Key Points
Lung neutrophils, which express unique surface proteins and genes differently from other neutrophils, are maintained by PGE2.
Tgm2 in lung neutrophils performs as an inflammatory checkpoint in the protection of the lungs against acute respiratory distress syndrome.
Abstract
Lung-resident neutrophils need to be tightly regulated to avoid degranulation- and cytokine-associated damage to fragile alveolar structures that can lead to fatal outcomes. Here we show that lung neutrophils (LNs) express distinct surface proteins and genes that distinguish LNs from bone marrow and blood neutrophils. Functionally, LNs show impaired migratory activity toward chemoattractants and produce high levels of interleukin-6 (IL-6) at steady state and low levels of tumor necrosis factor-α in response to lipopolysaccharide (LPS) challenge. Treating bone marrow neutrophils with bronchoalveolar lavage fluid or prostaglandin E2 induces LN-associated characteristics, including the expression of transglutaminase 2 (Tgm2) and reduced production of inflammatory cytokines upon LPS challenge. Neutrophils from Tgm2−/− mice release high levels of inflammatory cytokines in response to LPS. Lung damage is significantly exacerbated in Tgm2−/− mice in an LPS-induced acute respiratory distress syndrome model. Collectively, we demonstrate that prostaglandin E2 is a key factor for the generation of LNs with unique immune suppressive characteristics, acting through protein kinase A and Tgm2, and LNs play essential roles in protection of the lungs against pathogenic inflammation.
Introduction
Neutrophils make up the largest portion of white blood cells in the human circulatory system and are first-line defenders against infection.1 They rapidly eliminate invading pathogens with diverse bactericidal activities such as phagocytosis, generation of reactive oxygen species (ROS), and neutrophil extracellular traps without transcriptional activation.2 Neutrophils are homeostatically generated in the bone marrow.3 Upon infection or tissue injury, neutrophils migrate into the event area by sensing chemokine/chemoattractant gradients and play key roles in the regulation of innate immune defense.2,3 Accumulating evidence points to the existence of neutrophils in peripheral tissues at steady state that can regulate tissue specific homeostasis.4 Among different organs, the lung is a predominant site in which neutrophils are abundantly sustained at steady state.5 Compared with other neutrophils, lung neutrophils (LNs) have different phenotypic characteristics.6 Because alveolar structures are particularly sensitive to bystander damage associated with inflammation, it stands to reason that LNs need to be tightly regulated. Further detailed transcriptional, immunophenotypic, and functional characterization of LNs as to how this is achieved, however, remains to be determined.
Unlike other leukocytes, neutrophils have typically been considered to have low transcriptional activities because the cells are regarded as fully differentiated effectors.7 However, previous reports demonstrated that neutrophils are, in fact, transcriptionally active and express different surface markers and functional proteins for the maintenance of unique populations under homeostatic or pathological conditions.8-10 Gut microbiota, for example, affect the expression of neutrophil surface proteins through transcriptional activation, generating aged neutrophils leading to their homing to bone marrow.11 In addition, tissue-resident leukocytes are affected by tissue-associated environmental factors to have unique characteristics. Tissue-resident macrophages, for example, are located in many peripheral tissues, and their tissue-specific phenotypes are regulated by environmental factors via different transcriptional programs.12 However, it has not been investigated which pulmonary factors play essential roles in the generation or phenotype of lung-sustained neutrophils or how the transcriptional program of LNs may differ from neutrophils obtained from other tissues.
Prostaglandin E2 (PGE2) is a bioactive lipid with both pro- and antiinflammatory activities. It exacerbates ulcerative colitis13 and multiple sclerosis14 by modulating Th1 cell differentiation or Th17 expansion through EP receptors and protein kinase A (PKA). On the other hand, PGE2 has beneficial activity in lung inflammation and repair15 in bronchial asthma16 or lung fibrosis.17 The concentration of PGE2 in the human lung is 50- to 80-fold higher than that of plasma18 at steady state, and it is detected in the airways of steady-state mice.19 However, the roles of PGE2 in neutrophil homeostasis have not been well elucidated.
In this study, we discovered that lung-resident neutrophils express a unique pattern of surface markers and genes that distinguish them from bone marrow neutrophils (BMNs) or peripheral blood neutrophils (BNs). Pulmonary PGE2 is a key environmental driver of the LN phenotype. Downstream of PGE2/PKA signaling in LNs, we found that transglutaminase 2 (Tgm2) serves as a novel inflammatory checkpoint for lung-resident neutrophil homeostasis and immunosuppressant activity.
Materials and methods
Detailed information on materials and methods is available in the supplemental Materials (available on the Blood Web site).
Mice
C57BL/6 mice were purchased from Orient Bio (Seongnam, Korea). Tgm2−/− mice were generated as previously described.20 Animal protocols were approved by the Institutional Review Committee for Animal Care and Use at Sungkyunkwan University.
Isolation of single cells from tissues of mice
Bone marrow cells were collected by flushing the tibia and femur with HBSS (Sigma, St. Louis, MO). Peripheral blood was collected via the retroorbital venous sinus with heparinized capillary tubes (Kimble Chase, Vineland, NJ) into EDTA-coated microtainers (BD, Franklin Lakes, NJ). Lungs were first perfused with phosphate-buffered saline through the right ventricle to left atrium, minced, and further dissociated in digestion buffer (0.4 mg/mL of collagenase type IV from Clostridium histolyticum [Sigma, St. Louis, MO], 0.7 mg/mL of CaCl2·H2O, 12.5 U/mL of DNase I, 10 mM of HEPES [Gibco, Waltham, MA], and 5 mg/mL of bovine serum albumin [Sigma, St. Louis, MO] in HBSS [Sigma, St. Louis, MO]) for 30 minutes in 37°C shakers. After filtration through a 100-μm nylon strainer, residual red blood cells were lysed with red blood cell lysis buffer (Gibco, Waltham, MA). The samples were washed twice with phosphate-buffered saline, and single-cell suspensions were used for subsequent experiments.
Adoptive transfer of stained BMNs
BMNs from gradient isolation were stained with Cell Proliferation Dye eFluor 670 per manufacturer’s instructions. We transferred eF670+ 1 × 107 BMNs per head in 500 μL of HBSS with 0.5% BSA through retroorbital intravenous injection. After 16 hours, single cells from tissues were analyzed with Aurora staining (Cytek, Fremont, CA) with antibodies as indicated in the figure legends.
Results
LNs express a distinct pattern of surface markers compared with other neutrophils at steady state
We first questioned whether lungs could retain neutrophils at steady state after lung perfusion. We found that ∼11% of CD45+ leukocytes recovered from lungs were neutrophils (Figure 1A), consistent with a previous report.21 Intravenous injection of anti-Ly6G-FITC (supplemental Figure 1A) showed similar intensities of FITC between LNs and BNs after 5 minutes but showed lower levels in BMNs (supplemental Figure 1B), indicating that LNs might be located in intravascular regions.
LNs have a distinct immunophenotype and mRNA expression profile compared with other neutrophils. (A) Neutrophil proportions from total bone marrow, peripheral blood, lung, and spleen (n = 3) from 8-week-old C57BL/6 male mice were analyzed by flow cytometry. (B) Heatmap of Ly6G expression on bidimensional UMAP analysis of live CD45+ immune cells from bone marrow, peripheral blood, lung, and spleen (n = 3). (C) Three Ly6Ghigh clusters among 38 clusters from PARC analysis in bone marrow, blood, lung, and spleen. (D) Cluster proportion of each tissue neutrophil. Expression levels (MFI) of CD11b, CD62L, CXCR4, MHC II, and CD101 on Ly6G+ clusters (E) and tissue neutrophils (F). Correlation matrix (G) and up/down (log2FC ≥ 1/log2FC ≤ 1) (H) regulated count of genes from bulk RNA-seq data of BMNs, BNs, and LNs with log2FC ≥ 1, P < .05 from pooling 5 heads of 8-week-old C57BL/6 male mice. (I) Gene set enrichment analysis (GSEA) data of gene sets associated with translational activity in LNs compared with BNs. (J) Heatmap of surface protein mRNA expression on BMN, BN, and LN in RNA-seq. (K) Surface protein level (MFI) of Sca1, Plet1, and CD14 on tissue neutrophils (n = 4). Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. ns, not significant (one-way ANOVA) (A [right], E-F,K).
LNs have a distinct immunophenotype and mRNA expression profile compared with other neutrophils. (A) Neutrophil proportions from total bone marrow, peripheral blood, lung, and spleen (n = 3) from 8-week-old C57BL/6 male mice were analyzed by flow cytometry. (B) Heatmap of Ly6G expression on bidimensional UMAP analysis of live CD45+ immune cells from bone marrow, peripheral blood, lung, and spleen (n = 3). (C) Three Ly6Ghigh clusters among 38 clusters from PARC analysis in bone marrow, blood, lung, and spleen. (D) Cluster proportion of each tissue neutrophil. Expression levels (MFI) of CD11b, CD62L, CXCR4, MHC II, and CD101 on Ly6G+ clusters (E) and tissue neutrophils (F). Correlation matrix (G) and up/down (log2FC ≥ 1/log2FC ≤ 1) (H) regulated count of genes from bulk RNA-seq data of BMNs, BNs, and LNs with log2FC ≥ 1, P < .05 from pooling 5 heads of 8-week-old C57BL/6 male mice. (I) Gene set enrichment analysis (GSEA) data of gene sets associated with translational activity in LNs compared with BNs. (J) Heatmap of surface protein mRNA expression on BMN, BN, and LN in RNA-seq. (K) Surface protein level (MFI) of Sca1, Plet1, and CD14 on tissue neutrophils (n = 4). Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. ns, not significant (one-way ANOVA) (A [right], E-F,K).
Others have reported that neutrophils express different levels of surface markers such as CD101, CXCR2, CD62L, and CXCR4 based on their maturation and age status.22,23 To compare the expression patterns of multiple surface markers in LNs simultaneously vs BMNs, BNs, and splenic neutrophils (SPNs), we used 21-color spectral flow cytometry with a fixable viability dye (supplemental Figure 1C). Using uniform manifold approximation and projection (UMAP), we visualized global differences among various immune cells in a single bidimensional plot.24 CD11b+Ly6G+ neutrophils were located in the upper-right “big island” on the UMAP data (Figure 1B; supplemental Figure 1C). To determine whether neutrophils are composed of different marker-defined populations, we used an unsupervised clustering method, phenotyping by accelerated refined community-partitioning (PARC), which distinguishes small numbers of rare population with limited surface markers from other populations.25 PARC classified fixable viability dye−CD45+ live immune cells by 38 clusters with 19 surface markers (supplemental Figure 1D). Among the clusters, only 3 (PARC-03, -06, and -14) were CD11b+Ly6G+ neutrophils (Figure 1C). In UMAP data, PARC-03, -06 and -14 were neighboring in the same island, but PARC-14 was located in a relatively marginal region distinct from the major island with higher CD11b, CXCR4, and MHC II expression and lower CD62L expression (Figure 1C,E). CD101 expression of PARC-14 is higher than PARC-06 (Figure 1E). Approximately 97% of PARC-14 neutrophils were from the lungs, and almost all BNs and SPNs were similar in that >91% of the cells were PARC-03 (Figure 1D), and each neutrophil population has similar surface protein expression pattern to the corresponding cluster (Figure 1E-F). Therefore, unsupervised clustering using 19 markers effectively distinguished various immune cell subsets (supplemental Figure 1C-E), including an isolated neutrophil population comprising nearly all LNs immunophenotypically distinct from BNs, SPNs, and BMNs.
Unique characteristics of LNs arise from distinct mRNA expression and translation
To further examine characteristics of LNs from other neutrophils, we conducted RNA sequencing of sorted neutrophils. In multidimensional scaling, a dimension reduction analysis was performed for multiple factors.26 LNs, BNs, and BMNs were located in distinct regions (supplemental Figure 1F). Correlation scale scores between LNs and other neutrophils were lower than that between BMNs and BNs (Figure 1G). In addition, many genes were differentially expressed, despite both LNs and BNs likely being located in intravascular regions; 544 genes were significantly upregulated and 568 genes were significantly downregulated in LNs compared with BNs (Figure 1H). Taken together, differential mRNA expression patterns clearly distinguish LNs from other neutrophils.
Classically, translational activation of neutrophils is underestimated because major innate defense activities do not require de novo protein synthesis.8 However, aged neutrophils have activated characteristics of translation, and the microbiome can regulate neutrophil translational activity.23 Like aged neutrophils, LNs have a significantly positive enrichment score of translation-related gene sets in gene set enrichment analysis27 compared with BNs (Figure 1I). Therefore, we hypothesize that LNs might translate proteins de novo in response to lung environmental factors. In support of this, both mRNA and protein levels for Sca1/Ly6a, Cd14, and Plet1 were increased in LNs (Figure 1J-K).
LNs are impaired in chemotactic migration and produce different levels and types of inflammatory cytokines than BMNs
Neutrophil migration into infected or injured areas is a key initial step for immune defense.2 Several classical chemoattractants such as N-formylated peptides and C5a induce chemotactic migration of neutrophils via formyl peptide receptor (FPR) and C5a receptor,28 respectively. WKYMVm and MMK-1 are also strong chemotactic agonists for FPR members.28 Unlike BMNs, LNs do not migrate effectively toward FPR agonists (WKYMVm, MMK-1, fMLF) or C5a (Figure 2A). LNs also failed to migrate toward AcPGP, an agonist for CXCR2, a representative neutrophil chemokine receptor (Figure 2A). Transcriptome analysis showed that LNs express very low levels of chemoattractant/chemokine receptors (Figure 2B), consistent with their impaired migratory activity.
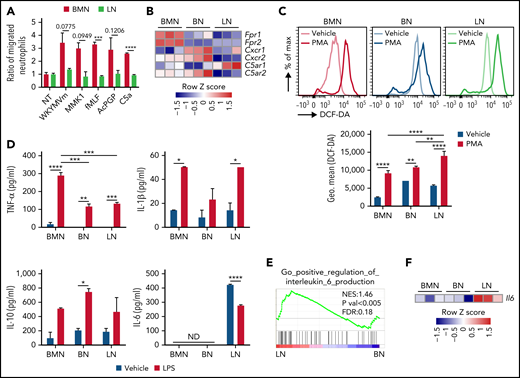
LNs have impaired migratory activity, reduced TNF-α, and increased IL-6 secretion compared with BMNs. (A) Ratio of migrated BMN and LN number by each chemoattractant compared with that of vehicle. (B) mRNA expression of chemotactic receptors in neutrophils. (C) ROS production of neutrophils 30 minutes after PMA stimulation. (D) Inflammatory cytokines from cultured media of sorted neutrophils after 24 hours of vehicle or LPS stimulation. ND, not detected. (E) GSEA of IL-6 production in LNs compared with BNs. (F) IL-6 mRNA expression in RNA-seq. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. % of max scale of each channel is presented as a percentage of maximum count (C).
LNs have impaired migratory activity, reduced TNF-α, and increased IL-6 secretion compared with BMNs. (A) Ratio of migrated BMN and LN number by each chemoattractant compared with that of vehicle. (B) mRNA expression of chemotactic receptors in neutrophils. (C) ROS production of neutrophils 30 minutes after PMA stimulation. (D) Inflammatory cytokines from cultured media of sorted neutrophils after 24 hours of vehicle or LPS stimulation. ND, not detected. (E) GSEA of IL-6 production in LNs compared with BNs. (F) IL-6 mRNA expression in RNA-seq. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. % of max scale of each channel is presented as a percentage of maximum count (C).
Recruited neutrophils perform innate defense activity through phagocytosis of invading pathogens.29 LNs effectively ingested AF488-conjugated ovalbumin (OVA) similar to BMNs and BNs (supplemental Figure 2A). Neutrophils produce ROS such as superoxide anion and hydrogen peroxide to kill pathogens.29 In response to phorbol 12-myristate 13-acetate (PMA), LNs produced somewhat higher levels of ROS than BMNs (Figure 2C). LNs showed similar neutrophil extracellular traps forming activity to BMNs and BNs in response to ionomycin (supplemental Figure 2B-C). Neutrophils play an important role in the regulation of inflammatory immune responses by releasing cytokines that modulate leukocyte activity.30 Unlike BMNs and BNs, only LNs basally produce high levels of IL-6 (Figure 2D). LNs also had a positive enrichment score for IL-6 production (Figure 2E), and high expression of IL-6 mRNA compared with other neutrophils (Figure 2F). The production of IL-1β and IL-10 was not significantly different in LNs compared with other neutrophils, but LNs produced reduced levels of tumor necrosis factor-α (TNF-α) vs BMNs in response to LPS (Figure 2D).
Pulmonary environmental factors are necessary to induce LN-like characteristics
To investigate whether LNs come from bone marrow through blood vessels, eF670-stained BMNs were transferred via intravenous injection (supplemental Figure 3A). After 16 hours, transferred eF670+ LNs showed similar surface marker expression pattern with those of endogenous LNs such as CD11bhigh, CD14high, Sca1high, and CD62Llow (Figure 3A-B; supplemental Figure 3B-C), but SPNs showed lower expression of CD101 (supplemental Figure 3D-E).
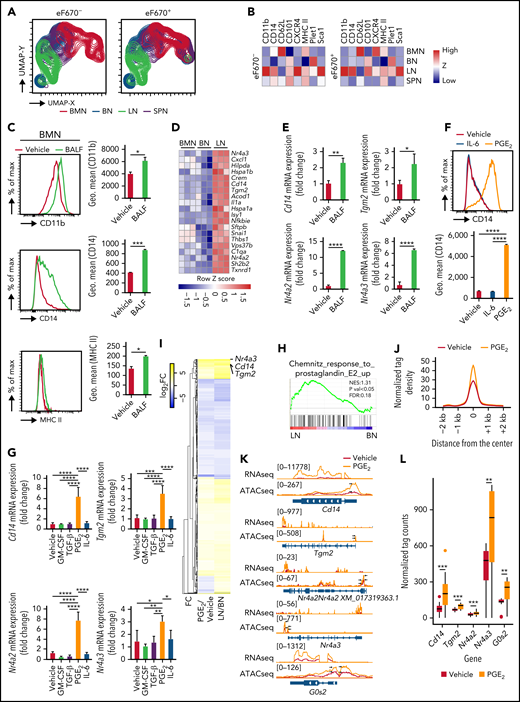
PGE2 and BALF induce LN-like characteristics in BMNs. (A) Bidimensional UMAP analysis of down sampled tissue neutrophils by CD11b, CD62L, CD101, MHC II, Plet1, Sca1 from Zombie NIR fixable viability dye negative adoptive eF670 stained BMNs transferred mice. (B) Heatmap of surface marker expression in transferred eF670+ and endogenous eF670− tissue neutrophils. (C) Surface protein expression of CD11b, MHC II, CD14, and CD101 on BMNs following 24 hours of incubation with BALF. (D) Top 20 most highly upregulated genes in LNs compared with BNs by RNA-seq. (E) mRNA expression of Cd14, Tgm2, Nr4a2, and Nr4a3 from vehicle- or BALF-treated BMNs. (F) Surface protein expression of CD14, Plet1, and CD11b following 24 hours of incubation with vehicle, IL-6, or PGE2. (G) mRNA expression of Tgm2, Cd14, Nr4a2, and Nr4a3 from vehicle-, GM-CSF-, TGF-β-, PGE2-, or IL-6–treated BMNs (H) GSEA of PGE2 signaling in LNs compared with BNs. (I) Heatmap of log2FC value comparing PGE2- vs vehicle-treated BMNs and LNs vs BNs. (J) Tag density plot of ATAC-seq in PGE2- (orange) or vehicle- (red) treated BMNs in 107 selected upregulated genes. (K) Representative genome browser snapshot showing signal intensity of ATAC-seq and RNA-seq in annotated sequences in PGE2- or vehicle-treated BMNs. Arrows indicate the peaks on the promotor regions of each gene. Upper-left number shows the range of read count. (L) Tag counts under ATAC-seq peak spanning the promotor regions of Cd14, Nr4a2, Nr4a3, Tgm2, and G0s2 genes. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. % of max scale of each channel is presented as a percentage of maximum count (C,F).
PGE2 and BALF induce LN-like characteristics in BMNs. (A) Bidimensional UMAP analysis of down sampled tissue neutrophils by CD11b, CD62L, CD101, MHC II, Plet1, Sca1 from Zombie NIR fixable viability dye negative adoptive eF670 stained BMNs transferred mice. (B) Heatmap of surface marker expression in transferred eF670+ and endogenous eF670− tissue neutrophils. (C) Surface protein expression of CD11b, MHC II, CD14, and CD101 on BMNs following 24 hours of incubation with BALF. (D) Top 20 most highly upregulated genes in LNs compared with BNs by RNA-seq. (E) mRNA expression of Cd14, Tgm2, Nr4a2, and Nr4a3 from vehicle- or BALF-treated BMNs. (F) Surface protein expression of CD14, Plet1, and CD11b following 24 hours of incubation with vehicle, IL-6, or PGE2. (G) mRNA expression of Tgm2, Cd14, Nr4a2, and Nr4a3 from vehicle-, GM-CSF-, TGF-β-, PGE2-, or IL-6–treated BMNs (H) GSEA of PGE2 signaling in LNs compared with BNs. (I) Heatmap of log2FC value comparing PGE2- vs vehicle-treated BMNs and LNs vs BNs. (J) Tag density plot of ATAC-seq in PGE2- (orange) or vehicle- (red) treated BMNs in 107 selected upregulated genes. (K) Representative genome browser snapshot showing signal intensity of ATAC-seq and RNA-seq in annotated sequences in PGE2- or vehicle-treated BMNs. Arrows indicate the peaks on the promotor regions of each gene. Upper-left number shows the range of read count. (L) Tag counts under ATAC-seq peak spanning the promotor regions of Cd14, Nr4a2, Nr4a3, Tgm2, and G0s2 genes. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. % of max scale of each channel is presented as a percentage of maximum count (C,F).
We next questioned whether soluble environmental factors in the lungs contribute to generation and/or maintenance of the steady-state LN phenotype. We first examined the effects of bronchoalveolar lavage fluid (BALF) on BMNs. Incubation of BMNs with BALF led to upregulation of CD11b, MHC II, and CD14 consistent with the phenotype of freshly isolated LNs (Figure 3C; supplemental Figure 4A). RNA-sequencing (RNA-seq) and quantitative reverse transcription polymerase chain reaction data from sorted neutrophils showed that LNs had the highest expression of Tgm2, Nr4a2, Nr4a3, and Cd14 among 3 different neutrophil populations (Figure 3D; supplemental Figure 4B), consistent with the results from Ballesteros et al6 (supplemental Figure 4C). Incubation of BMNs with BALF also induced expression of 4 defining markers of LNs: Tgm2, Nr4a2, Nr4a3, and Cd14 (Figure 3E). Collectively, BALF induced defining immunophenotypic and transcriptional characteristics of LNs.
Several pulmonary-related soluble factors, including cytokines and lipid mediators, regulate tissue-resident immune cells. Lung-resident alveolar macrophages sustain their population dependent on autocrine transforming growth factor-β (TGF-β) and lung environmental granulocyte-macrophage colony-stimulating factor (GM-CSF).31 It was previously reported that PGE2 level in the lungs is higher than that of blood.18,19 We therefore asked if several candidate pulmonary environmental factors such as PGE2, TGF-β, GM-CSF, and IL-6 were important for the generation of LNs from BMNs. Of these factors, only PGE2 upregulated the expression of surface CD14 (Figure 3F) but not Plet1 or CD11b (supplemental Figure 4D). mRNA expression of Tgm2, Cd14, Nr4a2, and Nr4a3 was induced only by PGE2 (Figure 3G), and the PGE2 pathway32 was activated in LNs (Figure 3H). To examine how PGE2 regulates characteristics of LNs, we performed RNA-seq and assay for transposase-accessible chromatin using sequencing (ATAC-seq) in vehicle- or PGE2-treated BMNs. PGE2-treated BMNs showed increased mRNA expression levels of Cd14, Tgm2, and Nr4a3 as well as G0s2, previously referred to as LN signature genes6 (supplemental Figure 5A). We then compared RNA-seq data from PGE2- vs vehicle-treated BMNs with RNA-seq data from LNs vs BNs (Figure 3I). We determined that 2032 differentially expressed genes (DEGs) were obtained in LNs compared with BNs, and 1359 DEGs were obtained in PGE2- compared with vehicle-treated BMNs with P < .05. There were 665 shared DEGs between each comparison, and the overall gene expression pattern in PGE2-treated BMNs was conserved with LNs. Specifically, Nr4a3, Cd14, and Tgm2 were significantly upregulated in PGE2-treated BMNs and LNs vs their respective comparators (Figure 3I). To examine whether chromatin structure could affect gene expressions, 107 upregulated genes (>2-fold change) in LNs and PGE2-treated BMNs were analyzed with ATAC-seq. ATAC signals in the promoters of these genes were increased in PGE2-treated compared with vehicle-treated BMNs (Figure 3J) and LNs compared with BNs (supplemental Figure 5B). Snapshot showed that accessibility and gene expression levels of Cd14, Tgm2, Nr4a2, Nr4a3, and G0s2 were significantly increased in PGE2-treated BMNs (Figure 3K-L; supplemental Figure 5A). Therefore, PGE2 can increase neutrophil chromatin openness and induce characteristics of LNs at the transcriptional level.
GM-CSF upregulated MHC II expression on BMNs (supplemental Figure 4E) such as LNs (Figure 1F), and previous reports showed that MHC II+ neutrophils act as antigen-presenting cells.33-36 However, LNs did not show antigen-presenting activity (supplemental Figure 6A-C). OVA-pulsed BMNs, BNs, and LNs failed to stimulate the proliferation of OT-II T cells compared with OVA-pulsed CD11c+ lung dendritic cells (DCs) (supplemental Figure 6A). LNs also failed to show antigen-presenting capacity in vivo. Injection of bone marrow–derived DCs but not BMNs or LNs pulsed with house dust mite extracts elicited strong production of representative Th2 cytokines (IL-4, IL-5, and IL-13) in experimental asthma models (supplemental Figure 6B-C). Collectively, our results suggest that among various pulmonary factors, PGE2 is a key player for the generation and/or maintenance of steady-state LNs.
The PGE2/PKA pathway regulates LN lifespan and production of inflammatory cytokines
Because neutrophils in peripheral tissues persist for >12 hours,37 we hypothesize that environmental factors may elongate the lifespan of LNs. Nr4a2 and Nr4a3, antiapoptotic factors for neutrophils, were upregulated by PGE237 and increased in LNs (Figure 3D) and in BALF-treated BMNs (Figure 3E). Thus, we first examined the effect of BALF on neutrophil apoptosis. Apoptosis was significantly decreased as determined by Annexin V and blocked by an EP2 antagonist (AH6809) but not by an EP4 antagonist (AH23848) in BALF-treated BMNs (Figure 4A). Furthermore, intratracheal (IT) instillation of NS398, a PGE2 synthase inhibitor, not only decreased PGE2 concentration in BALF but also increased apoptotic LNs in the lungs (Figure 4B) and decreased expression levels of Cd14, Nr4a3, and Tgm2 from sorted LNs (supplemental Figure 7A). Not only NS398-treated mice but also Tgm2−/− mice have lower LNs in the lung at steady states (supplemental Figure 7B-C). In addition, PGE2-treated BMNs produce higher levels of ROS in response to PMA (Figure 4C) such as LNs, as shown in Figure 2C. The results suggest that PGE2 may elongate the lifespan of LNs through EP2 and augment ROS-producing capacity.
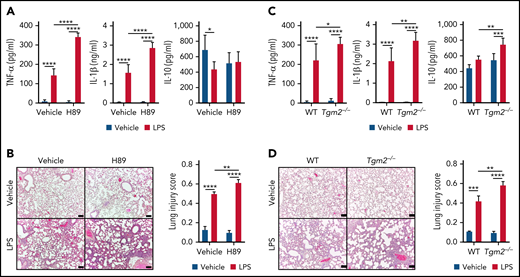
PGE2 and BALF have antiapoptotic and antiinflammatory effects on BMNs. (A) AnnexinV+ BMNs were measured by flow cytometry after addition of vehicle or BALF in the presence of AH6809 (an EP2 antagonist), AH23848 (an EP4 antagonist), or anti–IL-6 antibody. (B) PGE2 concentration of BALF (left) and % of annexin V+ cells among CD11b+ Ly6G+ neutrophils in lung (right) from 5 mg/kg NS398- or vehicle-treated mice via intratracheal instillation. (C) ROS production of BMNs with or without PGE2 pretreatment 30 minutes before PMA stimulation. Vehicles for PGE2 and PMA are distilled water (DW) and dimethyl sulfoxide (DMSO), respectively. (D-E) Inflammatory cytokine release following LPS stimulation of BMNs in the presence or absence of PGE2 (D) or BALF (E). (F) Tgm2 expression from BALF- or PGE2-treated BMNs with or without PKA inhibitor H89. (G) Inflammatory cytokine release from BALF- or PGE2-treated BMNs with or without H89 pretreatment 12 hours before LPS challenge. Dotted line indicates LPS only. (H) Cytokine release from BMNs from WT or Tgm2−/− mice following LPS stimulation. (I) Inflammatory cytokine release from BMNs from WT or Tgm2−/− mice pretreated with BALF from WT or Tgm2−/− mice. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. ns, not significant (two-way ANOVA) (A-B, C [right], D-I). % of max scale of each channel is presented as a percentage of maximum count (C).
PGE2 and BALF have antiapoptotic and antiinflammatory effects on BMNs. (A) AnnexinV+ BMNs were measured by flow cytometry after addition of vehicle or BALF in the presence of AH6809 (an EP2 antagonist), AH23848 (an EP4 antagonist), or anti–IL-6 antibody. (B) PGE2 concentration of BALF (left) and % of annexin V+ cells among CD11b+ Ly6G+ neutrophils in lung (right) from 5 mg/kg NS398- or vehicle-treated mice via intratracheal instillation. (C) ROS production of BMNs with or without PGE2 pretreatment 30 minutes before PMA stimulation. Vehicles for PGE2 and PMA are distilled water (DW) and dimethyl sulfoxide (DMSO), respectively. (D-E) Inflammatory cytokine release following LPS stimulation of BMNs in the presence or absence of PGE2 (D) or BALF (E). (F) Tgm2 expression from BALF- or PGE2-treated BMNs with or without PKA inhibitor H89. (G) Inflammatory cytokine release from BALF- or PGE2-treated BMNs with or without H89 pretreatment 12 hours before LPS challenge. Dotted line indicates LPS only. (H) Cytokine release from BMNs from WT or Tgm2−/− mice following LPS stimulation. (I) Inflammatory cytokine release from BMNs from WT or Tgm2−/− mice pretreated with BALF from WT or Tgm2−/− mice. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. ns, not significant (two-way ANOVA) (A-B, C [right], D-I). % of max scale of each channel is presented as a percentage of maximum count (C).
Because LNs produced lower levels of TNF-α upon LPS stimulation than BMNs (Figure 2D), we checked whether pulmonary environmental factors could regulate the production of inflammatory cytokines in BMNs. Addition of BALF or PGE2 to BMNs significantly decreased the production of inflammatory cytokines in response to LPS (Figure 4D-E). PGE2 also suppressed BN production of TNF-α and IL-10 (supplemental Figure 7D), further supporting the notion that PGE2 is important for generating LNs. PGE2 and BALF upregulated Tgm2 in BMNs (Figure 3E,G), and PGE2 is well known to activate PKA.38 Treatment of BMNs with PKA inhibitor H89 significantly blunted Tgm2 transcription (Figure 4F). Moreover, H89 treatment reversed inhibitory effects of BALF and PGE2 on cytokine production from LPS stimulation (Figure 4G). We next examined the role of Tgm2 on the production of inflammatory cytokines in response to LPS. LPS stimulated BMNs from Tgm2−/− mice showed higher levels of TNF-α, IL-1β, and IL-10 production compared with control BMNs from wild-type (WT) mice (Figure 4H). To specify whether Tgm2 in neutrophils or other pulmonary environmental cells are important for the regulation of inflammatory cytokine production, we treated BMNs from WT or Tgm2−/− mice with BALF from WT or Tgm2−/− mice (Figure 4I). BMNs from Tgm2−/− mice produced more TNF-α and IL-10 in WT BALF condition in response to LPS, and BALF from Tgm2−/− mice decreased TNF-α and IL-10 productions in Tgm2−/− BMNs by LPS (Figure 4I). The results suggest that Tgm2 deficiency in neutrophils increases inflammatory cytokine release, but pulmonary environmental deficiency of Tgm2 decreased inflammatory cytokine production. Taken together, a PGE2/PKA pathway in LNs modulates Tgm2 and decreases inflammatory cytokine release in response to LPS.
Tgm2 regulates neutrophilic inflammatory lung disease
Our finding that LNs produce lower levels of inflammatory cytokines in response to LPS compared with BMNs led us to examine whether LNs have protective roles in experimental neutrophilic acute respiratory distress syndrome (ARDS).39 Because H89 blocked the protective and antiinflammatory roles of BALF or PGE2 (Figure 4F-G), we first examined the effects of H89 in ARDS. H89 treatment 24 hours prior to ARDS induction via IT LPS instillation significantly increased the levels of airway inflammatory cytokines TNF-α and IL-1β but not IL-10 (Figure 5A) and exacerbated alveolar destruction (Figure 5B). Consistent with the in vitro data (Figure 4I), inflammatory cytokine levels in the airways and lung injury score were significantly higher in Tgm2−/− mice than WT mice (Figure 5C-D). The proportion of neutrophils in BALF was similar in WT, H89-treated, and Tgm2−/− mice (supplemental Figure 8A-B). The results suggest that the PKA/Tgm2 pathway plays a protective role as a novel neutrophil inflammatory checkpoint, diminishing inflammatory cytokine release in experimental ARDS and suppressing alveolar destruction.
Severe ARDS following intratracheal instillation of LPS in H89-treated or Tgm2−/− mice. (A) Inflammatory cytokines in BALF of mice pretreated with H89 24 hours before IT LPS instillation. (B) Histological evidence of lung inflammation by hematoxylin and eosin staining of lung tissues from ARDS mice with or without H89 pretreatment. (C) Inflammatory cytokines in BALF of WT or Tgm2−/− mice 24 hours after IT LPS instillation. (D) Hematoxylin and eosin staining of lung sections from WT or Tgm2−/− mice 24 hours after LPS IT instillation. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. Scale bars represent 100 μm (B,D).
Severe ARDS following intratracheal instillation of LPS in H89-treated or Tgm2−/− mice. (A) Inflammatory cytokines in BALF of mice pretreated with H89 24 hours before IT LPS instillation. (B) Histological evidence of lung inflammation by hematoxylin and eosin staining of lung tissues from ARDS mice with or without H89 pretreatment. (C) Inflammatory cytokines in BALF of WT or Tgm2−/− mice 24 hours after IT LPS instillation. (D) Hematoxylin and eosin staining of lung sections from WT or Tgm2−/− mice 24 hours after LPS IT instillation. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. Scale bars represent 100 μm (B,D).
LNs basally produce high levels of IL-6 (Figure 2D) with a positive enrichment score for IL-6 production (Figure 2E) and IL-6 signaling (supplemental Figure 8C). We therefore investigated the role of IL-6 on LNs at steady state. The IT instillation of neutralizing anti–IL-6 antibody significantly diminished LN numbers after 24 hours (supplemental Figure 8D). IL-6 neutralization also exacerbated inflammatory cytokine release but not alveolar destruction in experimental ARDS (supplemental Figure 8E-F). These data suggest that IL-6 may partly play an immunoprotective role in ARDS by maintaining the number of LNs with autocrine or paracrine activity.
Discussion
Despite reports on the existence of steady-state neutrophils in the lungs, a detailed gene expression analysis, functional phenotypic assessment, and identification of environmental factors and downstream signaling pathways necessary for the generation of LNs are limited. In this study, we used unsupervised UMAP analysis and transcriptome analysis to identify definitive surface marker expression and transcriptional profiles that distinguish LNs from BMNs and BNs. We also uncovered a novel functional role for PGE2/PKA/Tgm2 as an inflammatory checkpoint for the generation of LNs and regulation of inflammatory responses. These results indicate that LNs acquire unique characteristics and functions in the lung environment.
Almost all LNs are CD62Llow and have high expression of CXCR4 (Figure 1F), similar to aged neutrophils.23,40 Although LNs are present in intravascular regions (supplemental Figure 1B) such as BNs, LNs are transcriptionally and translationally active (Figure 1I). They express a unique gene expression pattern and markers such as CD14 that differentiate LNs from aged BNs (Figure 1B-H,J-K), consistent with a previous report.6 Previously, LNs have been reported to express high levels of CXCR4, a target receptor for CXCL12.4 Blocking of CXCR4 strongly detached neutrophils from intravascular sites in pulmonary capillary regions,5 indicating a crucial role for CXCR4 in the localization of LNs in the pulmonary environment. In this study, we found that LNs have impaired migratory activity toward chemoattractants for classical chemoattractant receptors (Fpr1, Fpr2, C5ar2) and chemokine receptors (CXCR1, CXCR2) (Figure 2A), with concomitant reduced receptor expression (Figure 2B). Thus, the intratissue positioning of LNs may be dictated largely by CXCL12, with little repositioning even upon challenge by bacterial infection or tissue injury.
Neutrophils in the lungs can be influenced by pulmonary environmental factors to express unique patterns of mRNA and proteins. Although neutrophil aging is known to be regulated by the microbiome,23 LNs were not affected by the microbiome,6 suggesting additional differences from aged neutrophils. LNs obtained from CXCR4 conditional knockout mice failed to express lung signature genes.6 Thus, high expression of CXCR4 might be important to migrate and sustain LNs in the lungs, where LNs may receive environmental signals in vivo. Because we found that transferred BMNs express different surface markers depending on environmental conditions (Figure 3A-B) and stimulation of BMNs with BALF upregulated some of surface expression of LN-associated markers (Figure 3C), we hypothesized that BALF contains crucial soluble environmental factors for the programming of the LN phenotype. Moreover, we found that GM-CSF, like BALF, induced the upregulation of CD11b and MHC II (supplemental Figure 4E; Figure 3C). Although transferred eF670+ BMNs show LN-like immunophenotype in the lungs (Figure 3A), stimulation of BMNs with BALF did not alter CXCR4, CD62L, Plet1, and Sca1 (supplemental Figure 4A), suggesting that additional factors may confer LN characteristics.
Because intravascular BNs could be affected by pulmonary soluble factors, we compared upregulated gene expression profiles of LNs vs BNs with PGE2-treated BMNs (Figure 3I). Among 203 upregulated genes from PGE2 treatment, 107 genes were also upregulated in LNs from BNs. We also found that PGE2 increased surface levels of CD14 on BMNs as well as mRNA expression of Tgm2, Cd14, Nr4a2, and Nr4a3 (Figure 3F-G), which are characteristics of LNs. Chromatin regions of these genes were also more accessible in PGE2-treated BMNs (Figure 3K-L). Expression and chromatin openness of these genes were also increased in LN data from Ballesteros et al6 (supplemental Figures 4C and 5B), supporting the notion that PGE2 could induce characteristics of LNs in neutrophils at a transcriptional level. Previously, others reported that the elongated lifespan of LNs is also regulated by PGE2/cAMP/PKA in Nr4a2 and Nr4a3-dependent ways.37 Our finding indicated that lung environmental PGE2 elicited antiapoptotic activity through EP2 but not EP4 receptor (Figure 4A) and that the percentage of apoptotic LNs increased by injection of PGE2 synthase inhibitor NS398 (Figure 4B). Decreased frequency and number of LNs by NS398 (supplemental Figure 7B) suggested that the elongated lifespan of LNs might be due, at least in part, to the action of PGE2.
Although lung-resident alveolar macrophages and DCs can regulate pulmonary diseases, the role of resident neutrophils in the pathomechanism of lung disease is unclear. In this study, we found that PGE2 or BALF suppressed inflammatory cytokine production following LPS challenge and that this effect was dependent on PKA/Tgm2 in neutrophils (Figure 4D-I). Tgm2 is known to activate NF-κB,41 but the detailed molecular mechanism involved in the process remains controversial41-44 as others have reported that overexpression of Tgm2 failed to phosphorylate NF-κB in gastric cancer cells.45 Tgm2 also could regulate immune response in macrophages,46 but its role in neutrophils is unclear. Here we showed that cytokine production was significantly elevated in neutrophils from Tgm2−/− mice (Figure 4H) and that inflammatory cytokine concentrations were significantly increased in BALF from Tgm2−/− mice in the ARDS model compared with WT mice (Figure 5D). LNs and BALF- or PGE2-treated neutrophils upregulated LPS coreceptor CD14, which can modulate TLR4-dependent cytokine production,47 but these LN-like neutrophils produced lower levels of cytokines than vehicle-treated cells (Figure 4D-E). Mice pretreated with PKA inhibitor H89 had worse histological lung inflammation in the ARDS model (Figure 5B). Additional studies are needed to elucidate the functional role of CD14 in LNs generated by the PGE2/PKA/Tgm2 pathway.
In conclusion, in this study we comprehensively characterized the gene expression profile and immunophenotype of LNs and discovered pulmonary environmental factors that drive the generation of LNs. Functionally, LNs have impaired migratory activity but produce high levels of IL-6 basally and produce lower inflammatory cytokines than other neutrophils upon LPS challenge. Pulmonary factors such as GM-CSF and PGE2 drive surface expression of CD11b, MHC II, and CD14. PGE2 elongates the lifespan of LNs via PKA and suppresses inflammatory cytokine production through a PKA/Tgm2-dependent axis. Using Tgm2−/− mice, we found that LNs negatively regulate lung inflammation to protect the lungs in response to infection or inflammatory stimuli. Thus, our findings provide new insight into LNs that offer novel avenues for the therapeutic control of pathologic inflammatory lung diseases.
Acknowledgments
This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and future Planning (NRF-2017R1A5A1014560, NRF-2018R1A2B3003868, 2020M3A9D3038435, and NRF-2021R1A2C3011228). A visual abstract was created with BioRender.com.
Authorship
Contribution: G.H.B. and Yoe-Sik Bae designed the experiments and wrote the draft manuscript; G.H.B., Y.S.K., J.Y.P., M.L., S.K.L., J.C.K., J.G.K., Y.J.S., and H.S.K. performed the research and analyzed data; H.L. and S.-Y.K. supplied Tgm2−/− mice; and G.H.B., B.A.Z., Yong-Soo Bae, H.-S.K., and Yoe-Sik Bae wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yoe-Sik Bae, Department of Biological Sciences, Sungkyunkwan University, Suwon 16419, Republic of Korea; e-mail: yoesik@skku.edu.
RNA sequencing data are available in the Gene Expression Omnibus (GEO) database accession numbers with GSE199773 for PGE2-treated neutrophils and GSE199774 for tissue neutrophils. ATAC-seq data of PGE2-treated neutrophils are available in the GEO database accession numbers with GSE199772.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![LNs have a distinct immunophenotype and mRNA expression profile compared with other neutrophils. (A) Neutrophil proportions from total bone marrow, peripheral blood, lung, and spleen (n = 3) from 8-week-old C57BL/6 male mice were analyzed by flow cytometry. (B) Heatmap of Ly6G expression on bidimensional UMAP analysis of live CD45+ immune cells from bone marrow, peripheral blood, lung, and spleen (n = 3). (C) Three Ly6Ghigh clusters among 38 clusters from PARC analysis in bone marrow, blood, lung, and spleen. (D) Cluster proportion of each tissue neutrophil. Expression levels (MFI) of CD11b, CD62L, CXCR4, MHC II, and CD101 on Ly6G+ clusters (E) and tissue neutrophils (F). Correlation matrix (G) and up/down (log2FC ≥ 1/log2FC ≤ 1) (H) regulated count of genes from bulk RNA-seq data of BMNs, BNs, and LNs with log2FC ≥ 1, P < .05 from pooling 5 heads of 8-week-old C57BL/6 male mice. (I) Gene set enrichment analysis (GSEA) data of gene sets associated with translational activity in LNs compared with BNs. (J) Heatmap of surface protein mRNA expression on BMN, BN, and LN in RNA-seq. (K) Surface protein level (MFI) of Sca1, Plet1, and CD14 on tissue neutrophils (n = 4). Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. ns, not significant (one-way ANOVA) (A [right], E-F,K).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/8/10.1182_blood.2021014283/3/m_bloodbld2021014283r2f1.png?Expires=1763554643&Signature=2cZpaWY6AGWYHnCtvEaKPId2hNlXNYetjg~cYdKctEzlQFuQA7Vwbfh5bZVOPWrjTG0NUqBPQ6mqeyunTp3Iptkf234XLi56P9X~NJC853uLizXC8KTnanlo-AkIzfWzFI3VOCYvxdwq0eSniECPQnBPI-I8pPnpuImS3Eezl9IP7Kd84geOmwzHrC5qiKc1CpI1woU8XW8hgcNUMpHc~3SWuPbM6iuk2YfNWAlnNy11tG1P~U7ZerLfHBHln-6zrASwTmTapP0geUOESAnW1GqCKIgHYBDucOfSYCOE-qgBX-yNSyEZYuAm4AwZJSxuSnKUAz9Q6jzCgLNko1L4Sg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![PGE2 and BALF have antiapoptotic and antiinflammatory effects on BMNs. (A) AnnexinV+ BMNs were measured by flow cytometry after addition of vehicle or BALF in the presence of AH6809 (an EP2 antagonist), AH23848 (an EP4 antagonist), or anti–IL-6 antibody. (B) PGE2 concentration of BALF (left) and % of annexin V+ cells among CD11b+ Ly6G+ neutrophils in lung (right) from 5 mg/kg NS398- or vehicle-treated mice via intratracheal instillation. (C) ROS production of BMNs with or without PGE2 pretreatment 30 minutes before PMA stimulation. Vehicles for PGE2 and PMA are distilled water (DW) and dimethyl sulfoxide (DMSO), respectively. (D-E) Inflammatory cytokine release following LPS stimulation of BMNs in the presence or absence of PGE2 (D) or BALF (E). (F) Tgm2 expression from BALF- or PGE2-treated BMNs with or without PKA inhibitor H89. (G) Inflammatory cytokine release from BALF- or PGE2-treated BMNs with or without H89 pretreatment 12 hours before LPS challenge. Dotted line indicates LPS only. (H) Cytokine release from BMNs from WT or Tgm2−/− mice following LPS stimulation. (I) Inflammatory cytokine release from BMNs from WT or Tgm2−/− mice pretreated with BALF from WT or Tgm2−/− mice. Data are shown as mean ± standard error of the mean. Significant differences are denoted as *P < .05; **P < .01; ***P < .001; ****P < .0001. ns, not significant (two-way ANOVA) (A-B, C [right], D-I). % of max scale of each channel is presented as a percentage of maximum count (C).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/8/10.1182_blood.2021014283/3/m_bloodbld2021014283r2f4.png?Expires=1763554643&Signature=M-YdKzTSMWLh9DJ7kelfnB-mBvOYKI~2xA3jnHbVtgLELwSF~XX267F8OWi~1Dvu-dn1OPeDChZnQoQGbluuzlLiUi678-L~UR7Cgl7EeMpfz-~UUF681r7wB4DT7S7fgcCaMYbN7rd6nM4ttYjhGihvpMjdKqICmjAa58zIqm86C4OzvoSo7UACC3KAcgACH8NQlvlUffDrk~~8CdaHO3ZiXDiwSb6h9o0xc8gw6fhlOHJQfXUKsRVNYHd3WF4oFM4JPHh4xix0bcbe709rs8c8OzYmBV9j01x2fQ0A277Gu9Cd8EDlELGTOB2aTQX52OnCfZ~hEWtjjbp51kZbMw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal