-length isoform into early-disruption and no-disruption. The early-disruption group showed intermediate outcome even though most of these patients express the full-length transcript, suggesting that the function of the resulting protein is not solely responsible for favorable or poor outcome. Other unidentified factors may be involved in regulating the expression and function of NSD2 within the 5' UTR and upstream promoter region. In contrast, the truncated fusion NSD2 transcript’s exclusive association with the HR group points to its critical role in the biology of cells containing the late-disruption translocation. The translated shorter NSD2 protein is predicted to lose the first PWWP domain, which is required for stabilizing the protein with chromatin and for enzymatic activity.22 It is likely that expression of this variant protein may lead to an altered gene expression prolife in the cells expressing this protein. Transcriptional analysis from patient samples is being planned to explore the key pathways and associated downstream targets and will be an interesting area for future research.
In conclusion, our analysis of the largest data set of NDMM patients with t(4;14) provides new genomic insights into this patient subset and provides a clinically actionable opportunity to identify HR t(4;14) patients at diagnosis to inform risk-adjusted treatment decisions. Our results could have practical application for the diagnosis of HR patients by using either a next generation sequencing- or PCR-based approach to identify HR t(4;14) patients in lieu of the current FISH-based methods. These data along with previous publications identifying HR del17p (CCF ≥ 0.55)23 and amp1q17,24 as high-risk features in MM make a scientific case to discuss modifications to the R-ISS criteria to redefine HR MM.
Acknowledgments
The authors thank Jonathan Keats for his review and comments on the analysis.
Funding for data processing and storage was provided by BMS Corporation.
Authorship
Contribution: The project was conceived and designed by A.T. and H.A.-L.; funding acquisition was conducted by E.F. and A.T.; project administration was conducted by M.O. and E.F.; oversight and management of resources (data generation, collection, transfer, infrastructure, data processing) were conducted by E.F., M.O., N.S., F.T., H.A.-L., M.S., N.M., J.C., and A.T.; analyses and interpretation were designed and performed by M.O., N.S., F.T., E.F., and A.T.; data visualization was performed by M.O. and N.S.; supervision and scientific direction were provided by A.T.; and the manuscript was written and critically reviewed by N.S., M.O., E.F., J.C., H.A.-L., M.S., N.M., A.A., and A.T.
Conflict-of-interest disclosure: N.S., M.O., F.T., E.F., and A.T. are employed by and have equity ownership in BMS Corporation. The remaining authors declare no competing financial interests.
Correspondence: Anjan Thakurta, Botnar Research Centre, Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, University of Oxford, Windmill Rd, Headington, Oxford OX3 7LD, United Kingdom; e-mail: anjan.thakurta@ndorms.ox.ac.uk.
References
Author notes
The current affiliation for A.T. is Nuffield Department of Orthopedics, Rheumatology and Musculoskeletal Sciences and Oxford Centre for Translational Myeloma Research, University of Oxford, Oxford, United Kingdom.
Whole-genome and RNA-sequencing data (FASTQ and VCF or quant.sf files) pertaining to the t(4;14) cohort are deposited on the Amazon Web Services (AWS) S3 cloud storage at http://s3/celgene-rnd-riku-prismm/DA0000715.
Data for academic/noncommercial purposes are available on request from Erin Flynt (erin.flynt@bms.com). Bristol Myers Squibb’s policy on data sharing can be found at https://www.bms.com/researchers-and-partners/clinical-trials-and-research/disclosure-commitment.html.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal