Key Points
AML/MDS defined by germline DDX41 CV represents a unique entity with favorable outcome.
Germline DDX41 CVs predisposing patients to MN are often associated with somatic DDX41 mutations.
Abstract
Germline DDX41 variants are the most common mutations predisposing to acute myeloid leukemia (AML)/myelodysplastic syndrome (MDS) in adults, but the causal variant (CV) landscape and clinical spectrum of hematologic malignancies (HMs) remain unexplored. Here, we analyzed the genomic profiles of 176 patients with HM carrying 82 distinct presumably germline DDX41 variants among a group of 9821 unrelated patients. Using our proposed DDX41-specific variant classification, we identified features distinguishing 116 patients with HM with CV from 60 patients with HM with variant of uncertain significance (VUS): an older age (median 69 years), male predominance (74% in CV vs 60% in VUS, P = .03), frequent concurrent somatic DDX41 variants (79% in CV vs 5% in VUS, P < .0001), a lower somatic mutation burden (1.4 ± 0.1 in CV vs 2.9 ± 0.04 in VUS, P = .012), near exclusion of canonical recurrent genetic abnormalities including mutations in NPM1, CEBPA, and FLT3 in AML, and favorable overall survival (OS) in patients with AML/MDS. This superior OS was determined independent of blast count, abnormal karyotypes, and concurrent variants, including TP53 in patients with AML/MDS, regardless of patient’s sex, age, or specific germline CV, suggesting that germline DDX41 variants define a distinct clinical entity. Furthermore, unrelated patients with myeloproliferative neoplasm and B-cell lymphoma were linked by DDX41 CV, thus expanding the known disease spectrum. This study outlines the CV landscape, expands the phenotypic spectrum in unrelated DDX41-mutated patients, and underscores the urgent need for gene-specific diagnostic and clinical management guidelines.
Introduction
Hereditary hematologic malignancies (HM) typically manifest at earlier ages than de novo disease,1 usually with substantial familial clustering.2,3 Inclusion of hereditary HM in the fourth edition of World Health Organization classification of hematopoietic and lymphoid tissues4 emphasizes the importance of germline evaluation in patients with myeloid neoplasm (MN). The National Comprehensive Cancer Network guidelines on next-generation sequencing (NGS) for patients with MN facilitate comprehensive large-scale screening in the general population for variants of interest, which has revealed the surprisingly high incidence of presumably germline mutations in genes predisposing to HM in children and adults. Approximately 8% of pediatric and adult patients have a pathogenic germline variant, and many patients lack a pertinent family history (FH).5,6 These recent studies have revealed that familial HM predisposition syndromes, previously thought to be rare diseases, are more common than anticipated.
Recently, our group and others identified DDX41 as 1 of the most common MN predisposition genes in adults.7-9 Unlike some other hereditary HMs that present in childhood or adolescence, DDX41 is associated with late-onset MN, at ages typical of sporadic acute myeloid leukemia (AML)/myelodysplastic syndrome (MDS),10 years after indolent and mild cytopenia,7,8,10-12 and these patients often lack FH.7,8,10-12 The subacute disease course of DDX41-associated AML is generally accompanied by bone marrow hypocellularity and a borderline increase in blasts with a near normal immunoprofile, and most patients have a normal karyotype. These features make the initial diagnosis of this inherited AML more challenging than other hereditary HM predisposition syndromes.8
Despite increasing integration of NGS assessment in clinical practice, the accurate diagnosis of DDX41-associated HM is further complicated by the currently limited ability to distinguish between causal and benign variants. The lack of FH, reduced penetrance and the long disease latency complicate the power of the familial segregation studies to accurately identify causal variants (CVs) among the increasing pool of novel missense variants identified by NGS. Identification of germline CVs can inform long-term patient management and prevent engraftment failure13 and donor-derived leukemia14-18 in some clinical contexts where allogeneic stem cell transplantation (HSCT) is necessary.7,8 Furthermore, family members also benefit from identification of a germline variant in informing their own risk of developing MN. Unfortunately, no consensus has been reached on guidelines of DDX41-specific diagnosis and patient management because of limited awareness of this disease and the inherent challenges in classification of novel missense germline variants. Thus, collaboration on variant curation among expert panels to develop gene-specific diagnostic guidance has become urgent as these novel variants are increasingly detected.
Beyond AML and MDS, germline DDX41 mutations appear to predispose to other MN, such as chronic myelomonocytic leukemia (CMML), chronic myeloid leukemia (CML), and myelodysplastic/myeloproliferative neoplasm (MDS/MPN), lymphoproliferative disorders (LPD), and potentially nonhematopoietic neoplasms.7,10,19 Previously reported cases with mixed germline CV and variant of uncertain significance (VUS) provide insufficient support to link germline DDX41 mutations to LPD and other MN. Moreover, the overall natural course and characteristic pathologic findings of each DDX41-associated entity remain unclear, and long-term surveillance for asymptomatic individuals with germline CV and guidelines for early intervention are also needed.
In this study, we identified 176 patients with HM with presumably germline DDX41 variants in an unselected and unrelated 9821 patient cohort from six institutions. Applying our proposed DDX41-specific classification criteria, we analyzed the genomic profiles, demographic characteristics, and clinical outcomes of each specific disease entity. The striking partitioning of these features with variants we deemed CV vs VUS indicates the variant classification criteria are warranted and supports the assertion that AML/MDS with DDX41 germline CV represent 1 distinct clinical entity with a favorable outcome. Building on the current literature, we further delineated the germline CV landscape, expanded the phenotypic spectrum of HM with germline DDX41 CV to include MPN and B-cell lymphoma to improve the recognition and refine the management of this HM predisposition syndrome.
Materials and methods
Case selection
Cases with at least 1 DDX41 variant (n = 195) were identified through retrospective search of the pathology archives from January 2015 to June 2021 at the University of Utah, ARUP Laboratories, Oregon Health and Science University, University of Kansas Medical Center, Emory University, Stanford University, and University of California San Diego in 9821 unrelated and unselected patients with HM (including 3583 AML, 2161 MDS, 1029 MPN, and 3048 other diagnoses) who underwent targeted panel testing by NGS (Figure 1). Nineteen patients (0.2%) exhibited somatic DDX41 variants without germline variants, whereas 176 patients (1.8%) had at least 1 presumed germline variant. Germline variants were further classified into CV or VUS according to the proposed DDX41-specific classification criteria (Figure 1) modified from the American College of Medical Genetics/the Association for Molecular Pathology (ACMG/AMP) guidelines (supplemental Table 3, available on the Blood Web site).20 Demographic data, clinical information, and molecular and cytogenetic profiles were further analyzed to test the proposed classification criteria. Among the 176 patients with HM carrying germline DDX41 variants, the World Health Organization entities of HM included 84 AML (66 CV and 18 VUS), 40 MDS (28 CV and 12 VUS), 15 MPN (4 CV and 11 VUS), and 37 others. The other category (Figure 2; Tables 1 and 2) included 32 clonal cytopenia of undetermined significance (CCUS, designated as cytopenia), 4 B-cell LPD, and 1 multiple myeloma (MM). Of note, 24 patients with AML with DDX41 CV have been documented in a previous study.8 A control cohort of 4307 patients without DDX41 variants (including 1365 AML, 1109 MDS, 479 MPN, and 1354 other, primarily CCUS) were identified by retrospective search of patients tested at ARUP. All were adult patients (age range, 18-97 years), and their demographic and genetic characteristics were compared with those with germline DDX41 variants (Figure 1A). Clinical follow-up information was available in 158 patients with AML and 87 patients with MDS and included in overall survival (OS) analysis as the controls (Figure 1A) as they were tested and treated at Huntsman Cancer Institute in the same period as DDX41 mutant patients. As ethnicity and clinical outcome data were not available for some samples tested at ARUP Laboratories (a national reference laboratory), these patients were excluded from ethnicity-specific and OS analysis. This study was approved by the institutional review boards at the participating institutions.
Flowchart of this multi-institutional study and graphical representation of DDX41 variants found in this study. (A) In this study, 195 (2%) patients with HM with at least 1 DDX41 variant (MAF < 0.1%) are identified in 9821 unrelated and unselected adult patients from 6 medical centers and at ARUP Laboratories. Among these patients with HM, 3583 are diagnosed with AML, 2160 with MDS, 1030 with MPN, and 3048 with others including cytopenia and other myeloid and lymphoid neoplasms. These DDX41 variants are further classified into somatic variants alone (variants with a VAF < 40% in isolation) and presumed germline variants (VAFs of 40% or above, with or without concurrent somatic DDX41 variants). The germline variants are further classified into CV (PV/LPV, n = 116) and VUS (n = 60), according to the proposed gene-specific diagnostic criteria, modified from the ACMG guidelines.20 Among the 116 patients with germline DDX41 CV, 66 are diagnosed with AML, 28 with MDS, 4 with MPN, and 18 with cytopenia (others). Similarly, among the 60 patients with germline VUS, 18 are diagnosed with AML, 12 with MDS, 11 with MPN, and 19 with others. Among others, 4 are diagnosed with B-cell LPD, 1 with MM, and 14 with cytopenia. In addition, we select 4307 adult patients with HM (age of 18 years or above) with wild-type DDX41 (DDX41−), confirmed by NGS testing at ARUP laboratories during the same time period. Among these control patients (DDX41−), 1365 have a documented AML diagnosis, and the remaining cases include 1109 MDS, 479 MPN, and 1354 others, most of which are cytopenia, similar to those in the cohort of 9821 patients described above. Patients’ age, sex, and cytogenetic and molecular profiles are summarized and sorted by each distinct MN entity and correlated with their DDX41 genotypes (short double-headed arrows indicate the epidemiologic and molecular profile comparisons in between DDX41+ CV, VUS, and DDX41− cohorts). Furthermore, we summarize the OS in patients with AML and MDS who were treated at Huntsman Cancer Institute and other medical centers in comparison with the age-matched cohorts (long double-headed arrows indicate the OS comparisons in between DDX41+ CV, VUS, and DDX41− cohorts). #Of note, 24 patients with AML with DDX41 CV have been documented in a previous study.8 (B) Graphic distribution of variants identified in this study, positioned on the protein sequence (NM_016222.4) with major functional domains (red, DEAD domain; green, helicase domain; orange, Znf, zinc finger domain; teal, NLS, nuclear localization signaling domain) is separated by germline (above-protein sequences) or somatic (below) variants. Each symbol in germline variants represents 1 patient. The underline indicates novel variants reported in this study. Red, DDX41 CV; blue, DDX41 VUS; orange, p.R164W, likely CV in lymphoma. *With specific exceptions (eg, p.M155I and p.P510S).
Flowchart of this multi-institutional study and graphical representation of DDX41 variants found in this study. (A) In this study, 195 (2%) patients with HM with at least 1 DDX41 variant (MAF < 0.1%) are identified in 9821 unrelated and unselected adult patients from 6 medical centers and at ARUP Laboratories. Among these patients with HM, 3583 are diagnosed with AML, 2160 with MDS, 1030 with MPN, and 3048 with others including cytopenia and other myeloid and lymphoid neoplasms. These DDX41 variants are further classified into somatic variants alone (variants with a VAF < 40% in isolation) and presumed germline variants (VAFs of 40% or above, with or without concurrent somatic DDX41 variants). The germline variants are further classified into CV (PV/LPV, n = 116) and VUS (n = 60), according to the proposed gene-specific diagnostic criteria, modified from the ACMG guidelines.20 Among the 116 patients with germline DDX41 CV, 66 are diagnosed with AML, 28 with MDS, 4 with MPN, and 18 with cytopenia (others). Similarly, among the 60 patients with germline VUS, 18 are diagnosed with AML, 12 with MDS, 11 with MPN, and 19 with others. Among others, 4 are diagnosed with B-cell LPD, 1 with MM, and 14 with cytopenia. In addition, we select 4307 adult patients with HM (age of 18 years or above) with wild-type DDX41 (DDX41−), confirmed by NGS testing at ARUP laboratories during the same time period. Among these control patients (DDX41−), 1365 have a documented AML diagnosis, and the remaining cases include 1109 MDS, 479 MPN, and 1354 others, most of which are cytopenia, similar to those in the cohort of 9821 patients described above. Patients’ age, sex, and cytogenetic and molecular profiles are summarized and sorted by each distinct MN entity and correlated with their DDX41 genotypes (short double-headed arrows indicate the epidemiologic and molecular profile comparisons in between DDX41+ CV, VUS, and DDX41− cohorts). Furthermore, we summarize the OS in patients with AML and MDS who were treated at Huntsman Cancer Institute and other medical centers in comparison with the age-matched cohorts (long double-headed arrows indicate the OS comparisons in between DDX41+ CV, VUS, and DDX41− cohorts). #Of note, 24 patients with AML with DDX41 CV have been documented in a previous study.8 (B) Graphic distribution of variants identified in this study, positioned on the protein sequence (NM_016222.4) with major functional domains (red, DEAD domain; green, helicase domain; orange, Znf, zinc finger domain; teal, NLS, nuclear localization signaling domain) is separated by germline (above-protein sequences) or somatic (below) variants. Each symbol in germline variants represents 1 patient. The underline indicates novel variants reported in this study. Red, DDX41 CV; blue, DDX41 VUS; orange, p.R164W, likely CV in lymphoma. *With specific exceptions (eg, p.M155I and p.P510S).
Summary of DDX41 variants and ethnic difference in germline CV identified in this study and literature. (A) Summary of DDX41 germline (above the protein sequence) and somatic (below the protein sequence) variants. The colors in the boxes above and the horizontal bars below the protein sequence are designated corresponding to the protein functional domains. Numbers in parentheses alone or before a slash indicate the total times of a certain variant was reported in literature including those reported in this study, whereas numbers after a slash represent variants seen in the current study. Red, CV; blue, VUS; orange, likely CV for lymphoma. (B-E) Ethnic difference in DDX41 CV as data combined in this study and collected and reanalyzed in literature.28,29 (B) Germline variants of p.M1I (98%, 39 White and 1 Asian patients) and p.D140fs (95%, 23 White and 1 African American patients) are the leading CVs in White patients. (C) Missense germline variants, although uncommon in Whites (15%), are seen in 49% of Asian patients with HM (P < .0001). (D) p.Y592C (92%, 11 Asians and 1 non-Asian) and p.A500fs (100%, 10 all in Asian) appear the most common germline CV in Asian patients. (E) Somatic DDX41 variants alone, in the absence of associated germline variants, appear more frequently in Asian than White patients (36% in Asian vs 15% in White, P = .0007). ***, P < .001; ****, P < .0001.
Summary of DDX41 variants and ethnic difference in germline CV identified in this study and literature. (A) Summary of DDX41 germline (above the protein sequence) and somatic (below the protein sequence) variants. The colors in the boxes above and the horizontal bars below the protein sequence are designated corresponding to the protein functional domains. Numbers in parentheses alone or before a slash indicate the total times of a certain variant was reported in literature including those reported in this study, whereas numbers after a slash represent variants seen in the current study. Red, CV; blue, VUS; orange, likely CV for lymphoma. (B-E) Ethnic difference in DDX41 CV as data combined in this study and collected and reanalyzed in literature.28,29 (B) Germline variants of p.M1I (98%, 39 White and 1 Asian patients) and p.D140fs (95%, 23 White and 1 African American patients) are the leading CVs in White patients. (C) Missense germline variants, although uncommon in Whites (15%), are seen in 49% of Asian patients with HM (P < .0001). (D) p.Y592C (92%, 11 Asians and 1 non-Asian) and p.A500fs (100%, 10 all in Asian) appear the most common germline CV in Asian patients. (E) Somatic DDX41 variants alone, in the absence of associated germline variants, appear more frequently in Asian than White patients (36% in Asian vs 15% in White, P = .0007). ***, P < .001; ****, P < .0001.
Molecular and cytogenetic profiles of 205 individuals with DDX41 variants
| Patient . | Diagnosis . | gl DDX41 . | gl DDX41 . | VAF . | Tier . | s DDX41 . | s DDX41 . | VAF . | Tiers . | Concomitant variants . | VAF (%) . | Tier . | Cytogenetics . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASXL1 c.1900_1922del, p.E635fs | 19 | 1 | |||||||||||
| 1* | AML | c.3G>A | p.M1I | 44 | 1 | c.1574G>A | p.R525H | 6 | 1 | PFH6 c.811G>A, p.E271K | 12 | 1 | 46,XX[20] |
| CUX1 c.607 + 1G>A, p.? | 3 | 1 | |||||||||||
| CUX1 c.2786del, p.P929fs | 1 | 1 | |||||||||||
| 2† | AML | c.3G>A | p.M1I | 46 | 1 | c.1574G>A | p.R525H | 6 | 1 | ASXL1 c.1900_1922del, p.E635fs | 6 | 1 | 46,XX[20] |
| PHF6 c.940A>T, p.I314F | 4 | 2 | |||||||||||
| CUX1 c.2485C>T, p.Q829* | 5 | 1 | |||||||||||
| 3 | AML | c.3G>A | p.M1I | 48 | 1 | c.1574G>A | p.R525H | 0 | 1 | ASXL1 c.1627G>T, p.E543* | 2 | 1 | NI |
| PHF6 c.880A>G, p.I294V | 3 | 2 | |||||||||||
| 4† | AML | c.3G>A | p.M1I | 48 | 1 | c.1574G>A | p.R525H | 16 | 1 | ASXL1 c.1779dup, p.C594fs | 8 | 1 | 45,X,-Y[6]/46,XY[14] |
| SF3B1 c.2098A>G, p.K700E | 4 | 1 | |||||||||||
| JAK2 c.1849G>T, p.V617F | 5 | 1 | |||||||||||
| 5 | AML | c.3G>A | p.M1I | 48 | 1 | c.1574G>A | p.R525H | 3 | 1 | ASXL1 c.2905_ 2926delinsTACTGTT, p.D969_N971delinsYC* | 5 | 1 | NI |
| TP53 c.850A> C, p.T284P | 5 | ||||||||||||
| KMT2A c.2830_2847dup, p.D944_T949dup | 33 | 2 | |||||||||||
| 6*,† | AML | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 1 | 1 | ASXL1 c.1761_1768del, p.Q588fs | 20 | 1 | 45,XY,del(5)(q15q35), der(7;16)(q10;q10), add(10)(p11.2), |
| BCORL1 c.1339C>T, p.Q447* | 1 | 1 | add(10)(q24),+20, -22[13]/45,XY,-4, del(5)(q15q35), | ||||||||||
| TP53 c.743G>A, p.R248Q | 2 | 1 | der(7)t(7;9)(p11.2;q13), add(12)(p12), -13,+14, | ||||||||||
| TP53 c.817C>T, p.R273C | 1 | 1 | add(14)(q32), add(18)(q21),-22,+mar[7] | ||||||||||
| 7 | AML | c.3G>A | p.M1I | 45 | 1 | c.1574G>A | p.R525H | 5 | 1 | ASXL1 c.3030_3031delinsTT, p.E1011* | 3 | 1 | 46,XY[20] |
| ASXL1 c.2122del, p.Q708fs | 3 | 1 | |||||||||||
| 8 | AML | c.3G>A | p.M1I | 51 | 1 | c.1574G>A | p.R525H | 7 | 1 | ASXL1 c.1960dup, p.A654fs | 6 | 1 | 46,XY[20] |
| 9† | AML | c.3G>A | p.M1I | 58 | 1 | c.1574G>A | p.R525H | 3 | 1 | ASXL1 c.3824C>G, p.S1275* | 11 | 1 | 46,XY[20] |
| ZRSR2 c.202_203del, p.R68fs | 5 | 1 | |||||||||||
| 10*,† | AML | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 1 | 1 | DNMT3A c.1015-1G>C, p.? | 5 | 1 | 46,XX[20] |
| SETBP1 c.1977T>A, p.D659G | 1 | 2 | |||||||||||
| 11 | AML | c.3G>A | p.M1I | 51 | 1 | c.1574G>A | p.R525H | 4 | 1 | RUNX1 c.385C>G, p.L129V | 6 | 2 | 46,XX[20] |
| JAK2 c.1849G>T, p.V617F | 4 | 1 | |||||||||||
| 12† | AML | c.3G>A | p.M1I | 52 | 1 | c.1574G>A | p.R525H | 5 | 1 | RUNX1 c.776_777del, p.F259* | 5 | 1 | 46,XX[20] |
| NF1 c.2033dupC, p.I679fs | 45 | 1 | |||||||||||
| 13 | AML | c.3G>A | p.M1I | 50 | 1 | c.1574G>A | p.R525H | 3 | 1 | 46,XX[20] | |||
| 14† | AML | c.3G>A | p.M1I | 46 | 1 | c.1574G>A | p.R525H | 2 | 1 | trisomy 8 | |||
| 15† | AML | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 6 | 1 | 46,XX[20] | |||
| 16† | AML | c.3G>A | p.M1I | 50 | 1 | c.1574G>A | p.R525H | 7 | 1 | NI | |||
| 17† | AML | c.3G>A | p.M1I | 43 | 1 | c.1574G>A | p.R525H | 5 | 1 | 46,XY[20] | |||
| 18† | AML | c.3G>A | p.M1I | 49 | 1 | c.1574G>A | p.R525H | 7 | 1 | 46,XY[19] | |||
| 19* | AML | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 11 | 1 | 46,XY[20] | |||
| 20*,† | AML | c.3G>A | p.M1I | 44 | 1 | c.971G>A | p.C264Y | 5 | 1 | 45,X,-Y[6]/46,XY[14] | |||
| 21† | AML | c.3G>A | p.M1I | 45 | 1 | c.1037C>T | p.A346P | 3 | 1 | 46,XY[20] | |||
| 22† | AML | c.3G>A | p.M1I | 50 | 1 | ASXL1 c.1934dup, p.G646fs | 13 | 1 | 46, XY[20] | ||||
| KRAS c.35G>T, p.G12V | 13 | 1 | |||||||||||
| 23 | AML | c.3G>A | p.M1I | 41 | 1 | ASXL1 c.1934dup, p.G646fs | 28 | 1 | 45, XY, −7[16]/46, XY, [4]. | ||||
| 24† | AML | c.3G>A | p.M1I | 49 | 1 | RUNX1 c.743dupA, p.N248fs | 21 | 1 | 46,XY, del(5)(q13q33)[2]/47, sl,+21[2]/ | ||||
| TP53 c.827C>A, p.A276D | 7 | 1 | 46∼48, sdl1,t(1;2) (p36.3;q31), t(1;6)(32P;q27)t(1;2), +mar[cp16] | ||||||||||
| 25 | AML | c.323del | p.K108fs | 51 | 1 | c.1574G>A | p.R525H | 9 | 1 | TET2 c.3965T>A, p.L1322Q | 9 | 1 | 46,XY[20] |
| SRSF2 c.284C>T, p.P95L | 13 | 1 | |||||||||||
| TP53 c.743G>A, p.R248Q | 5 | 1 | |||||||||||
| 26 | AML | c.415_418dup | p.D140fs | 49 | 1 | c.1574G>A | p.R525H | 22 | 1 | ASXL1 c.1924_1928del, p.G644fs | 4 | 1 | 46,XX[20] |
| TET2 c.2456dup, p.Y819* | 4 | 1 | |||||||||||
| TET2 c.2459G>A, p.S820N | 4 | 2 | |||||||||||
| SH2B3 c.1200dup, p.Y401fs | 36 | 1 | |||||||||||
| 27 | AML | c.415_418dup | p.D140fs | 43 | 1 | c.1574G>A | p.R525H | 3 | 1 | ASXL1 c.1900_1922del, p.E635fs | 2 | 1 | 46,XY[20] |
| DNMT3A c.976C>T, p.R326C | 3 | 12 | |||||||||||
| CSF3R c.1640G>A, p.W547* | 48 | ||||||||||||
| 28† | AML | c.415_418dup | p.D140fs | 43 | 1 | c.1574G>A | p.R525H | 5 | 1 | ASXL1 c.1919_1929del, p.A640fs | 7 | 1 | 46,XY[20] |
| PHF6 c.834G>T, p.M278I | 13 | 2 | |||||||||||
| 29 | AML | c.415_418dup | p.D140fs | 50 | 1 | c.1574G>A | p.R525H | 7 | 1 | ASXL1 c.2275_2284del, p.Gln760fs | 3 | 1 | 46,XY, inv(11)(q21q23)[20] |
| TP53 c.830G>A, p.C277Y | 6 | 1 | |||||||||||
| 30*,† | AML | c.415_418dup | p.D140fs | 45 | 1 | c.1574G>A | p.R525H | 16 | 1 | TET2 c.1847del, p.P616fs | 1 | 1 | 46,XY[20] |
| TET2 c.782_786del, p.S261* | 1 | 1 | |||||||||||
| 31 | AML | c.415_418dup | p.D140fs | 45 | 1 | c.1574G>A | p.R525H | 1 | 1 | TET2 c.5577_5578del, p.I1859fs | 2 | 1 | NI |
| 32† | AML | c.415_418dup | p.D140fs | 46 | 1 | c.1574G>A | p.R525H | 2 | 1 | TET2 c.2340dup, p.V781fs | 3 | 1 | NI |
| DDX41 c.138 + 5G>T, p.? | 47 | 2 | |||||||||||
| 33 | AML | c.415_418dup | p.D140fs | 46 | 1 | c.1574G>A | p.R525H | 1 | 1 | CUX1 c.2459G>A, p.W820* | 1 | 1 | NI |
| 34† | AML | c.415_418dup | p.D140fs | 46 | 1 | c.1574G>A | p.R525H | 2 | 1 | 46,XX[20] | |||
| 35 | AML | c.415_418dup | p.D140fs | 42 | 1 | c.1574G>A | p.R525H | 6 | 1 | 46,XY[20] | |||
| 36 | AML | c.415_418dup | p.D140fs | 43 | 1 | c.1589G>A | p.G530D | 35 | 1 | ASXL1 c.3001dup, p.T1001fs | 34 | 1 | NI |
| EZH2 c.349C>T, p.Q117* | 31 | 1 | |||||||||||
| SETBP1 c.2608G>A, p.G870S | 4 | 1 | |||||||||||
| 37 | AML | c.415_418dup | p.D140fs | 46 | 1 | NI | |||||||
| 38† | AML | c.415_418dup | p.D140fs | 49 | 1 | 46,X,-X,der1, (X;1)(p11.3;p36.3), inv9(p12q13)c, +14[4]/46, XX, inv9c[16] | |||||||
| 39† | AML | c.415_418dup | p.D140fs | 47 | 1 | 46,XY[20] | |||||||
| 40† | AML | c.415_418dup | p.D140fs | 46 | 1 | 46,XY[19] | |||||||
| 41 | AML | c.415_418dup | p.D140fs | 44 | 1 | NI | |||||||
| 42 | AML | c.415_418dup | p.D140fs | 46 | 1 | 46,XX[20] | |||||||
| 43 | AML | c.415_418dup | p.D140fs | 44 | 1 | 46,XY[20] | |||||||
| 44 | AML | c.668dup | p.I224fs | 45 | 1 | c.1574G>A | p.R525H | 16 | 1 | ASXL1 c.2541del, p.T848fs | 17 | 1 | NI |
| PHF6 c.138 + 1G>A, p.? | 5 | 1 | |||||||||||
| PHF6 c.255C>G, p.C85W | 9 | 2 | |||||||||||
| 45† | AML | c.847del | p.L283fs | 43 | 1 | c.1574G>A | p.R525H | 5 | 1 | NI | |||
| 46 | AML | c.946_947del | p.M316fs | 52 | 1 | c.1574G>A | p.R525H | 3 | 1 | CUX1 c.3855del, p.S1286fs | 1 | 1 | 46,XY[20] |
| 47 | AML | c.1394del | p.G465fs | 46 | 1 | c.1574G>A | p.R525H | 1 | 1 | DNMT3A c.2026C>T, p.R676W | 6 | 1 | 46,XY[20] |
| ASXL1 c.2423_2427del, p.P808fs | 5 | 1 | |||||||||||
| ASXL1 c.2060_2061del, p.C687fs | 3 | 1 | |||||||||||
| 48 | AML | c.121C>T | p.Q41* | 48 | 1 | c.1574G>A | p.R525H | 1 | 1 | ASXL1 c.1900_1922del, p.E635fs | 10 | 1 | N/A |
| DNMT3A c.2256_2263del, p.W753* | 15 | 1 | |||||||||||
| PHF c.820C>T, p.R274* | 22 | 1 | |||||||||||
| 49 | AML | c.121C>T | p.Q41* | 45 | 1 | TET2 c.4133G>A, p.C1378Y | 44 | 1 | NI | ||||
| GATA2 c.599dup, p.S201* | 2 | 1 | |||||||||||
| KDM6A c.3704 + 1G>C, p.? | 67 | 1 | |||||||||||
| ZRSR2 c.505C>T, p.R169* | 93 | 1 | |||||||||||
| NPM1 c.860_863dup, p.W288fs | 35 | 1 | |||||||||||
| 50 | AML | c.475C>T | p.R159* | 51 | 1 | c.1589G>A | p.G530D | 2 | 1 | U2AF1 c.101C>A, p.S34Y | 3 | 1 | 46,XY[20] |
| 51 | AML | c.931C>T | p.R311* | 49 | 1 | c.1589G>A | p.G530D | 5 | 1 | PHF6 c.730-1G>A, p.? | 3 | 1 | NI |
| DNMT3A c.2255_2257del, p.F752del | 1 | 1 | |||||||||||
| 52 | AML | c.1105C>T | p.R369* | 46 | 1 | c.1574G>A | p.R525H | 18 | 1 | ASXL1 c.1900_1922del, p.E635fs | 13 | 1 | 46,XY[20] |
| SETBP1 c.2602G>A, p.D868N | 5 | 1 | |||||||||||
| SETBP1 c.2608G>A, p.G870S | 9 | 1 | |||||||||||
| SETBP1 c.2612T>C, p.I871T | 5 | 1 | |||||||||||
| 53 | AML | c.1105C>T | p.R369* | 47 | 1 | c.1574G>A | p.R525H | 1 | 1 | TET2 c.3632G>A, p.C1211Y | 4 | 1 | NI |
| SH2B3 c.794G>A, p.R265Q | 48 | 2 | |||||||||||
| 54 | AML | c.1108C>T | p.Q370* | 48 | 1 | c.1588G>A | p.G530S | 25 | 1 | JAK2 c.1849G>T, p.V617F | 22 | 1 | 46,XY[20] |
| 55 | AML | c.1504C>T | p.Q502* | 49 | 1 | c.1035G>C | p.E345D | 10 | 1 | ASXL1 c.2693G>A, p.W898* | 9 | 1 | 45,X,-Y[14]/46,XY[6] |
| 56† | AML | c.645-1G>T | p.? | 45 | 1 | c.1574G>A | p.R525H | 1 | 1 | ASXL1 c.3824C>G, p.S1275* | 1 | 1 | 46,XY[20] |
| DNMT3A c.1572T>A, p.C524* | 1 | 1 | |||||||||||
| 57 | AML | c.992_994del | p.K331del | 48 | 1 | c.1035G>C | p.E345D | 14 | 1 | ASXL1 p.E635fs | 5 | 1 | 46,XY, der(7)add(7)(p13) add(7)(q11.2)[10]/45,XY, -der(7)[5]/46,XY[13] |
| EZH2 p.N546K | 7 | 2 | |||||||||||
| 58*,† | AML | c.646C>G | p.L216V | 51 | 1 | c.1035G>C | p.E345D | 30 | 1 | DNMT3A c.2656C>T, p.Q886* | 31 | 1 | NI |
| 59 | AML | c.653G>A | p.G218D | 50 | 1 | c.1589G>A | p.G530D | 4 | 1 | TP53 c.488A>G, p.Y163C | 6 | 1 | NI |
| RUNX1 c.288_291delinsAAA, p.N96fs | 3 | 1 | |||||||||||
| DNMT3A c.1627G>T, p.G543C | 5 | 1 | |||||||||||
| DNMT3A c.1578C>G, p.Y526* | 5 | 1 | |||||||||||
| 60 | AML; breast cancer | c.773C>T | p.P258L | 56 | 1 | c.1574G>A | p.R525H | 15 | 1 | 46,XY[20] | |||
| 61 | AML | c.967C>T | p.R323C | 45 | 1 | c.1574G>A | p.R525H | 3 | 1 | TET2 c.3585_3588delnsAG, p.A1196fs | 5 | 1 | NI |
| SRSF2 c.284C>A, p.P95H | 4 | 1 | |||||||||||
| 62 | AML | c.1046T>A | p.M349K | 45 | 1 | 46,XY[20] | |||||||
| 63 | AML | c.1046T>A | p.M349K | 50 | 1 | 46,XY[20] | |||||||
| 64 | AML | c.1105C>G | p.R369G | 48 | 1 | c.1574G>A | p.R525H | 5 | 1 | 46,XX[20] | |||
| 65 | AML | c.1399G>T | p.D467Y | 45 | 1 | c.1589G>A | p.G530D | 13 | 1 | ASXL1 c.1934dup, p.G646fs | 13 | 1 | NI |
| ASXL1 c.1900_1922del, p.E635fs | 2 | 1 | |||||||||||
| EZH2 c.2022G>C, p.L674F | 3 | 1 | |||||||||||
| SETBP1 c.2608G>A, p.G870S | 4 | 1 | |||||||||||
| EZH2 c.2197T>A, p.Y733N | 10 | 1 | |||||||||||
| SETBP1 c.2612T>C, p.I871T | 8 | 1 | |||||||||||
| 66 | AML | c.1574G>A | p.R525H | 54 | 1 | NI | |||||||
| 67 | MDS | c.3G>A | p.M1I | 45 | 1 | c.1574G>A | p.R525H | 2 | 1 | DNMT3A c.1010C>G, p.S337* | 1 | 1 | NI |
| 68 | MDS | c.3G>A | p.M1I | 49 | 1 | c.1574G>A | p.R525H | 6 | 1 | 46,XY[20] | |||
| 69 | MDS | c.3G>A | p.M1I | 43 | 1 | c.1574G>A | p.R525H | 4 | 1 | 46,XX[20] | |||
| 70 | MDS | c.3G>A | p.M1I | 53 | 1 | c.1574G>A | p.R525H | 2 | 1 | 46,XY[20] | |||
| 71 | MDS | c.3G>A | p.M1I | 50 | 1 | c.694A>G | p.T232A | 3 | 1 | 46,XY[20] | |||
| 72 | MDS | c.3G>A | p.M1I | 49 | 1 | c.962C>T | p.P321L | 14 | 1 | DNMT3A c.1792C>T, p.598* | 14 | 1 | 46,XY[20] |
| 73 | MDS | c.3G>A | p.M1I | 49 | 1 | c.962C>T | p.P321L | 14 | 1 | 46,XY[20] | |||
| 74 | MDS | c.3G>A | p.M1I | 48 | 1 | c.1037C>T | p.A346P | 3 | 1 | 46,XY[20] | |||
| 75 | MDS | c.3G>A | p.M1I | 46 | 1 | c.1643T>A | p.L548H | 4 | 1 | 46,XX[20] | |||
| 76 | MDS; MM; MBL | c.3G>A | p.M1I | 50 | 1 | DNMT3A c.2645G>A, p.R882H | 15 | 1 | 46,XX[20] | ||||
| 77 | MDS | c.3G>A | p.M1I | 46 | 1 | PTPN11 c.215C>T, p.A72V | 42 | 1 | 47,XY,del(5)(q13q33), +21[2]/48,sl,t(9;21) (q10;q10), +21[12]/ | ||||
| RUNX1 c.1036dup, p.R346fs | 32 | 1 | |||||||||||
| 78 | MDS | c.3G>A | p.M1I | 49 | 1 | JAK2 c.1849G>T, p.V617F | 1 | 1 | 46,XY[20] | ||||
| 79 | MDS | c.3G>A | p.M1I | 49 | 1 | 45,X,-Y[6]/46,XY[14] | |||||||
| 80 | MDS | c.415_418dup | p.D140fs | 44 | 1 | c.1574G>A | p.R525H | 10 | 1 | ASXL1 c.4127dup, p.P1377fs | 12 | 1 | NI |
| 81 | MDS | c.415_418dup | p.D140fs | 45 | 1 | c.794C>T | p.P265L | 27 | 1 | DNMT3A c.1579C>T, p.Q527* | 9 | 1 | 46,XY[20] |
| 82 | MDS | c.1496dup | p.A500fs | 47 | 1 | c.1660G>A | p.E554K | 2 | 1 | 46,XY[20] | |||
| 83 | MDS | c.130C>T | p.Q44* | 60 | 1 | c.1126C>T | p.A376T | 2 | 1 | DNMT3A c.2207G>A, p.R736H | 7 | 1 | 46,XX[20] |
| 84 | MDS | c.475C>T | p.R159* | 47 | 1 | c.1589G>A | p.G530D | 6 | 1 | NI | |||
| c.1574G>A | p.R525H | 3 | 1 | ||||||||||
| c.1588G>A | p.G530S | 1 | 2 | ||||||||||
| 85 | MDS | c.931C>T | p.R311* | 51 | 1 | c.1574G>A | p.R525H | 6 | 1 | SRSF2 c.284C>T, p.P95L | 3 | 1 | NI |
| 86 | MDS | c.931C>T | p.R311* | 50 | 1 | c.1574G>A | p.R525H | 7 | 1 | NI | |||
| 87 | MDS | c.992_994del | p.K331del | 43 | 1 | c.1574G>A | p.R525H | 14 | 1 | TET2 c.3866G>T, p.C1289F | 3 | 1 | 46,XY[20] |
| 88 | MDS | c.992_994del | p.K331del | 48 | 1 | 46,XY[20] | |||||||
| 89 | MDS | c.566C>T | p.P189L | 49 | 1 | c.1574G>A | p.R525H | 4 | 1 | EZH2 c.434T>C, p.F145S | 3 | 1 | NI |
| 90 | MDS | c.710T>G | p.L237W | 49 | 1 | c.1589G>A | p.G530D | 13 | 1 | 46,XY[20] | |||
| 91 | MDS | c.1016G>A | p.R339H | 51 | 1 | IDH1 c.605del, p.S202fs | 31 | 2 | 45,X,-Y/46,XY[15] | ||||
| 92 | MDS | c.1015C>T | p.R339C | 49 | 1 | c.680C>T | p.T227M | 13 | 1 | TET2 c.1793del, p.N598fs | 2 | 1 | 46,XY[20] |
| 93 | MDS | c.1018T>A | p.Y340N | 47 | 1 | c.1574G>A | p.R525H | 7 | 1 | CUX1, c.2389del, p.Q797fs | 13 | 1 | 46,XY[20] |
| EZH2 c.371A>G, p.D124G | 15 | 2 | |||||||||||
| 94* | MDS | c.1105C>G | p.R369G | 47 | 1 | c.645-1G>A | p.? | 1 | 1 | TET2 c.1263del; p.G422fs | 31 | 1 | NI |
| TET2 c.3860_3869del, p.F1287fs | 1 | 1 | |||||||||||
| 95 | Pancytopenia | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 14 | 1 | JAK2 c.1849G>T, p.V617F | 15 | 1 | NI |
| TET2 c.1648C>T, p.R550* | 7 | 1 | |||||||||||
| 96 | Pancytopenia | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 2 | 1 | NI | |||
| 97 | Pancytopenia | c.415_418dup | p.D140fs | 46 | 1 | c.1574G>A | p.R525H | 9 | 1 | 46,XY, del(20)(q11.2q13.1) [1]/46,XY[20] | |||
| 98 | Pancytopenia | c.430del | p.T144fs | 45 | 1 | c.1574G>A | p.R525H | 4 | 1 | ASXL1c.4002del, p.S1335fs | 21 | 1 | NI |
| TP53 c.586C>T, p.R196* | 4 | 1 | |||||||||||
| TP53 c.916C>T, p.R306* | 3 | 1 | |||||||||||
| IDH2 c.419G>A, p.R140Q | 2 | 1 | |||||||||||
| 99 | Pancytopenia | c.946_947del | p.M316fs | 47 | 1 | c.1574G>A | p.R525H | 5 | 1 | ASXL1 c.2644C>T, p.Q882* | 6 | 1 | 46,XY[20] |
| PHF6 c.941T>C, p.I314T | 11 | 2 | |||||||||||
| 100 | Pancytopenia | c.1354del | p.L452fs | 48 | 1 | c.1574G>A | p.R525H | 8 | 1 | DNMT3A c.929T>C, p.I310T | 5 | 1 | 46,XY[20] |
| 101 | Pancytopenia | c.475C>T | p.R159* | 48 | 1 | c.1574G>A | p.R525H | 12 | 1 | SRSF2 c.284C>A, p.P95H | 10 | 1 | NI |
| STAG2 c.1243C>T, p.H415Y | 16 | 2 | |||||||||||
| 102 | Pancytopenia | c.1628C>G | p.S543* | 50 | 1 | c.1574G>A | p.R525H | 4 | 1 | ASXL1 c.1934dup, p.G646fs | 3 | 1 | NI |
| RUNX1 c.540del, p.F180fs | 3 | 1 | |||||||||||
| PHF6 c.941T>C, p.I314T | 7 | 1 | |||||||||||
| KDM6A c.2665del, p.T889fs | 1 | 1 | |||||||||||
| 103 | Pancytopenia | c.649T>C | p.S217P | 48 | 1 | c.1589G>A | p.G530D | 9 | 1 | ASXL1 c.2467_2468insA, p.L823fs | 7 | 1 | NI |
| KRAS c.437C>T, p.A146V | 3 | 1 | |||||||||||
| SRSF2 c.47T>A, p.L16H | 7 | 2 | |||||||||||
| 104 | Pancytopenia | c.773C>T | p.P258L | 47 | 1 | c.1574G>A | p.R525H | 4 | 1 | KDM6A c.3107delT, p.F1036fs | 2 | 1 | NI |
| SRSF2 c.284C>A, p.P95H | 7 | 1 | |||||||||||
| 105 | Pancytopenia | c.776A>G | p.Y259C | 48 | 1 | c.1574G>A | p.R525H | 6 | 1 | FBXW7 c.62G>A, p.G21D | 19 | 2 | 46,XY[20] |
| 106 | Pancytopenia | c.1016G>T | p.R339L | 50 | 1 | c.1574G>A | p.R525H | 1 | 1 | NI | |||
| 107 | Thrombocytopenia | c.415_418dup | p.D140fs | 45 | 1 | c.1574G>A | p.R525H | 10 | 1 | ASXL1 c.1900_1922del, p.E635fs | 2 | 1 | NI |
| CUX1 c.2161C>T, p.Q721* | 8 | 1 | |||||||||||
| SMC1A c.2132G>A, p.R711Q | 4 | 2 | |||||||||||
| 108 | Thrombocytopenia | c.1586_ 1587delCA | p.T529fs | 52 | 1 | TP53 c.1024C>T, p.R342* | 40 | 1 | 46,XX[20] | ||||
| 109 | Thrombocytopenia | c.3G>A | p.M1I | 45 | 1 | c.962C>T | p.P321L | 23 | 1 | RUNX1 c.281G>A, p.S94N | 30 | 1 | NI |
| TP53 c.742C>T, p.R248W | 35 | 1 | |||||||||||
| SRSF2 c.284C>G, p.P95R | 26 | 1 | |||||||||||
| 110 | Neutropenia | c.3G>A | p.M1I | 48 | 1 | c.1574G>A | p.R525H | 10 | 1 | NI | |||
| 111 | Neutropenia | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 1 | 1 | NI | |||
| 112 | Anemia | c.1105C>T | p.R369* | 45 | 1 | c.1574G>A | p.R525H | 5 | 1 | ASXL1 c.1934dup, p.G646fs | 7 | 1 | 46,XX,+1, der(1;7)(q10;p10) [16]/46,XX[4] |
| EZH2 c.786dup, p.N263fs | 4 | 1 | |||||||||||
| SETBP1 c.2613_2614delinsGC, p.I871_G872delinsMR | 4 | 2 | |||||||||||
| 113 | MPN | c.415_418dup | p.D140fs | 49 | 1 | 46,XX[20] | |||||||
| 114 | MPN | c.946_947del | p.M316fs | 48 | 1 | NI | |||||||
| 115 | MPN | c.916C>T | p.Q306* | 48 | 1 | 46,XX[20] | |||||||
| 116 | MPN | c.1141A>T | p.K381* | 44 | 1 | c.1574G>A | p.R525H | 7 | 1 | CUX1 c.988C>T, p.Q330* | 7 | 1 | 46,XY[20] |
| 117 | AML | c.6G>T | p.E2D | 48 | 2 | FLT3 c.1805_1806ins42, p.K602_W603ins14 | n/a | 1 | 46,XX[20] | ||||
| NPM1 c.863_864insCCTG, p.W288fs | 35 | 1 | |||||||||||
| WT1 c.1141_1144dup, p.A382fs | 8 | 1 | |||||||||||
| 118* | AML | c.55G>A | p.G19R | 48 | 2 | FLT3 ITD c.1837 + 11_1837 + 12ins114, p.? | n/a | 1 | NI | ||||
| NPM1 c.863_864insCCTG, p.W288fs | 43 | 1 | |||||||||||
| IDH1 c.395G>A, p.R132H | 41 | 1 | |||||||||||
| DNMT3A c.1627G>T, p.G543C | 42 | 1 | |||||||||||
| 119* | AML | c.97T>C | p.Y33H | 45 | 2 | FLT3 c.1770_1811dup42, p.W603_E604ins14 | n/a | 1 | NI | ||||
| NPM1 c.860_863dup, p.W288fs | 18 | 1 | |||||||||||
| DNMT3A c.860_863dup, p.W288fs | 27 | 1 | |||||||||||
| TET2 c.2490dup, p.Q831fs | 23 | 1 | |||||||||||
| 120 | AML | c.465G>A | p.M155I | 47 | 2 | NPM1 c.860_863dup, p.W288fs | 7 | 1 | 46,XY[20] | ||||
| SRSF2 c.284C>G, p.P95R | 28 | 1 | |||||||||||
| TET2 c.2244dup, p.Q749fs | 44 | 1 | |||||||||||
| 121 | AML | c.465G>A | p.M155I | 47 | 2 | NPM1 c.863_864insCTTG, p.W288Cfs | 33 | 1 | 46,XY[20] | ||||
| GATA2 c.599dup, p.S201* | 2 | 1 | |||||||||||
| 122* | AML | c.491G>A | p.R164N | 48 | 2 | c.1774A>T | p.I592F | 35 | 2 | NPM1 c.863_864insCTTG, p.W288Cfs | 33 | 1 | 46,XX[20] |
| 123 | AML | c.1528C>T | p.P510S | 48 | 2 | NPM1 c.860_863dup, p.W288fs | 34 | 1 | 46,XY[20] | ||||
| SRSF2 c.284C>T, p.P95L | 42 | 1 | |||||||||||
| KRAS c.35G>A, p.G12D | 11 | 1 | |||||||||||
| 124 | AML | c.380T>A | p.M127K | 49 | 2 | FLT3 c.2505T>G, p.D835E | 3 | 1 | 46,XY,+12, der(17)t(17;18) (p10;q10),-18[8] | ||||
| TP53 c.400T>A, p.F134I | 46 | 1 | |||||||||||
| TP53 c.458_462del, p.P153fs | 3 | 1 | |||||||||||
| U2AF1 c.101C>A, p.S34Y | 6 | 1 | |||||||||||
| KRAS c.351A>T, p.K117N | 27 | 1 | |||||||||||
| 125 | AML | c.465G>A | p.M155I | 47 | 2 | FLT3 p.N841K | 7 | 1 | 46,XY[20] | ||||
| KRAS c.35G>T, p.G12V | 3 | 1 | |||||||||||
| 126 | AML | c.199G>C | p.G67R | 50 | 2 | ASXL1 c.2959G>T, p.G987* | 47 | 1 | 46,XY[20] | ||||
| CEBPA c.985_986insCC, E329fs | 42 | 1 | |||||||||||
| CEBPA c.68del, p.P23fs | 46 | 1 | |||||||||||
| IDH2 c.418C>T, p.R140W | 48 | 1 | |||||||||||
| JAK2 c.1849G>T, p.V617F | 6 | 1 | |||||||||||
| 127 | AML | c.199G>C | p.G67R | 48 | 2 | ASXL1 c.2959G>T, p.G987* | 22 | 1 | 46,XY[20] | ||||
| IDH2 c.418C>T, p.R140W | 25 | 1 | |||||||||||
| SRSF2 c.284C>A, p.P95H | 28 | 1 | |||||||||||
| STAG2 c.1196 + 1G>A, p.? | 4 | 1 | |||||||||||
| STAG2 c.1999del, p.R667fs | 15 | 1 | |||||||||||
| 128* | AML | c.465G>A | p.M155I | 51 | 2 | 48,XY,+8,+22[20] | |||||||
| 129 | AML | c.883G>A | p.A295T | 53 | 2 | TP53 c.743G>A, p.R248Q | 35 | 1 | 44,XY,-3,add(5) (q11.2),+8, add(8)(q22), der(12;17)(q10;q10), | ||||
| TP53 c.818G>A, p.R273H | 15 | 1 | −14,i(14)(q10), i(21)(q10)[12]/ 43-45, sl,-add(5)(q11.2), i(5)(q10)[cp4]/88 < 4n>,slx2[1] | ||||||||||
| 130 | AML | c.893T>C | p.I298T | 49 | 2 | DNMT3A c.989G>A, p.W330* | 2 | 1 | NI | ||||
| TP53 c.844C>T, p.R282W | 61 | 1 | |||||||||||
| TP53 c.535C>T, p.H179Y | 4 | 1 | |||||||||||
| PTPN11 c.1504T>C, p.S502P | 2 | 1 | |||||||||||
| 131 | AML | c.1063G>A | p.E355K | 47 | 2 | PHF6 c.418 + 2T>C, p.? | 21 | 1 | 46,XX[7] | ||||
| KMT2A c.10462C>T, p.Gln3488* | 25 | 2 | |||||||||||
| 132 | AML | c.465G>A | p.M155I | 48 | 2 | NRAS c.181C>A, p.Q61K | 15 | 1 | 46,XY,inv(16) (p13.1q22)[20] | ||||
| 133 | AML | c.465G>A | p.M155I | 47 | 2 | CSF3R c.1843A>G, p.T615A CSF3R c.1853C>T, p.T618I | 22 4 | 1 1 | 46,XX, der(8)t(8;21) (q22;q22), der(8)?(8pter-> 8p22::?:: 8p11.2-> 8q?13::8q22 ->8qter), der(21)(21pter-> 21q22::8?q13-> 8q?22:: 8q22-> 8qter)[7]/45, sl,-X[8]/46,XX[5] | ||||
| 134 | AML | c.138 + 5G>A | p.? | 50 | 2 | NRAS c.181C>A, p.Q61Lys | 39 | 1 | 47,XY, +8,inv(16) (p13.1q22)[20] | ||||
| 135 | MDS | c.367G>A | p.G123S | 48 | 2 | ASXL1 c.2239_2244delinsCC, p.S747fs | 42 | 1 | 46,XX[20] | ||||
| ASXL1 c.1934dup, p.G646fs | 2 | 1 | |||||||||||
| TET2 c.5543C>G, p.S1848* | 1 | 1 | |||||||||||
| EZH2 c.1119dup, p.T374fs | 2 | 1 | |||||||||||
| IDH2 c.419G>A, p.R140Q | 31 | 1 | |||||||||||
| SF3B1 c.2347G>A, p.E783K | 2 | 2 | |||||||||||
| JAK2 c.1849G>T, p.V617F | 1 | 1 | |||||||||||
| 136 | MDS | c.465G>A | p.M155I | 43 | 2 | JAK2 c.1849G>T, p.V617F | 27 | 1 | NI | ||||
| DNMT1 c.4663G>A, p.V1555M | 32 | 2 | |||||||||||
| SH2B3 c.127C>T, p.R43C | 35 | 2 | |||||||||||
| 137 | MDS | c.644 + 5G>C | p.? | 43 | 2 | SH2B3 c.947_953del, p.E316fs | 21 | 1 | 47,XY, +9,del(20) (q11.2q13.1)[19]/46,XY[1] | ||||
| JAK2 c.1849G>T, p.V617F | 40 | 1 | |||||||||||
| 138 | MDS | c.523G>A | p.G175S | 48 | 2 | NRAS c.34G>A, p.G12S | 44 | 1 | NI | ||||
| TET2 c.2290dup, p.Q764fs | 50 | 1 | |||||||||||
| GATA2 c.599del, p.G200fs | 1 | 1 | |||||||||||
| SRSF2 c.284C>A, p.P95H | 47 | 1 | |||||||||||
| 139 | MDS | c.529C>T | p.P177S | 45 | 2 | SMC1A c.197A>G, p.H66R | 9 | 2 | NI | ||||
| BCOR c.441dup, p.Ile148fs | 50 | 1 | |||||||||||
| U2AF1 c.101C>T, p.S34F | 37 | 1 | |||||||||||
| RUNX1 c.601del, p.R201fs | 10 | 1 | |||||||||||
| 140 | MDS | c.1301C>T | p.P434L | 49 | 2 | GATA2 c.599dup; p.S201* | 31 | 1 | 46,XY[20] | ||||
| NPM1 c.867_868insAGGA, p.W290fs | 48 | 1 | |||||||||||
| 141 | MDS | c.1528C>T | p.P510S | 48 | 2 | ASXL1 c.1934dup, p.G646fs | 35 | 1 | NI | ||||
| CBL c.800G>A, p.G267D | 2 | 2 | |||||||||||
| 142 | MDS | c.1704C>G | p.C568W | 48 | 2 | KIT c.2446_2447delinsAT, p.D816I | 10 | 1 | NI | ||||
| KIT c.2447A>T, p.D816V | 1 | 1 | |||||||||||
| PTPN11 c.154A>G, p.T52A | 5 | 1 | |||||||||||
| PTPN11 c.1508G>A, p.G503E | 3 | 2 | |||||||||||
| 143 | MDS/MPN | c.1760G>C | p.G587A | 47 | 2 | CSF3R c.1853C>T, p.T618I | 3 | 1 | 47,XY,+8[20] | ||||
| KRAS c.436G>C, p.A146P | 37 | 1 | |||||||||||
| ASXL1 c.1900_1922del, p.E635fs | 44 | 1 | |||||||||||
| STAG2 c.1191dup, p.Q399fs | 92 | 1 | |||||||||||
| 144 | MDS | c.1276G>A | p.E426K | 49 | 2 | SRSF2 c.284C>A, p.P95H | 46 | 1 | NI | ||||
| IDH2 c.419G>A, p.R140Q | 45 | 1 | |||||||||||
| 145 | MDS | c.1528C>T | p.P510S | 48 | 2 | TP53 c.734G>A, p.G45D | 20 | 1 | NI | ||||
| 146 | MDS | c.1663G>A | p.A555T | 43 | 2 | TP53 c.706dup, p.Y236fs | 39 | 1 | 44-47,XY, del(5)(q22q35), -7,der(11)t(11;13) (p15;q14), | ||||
| TP53 c.713G>A, p.C238Y | 37 | 1 | |||||||||||
| 147 | Pancytopenia | c.27G>A | p.K9K | 43 | 2 | NI | |||||||
| 148 | Pancytopenia | c.465G>A | p.M155I | 50 | 2 | SF3B1 c.1996A>G, p.K666E | 19 | 1 | 45,X,-Y[14]/46,XY[6] | ||||
| U2AF1 c.101C>T, p.S34F | 5 | 1 | |||||||||||
| 149 | Pancytopenia | c.556A>T | p.M186L | 47 | 2 | ETNK1 c.731A>G, p.N244S | 24 | 1 | NI | ||||
| Small cell carcinoma | KRAS c.468C>G, p.F156L | 18 | 2 | ||||||||||
| 150 | Pancytopenia | c.656G>A | p.R219H | 49 | 2 | c.679A>G | p.T227A | 30 | 2 | 46,XY[20] | |||
| 151 | Pancytopenia | c.845G>A | p.R282H | 40 | 2 | TP53 c.377A>G, p.Y126C | 67 | 1 | NI | ||||
| BRAF c.1391G>T, p.G464V | 38 | 1 | |||||||||||
| 152 | Pancytopenia | c.881G>T | p.C294F | 48 | 2 | CBL c.1211G>A, p.C404Y | 2 | 1 | NI | ||||
| 153 | Pancytopenia | c.1477T>G | p.S493A | 47 | 2 | ASXL1 c.1900_1922del, p.E635fs | 15 | 1 | NI | ||||
| ASXL1 c.1934dup, p.G646fs | 3 | 1 | |||||||||||
| ASXL1 c.2295del, p.S766fs | 1 | 1 | |||||||||||
| SH2B3 c.703C>G, p.R235G | 16 | 1 | |||||||||||
| 154 | Thrombocytopenia | c.465G>A | p.M155I | 50 | 2 | 46,XY[20] | |||||||
| 155 | Thrombocytopenia | c.707C>T | p.T236M | 47 | 2 | NI | |||||||
| 156 | Thrombocytopenia | c.138 + 5G>A | p.? | 54 | 2 | NI | |||||||
| 157 | Thrombocytopenia | c.644 + 5G>C | p.? | 47 | 2 | RAD21 c.507_508del, p.E169fs | 38 | 1 | NI | ||||
| NRAS c.35G>A, p.G12D | 20 | 1 | |||||||||||
| TP53 c.818G>A, p.R273H | 31 | 1 | |||||||||||
| TP53 c.559 + 1G>A, p.? | 42 | 1 | |||||||||||
| 158 | Neutropenia | c.138 + 5G>A | p.? | 50 | 2 | NI | |||||||
| 159 | Anemia | c.751C>T | p.P251S | 46 | 2 | 46,XX[20] | |||||||
| 160 | Anemia | c.1748C>T | p.A583V | 47 | 2 | PTPN11 c.227A>T, p.E76V | 45 | 1 | NI | ||||
| 161 | MPN, PV | c.23G>A | p.R8Q | 47 | 2 | JAK2 c.1849G>T, p.V617F | 45 | 1 | 46,XY[20] | ||||
| 162 | MPN, PV | c.94G>A | p.D32N | 48 | 2 | JAK2 c.1849G>T, p.V617F | 85 | 1 | 46,XY[20] | ||||
| 163 | MPN, ET | c.398C>T | p.A133V | 44 | 2 | CALR c.1122_1125del, p.K374fs | 44 | 1 | NI | ||||
| U2AF1 c.470A>G, p.Q157R | 45 | 1 | |||||||||||
| ZRSR2 c.236_237del, p.E79Afs | 87 | 1 | |||||||||||
| 164 | MPN | c.644T>C | p.I215T | 48 | 2 | JAK2 c.1849G>T, p.V617F | 26 | 1 | NI | ||||
| ASXL1 c.1934dup, p.G646fs | 1 | 1 | |||||||||||
| 165 | MPN | c.647T>C | p.L216P | 48 | 2 | c.1655T>G | p.L552R | 14 | 2 | JAK2 c.1849G>T, p.V617F | 12 | 1 | NI |
| ASXL1 c.1900_1922del, p.E635fs | 8 | 1 | |||||||||||
| 166 | MPN, ET | c.707C>T | p.T236M | 50 | 2 | JAK2 c.1849G>T, p.V617F | 18 | 1 | 46,XY[20] | ||||
| SF3B1 c.2110A>T, p.I704F | 15 | 1 | |||||||||||
| 167 | MPN | c.1471G>A | p.V491I | 52 | 2 | JAK2 c.1849G>T, p.V617F | 47 | 1 | 46,XY[20] | ||||
| TP53 c.743G>A, p.R248Q | 6 | 1 | |||||||||||
| TP53 c.742C>T, p.R248W | 5 | 1 | |||||||||||
| NF1 c.2035del, p.I679fs | 15 | 1 | |||||||||||
| 168 | MPN, ET | c.1630G>T | p.V544L | 49 | 2 | JAK2 c.1849G>T, p.V617F | 26 | 1 | 46,XY[20] | ||||
| SF3B1 c.1997A>C, p.K666T | 16 | 1 | |||||||||||
| 169 | MPN, ET | c.138 + 5G>T | p.? | 49 | 2 | NI | |||||||
| 170 | MPN, ET | c.465G>A | p.M155T | 49 | 2 | 46,XX[20] | |||||||
| 171 | MPN | c.560A>G | p.K187R | 46 | 2 | NI | |||||||
| 172 | LPL with pancytopenia | c.490C>T | p.R164W | 50 | 2 | SF3B1 c.2098A>G, p.K700E | 41 | 1 | 46,XY[20] | ||||
| 173 | γ heavy chain disease | c.490C>T | p.R164W | 49 | 2 | 46,XY[20] | |||||||
| MYD88 negative LPL | |||||||||||||
| 174 | CLL; breast cancer | c.490C>T | p.R164W | 47 | 2 | 46,XY[20] | |||||||
| 175 | CLL | c.751C>T | p.P251S | 50 | 2 | 46,XY, del(13)(q12;q22), add(18)(p11.20[4]/46,XY[16] | |||||||
| 176 | MM | c.29G>A | p.R10Q | 46 | 2 | TP53 c.376T>G, p.Y126D | 24 | 1 | 44∼46,X, add(X)(p10), del(6)(q21q23), i(8)(q10), | ||||
| DNMT3A c.2339T>C, p.I780T | 18 | 2 | t(11;14)(q13;q32), del(13)(q12q22),- 14,-17,+1∼ 2mar[cp4]/46,XX[16] | ||||||||||
| KDM6A c.1354_1355del, p.G452fs | 26 | 1 | |||||||||||
| 177 | AML | c.1574G>A | p.R525H | 12 | 1 | ASXL1 c.1774C>T, p.Q592* | 4 | 1 | 46,XY,t(1;4)? (q21;q31)[2]/46,XY[6] | ||||
| PHF6 c.58del, p.C20fs | 17 | 1 | |||||||||||
| 178 | AML | c.1574G>A | p.R525H | 25 | 1 | ASXL1 c.2077C>T, p.R693* | 2 | 1 | 46,XY[21] | ||||
| 179 | AML | c.1574G>A | p.R525H | 1 | 1 | CBL c.1211G>A, p.C404Y | 15 | 1 | 46,XX[20] | ||||
| U2AF2 c.766G>A, p.D256G | 16 | 2 | |||||||||||
| 180 | AML | c.1574G>A | p.R525H | 4 | 1 | SH2B3 c.519_523del, p.R175fs | 3 | 1 | 45,X,-Y[5]/46,XY[15] | ||||
| PHF6 c.635G>T, p.C212F | 8 | 2 | |||||||||||
| 181 | AML | c.1574G>A | p.R525H | 16 | 1 | SRSF2 c.284C>A, p.P95H | 17 | 1 | 46,X,del(X)?(q22q26)[2]/46,XX[18] | ||||
| 182 | AML | c.1574G>A | p.R525H | 9 | 1 | SETBP1 c.2602G>C, p.D868H | 2 | 1 | NI | ||||
| c.944A>G | p.H315R | 8 | 2 | CUX1 c.439C>T, p.R147* | 7 | 1 | |||||||
| STAG2 1693G>T, p.E565* | 5 | 1 | |||||||||||
| EZH2 c.1967C>T, p.A656V | 4 | 1 | |||||||||||
| ASXL1 c.2156del, p.E719fs | 8 | 1 | |||||||||||
| 183 | AML | c.1589G>A | p.G530D | 15 | 1 | ASXL1 c.2985del, p.H995fs | 5 | 1 | 46,XY[20] | ||||
| c.629A>G | p.Q210R | 15 | 2 | ASXL1 c.2083C>T, p.R695* | 3 | 1 | |||||||
| ASXL1 c.2077C>T, p.H963* | 5 | 1 | |||||||||||
| EZH2 c.2213del, p.A738fs | 15 | 1 | |||||||||||
| NF1 c.3774G>C, p.W1258C | 12 | 1 | |||||||||||
| 184 | AML; DLBCL | c.1589G>A | p.G530D | 6 | 1 | 46,XY[20] | |||||||
| 185 | MDS | c.1574G>A | p.R525H | 4 | 1 | DNMT3A c.2645G>A, p.R882H | 3 | 1 | 46,XX[11] | ||||
| 186 | MDS | c.1574G>A | p.R525H | 9 | 1 | 45,X,-Y[4]/46,XY[16] | |||||||
| 187 | MDS | c.1574G>A | p.R525H | 10 | 1 | 46,XX[20] | |||||||
| 188 | MDS | c.1574G>A | p.R525H | 1 | 1 | 46,XY[20] | |||||||
| 189 | MDS | c.1574G>A | p.R525H | 4 | 1 | 46,XY[20] | |||||||
| 190 | AML | c.157C>G | p.R53G | 5 | 2 | SF3B1 c.2098A>G, p.K700E | 32 | 1 | NI | ||||
| PTPN11 c.214G>A, p.A72T | 37 | 1 | |||||||||||
| 191 | AML | c.1760_ 1761TT | p.G587V | 4 | 2 | DNMT3A c.2645G>A, p.R882H | 7 | 1 | 46,XY[20] | ||||
| DNMT3A c.2095G>A, p.G699R | 3 | 2 | |||||||||||
| 192 | MDS | c.622C>G | p.Q208E | 14 | 2 | SETBP1 c.2608G>A, p.G870S | 6 | 1 | 46,XY,del(7)(q22)[6]/46,XY[14] | ||||
| SETBP1 c.2602G>A, p.D868N | 2 | 1 | |||||||||||
| DNMT3A c.2645G>A, p.R882H12 | 12 | 1 | |||||||||||
| EVT6 c.313C>G, p.R105G | 2 | 2 | |||||||||||
| TET2 c.4079T>C, p.L1360P | 1 | 2 | |||||||||||
| 193 | MDS | c.1199G>T | p.R400L | 3 | 2 | ASXL1 c.2324T>G, p.L775* | 20 | 1 | 46,XY, del(5)(q31q33) [13]/46,XX[7] | ||||
| 194 | MDS | c.1369G>C | p.V457L | 2 | 2 | TP53 c.818G>A, p.R273H | 30 | 1 | 46,XY, del(5)(q31q33) [14]/46,XX[6] | ||||
| TP53 c.578A>C, p.H193P | 2 | 2 | |||||||||||
| 195 | Neutropenia | c.1775T>A | p.I592N | 4 | 2 | SRSF2 c.284C>T, p.P95L | 4 | 1 | NI | ||||
| RUNX1 c.606dup, p.P203fs | 2 | 1 | |||||||||||
| TET2 c.330G>C, p.Lys110N | 3 | 2 | |||||||||||
| 196* | Normal | c.3G>A | p.M1I | 53 | 1 | 46,XY[20] | |||||||
| 197* | Normal | c.3G>A | p.M1I | 50 | 1 | 46,XX[20] | |||||||
| 198* | Normal | c.415_418dup | p.D140fs | 46 | 1 | Not done | |||||||
| 199* | Normal | c.415_418dup | p.D140fs | 45 | 1 | NI | |||||||
| 200* | Normal | c.415_418dup | p.D140fs | 56 | 1 | 46,XY[20] | |||||||
| 201* | Normal | c.992_994del | p.K331del | 47 | 1 | Not done | |||||||
| 202 | Normal∼(donor) | c.931C>T | p.311* | 50 | 1 | 47,XX,+8[19] | |||||||
| 203 | Normal∼(donor) | c.465G>A | p.M155I | 46 | 2 | 46,XY[20] | |||||||
| 204 | Normal∼(donor) | c.1693G>A | p.V565M | 50 | 2 | 46,XY[20] | |||||||
| 205 | Normal∼(donor) | c.1585dup | p.T529fs | 46 | 1 | 44,XY,inc[1]//46,XX[16] |
| Patient . | Diagnosis . | gl DDX41 . | gl DDX41 . | VAF . | Tier . | s DDX41 . | s DDX41 . | VAF . | Tiers . | Concomitant variants . | VAF (%) . | Tier . | Cytogenetics . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASXL1 c.1900_1922del, p.E635fs | 19 | 1 | |||||||||||
| 1* | AML | c.3G>A | p.M1I | 44 | 1 | c.1574G>A | p.R525H | 6 | 1 | PFH6 c.811G>A, p.E271K | 12 | 1 | 46,XX[20] |
| CUX1 c.607 + 1G>A, p.? | 3 | 1 | |||||||||||
| CUX1 c.2786del, p.P929fs | 1 | 1 | |||||||||||
| 2† | AML | c.3G>A | p.M1I | 46 | 1 | c.1574G>A | p.R525H | 6 | 1 | ASXL1 c.1900_1922del, p.E635fs | 6 | 1 | 46,XX[20] |
| PHF6 c.940A>T, p.I314F | 4 | 2 | |||||||||||
| CUX1 c.2485C>T, p.Q829* | 5 | 1 | |||||||||||
| 3 | AML | c.3G>A | p.M1I | 48 | 1 | c.1574G>A | p.R525H | 0 | 1 | ASXL1 c.1627G>T, p.E543* | 2 | 1 | NI |
| PHF6 c.880A>G, p.I294V | 3 | 2 | |||||||||||
| 4† | AML | c.3G>A | p.M1I | 48 | 1 | c.1574G>A | p.R525H | 16 | 1 | ASXL1 c.1779dup, p.C594fs | 8 | 1 | 45,X,-Y[6]/46,XY[14] |
| SF3B1 c.2098A>G, p.K700E | 4 | 1 | |||||||||||
| JAK2 c.1849G>T, p.V617F | 5 | 1 | |||||||||||
| 5 | AML | c.3G>A | p.M1I | 48 | 1 | c.1574G>A | p.R525H | 3 | 1 | ASXL1 c.2905_ 2926delinsTACTGTT, p.D969_N971delinsYC* | 5 | 1 | NI |
| TP53 c.850A> C, p.T284P | 5 | ||||||||||||
| KMT2A c.2830_2847dup, p.D944_T949dup | 33 | 2 | |||||||||||
| 6*,† | AML | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 1 | 1 | ASXL1 c.1761_1768del, p.Q588fs | 20 | 1 | 45,XY,del(5)(q15q35), der(7;16)(q10;q10), add(10)(p11.2), |
| BCORL1 c.1339C>T, p.Q447* | 1 | 1 | add(10)(q24),+20, -22[13]/45,XY,-4, del(5)(q15q35), | ||||||||||
| TP53 c.743G>A, p.R248Q | 2 | 1 | der(7)t(7;9)(p11.2;q13), add(12)(p12), -13,+14, | ||||||||||
| TP53 c.817C>T, p.R273C | 1 | 1 | add(14)(q32), add(18)(q21),-22,+mar[7] | ||||||||||
| 7 | AML | c.3G>A | p.M1I | 45 | 1 | c.1574G>A | p.R525H | 5 | 1 | ASXL1 c.3030_3031delinsTT, p.E1011* | 3 | 1 | 46,XY[20] |
| ASXL1 c.2122del, p.Q708fs | 3 | 1 | |||||||||||
| 8 | AML | c.3G>A | p.M1I | 51 | 1 | c.1574G>A | p.R525H | 7 | 1 | ASXL1 c.1960dup, p.A654fs | 6 | 1 | 46,XY[20] |
| 9† | AML | c.3G>A | p.M1I | 58 | 1 | c.1574G>A | p.R525H | 3 | 1 | ASXL1 c.3824C>G, p.S1275* | 11 | 1 | 46,XY[20] |
| ZRSR2 c.202_203del, p.R68fs | 5 | 1 | |||||||||||
| 10*,† | AML | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 1 | 1 | DNMT3A c.1015-1G>C, p.? | 5 | 1 | 46,XX[20] |
| SETBP1 c.1977T>A, p.D659G | 1 | 2 | |||||||||||
| 11 | AML | c.3G>A | p.M1I | 51 | 1 | c.1574G>A | p.R525H | 4 | 1 | RUNX1 c.385C>G, p.L129V | 6 | 2 | 46,XX[20] |
| JAK2 c.1849G>T, p.V617F | 4 | 1 | |||||||||||
| 12† | AML | c.3G>A | p.M1I | 52 | 1 | c.1574G>A | p.R525H | 5 | 1 | RUNX1 c.776_777del, p.F259* | 5 | 1 | 46,XX[20] |
| NF1 c.2033dupC, p.I679fs | 45 | 1 | |||||||||||
| 13 | AML | c.3G>A | p.M1I | 50 | 1 | c.1574G>A | p.R525H | 3 | 1 | 46,XX[20] | |||
| 14† | AML | c.3G>A | p.M1I | 46 | 1 | c.1574G>A | p.R525H | 2 | 1 | trisomy 8 | |||
| 15† | AML | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 6 | 1 | 46,XX[20] | |||
| 16† | AML | c.3G>A | p.M1I | 50 | 1 | c.1574G>A | p.R525H | 7 | 1 | NI | |||
| 17† | AML | c.3G>A | p.M1I | 43 | 1 | c.1574G>A | p.R525H | 5 | 1 | 46,XY[20] | |||
| 18† | AML | c.3G>A | p.M1I | 49 | 1 | c.1574G>A | p.R525H | 7 | 1 | 46,XY[19] | |||
| 19* | AML | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 11 | 1 | 46,XY[20] | |||
| 20*,† | AML | c.3G>A | p.M1I | 44 | 1 | c.971G>A | p.C264Y | 5 | 1 | 45,X,-Y[6]/46,XY[14] | |||
| 21† | AML | c.3G>A | p.M1I | 45 | 1 | c.1037C>T | p.A346P | 3 | 1 | 46,XY[20] | |||
| 22† | AML | c.3G>A | p.M1I | 50 | 1 | ASXL1 c.1934dup, p.G646fs | 13 | 1 | 46, XY[20] | ||||
| KRAS c.35G>T, p.G12V | 13 | 1 | |||||||||||
| 23 | AML | c.3G>A | p.M1I | 41 | 1 | ASXL1 c.1934dup, p.G646fs | 28 | 1 | 45, XY, −7[16]/46, XY, [4]. | ||||
| 24† | AML | c.3G>A | p.M1I | 49 | 1 | RUNX1 c.743dupA, p.N248fs | 21 | 1 | 46,XY, del(5)(q13q33)[2]/47, sl,+21[2]/ | ||||
| TP53 c.827C>A, p.A276D | 7 | 1 | 46∼48, sdl1,t(1;2) (p36.3;q31), t(1;6)(32P;q27)t(1;2), +mar[cp16] | ||||||||||
| 25 | AML | c.323del | p.K108fs | 51 | 1 | c.1574G>A | p.R525H | 9 | 1 | TET2 c.3965T>A, p.L1322Q | 9 | 1 | 46,XY[20] |
| SRSF2 c.284C>T, p.P95L | 13 | 1 | |||||||||||
| TP53 c.743G>A, p.R248Q | 5 | 1 | |||||||||||
| 26 | AML | c.415_418dup | p.D140fs | 49 | 1 | c.1574G>A | p.R525H | 22 | 1 | ASXL1 c.1924_1928del, p.G644fs | 4 | 1 | 46,XX[20] |
| TET2 c.2456dup, p.Y819* | 4 | 1 | |||||||||||
| TET2 c.2459G>A, p.S820N | 4 | 2 | |||||||||||
| SH2B3 c.1200dup, p.Y401fs | 36 | 1 | |||||||||||
| 27 | AML | c.415_418dup | p.D140fs | 43 | 1 | c.1574G>A | p.R525H | 3 | 1 | ASXL1 c.1900_1922del, p.E635fs | 2 | 1 | 46,XY[20] |
| DNMT3A c.976C>T, p.R326C | 3 | 12 | |||||||||||
| CSF3R c.1640G>A, p.W547* | 48 | ||||||||||||
| 28† | AML | c.415_418dup | p.D140fs | 43 | 1 | c.1574G>A | p.R525H | 5 | 1 | ASXL1 c.1919_1929del, p.A640fs | 7 | 1 | 46,XY[20] |
| PHF6 c.834G>T, p.M278I | 13 | 2 | |||||||||||
| 29 | AML | c.415_418dup | p.D140fs | 50 | 1 | c.1574G>A | p.R525H | 7 | 1 | ASXL1 c.2275_2284del, p.Gln760fs | 3 | 1 | 46,XY, inv(11)(q21q23)[20] |
| TP53 c.830G>A, p.C277Y | 6 | 1 | |||||||||||
| 30*,† | AML | c.415_418dup | p.D140fs | 45 | 1 | c.1574G>A | p.R525H | 16 | 1 | TET2 c.1847del, p.P616fs | 1 | 1 | 46,XY[20] |
| TET2 c.782_786del, p.S261* | 1 | 1 | |||||||||||
| 31 | AML | c.415_418dup | p.D140fs | 45 | 1 | c.1574G>A | p.R525H | 1 | 1 | TET2 c.5577_5578del, p.I1859fs | 2 | 1 | NI |
| 32† | AML | c.415_418dup | p.D140fs | 46 | 1 | c.1574G>A | p.R525H | 2 | 1 | TET2 c.2340dup, p.V781fs | 3 | 1 | NI |
| DDX41 c.138 + 5G>T, p.? | 47 | 2 | |||||||||||
| 33 | AML | c.415_418dup | p.D140fs | 46 | 1 | c.1574G>A | p.R525H | 1 | 1 | CUX1 c.2459G>A, p.W820* | 1 | 1 | NI |
| 34† | AML | c.415_418dup | p.D140fs | 46 | 1 | c.1574G>A | p.R525H | 2 | 1 | 46,XX[20] | |||
| 35 | AML | c.415_418dup | p.D140fs | 42 | 1 | c.1574G>A | p.R525H | 6 | 1 | 46,XY[20] | |||
| 36 | AML | c.415_418dup | p.D140fs | 43 | 1 | c.1589G>A | p.G530D | 35 | 1 | ASXL1 c.3001dup, p.T1001fs | 34 | 1 | NI |
| EZH2 c.349C>T, p.Q117* | 31 | 1 | |||||||||||
| SETBP1 c.2608G>A, p.G870S | 4 | 1 | |||||||||||
| 37 | AML | c.415_418dup | p.D140fs | 46 | 1 | NI | |||||||
| 38† | AML | c.415_418dup | p.D140fs | 49 | 1 | 46,X,-X,der1, (X;1)(p11.3;p36.3), inv9(p12q13)c, +14[4]/46, XX, inv9c[16] | |||||||
| 39† | AML | c.415_418dup | p.D140fs | 47 | 1 | 46,XY[20] | |||||||
| 40† | AML | c.415_418dup | p.D140fs | 46 | 1 | 46,XY[19] | |||||||
| 41 | AML | c.415_418dup | p.D140fs | 44 | 1 | NI | |||||||
| 42 | AML | c.415_418dup | p.D140fs | 46 | 1 | 46,XX[20] | |||||||
| 43 | AML | c.415_418dup | p.D140fs | 44 | 1 | 46,XY[20] | |||||||
| 44 | AML | c.668dup | p.I224fs | 45 | 1 | c.1574G>A | p.R525H | 16 | 1 | ASXL1 c.2541del, p.T848fs | 17 | 1 | NI |
| PHF6 c.138 + 1G>A, p.? | 5 | 1 | |||||||||||
| PHF6 c.255C>G, p.C85W | 9 | 2 | |||||||||||
| 45† | AML | c.847del | p.L283fs | 43 | 1 | c.1574G>A | p.R525H | 5 | 1 | NI | |||
| 46 | AML | c.946_947del | p.M316fs | 52 | 1 | c.1574G>A | p.R525H | 3 | 1 | CUX1 c.3855del, p.S1286fs | 1 | 1 | 46,XY[20] |
| 47 | AML | c.1394del | p.G465fs | 46 | 1 | c.1574G>A | p.R525H | 1 | 1 | DNMT3A c.2026C>T, p.R676W | 6 | 1 | 46,XY[20] |
| ASXL1 c.2423_2427del, p.P808fs | 5 | 1 | |||||||||||
| ASXL1 c.2060_2061del, p.C687fs | 3 | 1 | |||||||||||
| 48 | AML | c.121C>T | p.Q41* | 48 | 1 | c.1574G>A | p.R525H | 1 | 1 | ASXL1 c.1900_1922del, p.E635fs | 10 | 1 | N/A |
| DNMT3A c.2256_2263del, p.W753* | 15 | 1 | |||||||||||
| PHF c.820C>T, p.R274* | 22 | 1 | |||||||||||
| 49 | AML | c.121C>T | p.Q41* | 45 | 1 | TET2 c.4133G>A, p.C1378Y | 44 | 1 | NI | ||||
| GATA2 c.599dup, p.S201* | 2 | 1 | |||||||||||
| KDM6A c.3704 + 1G>C, p.? | 67 | 1 | |||||||||||
| ZRSR2 c.505C>T, p.R169* | 93 | 1 | |||||||||||
| NPM1 c.860_863dup, p.W288fs | 35 | 1 | |||||||||||
| 50 | AML | c.475C>T | p.R159* | 51 | 1 | c.1589G>A | p.G530D | 2 | 1 | U2AF1 c.101C>A, p.S34Y | 3 | 1 | 46,XY[20] |
| 51 | AML | c.931C>T | p.R311* | 49 | 1 | c.1589G>A | p.G530D | 5 | 1 | PHF6 c.730-1G>A, p.? | 3 | 1 | NI |
| DNMT3A c.2255_2257del, p.F752del | 1 | 1 | |||||||||||
| 52 | AML | c.1105C>T | p.R369* | 46 | 1 | c.1574G>A | p.R525H | 18 | 1 | ASXL1 c.1900_1922del, p.E635fs | 13 | 1 | 46,XY[20] |
| SETBP1 c.2602G>A, p.D868N | 5 | 1 | |||||||||||
| SETBP1 c.2608G>A, p.G870S | 9 | 1 | |||||||||||
| SETBP1 c.2612T>C, p.I871T | 5 | 1 | |||||||||||
| 53 | AML | c.1105C>T | p.R369* | 47 | 1 | c.1574G>A | p.R525H | 1 | 1 | TET2 c.3632G>A, p.C1211Y | 4 | 1 | NI |
| SH2B3 c.794G>A, p.R265Q | 48 | 2 | |||||||||||
| 54 | AML | c.1108C>T | p.Q370* | 48 | 1 | c.1588G>A | p.G530S | 25 | 1 | JAK2 c.1849G>T, p.V617F | 22 | 1 | 46,XY[20] |
| 55 | AML | c.1504C>T | p.Q502* | 49 | 1 | c.1035G>C | p.E345D | 10 | 1 | ASXL1 c.2693G>A, p.W898* | 9 | 1 | 45,X,-Y[14]/46,XY[6] |
| 56† | AML | c.645-1G>T | p.? | 45 | 1 | c.1574G>A | p.R525H | 1 | 1 | ASXL1 c.3824C>G, p.S1275* | 1 | 1 | 46,XY[20] |
| DNMT3A c.1572T>A, p.C524* | 1 | 1 | |||||||||||
| 57 | AML | c.992_994del | p.K331del | 48 | 1 | c.1035G>C | p.E345D | 14 | 1 | ASXL1 p.E635fs | 5 | 1 | 46,XY, der(7)add(7)(p13) add(7)(q11.2)[10]/45,XY, -der(7)[5]/46,XY[13] |
| EZH2 p.N546K | 7 | 2 | |||||||||||
| 58*,† | AML | c.646C>G | p.L216V | 51 | 1 | c.1035G>C | p.E345D | 30 | 1 | DNMT3A c.2656C>T, p.Q886* | 31 | 1 | NI |
| 59 | AML | c.653G>A | p.G218D | 50 | 1 | c.1589G>A | p.G530D | 4 | 1 | TP53 c.488A>G, p.Y163C | 6 | 1 | NI |
| RUNX1 c.288_291delinsAAA, p.N96fs | 3 | 1 | |||||||||||
| DNMT3A c.1627G>T, p.G543C | 5 | 1 | |||||||||||
| DNMT3A c.1578C>G, p.Y526* | 5 | 1 | |||||||||||
| 60 | AML; breast cancer | c.773C>T | p.P258L | 56 | 1 | c.1574G>A | p.R525H | 15 | 1 | 46,XY[20] | |||
| 61 | AML | c.967C>T | p.R323C | 45 | 1 | c.1574G>A | p.R525H | 3 | 1 | TET2 c.3585_3588delnsAG, p.A1196fs | 5 | 1 | NI |
| SRSF2 c.284C>A, p.P95H | 4 | 1 | |||||||||||
| 62 | AML | c.1046T>A | p.M349K | 45 | 1 | 46,XY[20] | |||||||
| 63 | AML | c.1046T>A | p.M349K | 50 | 1 | 46,XY[20] | |||||||
| 64 | AML | c.1105C>G | p.R369G | 48 | 1 | c.1574G>A | p.R525H | 5 | 1 | 46,XX[20] | |||
| 65 | AML | c.1399G>T | p.D467Y | 45 | 1 | c.1589G>A | p.G530D | 13 | 1 | ASXL1 c.1934dup, p.G646fs | 13 | 1 | NI |
| ASXL1 c.1900_1922del, p.E635fs | 2 | 1 | |||||||||||
| EZH2 c.2022G>C, p.L674F | 3 | 1 | |||||||||||
| SETBP1 c.2608G>A, p.G870S | 4 | 1 | |||||||||||
| EZH2 c.2197T>A, p.Y733N | 10 | 1 | |||||||||||
| SETBP1 c.2612T>C, p.I871T | 8 | 1 | |||||||||||
| 66 | AML | c.1574G>A | p.R525H | 54 | 1 | NI | |||||||
| 67 | MDS | c.3G>A | p.M1I | 45 | 1 | c.1574G>A | p.R525H | 2 | 1 | DNMT3A c.1010C>G, p.S337* | 1 | 1 | NI |
| 68 | MDS | c.3G>A | p.M1I | 49 | 1 | c.1574G>A | p.R525H | 6 | 1 | 46,XY[20] | |||
| 69 | MDS | c.3G>A | p.M1I | 43 | 1 | c.1574G>A | p.R525H | 4 | 1 | 46,XX[20] | |||
| 70 | MDS | c.3G>A | p.M1I | 53 | 1 | c.1574G>A | p.R525H | 2 | 1 | 46,XY[20] | |||
| 71 | MDS | c.3G>A | p.M1I | 50 | 1 | c.694A>G | p.T232A | 3 | 1 | 46,XY[20] | |||
| 72 | MDS | c.3G>A | p.M1I | 49 | 1 | c.962C>T | p.P321L | 14 | 1 | DNMT3A c.1792C>T, p.598* | 14 | 1 | 46,XY[20] |
| 73 | MDS | c.3G>A | p.M1I | 49 | 1 | c.962C>T | p.P321L | 14 | 1 | 46,XY[20] | |||
| 74 | MDS | c.3G>A | p.M1I | 48 | 1 | c.1037C>T | p.A346P | 3 | 1 | 46,XY[20] | |||
| 75 | MDS | c.3G>A | p.M1I | 46 | 1 | c.1643T>A | p.L548H | 4 | 1 | 46,XX[20] | |||
| 76 | MDS; MM; MBL | c.3G>A | p.M1I | 50 | 1 | DNMT3A c.2645G>A, p.R882H | 15 | 1 | 46,XX[20] | ||||
| 77 | MDS | c.3G>A | p.M1I | 46 | 1 | PTPN11 c.215C>T, p.A72V | 42 | 1 | 47,XY,del(5)(q13q33), +21[2]/48,sl,t(9;21) (q10;q10), +21[12]/ | ||||
| RUNX1 c.1036dup, p.R346fs | 32 | 1 | |||||||||||
| 78 | MDS | c.3G>A | p.M1I | 49 | 1 | JAK2 c.1849G>T, p.V617F | 1 | 1 | 46,XY[20] | ||||
| 79 | MDS | c.3G>A | p.M1I | 49 | 1 | 45,X,-Y[6]/46,XY[14] | |||||||
| 80 | MDS | c.415_418dup | p.D140fs | 44 | 1 | c.1574G>A | p.R525H | 10 | 1 | ASXL1 c.4127dup, p.P1377fs | 12 | 1 | NI |
| 81 | MDS | c.415_418dup | p.D140fs | 45 | 1 | c.794C>T | p.P265L | 27 | 1 | DNMT3A c.1579C>T, p.Q527* | 9 | 1 | 46,XY[20] |
| 82 | MDS | c.1496dup | p.A500fs | 47 | 1 | c.1660G>A | p.E554K | 2 | 1 | 46,XY[20] | |||
| 83 | MDS | c.130C>T | p.Q44* | 60 | 1 | c.1126C>T | p.A376T | 2 | 1 | DNMT3A c.2207G>A, p.R736H | 7 | 1 | 46,XX[20] |
| 84 | MDS | c.475C>T | p.R159* | 47 | 1 | c.1589G>A | p.G530D | 6 | 1 | NI | |||
| c.1574G>A | p.R525H | 3 | 1 | ||||||||||
| c.1588G>A | p.G530S | 1 | 2 | ||||||||||
| 85 | MDS | c.931C>T | p.R311* | 51 | 1 | c.1574G>A | p.R525H | 6 | 1 | SRSF2 c.284C>T, p.P95L | 3 | 1 | NI |
| 86 | MDS | c.931C>T | p.R311* | 50 | 1 | c.1574G>A | p.R525H | 7 | 1 | NI | |||
| 87 | MDS | c.992_994del | p.K331del | 43 | 1 | c.1574G>A | p.R525H | 14 | 1 | TET2 c.3866G>T, p.C1289F | 3 | 1 | 46,XY[20] |
| 88 | MDS | c.992_994del | p.K331del | 48 | 1 | 46,XY[20] | |||||||
| 89 | MDS | c.566C>T | p.P189L | 49 | 1 | c.1574G>A | p.R525H | 4 | 1 | EZH2 c.434T>C, p.F145S | 3 | 1 | NI |
| 90 | MDS | c.710T>G | p.L237W | 49 | 1 | c.1589G>A | p.G530D | 13 | 1 | 46,XY[20] | |||
| 91 | MDS | c.1016G>A | p.R339H | 51 | 1 | IDH1 c.605del, p.S202fs | 31 | 2 | 45,X,-Y/46,XY[15] | ||||
| 92 | MDS | c.1015C>T | p.R339C | 49 | 1 | c.680C>T | p.T227M | 13 | 1 | TET2 c.1793del, p.N598fs | 2 | 1 | 46,XY[20] |
| 93 | MDS | c.1018T>A | p.Y340N | 47 | 1 | c.1574G>A | p.R525H | 7 | 1 | CUX1, c.2389del, p.Q797fs | 13 | 1 | 46,XY[20] |
| EZH2 c.371A>G, p.D124G | 15 | 2 | |||||||||||
| 94* | MDS | c.1105C>G | p.R369G | 47 | 1 | c.645-1G>A | p.? | 1 | 1 | TET2 c.1263del; p.G422fs | 31 | 1 | NI |
| TET2 c.3860_3869del, p.F1287fs | 1 | 1 | |||||||||||
| 95 | Pancytopenia | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 14 | 1 | JAK2 c.1849G>T, p.V617F | 15 | 1 | NI |
| TET2 c.1648C>T, p.R550* | 7 | 1 | |||||||||||
| 96 | Pancytopenia | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 2 | 1 | NI | |||
| 97 | Pancytopenia | c.415_418dup | p.D140fs | 46 | 1 | c.1574G>A | p.R525H | 9 | 1 | 46,XY, del(20)(q11.2q13.1) [1]/46,XY[20] | |||
| 98 | Pancytopenia | c.430del | p.T144fs | 45 | 1 | c.1574G>A | p.R525H | 4 | 1 | ASXL1c.4002del, p.S1335fs | 21 | 1 | NI |
| TP53 c.586C>T, p.R196* | 4 | 1 | |||||||||||
| TP53 c.916C>T, p.R306* | 3 | 1 | |||||||||||
| IDH2 c.419G>A, p.R140Q | 2 | 1 | |||||||||||
| 99 | Pancytopenia | c.946_947del | p.M316fs | 47 | 1 | c.1574G>A | p.R525H | 5 | 1 | ASXL1 c.2644C>T, p.Q882* | 6 | 1 | 46,XY[20] |
| PHF6 c.941T>C, p.I314T | 11 | 2 | |||||||||||
| 100 | Pancytopenia | c.1354del | p.L452fs | 48 | 1 | c.1574G>A | p.R525H | 8 | 1 | DNMT3A c.929T>C, p.I310T | 5 | 1 | 46,XY[20] |
| 101 | Pancytopenia | c.475C>T | p.R159* | 48 | 1 | c.1574G>A | p.R525H | 12 | 1 | SRSF2 c.284C>A, p.P95H | 10 | 1 | NI |
| STAG2 c.1243C>T, p.H415Y | 16 | 2 | |||||||||||
| 102 | Pancytopenia | c.1628C>G | p.S543* | 50 | 1 | c.1574G>A | p.R525H | 4 | 1 | ASXL1 c.1934dup, p.G646fs | 3 | 1 | NI |
| RUNX1 c.540del, p.F180fs | 3 | 1 | |||||||||||
| PHF6 c.941T>C, p.I314T | 7 | 1 | |||||||||||
| KDM6A c.2665del, p.T889fs | 1 | 1 | |||||||||||
| 103 | Pancytopenia | c.649T>C | p.S217P | 48 | 1 | c.1589G>A | p.G530D | 9 | 1 | ASXL1 c.2467_2468insA, p.L823fs | 7 | 1 | NI |
| KRAS c.437C>T, p.A146V | 3 | 1 | |||||||||||
| SRSF2 c.47T>A, p.L16H | 7 | 2 | |||||||||||
| 104 | Pancytopenia | c.773C>T | p.P258L | 47 | 1 | c.1574G>A | p.R525H | 4 | 1 | KDM6A c.3107delT, p.F1036fs | 2 | 1 | NI |
| SRSF2 c.284C>A, p.P95H | 7 | 1 | |||||||||||
| 105 | Pancytopenia | c.776A>G | p.Y259C | 48 | 1 | c.1574G>A | p.R525H | 6 | 1 | FBXW7 c.62G>A, p.G21D | 19 | 2 | 46,XY[20] |
| 106 | Pancytopenia | c.1016G>T | p.R339L | 50 | 1 | c.1574G>A | p.R525H | 1 | 1 | NI | |||
| 107 | Thrombocytopenia | c.415_418dup | p.D140fs | 45 | 1 | c.1574G>A | p.R525H | 10 | 1 | ASXL1 c.1900_1922del, p.E635fs | 2 | 1 | NI |
| CUX1 c.2161C>T, p.Q721* | 8 | 1 | |||||||||||
| SMC1A c.2132G>A, p.R711Q | 4 | 2 | |||||||||||
| 108 | Thrombocytopenia | c.1586_ 1587delCA | p.T529fs | 52 | 1 | TP53 c.1024C>T, p.R342* | 40 | 1 | 46,XX[20] | ||||
| 109 | Thrombocytopenia | c.3G>A | p.M1I | 45 | 1 | c.962C>T | p.P321L | 23 | 1 | RUNX1 c.281G>A, p.S94N | 30 | 1 | NI |
| TP53 c.742C>T, p.R248W | 35 | 1 | |||||||||||
| SRSF2 c.284C>G, p.P95R | 26 | 1 | |||||||||||
| 110 | Neutropenia | c.3G>A | p.M1I | 48 | 1 | c.1574G>A | p.R525H | 10 | 1 | NI | |||
| 111 | Neutropenia | c.3G>A | p.M1I | 47 | 1 | c.1574G>A | p.R525H | 1 | 1 | NI | |||
| 112 | Anemia | c.1105C>T | p.R369* | 45 | 1 | c.1574G>A | p.R525H | 5 | 1 | ASXL1 c.1934dup, p.G646fs | 7 | 1 | 46,XX,+1, der(1;7)(q10;p10) [16]/46,XX[4] |
| EZH2 c.786dup, p.N263fs | 4 | 1 | |||||||||||
| SETBP1 c.2613_2614delinsGC, p.I871_G872delinsMR | 4 | 2 | |||||||||||
| 113 | MPN | c.415_418dup | p.D140fs | 49 | 1 | 46,XX[20] | |||||||
| 114 | MPN | c.946_947del | p.M316fs | 48 | 1 | NI | |||||||
| 115 | MPN | c.916C>T | p.Q306* | 48 | 1 | 46,XX[20] | |||||||
| 116 | MPN | c.1141A>T | p.K381* | 44 | 1 | c.1574G>A | p.R525H | 7 | 1 | CUX1 c.988C>T, p.Q330* | 7 | 1 | 46,XY[20] |
| 117 | AML | c.6G>T | p.E2D | 48 | 2 | FLT3 c.1805_1806ins42, p.K602_W603ins14 | n/a | 1 | 46,XX[20] | ||||
| NPM1 c.863_864insCCTG, p.W288fs | 35 | 1 | |||||||||||
| WT1 c.1141_1144dup, p.A382fs | 8 | 1 | |||||||||||
| 118* | AML | c.55G>A | p.G19R | 48 | 2 | FLT3 ITD c.1837 + 11_1837 + 12ins114, p.? | n/a | 1 | NI | ||||
| NPM1 c.863_864insCCTG, p.W288fs | 43 | 1 | |||||||||||
| IDH1 c.395G>A, p.R132H | 41 | 1 | |||||||||||
| DNMT3A c.1627G>T, p.G543C | 42 | 1 | |||||||||||
| 119* | AML | c.97T>C | p.Y33H | 45 | 2 | FLT3 c.1770_1811dup42, p.W603_E604ins14 | n/a | 1 | NI | ||||
| NPM1 c.860_863dup, p.W288fs | 18 | 1 | |||||||||||
| DNMT3A c.860_863dup, p.W288fs | 27 | 1 | |||||||||||
| TET2 c.2490dup, p.Q831fs | 23 | 1 | |||||||||||
| 120 | AML | c.465G>A | p.M155I | 47 | 2 | NPM1 c.860_863dup, p.W288fs | 7 | 1 | 46,XY[20] | ||||
| SRSF2 c.284C>G, p.P95R | 28 | 1 | |||||||||||
| TET2 c.2244dup, p.Q749fs | 44 | 1 | |||||||||||
| 121 | AML | c.465G>A | p.M155I | 47 | 2 | NPM1 c.863_864insCTTG, p.W288Cfs | 33 | 1 | 46,XY[20] | ||||
| GATA2 c.599dup, p.S201* | 2 | 1 | |||||||||||
| 122* | AML | c.491G>A | p.R164N | 48 | 2 | c.1774A>T | p.I592F | 35 | 2 | NPM1 c.863_864insCTTG, p.W288Cfs | 33 | 1 | 46,XX[20] |
| 123 | AML | c.1528C>T | p.P510S | 48 | 2 | NPM1 c.860_863dup, p.W288fs | 34 | 1 | 46,XY[20] | ||||
| SRSF2 c.284C>T, p.P95L | 42 | 1 | |||||||||||
| KRAS c.35G>A, p.G12D | 11 | 1 | |||||||||||
| 124 | AML | c.380T>A | p.M127K | 49 | 2 | FLT3 c.2505T>G, p.D835E | 3 | 1 | 46,XY,+12, der(17)t(17;18) (p10;q10),-18[8] | ||||
| TP53 c.400T>A, p.F134I | 46 | 1 | |||||||||||
| TP53 c.458_462del, p.P153fs | 3 | 1 | |||||||||||
| U2AF1 c.101C>A, p.S34Y | 6 | 1 | |||||||||||
| KRAS c.351A>T, p.K117N | 27 | 1 | |||||||||||
| 125 | AML | c.465G>A | p.M155I | 47 | 2 | FLT3 p.N841K | 7 | 1 | 46,XY[20] | ||||
| KRAS c.35G>T, p.G12V | 3 | 1 | |||||||||||
| 126 | AML | c.199G>C | p.G67R | 50 | 2 | ASXL1 c.2959G>T, p.G987* | 47 | 1 | 46,XY[20] | ||||
| CEBPA c.985_986insCC, E329fs | 42 | 1 | |||||||||||
| CEBPA c.68del, p.P23fs | 46 | 1 | |||||||||||
| IDH2 c.418C>T, p.R140W | 48 | 1 | |||||||||||
| JAK2 c.1849G>T, p.V617F | 6 | 1 | |||||||||||
| 127 | AML | c.199G>C | p.G67R | 48 | 2 | ASXL1 c.2959G>T, p.G987* | 22 | 1 | 46,XY[20] | ||||
| IDH2 c.418C>T, p.R140W | 25 | 1 | |||||||||||
| SRSF2 c.284C>A, p.P95H | 28 | 1 | |||||||||||
| STAG2 c.1196 + 1G>A, p.? | 4 | 1 | |||||||||||
| STAG2 c.1999del, p.R667fs | 15 | 1 | |||||||||||
| 128* | AML | c.465G>A | p.M155I | 51 | 2 | 48,XY,+8,+22[20] | |||||||
| 129 | AML | c.883G>A | p.A295T | 53 | 2 | TP53 c.743G>A, p.R248Q | 35 | 1 | 44,XY,-3,add(5) (q11.2),+8, add(8)(q22), der(12;17)(q10;q10), | ||||
| TP53 c.818G>A, p.R273H | 15 | 1 | −14,i(14)(q10), i(21)(q10)[12]/ 43-45, sl,-add(5)(q11.2), i(5)(q10)[cp4]/88 < 4n>,slx2[1] | ||||||||||
| 130 | AML | c.893T>C | p.I298T | 49 | 2 | DNMT3A c.989G>A, p.W330* | 2 | 1 | NI | ||||
| TP53 c.844C>T, p.R282W | 61 | 1 | |||||||||||
| TP53 c.535C>T, p.H179Y | 4 | 1 | |||||||||||
| PTPN11 c.1504T>C, p.S502P | 2 | 1 | |||||||||||
| 131 | AML | c.1063G>A | p.E355K | 47 | 2 | PHF6 c.418 + 2T>C, p.? | 21 | 1 | 46,XX[7] | ||||
| KMT2A c.10462C>T, p.Gln3488* | 25 | 2 | |||||||||||
| 132 | AML | c.465G>A | p.M155I | 48 | 2 | NRAS c.181C>A, p.Q61K | 15 | 1 | 46,XY,inv(16) (p13.1q22)[20] | ||||
| 133 | AML | c.465G>A | p.M155I | 47 | 2 | CSF3R c.1843A>G, p.T615A CSF3R c.1853C>T, p.T618I | 22 4 | 1 1 | 46,XX, der(8)t(8;21) (q22;q22), der(8)?(8pter-> 8p22::?:: 8p11.2-> 8q?13::8q22 ->8qter), der(21)(21pter-> 21q22::8?q13-> 8q?22:: 8q22-> 8qter)[7]/45, sl,-X[8]/46,XX[5] | ||||
| 134 | AML | c.138 + 5G>A | p.? | 50 | 2 | NRAS c.181C>A, p.Q61Lys | 39 | 1 | 47,XY, +8,inv(16) (p13.1q22)[20] | ||||
| 135 | MDS | c.367G>A | p.G123S | 48 | 2 | ASXL1 c.2239_2244delinsCC, p.S747fs | 42 | 1 | 46,XX[20] | ||||
| ASXL1 c.1934dup, p.G646fs | 2 | 1 | |||||||||||
| TET2 c.5543C>G, p.S1848* | 1 | 1 | |||||||||||
| EZH2 c.1119dup, p.T374fs | 2 | 1 | |||||||||||
| IDH2 c.419G>A, p.R140Q | 31 | 1 | |||||||||||
| SF3B1 c.2347G>A, p.E783K | 2 | 2 | |||||||||||
| JAK2 c.1849G>T, p.V617F | 1 | 1 | |||||||||||
| 136 | MDS | c.465G>A | p.M155I | 43 | 2 | JAK2 c.1849G>T, p.V617F | 27 | 1 | NI | ||||
| DNMT1 c.4663G>A, p.V1555M | 32 | 2 | |||||||||||
| SH2B3 c.127C>T, p.R43C | 35 | 2 | |||||||||||
| 137 | MDS | c.644 + 5G>C | p.? | 43 | 2 | SH2B3 c.947_953del, p.E316fs | 21 | 1 | 47,XY, +9,del(20) (q11.2q13.1)[19]/46,XY[1] | ||||
| JAK2 c.1849G>T, p.V617F | 40 | 1 | |||||||||||
| 138 | MDS | c.523G>A | p.G175S | 48 | 2 | NRAS c.34G>A, p.G12S | 44 | 1 | NI | ||||
| TET2 c.2290dup, p.Q764fs | 50 | 1 | |||||||||||
| GATA2 c.599del, p.G200fs | 1 | 1 | |||||||||||
| SRSF2 c.284C>A, p.P95H | 47 | 1 | |||||||||||
| 139 | MDS | c.529C>T | p.P177S | 45 | 2 | SMC1A c.197A>G, p.H66R | 9 | 2 | NI | ||||
| BCOR c.441dup, p.Ile148fs | 50 | 1 | |||||||||||
| U2AF1 c.101C>T, p.S34F | 37 | 1 | |||||||||||
| RUNX1 c.601del, p.R201fs | 10 | 1 | |||||||||||
| 140 | MDS | c.1301C>T | p.P434L | 49 | 2 | GATA2 c.599dup; p.S201* | 31 | 1 | 46,XY[20] | ||||
| NPM1 c.867_868insAGGA, p.W290fs | 48 | 1 | |||||||||||
| 141 | MDS | c.1528C>T | p.P510S | 48 | 2 | ASXL1 c.1934dup, p.G646fs | 35 | 1 | NI | ||||
| CBL c.800G>A, p.G267D | 2 | 2 | |||||||||||
| 142 | MDS | c.1704C>G | p.C568W | 48 | 2 | KIT c.2446_2447delinsAT, p.D816I | 10 | 1 | NI | ||||
| KIT c.2447A>T, p.D816V | 1 | 1 | |||||||||||
| PTPN11 c.154A>G, p.T52A | 5 | 1 | |||||||||||
| PTPN11 c.1508G>A, p.G503E | 3 | 2 | |||||||||||
| 143 | MDS/MPN | c.1760G>C | p.G587A | 47 | 2 | CSF3R c.1853C>T, p.T618I | 3 | 1 | 47,XY,+8[20] | ||||
| KRAS c.436G>C, p.A146P | 37 | 1 | |||||||||||
| ASXL1 c.1900_1922del, p.E635fs | 44 | 1 | |||||||||||
| STAG2 c.1191dup, p.Q399fs | 92 | 1 | |||||||||||
| 144 | MDS | c.1276G>A | p.E426K | 49 | 2 | SRSF2 c.284C>A, p.P95H | 46 | 1 | NI | ||||
| IDH2 c.419G>A, p.R140Q | 45 | 1 | |||||||||||
| 145 | MDS | c.1528C>T | p.P510S | 48 | 2 | TP53 c.734G>A, p.G45D | 20 | 1 | NI | ||||
| 146 | MDS | c.1663G>A | p.A555T | 43 | 2 | TP53 c.706dup, p.Y236fs | 39 | 1 | 44-47,XY, del(5)(q22q35), -7,der(11)t(11;13) (p15;q14), | ||||
| TP53 c.713G>A, p.C238Y | 37 | 1 | |||||||||||
| 147 | Pancytopenia | c.27G>A | p.K9K | 43 | 2 | NI | |||||||
| 148 | Pancytopenia | c.465G>A | p.M155I | 50 | 2 | SF3B1 c.1996A>G, p.K666E | 19 | 1 | 45,X,-Y[14]/46,XY[6] | ||||
| U2AF1 c.101C>T, p.S34F | 5 | 1 | |||||||||||
| 149 | Pancytopenia | c.556A>T | p.M186L | 47 | 2 | ETNK1 c.731A>G, p.N244S | 24 | 1 | NI | ||||
| Small cell carcinoma | KRAS c.468C>G, p.F156L | 18 | 2 | ||||||||||
| 150 | Pancytopenia | c.656G>A | p.R219H | 49 | 2 | c.679A>G | p.T227A | 30 | 2 | 46,XY[20] | |||
| 151 | Pancytopenia | c.845G>A | p.R282H | 40 | 2 | TP53 c.377A>G, p.Y126C | 67 | 1 | NI | ||||
| BRAF c.1391G>T, p.G464V | 38 | 1 | |||||||||||
| 152 | Pancytopenia | c.881G>T | p.C294F | 48 | 2 | CBL c.1211G>A, p.C404Y | 2 | 1 | NI | ||||
| 153 | Pancytopenia | c.1477T>G | p.S493A | 47 | 2 | ASXL1 c.1900_1922del, p.E635fs | 15 | 1 | NI | ||||
| ASXL1 c.1934dup, p.G646fs | 3 | 1 | |||||||||||
| ASXL1 c.2295del, p.S766fs | 1 | 1 | |||||||||||
| SH2B3 c.703C>G, p.R235G | 16 | 1 | |||||||||||
| 154 | Thrombocytopenia | c.465G>A | p.M155I | 50 | 2 | 46,XY[20] | |||||||
| 155 | Thrombocytopenia | c.707C>T | p.T236M | 47 | 2 | NI | |||||||
| 156 | Thrombocytopenia | c.138 + 5G>A | p.? | 54 | 2 | NI | |||||||
| 157 | Thrombocytopenia | c.644 + 5G>C | p.? | 47 | 2 | RAD21 c.507_508del, p.E169fs | 38 | 1 | NI | ||||
| NRAS c.35G>A, p.G12D | 20 | 1 | |||||||||||
| TP53 c.818G>A, p.R273H | 31 | 1 | |||||||||||
| TP53 c.559 + 1G>A, p.? | 42 | 1 | |||||||||||
| 158 | Neutropenia | c.138 + 5G>A | p.? | 50 | 2 | NI | |||||||
| 159 | Anemia | c.751C>T | p.P251S | 46 | 2 | 46,XX[20] | |||||||
| 160 | Anemia | c.1748C>T | p.A583V | 47 | 2 | PTPN11 c.227A>T, p.E76V | 45 | 1 | NI | ||||
| 161 | MPN, PV | c.23G>A | p.R8Q | 47 | 2 | JAK2 c.1849G>T, p.V617F | 45 | 1 | 46,XY[20] | ||||
| 162 | MPN, PV | c.94G>A | p.D32N | 48 | 2 | JAK2 c.1849G>T, p.V617F | 85 | 1 | 46,XY[20] | ||||
| 163 | MPN, ET | c.398C>T | p.A133V | 44 | 2 | CALR c.1122_1125del, p.K374fs | 44 | 1 | NI | ||||
| U2AF1 c.470A>G, p.Q157R | 45 | 1 | |||||||||||
| ZRSR2 c.236_237del, p.E79Afs | 87 | 1 | |||||||||||
| 164 | MPN | c.644T>C | p.I215T | 48 | 2 | JAK2 c.1849G>T, p.V617F | 26 | 1 | NI | ||||
| ASXL1 c.1934dup, p.G646fs | 1 | 1 | |||||||||||
| 165 | MPN | c.647T>C | p.L216P | 48 | 2 | c.1655T>G | p.L552R | 14 | 2 | JAK2 c.1849G>T, p.V617F | 12 | 1 | NI |
| ASXL1 c.1900_1922del, p.E635fs | 8 | 1 | |||||||||||
| 166 | MPN, ET | c.707C>T | p.T236M | 50 | 2 | JAK2 c.1849G>T, p.V617F | 18 | 1 | 46,XY[20] | ||||
| SF3B1 c.2110A>T, p.I704F | 15 | 1 | |||||||||||
| 167 | MPN | c.1471G>A | p.V491I | 52 | 2 | JAK2 c.1849G>T, p.V617F | 47 | 1 | 46,XY[20] | ||||
| TP53 c.743G>A, p.R248Q | 6 | 1 | |||||||||||
| TP53 c.742C>T, p.R248W | 5 | 1 | |||||||||||
| NF1 c.2035del, p.I679fs | 15 | 1 | |||||||||||
| 168 | MPN, ET | c.1630G>T | p.V544L | 49 | 2 | JAK2 c.1849G>T, p.V617F | 26 | 1 | 46,XY[20] | ||||
| SF3B1 c.1997A>C, p.K666T | 16 | 1 | |||||||||||
| 169 | MPN, ET | c.138 + 5G>T | p.? | 49 | 2 | NI | |||||||
| 170 | MPN, ET | c.465G>A | p.M155T | 49 | 2 | 46,XX[20] | |||||||
| 171 | MPN | c.560A>G | p.K187R | 46 | 2 | NI | |||||||
| 172 | LPL with pancytopenia | c.490C>T | p.R164W | 50 | 2 | SF3B1 c.2098A>G, p.K700E | 41 | 1 | 46,XY[20] | ||||
| 173 | γ heavy chain disease | c.490C>T | p.R164W | 49 | 2 | 46,XY[20] | |||||||
| MYD88 negative LPL | |||||||||||||
| 174 | CLL; breast cancer | c.490C>T | p.R164W | 47 | 2 | 46,XY[20] | |||||||
| 175 | CLL | c.751C>T | p.P251S | 50 | 2 | 46,XY, del(13)(q12;q22), add(18)(p11.20[4]/46,XY[16] | |||||||
| 176 | MM | c.29G>A | p.R10Q | 46 | 2 | TP53 c.376T>G, p.Y126D | 24 | 1 | 44∼46,X, add(X)(p10), del(6)(q21q23), i(8)(q10), | ||||
| DNMT3A c.2339T>C, p.I780T | 18 | 2 | t(11;14)(q13;q32), del(13)(q12q22),- 14,-17,+1∼ 2mar[cp4]/46,XX[16] | ||||||||||
| KDM6A c.1354_1355del, p.G452fs | 26 | 1 | |||||||||||
| 177 | AML | c.1574G>A | p.R525H | 12 | 1 | ASXL1 c.1774C>T, p.Q592* | 4 | 1 | 46,XY,t(1;4)? (q21;q31)[2]/46,XY[6] | ||||
| PHF6 c.58del, p.C20fs | 17 | 1 | |||||||||||
| 178 | AML | c.1574G>A | p.R525H | 25 | 1 | ASXL1 c.2077C>T, p.R693* | 2 | 1 | 46,XY[21] | ||||
| 179 | AML | c.1574G>A | p.R525H | 1 | 1 | CBL c.1211G>A, p.C404Y | 15 | 1 | 46,XX[20] | ||||
| U2AF2 c.766G>A, p.D256G | 16 | 2 | |||||||||||
| 180 | AML | c.1574G>A | p.R525H | 4 | 1 | SH2B3 c.519_523del, p.R175fs | 3 | 1 | 45,X,-Y[5]/46,XY[15] | ||||
| PHF6 c.635G>T, p.C212F | 8 | 2 | |||||||||||
| 181 | AML | c.1574G>A | p.R525H | 16 | 1 | SRSF2 c.284C>A, p.P95H | 17 | 1 | 46,X,del(X)?(q22q26)[2]/46,XX[18] | ||||
| 182 | AML | c.1574G>A | p.R525H | 9 | 1 | SETBP1 c.2602G>C, p.D868H | 2 | 1 | NI | ||||
| c.944A>G | p.H315R | 8 | 2 | CUX1 c.439C>T, p.R147* | 7 | 1 | |||||||
| STAG2 1693G>T, p.E565* | 5 | 1 | |||||||||||
| EZH2 c.1967C>T, p.A656V | 4 | 1 | |||||||||||
| ASXL1 c.2156del, p.E719fs | 8 | 1 | |||||||||||
| 183 | AML | c.1589G>A | p.G530D | 15 | 1 | ASXL1 c.2985del, p.H995fs | 5 | 1 | 46,XY[20] | ||||
| c.629A>G | p.Q210R | 15 | 2 | ASXL1 c.2083C>T, p.R695* | 3 | 1 | |||||||
| ASXL1 c.2077C>T, p.H963* | 5 | 1 | |||||||||||
| EZH2 c.2213del, p.A738fs | 15 | 1 | |||||||||||
| NF1 c.3774G>C, p.W1258C | 12 | 1 | |||||||||||
| 184 | AML; DLBCL | c.1589G>A | p.G530D | 6 | 1 | 46,XY[20] | |||||||
| 185 | MDS | c.1574G>A | p.R525H | 4 | 1 | DNMT3A c.2645G>A, p.R882H | 3 | 1 | 46,XX[11] | ||||
| 186 | MDS | c.1574G>A | p.R525H | 9 | 1 | 45,X,-Y[4]/46,XY[16] | |||||||
| 187 | MDS | c.1574G>A | p.R525H | 10 | 1 | 46,XX[20] | |||||||
| 188 | MDS | c.1574G>A | p.R525H | 1 | 1 | 46,XY[20] | |||||||
| 189 | MDS | c.1574G>A | p.R525H | 4 | 1 | 46,XY[20] | |||||||
| 190 | AML | c.157C>G | p.R53G | 5 | 2 | SF3B1 c.2098A>G, p.K700E | 32 | 1 | NI | ||||
| PTPN11 c.214G>A, p.A72T | 37 | 1 | |||||||||||
| 191 | AML | c.1760_ 1761TT | p.G587V | 4 | 2 | DNMT3A c.2645G>A, p.R882H | 7 | 1 | 46,XY[20] | ||||
| DNMT3A c.2095G>A, p.G699R | 3 | 2 | |||||||||||
| 192 | MDS | c.622C>G | p.Q208E | 14 | 2 | SETBP1 c.2608G>A, p.G870S | 6 | 1 | 46,XY,del(7)(q22)[6]/46,XY[14] | ||||
| SETBP1 c.2602G>A, p.D868N | 2 | 1 | |||||||||||
| DNMT3A c.2645G>A, p.R882H12 | 12 | 1 | |||||||||||
| EVT6 c.313C>G, p.R105G | 2 | 2 | |||||||||||
| TET2 c.4079T>C, p.L1360P | 1 | 2 | |||||||||||
| 193 | MDS | c.1199G>T | p.R400L | 3 | 2 | ASXL1 c.2324T>G, p.L775* | 20 | 1 | 46,XY, del(5)(q31q33) [13]/46,XX[7] | ||||
| 194 | MDS | c.1369G>C | p.V457L | 2 | 2 | TP53 c.818G>A, p.R273H | 30 | 1 | 46,XY, del(5)(q31q33) [14]/46,XX[6] | ||||
| TP53 c.578A>C, p.H193P | 2 | 2 | |||||||||||
| 195 | Neutropenia | c.1775T>A | p.I592N | 4 | 2 | SRSF2 c.284C>T, p.P95L | 4 | 1 | NI | ||||
| RUNX1 c.606dup, p.P203fs | 2 | 1 | |||||||||||
| TET2 c.330G>C, p.Lys110N | 3 | 2 | |||||||||||
| 196* | Normal | c.3G>A | p.M1I | 53 | 1 | 46,XY[20] | |||||||
| 197* | Normal | c.3G>A | p.M1I | 50 | 1 | 46,XX[20] | |||||||
| 198* | Normal | c.415_418dup | p.D140fs | 46 | 1 | Not done | |||||||
| 199* | Normal | c.415_418dup | p.D140fs | 45 | 1 | NI | |||||||
| 200* | Normal | c.415_418dup | p.D140fs | 56 | 1 | 46,XY[20] | |||||||
| 201* | Normal | c.992_994del | p.K331del | 47 | 1 | Not done | |||||||
| 202 | Normal∼(donor) | c.931C>T | p.311* | 50 | 1 | 47,XX,+8[19] | |||||||
| 203 | Normal∼(donor) | c.465G>A | p.M155I | 46 | 2 | 46,XY[20] | |||||||
| 204 | Normal∼(donor) | c.1693G>A | p.V565M | 50 | 2 | 46,XY[20] | |||||||
| 205 | Normal∼(donor) | c.1585dup | p.T529fs | 46 | 1 | 44,XY,inc[1]//46,XX[16] |
AF, variant allele frequency by %; ET, essential thrombocythemia; gl, germline; NI, no information; PMF, primary myelofibrosis; s, somatic; Tier 1, CV; Tier 2, VUS.
Germline variants confirmed by skin biopsies.
Patients reported in a prior study.9
The demographic features, family history, and overall survival of 205 individuals with DDX41 variants
| Patient . | Diagnosis . | Age . | Sex . | Ethnicity . | gl DDX41 . | FU (d) . | Survival (Y/N) . | FH_MN . | FH_LN . | FH_Other . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1* | AML | 62 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 2† | AML | 78 | F | Caucasian | p.M1I | 365 | Y | No | No | No |
| 3 | AML | 77 | M | Caucasian | p.M1I | 122 | Y | NI | NI | NI |
| 4† | AML | 64 | M | Caucasian | p.M1I | 519 | Y | No | No | Breast cancer (mother) |
| 5 | AML | 80 | M | Caucasian | p.M1I | 336 | Y | NI | NI | NI |
| 6*,† | AML | 77 | M | Caucasian | p.M1I | 2161 | Y | No | No | No |
| 7 | AML | 62 | M | Caucasian | p.M1I | 151 | Y | Leukemia (father) | No | No |
| 8 | AML | 76 | M | Caucasian | p.M1I | 1214 | Y | No | No | No |
| 9† | AML | 68 | M | Caucasian | p.M1I | 822 | Y | No | No | No |
| 10*,† | AML | 76 | F | Caucasian | p.M1I | 2161 | Y | No | No | Breast cancer (mother and sister); |
| pituitary tumor (brother); rhabdomyosarcoma (son) | ||||||||||
| 11 | AML | 88 | F | Caucasian | p.M1I | 98 | Y | No | No | No |
| 12† | AML | 48 | F | Caucasian | p.M1I | 700 | Y | No | No | No |
| 13 | AML | 63 | F | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 14† | AML | 78 | M | Caucasian | p.M1I | 1034 | Y | No | No | No |
| 15† | AML | 65 | M | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 16† | AML | 73 | M | Caucasian | p.M1I | 91 | Y | No | No | Cancer of unknown origin (mother) |
| 17† | AML | 72 | M | Caucasian | p.M1I | 822 | N | No | No | No |
| 18† | AML | 74 | M | Caucasian | p.M1I | 396 | Y | No | No | Cancer of unknown origin (brother) |
| 19* | AML | 75 | M | Caucasian | p.M1I | 608 | Y | NI | NI | NI |
| 20*# | AML | 60 | M | Caucasian | p.M1I | 639 | Y | No | No | No |
| 21# | AML | 65 | M | Caucasian | p.M1I | 365 | Y | No | No | Cancer of unknown origin (sister) |
| 22# | AML | 64 | M | Caucasian | p.M1I | 304 | Y | No | No | Lung cancer (father); cervical cancer (daughter) |
| 23 | AML | 68 | M | Caucasian | p.M1I | 400 | Y | No | No | Lung cancer (mother) |
| 24† | AML | 69 | M | Caucasian | p.M1I | 1216 | Y | NI | NI | NI |
| 25 | AML | 71 | F | Caucasian | p.K108fs | NI | NI | NI | NI | NI |
| 26 | AML | 71 | F | Caucasian | p.D140fs | NI | NI | NI | NI | NI |
| 27 | AML | 57 | M | Caucasian | p.D140fs | 561 | Y | NI | NI | NI |
| 28† | AML | 57 | M | Caucasian | p.D140fs | 403 | N | No | No | No |
| 29 | AML | 57 | M | Caucasian | p.D140fs | 1080 | Y | No | Lymphoma (father) | Prostatic cancer (paternal uncle) |
| 30*,† | AML | 59 | M | Caucasian | p.D140fs | 639 | Y | No | No | Pancreatic cancer (brother) |
| 31 | AML | 78 | M | Caucasian | p.D140fs | NI | NI | NI | NI | NI |
| 32 | AML | 81 | M | Caucasian | p.D140fs | 260 | Y | NI | NI | NI |
| 33 | AML | 63 | M | Caucasian | p.D140fs | 61 | Y | NI | NI | NI |
| 34† | AML | 90 | F | Caucasian | p.D140fs | 176 | N | No | No | No |
| 35 | AML | 66 | M | Caucasian | p.D140fs | 913 | Y | MDS (father and paternal uncle) | No | No |
| 36 | AML | 67 | M | NI | p.D140fs | 580 | Y | NI | NI | NI |
| 37 | AML | 50 | M | Caucasian | p.D140fs | 245 | Y | NI | NI | NI |
| 38† | AML | 70 | F | AA | p.D140fs | 183 | Y | No | No | Brain cancer (sister) |
| 39† | AML | 53 | M | Caucasian | p.D140fs | 945 | Y | No | No | Lung cancer (mother); colon cancer (father) |
| 40† | AML | 70 | M | Caucasian | p.D140fs | 488 | Y | No | No | Cancer of mouth and throat (brother) |
| 41 | AML | 57 | M | NI | p.D140fs | 675 | Y | NI | NI | NI |
| 42 | AML | 54 | F | Caucasian | p.D140fs | 300 | Y | No | No | No |
| 43 | AML | 61 | M | Caucasian | p.D140fs | 145 | Y | No | MM (father) | No |
| 44 | AML | 73 | M | Asian | p.I224fs | 731 | Y | NI | NI | NI |
| 45† | AML | 54 | F | Caucasian | p.L283fs | 580 | Y | No | No | No |
| 46 | AML | 58 | F | Caucasian | p.M316fs | 1071 | Y | No | No | No |
| 47 | AML | 76 | F | NI | p.G465fs | 640 | Y | NI | NI | NI |
| 48 | AML | 63 | M | Caucasian | p.Q41* | 212 | Y | No | No | No |
| 49 | AML | 70 | M | Caucasian | p.Q41* | NI | NI | NI | NI | NI |
| 50 | AML | 71 | M | Caucasian | p.R159* | 31 | Y | NI | NI | No |
| 51 | AML | 61 | M | Caucasian | p.R311* | NI | NI | NI | NI | NI |
| 52 | AML | 68 | M | Caucasian | p.R369* | 548 | N | No | No | No |
| 53 | AML | 78 | M | NI | p.R369* | 120 | Y | NI | NI | NI |
| 54 | AML | 70 | M | Caucasian | p.Q370* | 408 | Y | Leukemia (mother) | No | No |
| 55 | AML | 82 | M | Caucasian | p.Q502* | NI | NI | NI | NI | NI |
| 56† | AML | 68 | F | Caucasian | p.? | 905 | Y | Leukemia (maternal aunt) | No | No |
| 57 | AML | 74 | M | Caucasian | p.K331del | 232 | N | No | No | Thyroid and colon cancer (mother) |
| 58† | AML | 65 | F | Caucasian | p.L216V | 275 | Y | MDS (brother) | No | No |
| 59 | AML | 66 | M | Caucasian | p.G218D | 31 | Y | NI | NI | NI |
| 60 | AML; breast cancer | 47 | F | Caucasian | p.P258L | 653 | Y | No | No | No |
| 61 | AML | 60 | M | NI | p.R323C | 956 | Y | NI | NI | NI |
| 62 | AML | 69 | M | Caucasian | p.M349K | 365 | Y | NI | NI | NI |
| 63 | AML | 69 | M | Caucasian | p.M349K | 1134 | Y | No | No | Prostatic cancer (father) |
| 64 | AML | 81 | F | Caucasian | p.R369G | 151 | Y | AML (paternal cousin) | No | No |
| 65 | AML | 85 | M | Asian | p.D467Y | 458 | Y | NI | NI | NI |
| 66 | AML | 61 | M | Asian | p.R525H | 243 | N | MDS (father); Leukemia (paternal grandma) | No | No |
| Hematologic cancer (paternal uncle) | ||||||||||
| 67 | MDS | 72 | M | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 68 | MDS | 73 | M | Caucasian | p.M1I | 539 | Y | No | No | No |
| 69 | MDS | 81 | F | Caucasian | p.M1I | 670 | Y | No | No | No |
| 70 | MDS | 76 | M | Caucasian | p.M1I | 362 | Y | No | No | No |
| 71 | MDS | 79 | M | Caucasian | p.M1I | 763 | Y | No | No | Prostatic cancer (brother) |
| 72 | MDS | 61 | F | Caucasian | p.M1I | 192 | Y | No | No | Breast cancer (sister) |
| 73 | MDS | 60 | M | Caucasian | p.M1I | 1795 | Y | No | No | Throat cancer (father); melanoma (sister) |
| 74 | MDS | 65 | M | Caucasian | p.M1I | 153 | Y | NI | NI | No |
| 75 | MDS | 63 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 76 | MDS; MM; MBL | 67 | F | Caucasian | p.M1I | 98 | Y | No | No | No |
| 77 | MDS | 69 | M | Caucasian | p.M1I | 92 | Y | No | No | No |
| 78 | MDS | 72 | M | Caucasian | p.M1I | 2722 | Y | No | No | No |
| 79 | MDS | 60 | M | Caucasian | p.M1I | 395 | Y | NI | NI | No |
| 80 | MDS | 88 | F | Caucasian | p.D140fs | 476 | Y | NI | NI | NI |
| 81 | MDS | 69 | M | Caucasian | p.D140fs | 1078 | Y | NI | NI | NI |
| 82 | MDS | 63 | M | Asian | p.A500fs | 761 | Y | NI | NI | No |
| 83 | MDS | 76 | F | Asian | p.Q44* | 701 | Y | No | No | No |
| 84 | MDS | 77 | M | Caucasian | p.R159* | 876 | N | NI | NI | NI |
| 85 | MDS | 85 | F | Caucasian | p.R311* | NI | NI | NI | NI | NI |
| 86 | MDS | 84 | M | Caucasian | p.R311* | NI | NI | NI | NI | NI |
| 87 | MDS | 77 | M | Caucasian | p.K331del | 1793 | Y | Leukemia (father) | No | No |
| 88 | MDS | 62 | M | Caucasian | p.K331del | 900 | Y | No | No | Prostatic cancer, melanoma and stomach cancer (paternal uncles); |
| Pancreatic cancer (maternal aunt); lung cancer (maternal cousins); | ||||||||||
| Bile duct cancer (maternal cousin); breast cancer (maternal great aunt) | ||||||||||
| 89 | MDS | 84 | F | Caucasian | p.P189L | 2015 | Y | No | No | No |
| 90 | MDS | 73 | M | Caucasian | p.L237W | 134 | Y | MDS (father) | No | No |
| 91 | MDS | 74 | M | Caucasian | p.R339H | 2023 | Y | No | No | No |
| 92 | MDS | 76 | M | Caucasian | p.R339C | 305 | Y | NI | NI | NI |
| 93 | MDS | 63 | M | Caucasian | p.Y340N | 90 | Y | NI | NI | NI |
| 94* | MDS | 76 | M | Caucasian | p.R369G | 864 | Y | NI | NI | NI |
| 95 | Pancytopenia | 85 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 96 | Pancytopenia | 56 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 97 | Pancytopenia | 82 | M | Caucasian | p.D140fs | NI | NI | NI | NI | NI |
| 98 | Pancytopenia | 80 | M | Caucasian | p.T144fs | NI | NI | NI | NI | NI |
| 99 | Pancytopenia | 79 | M | NI | p.M316fs | NI | NI | NI | NI | NI |
| 100 | Pancytopenia | 83 | M | Caucasian | p.L452fs | NI | NI | NI | NI | NI |
| 101 | Pancytopenia | 65 | M | Caucasian | p.R159* | 61 | Y | NI | NI | NI |
| 102 | Pancytopenia | 56 | M | NI | p.S543* | NI | NI | NI | NI | NI |
| 103 | Pancytopenia | 55 | M | NI | p.S217P | NI | NI | NI | NI | NI |
| 104 | Pancytopenia | 67 | M | NI | p.P258L | NI | NI | NI | NI | NI |
| 105 | Pancytopenia | 77 | M | Asian | p.Y259C | NI | NI | NI | NI | NI |
| 106 | Pancytopenia | 58 | M | Asian | p.R339L | NI | NI | NI | NI | NI |
| 107 | Thrombocytopenia | 81 | M | NI | p.D140fs | NI | NI | NI | NI | NI |
| 108 | Thrombocytopenia | 37 | F | Caucasian | p.T529fs | 423 | Y | AML (father) | No | No |
| 109 | Thrombocytopenia | 68 | M | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 110 | Neutropenia | 64 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 111 | Neutropenia | 78 | F | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 112 | Anemia | 70 | F | NI | p.R369* | NI | NI | NI | NI | NI |
| 113 | MPN | 40 | F | Caucasian | p.D140fs | 278 | Y | Leukemia (family members, not specified) | No | Breast and uterine cancers (mother) |
| 114 | MPN | 76 | F | Caucasian | p.M316fs | NI | NI | NI | NI | NI |
| 115 | MPN, ET | 41 | F | NI | p.Q306* | NI | NI | NI | NI | NI |
| 116 | MPN | 85 | M | NI | p.K381* | NI | NI | NI | NI | NI |
| 117 | AML | 48 | F | Caucasian | p.E2D | 96 | NI | NI | NI | NI |
| 118* | AML | 54 | F | NI | p.G19R | 61 | Y | NI | NI | NI |
| 119* | AML | 62 | F | NI | p.Y33H | 516 | Y | NI | NI | NI |
| 120 | AML | 76 | M | Caucasian | p.M155I | NI | NI | NI | NI | NI |
| 121 | AML | 56 | F | NI | p.M155I | 613 | N | No | No | No |
| 122* | AML | 63 | F | Caucasian | p.R164N | 365 | Y | No | No | No |
| 123 | AML | 84 | M | Caucasian | p.P510S | 216 | Y | NI | NI | NI |
| 124 | AML | 81 | M | Caucasian | p.M127K | 15 | N | No | No | No |
| 125 | AML | 47 | F | Caucasian | p.M155I | NI | NI | NI | NI | NI |
| 126 | AML | 72 | M | Caucasian | p.G67R | 180 | Y | No | No | No |
| 127 | AML | 72 | M | Caucasian | p.G67R | 150 | Y | No | No | No |
| 128* | AML | 46 | M | Caucasian | p.M155I | 146 | Y | No | No | Cancer of unknown origin (mother) |
| 129 | AML | 79 | M | Caucasian | p.A295T | 241 | Y | No | No | Cancer of unknown origin (mother and father) |
| 130 | AML | 55 | M | NI | p.I298T | NI | NI | NI | NI | NI |
| 131 | AML | 47 | F | Caucasian | p.E355K | 19 | N | No | No | No |
| 132 | AML | 67 | M | Caucasian | p.M155I | 1056 | Y | No | No | No |
| 133 | AML | 7 | F | NI | p.M155I | NI | NI | NI | NI | NI |
| 134 | AML | 64 | M | NI | p.? | 28 | Y | NI | NI | NI |
| 135 | MDS | 79 | F | Caucasian | p.G123S | NI | NI | NI | NI | NI |
| 136 | MDS | 63 | M | NI | p.M155I | NI | NI | NI | NI | NI |
| 137 | MDS | 73 | M | Caucasian | p.? | 420 | N | NI | NI | NI |
| 138 | MDS | 50 | M | Caucasian | p.G175S | NI | NI | NI | NI | NI |
| 139 | MDS | 84 | M | Caucasian | p.P177S | NI | NI | NI | NI | NI |
| 140 | MDS | 60 | M | Caucasian | p.P434L | 303 | Y | NI | NI | NI |
| 141 | MDS | 83 | F | Caucasian | p.P510S | NI | NI | NI | NI | NI |
| 142 | MDS | 78 | M | Caucasian | p.C568W | 516 | N | No | No | No |
| 143 | MDS/MPN | 56 | M | Caucasian | p.G587A | 183 | Y | NI | NI | No |
| 144 | MDS | 92 | M | Caucasian | p.E426K | NI | NI | NI | NI | NI |
| 145 | MDS | 65 | F | NI | p.P510S | 456 | Y | NI | NI | NI |
| 146 | MDS | 62 | M | Caucasian | p.A555T | 255 | Y | NI | NI | NI |
| 147 | Pancytopenia | 63 | M | NI | p.K9K | NI | NI | NI | NI | NI |
| 148 | Pancytopenia | 81 | M | Caucasian | p.M155I | NI | NI | NI | NI | NI |
| 149 | Pancytopenia; SCC | 77 | M | NI | p.M186L | NI | NI | NI | NI | NI |
| 150 | Pancytopenia | 85 | M | NI | p.R219H | NI | NI | NI | NI | NI |
| 151 | Pancytopenia | 84 | F | NI | p.R282H | NI | NI | NI | NI | NI |
| 152 | Pancytopenia | 85 | F | NI | p.C294F | 550 | Y | NI | NI | NI |
| 153 | Pancytopenia | 70 | M | NI | p.S493A | NI | NI | NI | NI | NI |
| 154 | Thrombocytopenia | 88 | M | Caucasian | p.M155I | 37 | Y | No | No | No |
| 155 | Thrombocytopenia | 78 | F | NI | p.T236M | NI | NI | NI | NI | NI |
| 156 | Thrombocytopenia | 39 | M | Caucasian | p.? | NI | NI | NI | NI | NI |
| 157 | Thrombocytopenia | 73 | M | Caucasian | p.? | 31 | Y | NI | NI | NI |
| 158 | Neutropenia | 43 | F | Caucasian | p.? | NI | NI | NI | NI | NI |
| 159 | Anemia | 69 | F | Caucasian | p.P251S | 1777 | Y | No | No | No |
| 160 | Anemia | 78 | M | Caucasian | p.A583V | NI | NI | NI | NI | NI |
| 161 | MPN, PV | 53 | F | Caucasian | p.R8Q | 87 | Y | No | No | Cancer of unknown origin (brother) |
| 162 | MPN, PV | 61 | M | Caucasian | p.D32N | 395 | Y | NI | NI | NI |
| 163 | MPN, ET | 72 | M | NI | p.A133V | NI | NI | NI | NI | NI |
| 164 | MPN | 49 | F | NI | p.I215T | 943 | Y | NI | NI | NI |
| 165 | MPN | 85 | F | NI | p.L216P | NI | NI | NI | NI | NI |
| 166 | MPN, ET | 69 | M | Caucasian | p.T236M | 918 | Y | No | No | No |
| 167 | MPN | 66 | M | Caucasian | p.V491I | 3839 | Y | No | No | No |
| 168 | MPN, ET | 54 | M | Caucasian | p.V544L | 664 | Y | NI | NI | NI |
| 169 | MPN, ET | 39 | F | NI | p.? | NI | NI | NI | NI | NI |
| 170 | MPN, ET | 7 | F | Asian | p.M155T | NI | NI | NI | NI | NI |
| 171 | MPN | 46 | F | Caucasian | p.K187R | NI | NI | NI | NI | NI |
| 172 | Pancytopenia; LPL | 77 | M | AA | p.R164W | 10 | Y | No | No | No |
| 173 | Y heavy chain disease; | 52 | M | NI | p.R164W | 7955 | Y | Myelofibrosis (mother) | No | Lung cancer (father) |
| MYD88 negative LPL | ||||||||||
| 174 | CLL; breast cancer | 51 | F | NI | p.R164W | 1186 | Y | No | FL (mother) | Bladder & prostatic cancer (father) |
| 175 | CLL | 70 | M | Caucasian | p.P251S | 1004 | Y | No | No | No |
| 176 | MM | 82 | F | Caucasian | p.R10Q | 471 | N | No | No | No |
| 177 | AML | 64 | M | Asian | 1703 | Y | Leukemia (maternal uncle) | Gastric cancer (paternal grandfather) | ||
| 178 | AML | 74 | M | Caucasian | 578 | Y | No | No | Sarcoma (father) | |
| 179 | AML | 69 | F | Caucasian | 974 | Y | NI | NI | No | |
| 180 | AML | 54 | M | Caucasian | 42282 | N | No | No | Colon cancer (father) | |
| 181 | AML | 70 | F | Caucasian | 455 | Y | No | No | Breast cancer (paternal aunt) | |
| 182 | AML | 77 | M | Caucasian | NI | NI | NI | NI | NI | |
| 183 | AML | 70 | M | Caucasian | 81 | Y | No | No | No | |
| 184 | AML; DLBCL | 74 | M | Caucasian | NI | Y | No | No | Breast cancer (sisters × 2) | |
| 185 | MDS | 75 | M | Caucasian | 1127 | Y | No | No | No | |
| 186 | MDS | 71 | M | Caucasian | 2053 | Y | No | No | Cancer of unknown origin (mother and sister) | |
| 187 | MDS | 65 | F | Caucasian | 2466 | Y | No | No | Lung cancer (father); breast cancer (father's sister) | |
| 188 | MDS | 54 | M | Caucasian | 151 | Y | No | No | No | |
| 189 | MDS | 69 | M | Caucasian | 689 | N | No | No | Prostatic cancer (father) | |
| 190 | AML | 30 | M | NI | 304 | Y | NI | NI | NI | |
| 191 | AML | 63 | M | Caucasian | 21 | N | No | No | No | |
| 192 | MDS | 81 | M | Caucasian | NI | NI | NI | NI | NI | |
| 193 | MDS | 62 | M | Caucasian | 212 | Y | No | No | Yes | |
| 194 | MDS | 80 | F | Caucasian | NI | NI | NI | NI | NI | |
| 195 | Neutropenia | 86 | M | Caucasian | NI | NI | NI | NI | NI | |
| 196* | Normal | 43 | M | Caucasian | p.M1I | 65 | Y | MDS (father) | No | No |
| 197* | Normal | 69 | F | Caucasian | p.M1I | 496 | Y | Myeloid neoplasm (mother) | No | No |
| 198* | Normal | 28 | M | Caucasian | p.D140fs | 1044 | Y | AML (father) | No | No |
| 199* | Normal | 59 | F | Caucasian | p.D140fs | NI | Y | Leukemia (father) | NI | NI |
| 200* | Normal | 66 | M | Caucasian | p.D140fs | NI | Y | MDS (brother); AML (brothers); ALL (daughter) | No | Lung cancer (father) |
| 201* | Normal | 33 | Caucasian | p.K331del | 614 | Y | MDS (father) | No | Prostatic cancer (paternal uncle); pancreatic cancer (maternal aunt); lung cancer (maternal cousins); bile duct cancer (maternal cousin); melanoma and stomach cancer (paternal uncle); breast cancer (maternal great aunt) | |
| 202 | Normal∼(donor) | 71 | F | Caucasian | p.311* | 367/PHSCT | Y | No | No | |
| 203 | Normal∼(donor) | 59 | M | Caucasian | p.M155I | 2061/PHSCT | Y | No | No | No |
| 204 | Normal∼(donor) | 60 | M | Caucasian | p.V565M | 725/PHSCT | Y | NI | NI | NI |
| 205 | Normal∼(donor) | 71 | M | Caucasian | p.T529fs | 459/PHSCT | Y | No | No | No |
| Patient . | Diagnosis . | Age . | Sex . | Ethnicity . | gl DDX41 . | FU (d) . | Survival (Y/N) . | FH_MN . | FH_LN . | FH_Other . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1* | AML | 62 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 2† | AML | 78 | F | Caucasian | p.M1I | 365 | Y | No | No | No |
| 3 | AML | 77 | M | Caucasian | p.M1I | 122 | Y | NI | NI | NI |
| 4† | AML | 64 | M | Caucasian | p.M1I | 519 | Y | No | No | Breast cancer (mother) |
| 5 | AML | 80 | M | Caucasian | p.M1I | 336 | Y | NI | NI | NI |
| 6*,† | AML | 77 | M | Caucasian | p.M1I | 2161 | Y | No | No | No |
| 7 | AML | 62 | M | Caucasian | p.M1I | 151 | Y | Leukemia (father) | No | No |
| 8 | AML | 76 | M | Caucasian | p.M1I | 1214 | Y | No | No | No |
| 9† | AML | 68 | M | Caucasian | p.M1I | 822 | Y | No | No | No |
| 10*,† | AML | 76 | F | Caucasian | p.M1I | 2161 | Y | No | No | Breast cancer (mother and sister); |
| pituitary tumor (brother); rhabdomyosarcoma (son) | ||||||||||
| 11 | AML | 88 | F | Caucasian | p.M1I | 98 | Y | No | No | No |
| 12† | AML | 48 | F | Caucasian | p.M1I | 700 | Y | No | No | No |
| 13 | AML | 63 | F | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 14† | AML | 78 | M | Caucasian | p.M1I | 1034 | Y | No | No | No |
| 15† | AML | 65 | M | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 16† | AML | 73 | M | Caucasian | p.M1I | 91 | Y | No | No | Cancer of unknown origin (mother) |
| 17† | AML | 72 | M | Caucasian | p.M1I | 822 | N | No | No | No |
| 18† | AML | 74 | M | Caucasian | p.M1I | 396 | Y | No | No | Cancer of unknown origin (brother) |
| 19* | AML | 75 | M | Caucasian | p.M1I | 608 | Y | NI | NI | NI |
| 20*# | AML | 60 | M | Caucasian | p.M1I | 639 | Y | No | No | No |
| 21# | AML | 65 | M | Caucasian | p.M1I | 365 | Y | No | No | Cancer of unknown origin (sister) |
| 22# | AML | 64 | M | Caucasian | p.M1I | 304 | Y | No | No | Lung cancer (father); cervical cancer (daughter) |
| 23 | AML | 68 | M | Caucasian | p.M1I | 400 | Y | No | No | Lung cancer (mother) |
| 24† | AML | 69 | M | Caucasian | p.M1I | 1216 | Y | NI | NI | NI |
| 25 | AML | 71 | F | Caucasian | p.K108fs | NI | NI | NI | NI | NI |
| 26 | AML | 71 | F | Caucasian | p.D140fs | NI | NI | NI | NI | NI |
| 27 | AML | 57 | M | Caucasian | p.D140fs | 561 | Y | NI | NI | NI |
| 28† | AML | 57 | M | Caucasian | p.D140fs | 403 | N | No | No | No |
| 29 | AML | 57 | M | Caucasian | p.D140fs | 1080 | Y | No | Lymphoma (father) | Prostatic cancer (paternal uncle) |
| 30*,† | AML | 59 | M | Caucasian | p.D140fs | 639 | Y | No | No | Pancreatic cancer (brother) |
| 31 | AML | 78 | M | Caucasian | p.D140fs | NI | NI | NI | NI | NI |
| 32 | AML | 81 | M | Caucasian | p.D140fs | 260 | Y | NI | NI | NI |
| 33 | AML | 63 | M | Caucasian | p.D140fs | 61 | Y | NI | NI | NI |
| 34† | AML | 90 | F | Caucasian | p.D140fs | 176 | N | No | No | No |
| 35 | AML | 66 | M | Caucasian | p.D140fs | 913 | Y | MDS (father and paternal uncle) | No | No |
| 36 | AML | 67 | M | NI | p.D140fs | 580 | Y | NI | NI | NI |
| 37 | AML | 50 | M | Caucasian | p.D140fs | 245 | Y | NI | NI | NI |
| 38† | AML | 70 | F | AA | p.D140fs | 183 | Y | No | No | Brain cancer (sister) |
| 39† | AML | 53 | M | Caucasian | p.D140fs | 945 | Y | No | No | Lung cancer (mother); colon cancer (father) |
| 40† | AML | 70 | M | Caucasian | p.D140fs | 488 | Y | No | No | Cancer of mouth and throat (brother) |
| 41 | AML | 57 | M | NI | p.D140fs | 675 | Y | NI | NI | NI |
| 42 | AML | 54 | F | Caucasian | p.D140fs | 300 | Y | No | No | No |
| 43 | AML | 61 | M | Caucasian | p.D140fs | 145 | Y | No | MM (father) | No |
| 44 | AML | 73 | M | Asian | p.I224fs | 731 | Y | NI | NI | NI |
| 45† | AML | 54 | F | Caucasian | p.L283fs | 580 | Y | No | No | No |
| 46 | AML | 58 | F | Caucasian | p.M316fs | 1071 | Y | No | No | No |
| 47 | AML | 76 | F | NI | p.G465fs | 640 | Y | NI | NI | NI |
| 48 | AML | 63 | M | Caucasian | p.Q41* | 212 | Y | No | No | No |
| 49 | AML | 70 | M | Caucasian | p.Q41* | NI | NI | NI | NI | NI |
| 50 | AML | 71 | M | Caucasian | p.R159* | 31 | Y | NI | NI | No |
| 51 | AML | 61 | M | Caucasian | p.R311* | NI | NI | NI | NI | NI |
| 52 | AML | 68 | M | Caucasian | p.R369* | 548 | N | No | No | No |
| 53 | AML | 78 | M | NI | p.R369* | 120 | Y | NI | NI | NI |
| 54 | AML | 70 | M | Caucasian | p.Q370* | 408 | Y | Leukemia (mother) | No | No |
| 55 | AML | 82 | M | Caucasian | p.Q502* | NI | NI | NI | NI | NI |
| 56† | AML | 68 | F | Caucasian | p.? | 905 | Y | Leukemia (maternal aunt) | No | No |
| 57 | AML | 74 | M | Caucasian | p.K331del | 232 | N | No | No | Thyroid and colon cancer (mother) |
| 58† | AML | 65 | F | Caucasian | p.L216V | 275 | Y | MDS (brother) | No | No |
| 59 | AML | 66 | M | Caucasian | p.G218D | 31 | Y | NI | NI | NI |
| 60 | AML; breast cancer | 47 | F | Caucasian | p.P258L | 653 | Y | No | No | No |
| 61 | AML | 60 | M | NI | p.R323C | 956 | Y | NI | NI | NI |
| 62 | AML | 69 | M | Caucasian | p.M349K | 365 | Y | NI | NI | NI |
| 63 | AML | 69 | M | Caucasian | p.M349K | 1134 | Y | No | No | Prostatic cancer (father) |
| 64 | AML | 81 | F | Caucasian | p.R369G | 151 | Y | AML (paternal cousin) | No | No |
| 65 | AML | 85 | M | Asian | p.D467Y | 458 | Y | NI | NI | NI |
| 66 | AML | 61 | M | Asian | p.R525H | 243 | N | MDS (father); Leukemia (paternal grandma) | No | No |
| Hematologic cancer (paternal uncle) | ||||||||||
| 67 | MDS | 72 | M | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 68 | MDS | 73 | M | Caucasian | p.M1I | 539 | Y | No | No | No |
| 69 | MDS | 81 | F | Caucasian | p.M1I | 670 | Y | No | No | No |
| 70 | MDS | 76 | M | Caucasian | p.M1I | 362 | Y | No | No | No |
| 71 | MDS | 79 | M | Caucasian | p.M1I | 763 | Y | No | No | Prostatic cancer (brother) |
| 72 | MDS | 61 | F | Caucasian | p.M1I | 192 | Y | No | No | Breast cancer (sister) |
| 73 | MDS | 60 | M | Caucasian | p.M1I | 1795 | Y | No | No | Throat cancer (father); melanoma (sister) |
| 74 | MDS | 65 | M | Caucasian | p.M1I | 153 | Y | NI | NI | No |
| 75 | MDS | 63 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 76 | MDS; MM; MBL | 67 | F | Caucasian | p.M1I | 98 | Y | No | No | No |
| 77 | MDS | 69 | M | Caucasian | p.M1I | 92 | Y | No | No | No |
| 78 | MDS | 72 | M | Caucasian | p.M1I | 2722 | Y | No | No | No |
| 79 | MDS | 60 | M | Caucasian | p.M1I | 395 | Y | NI | NI | No |
| 80 | MDS | 88 | F | Caucasian | p.D140fs | 476 | Y | NI | NI | NI |
| 81 | MDS | 69 | M | Caucasian | p.D140fs | 1078 | Y | NI | NI | NI |
| 82 | MDS | 63 | M | Asian | p.A500fs | 761 | Y | NI | NI | No |
| 83 | MDS | 76 | F | Asian | p.Q44* | 701 | Y | No | No | No |
| 84 | MDS | 77 | M | Caucasian | p.R159* | 876 | N | NI | NI | NI |
| 85 | MDS | 85 | F | Caucasian | p.R311* | NI | NI | NI | NI | NI |
| 86 | MDS | 84 | M | Caucasian | p.R311* | NI | NI | NI | NI | NI |
| 87 | MDS | 77 | M | Caucasian | p.K331del | 1793 | Y | Leukemia (father) | No | No |
| 88 | MDS | 62 | M | Caucasian | p.K331del | 900 | Y | No | No | Prostatic cancer, melanoma and stomach cancer (paternal uncles); |
| Pancreatic cancer (maternal aunt); lung cancer (maternal cousins); | ||||||||||
| Bile duct cancer (maternal cousin); breast cancer (maternal great aunt) | ||||||||||
| 89 | MDS | 84 | F | Caucasian | p.P189L | 2015 | Y | No | No | No |
| 90 | MDS | 73 | M | Caucasian | p.L237W | 134 | Y | MDS (father) | No | No |
| 91 | MDS | 74 | M | Caucasian | p.R339H | 2023 | Y | No | No | No |
| 92 | MDS | 76 | M | Caucasian | p.R339C | 305 | Y | NI | NI | NI |
| 93 | MDS | 63 | M | Caucasian | p.Y340N | 90 | Y | NI | NI | NI |
| 94* | MDS | 76 | M | Caucasian | p.R369G | 864 | Y | NI | NI | NI |
| 95 | Pancytopenia | 85 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 96 | Pancytopenia | 56 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 97 | Pancytopenia | 82 | M | Caucasian | p.D140fs | NI | NI | NI | NI | NI |
| 98 | Pancytopenia | 80 | M | Caucasian | p.T144fs | NI | NI | NI | NI | NI |
| 99 | Pancytopenia | 79 | M | NI | p.M316fs | NI | NI | NI | NI | NI |
| 100 | Pancytopenia | 83 | M | Caucasian | p.L452fs | NI | NI | NI | NI | NI |
| 101 | Pancytopenia | 65 | M | Caucasian | p.R159* | 61 | Y | NI | NI | NI |
| 102 | Pancytopenia | 56 | M | NI | p.S543* | NI | NI | NI | NI | NI |
| 103 | Pancytopenia | 55 | M | NI | p.S217P | NI | NI | NI | NI | NI |
| 104 | Pancytopenia | 67 | M | NI | p.P258L | NI | NI | NI | NI | NI |
| 105 | Pancytopenia | 77 | M | Asian | p.Y259C | NI | NI | NI | NI | NI |
| 106 | Pancytopenia | 58 | M | Asian | p.R339L | NI | NI | NI | NI | NI |
| 107 | Thrombocytopenia | 81 | M | NI | p.D140fs | NI | NI | NI | NI | NI |
| 108 | Thrombocytopenia | 37 | F | Caucasian | p.T529fs | 423 | Y | AML (father) | No | No |
| 109 | Thrombocytopenia | 68 | M | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 110 | Neutropenia | 64 | M | NI | p.M1I | NI | NI | NI | NI | NI |
| 111 | Neutropenia | 78 | F | Caucasian | p.M1I | NI | NI | NI | NI | NI |
| 112 | Anemia | 70 | F | NI | p.R369* | NI | NI | NI | NI | NI |
| 113 | MPN | 40 | F | Caucasian | p.D140fs | 278 | Y | Leukemia (family members, not specified) | No | Breast and uterine cancers (mother) |
| 114 | MPN | 76 | F | Caucasian | p.M316fs | NI | NI | NI | NI | NI |
| 115 | MPN, ET | 41 | F | NI | p.Q306* | NI | NI | NI | NI | NI |
| 116 | MPN | 85 | M | NI | p.K381* | NI | NI | NI | NI | NI |
| 117 | AML | 48 | F | Caucasian | p.E2D | 96 | NI | NI | NI | NI |
| 118* | AML | 54 | F | NI | p.G19R | 61 | Y | NI | NI | NI |
| 119* | AML | 62 | F | NI | p.Y33H | 516 | Y | NI | NI | NI |
| 120 | AML | 76 | M | Caucasian | p.M155I | NI | NI | NI | NI | NI |
| 121 | AML | 56 | F | NI | p.M155I | 613 | N | No | No | No |
| 122* | AML | 63 | F | Caucasian | p.R164N | 365 | Y | No | No | No |
| 123 | AML | 84 | M | Caucasian | p.P510S | 216 | Y | NI | NI | NI |
| 124 | AML | 81 | M | Caucasian | p.M127K | 15 | N | No | No | No |
| 125 | AML | 47 | F | Caucasian | p.M155I | NI | NI | NI | NI | NI |
| 126 | AML | 72 | M | Caucasian | p.G67R | 180 | Y | No | No | No |
| 127 | AML | 72 | M | Caucasian | p.G67R | 150 | Y | No | No | No |
| 128* | AML | 46 | M | Caucasian | p.M155I | 146 | Y | No | No | Cancer of unknown origin (mother) |
| 129 | AML | 79 | M | Caucasian | p.A295T | 241 | Y | No | No | Cancer of unknown origin (mother and father) |
| 130 | AML | 55 | M | NI | p.I298T | NI | NI | NI | NI | NI |
| 131 | AML | 47 | F | Caucasian | p.E355K | 19 | N | No | No | No |
| 132 | AML | 67 | M | Caucasian | p.M155I | 1056 | Y | No | No | No |
| 133 | AML | 7 | F | NI | p.M155I | NI | NI | NI | NI | NI |
| 134 | AML | 64 | M | NI | p.? | 28 | Y | NI | NI | NI |
| 135 | MDS | 79 | F | Caucasian | p.G123S | NI | NI | NI | NI | NI |
| 136 | MDS | 63 | M | NI | p.M155I | NI | NI | NI | NI | NI |
| 137 | MDS | 73 | M | Caucasian | p.? | 420 | N | NI | NI | NI |
| 138 | MDS | 50 | M | Caucasian | p.G175S | NI | NI | NI | NI | NI |
| 139 | MDS | 84 | M | Caucasian | p.P177S | NI | NI | NI | NI | NI |
| 140 | MDS | 60 | M | Caucasian | p.P434L | 303 | Y | NI | NI | NI |
| 141 | MDS | 83 | F | Caucasian | p.P510S | NI | NI | NI | NI | NI |
| 142 | MDS | 78 | M | Caucasian | p.C568W | 516 | N | No | No | No |
| 143 | MDS/MPN | 56 | M | Caucasian | p.G587A | 183 | Y | NI | NI | No |
| 144 | MDS | 92 | M | Caucasian | p.E426K | NI | NI | NI | NI | NI |
| 145 | MDS | 65 | F | NI | p.P510S | 456 | Y | NI | NI | NI |
| 146 | MDS | 62 | M | Caucasian | p.A555T | 255 | Y | NI | NI | NI |
| 147 | Pancytopenia | 63 | M | NI | p.K9K | NI | NI | NI | NI | NI |
| 148 | Pancytopenia | 81 | M | Caucasian | p.M155I | NI | NI | NI | NI | NI |
| 149 | Pancytopenia; SCC | 77 | M | NI | p.M186L | NI | NI | NI | NI | NI |
| 150 | Pancytopenia | 85 | M | NI | p.R219H | NI | NI | NI | NI | NI |
| 151 | Pancytopenia | 84 | F | NI | p.R282H | NI | NI | NI | NI | NI |
| 152 | Pancytopenia | 85 | F | NI | p.C294F | 550 | Y | NI | NI | NI |
| 153 | Pancytopenia | 70 | M | NI | p.S493A | NI | NI | NI | NI | NI |
| 154 | Thrombocytopenia | 88 | M | Caucasian | p.M155I | 37 | Y | No | No | No |
| 155 | Thrombocytopenia | 78 | F | NI | p.T236M | NI | NI | NI | NI | NI |
| 156 | Thrombocytopenia | 39 | M | Caucasian | p.? | NI | NI | NI | NI | NI |
| 157 | Thrombocytopenia | 73 | M | Caucasian | p.? | 31 | Y | NI | NI | NI |
| 158 | Neutropenia | 43 | F | Caucasian | p.? | NI | NI | NI | NI | NI |
| 159 | Anemia | 69 | F | Caucasian | p.P251S | 1777 | Y | No | No | No |
| 160 | Anemia | 78 | M | Caucasian | p.A583V | NI | NI | NI | NI | NI |
| 161 | MPN, PV | 53 | F | Caucasian | p.R8Q | 87 | Y | No | No | Cancer of unknown origin (brother) |
| 162 | MPN, PV | 61 | M | Caucasian | p.D32N | 395 | Y | NI | NI | NI |
| 163 | MPN, ET | 72 | M | NI | p.A133V | NI | NI | NI | NI | NI |
| 164 | MPN | 49 | F | NI | p.I215T | 943 | Y | NI | NI | NI |
| 165 | MPN | 85 | F | NI | p.L216P | NI | NI | NI | NI | NI |
| 166 | MPN, ET | 69 | M | Caucasian | p.T236M | 918 | Y | No | No | No |
| 167 | MPN | 66 | M | Caucasian | p.V491I | 3839 | Y | No | No | No |
| 168 | MPN, ET | 54 | M | Caucasian | p.V544L | 664 | Y | NI | NI | NI |
| 169 | MPN, ET | 39 | F | NI | p.? | NI | NI | NI | NI | NI |
| 170 | MPN, ET | 7 | F | Asian | p.M155T | NI | NI | NI | NI | NI |
| 171 | MPN | 46 | F | Caucasian | p.K187R | NI | NI | NI | NI | NI |
| 172 | Pancytopenia; LPL | 77 | M | AA | p.R164W | 10 | Y | No | No | No |
| 173 | Y heavy chain disease; | 52 | M | NI | p.R164W | 7955 | Y | Myelofibrosis (mother) | No | Lung cancer (father) |
| MYD88 negative LPL | ||||||||||
| 174 | CLL; breast cancer | 51 | F | NI | p.R164W | 1186 | Y | No | FL (mother) | Bladder & prostatic cancer (father) |
| 175 | CLL | 70 | M | Caucasian | p.P251S | 1004 | Y | No | No | No |
| 176 | MM | 82 | F | Caucasian | p.R10Q | 471 | N | No | No | No |
| 177 | AML | 64 | M | Asian | 1703 | Y | Leukemia (maternal uncle) | Gastric cancer (paternal grandfather) | ||
| 178 | AML | 74 | M | Caucasian | 578 | Y | No | No | Sarcoma (father) | |
| 179 | AML | 69 | F | Caucasian | 974 | Y | NI | NI | No | |
| 180 | AML | 54 | M | Caucasian | 42282 | N | No | No | Colon cancer (father) | |
| 181 | AML | 70 | F | Caucasian | 455 | Y | No | No | Breast cancer (paternal aunt) | |
| 182 | AML | 77 | M | Caucasian | NI | NI | NI | NI | NI | |
| 183 | AML | 70 | M | Caucasian | 81 | Y | No | No | No | |
| 184 | AML; DLBCL | 74 | M | Caucasian | NI | Y | No | No | Breast cancer (sisters × 2) | |
| 185 | MDS | 75 | M | Caucasian | 1127 | Y | No | No | No | |
| 186 | MDS | 71 | M | Caucasian | 2053 | Y | No | No | Cancer of unknown origin (mother and sister) | |
| 187 | MDS | 65 | F | Caucasian | 2466 | Y | No | No | Lung cancer (father); breast cancer (father's sister) | |
| 188 | MDS | 54 | M | Caucasian | 151 | Y | No | No | No | |
| 189 | MDS | 69 | M | Caucasian | 689 | N | No | No | Prostatic cancer (father) | |
| 190 | AML | 30 | M | NI | 304 | Y | NI | NI | NI | |
| 191 | AML | 63 | M | Caucasian | 21 | N | No | No | No | |
| 192 | MDS | 81 | M | Caucasian | NI | NI | NI | NI | NI | |
| 193 | MDS | 62 | M | Caucasian | 212 | Y | No | No | Yes | |
| 194 | MDS | 80 | F | Caucasian | NI | NI | NI | NI | NI | |
| 195 | Neutropenia | 86 | M | Caucasian | NI | NI | NI | NI | NI | |
| 196* | Normal | 43 | M | Caucasian | p.M1I | 65 | Y | MDS (father) | No | No |
| 197* | Normal | 69 | F | Caucasian | p.M1I | 496 | Y | Myeloid neoplasm (mother) | No | No |
| 198* | Normal | 28 | M | Caucasian | p.D140fs | 1044 | Y | AML (father) | No | No |
| 199* | Normal | 59 | F | Caucasian | p.D140fs | NI | Y | Leukemia (father) | NI | NI |
| 200* | Normal | 66 | M | Caucasian | p.D140fs | NI | Y | MDS (brother); AML (brothers); ALL (daughter) | No | Lung cancer (father) |
| 201* | Normal | 33 | Caucasian | p.K331del | 614 | Y | MDS (father) | No | Prostatic cancer (paternal uncle); pancreatic cancer (maternal aunt); lung cancer (maternal cousins); bile duct cancer (maternal cousin); melanoma and stomach cancer (paternal uncle); breast cancer (maternal great aunt) | |
| 202 | Normal∼(donor) | 71 | F | Caucasian | p.311* | 367/PHSCT | Y | No | No | |
| 203 | Normal∼(donor) | 59 | M | Caucasian | p.M155I | 2061/PHSCT | Y | No | No | No |
| 204 | Normal∼(donor) | 60 | M | Caucasian | p.V565M | 725/PHSCT | Y | NI | NI | NI |
| 205 | Normal∼(donor) | 71 | M | Caucasian | p.T529fs | 459/PHSCT | Y | No | No | No |
AA, African American; F, female; M, male; N, deceased; NI, no information; PHSCT, post-HSCT; Y, survived.
Germline variants confirmed by skin biopsies.
Reported in a prior study.9
Targeted NGS
DNA was extracted from fresh bone marrow aspirates and NGS testing was performed using a targeted NGS panel at each institution. The ARUP myeloid malignancy NGS panel included 65 genes (supplemental Table 1), and targeted hybrid‐capture sequencing was performed using the SureselectXTHS kit (Agilent Technologies, Santa Clara, CA) following the manufacturer's protocol as described previously.7,8 The genes listed in NGS panels at each institution and the 53 common genes tested are summarized in supplemental Tables 1 and 2.
DDX41-specific variant classification and interpretation
Variants with a variant allele frequency (VAF) of 40% or above were presumed to be germline variants.7,8 Germline variants were classified as pathogenic/likely pathogenic variants (PV/LPV), described herein as CV or VUS according to ACMG/AMP guidelines (supplemental Table 3) with the following specific considerations.20 A pathogenic moderate criterion (PM2) was applied to variants with a Genome Aggregation Database (gnomAD) population frequency less than that of the 2 most frequent known pathogenic DDX41 variants: p.M1I and p.D140fs (both with gnomAD frequency of 0.008%). Another pathogenic moderate criterion (PM3) was used in a modified manner to account for the known mechanism of DDX41 second hits in affected individuals. This criterion was applied to the germline variant when a second pathogenic variant (presumed somatic) was also present in an affected patient with this variant in our study or reported in the literature. A pathogenic supporting criterion (PP3) was applied when the REVEL score for the variant was greater than 0.7.
Germline confirmation
Germline testing was performed prospectively on 12 patients (8 with CV and 4 with VUS) and 6 asymptomatic relatives who were referred to the genetics clinic based on the persistent presence of a DDX41 variant at near-heterozygous VAF and suspicious FH. Germline confirmation was performed as previously described using skin biopsies.7 The remaining patients either were not referred for genetic counseling or declined further testing.
Asymptomatic individuals with germline DDX41 variants under surveillance
Six asymptomatic relatives of the patients with HM with confirmed germline DDX41 CV underwent cancer surveillance (Tables 1 and 2, patients 196-201, all with CV) with bone marrow biopsies in conjunction with flow cytometric, cytogenetic, and NGS studies to establish a baseline. Bone marrow examination in all cases showed essentially normal trilineage hematopoiesis without evidence of malignancies, as reviewed by P.L. and M.W. independently. Furthermore, 4 patients with donor-derived DDX41 variants (2 CV and 2 VUS) after HSCT for previously diagnosed AML were included (Tables 1 and 2, patients 202-205), and all had unremarkable complete blood count (CBC) and 100% donor chimerism, confirmed by short tandem repeat testing at post-transplant surveillance.
Statistics
Descriptive statistics were used for patient epidemiologic characteristics and the number of somatic variants per case, and the results are summarized in figures as appropriate. Unpaired t test was used for all quantitative data, and Fisher exact test or χ2 test was used for qualitative data.8 OS was analyzed as a time-to-event date point using the Kaplan-Meier method.21-24 Time-to-event data were also analyzed with Cox proportional hazards regression to calculate hazard ratios (HR) by multivariate analysis.25,26
Literature review and gnomAD database search
A PubMed search for cases of sporadic and familial HM with germline DDX41 variant was performed. Individual studies were reviewed, and the variants were reclassified and summarized in Figure 2A. Clinical outcome information from 18 additional AML and high-grade MDS cases with germline DDX41 variants in association with 186 age-matched patients with DDX41 wild-type (WT) AML from a previous study7 was collected. Furthermore, additional data on OS of 3128 age-matched patients with MDS in the literature27 and 1040 patients with AML in cBioPortal (median age, 68 years; range, 47-99 years; access date, 28 February 2022) were retrieved and reanalyzed to extend OS analysis (supplemental Figure 2A). Where available in publications,15,28,29 ethnicity was reanalyzed in combination with the current study population and summarized in Figure 2B-E. GnomAD was searched to acquire the minor allele frequencies (MAF) of variants and was incorporated in supplemental Table 3.
Results
DDX41 variant landscape, genetic profiles, and ethnic differences of patients with HM
Among the 9821 unrelated and unselected patients with HM, 195 (2%) were found to have at least 1 DDX41 variant; of those, 176 (1.8%) patients had a putative germline DDX41 variant and 19 (0.2%) had somatic variants alone (Figure 1A). The 176 HM cases with germline variants (Tables 1 and 2, patients 1-176) included 84 AML, 40 MDS, 15 MPN, 32 cases of cytopenia, 4 B-cell lymphoma, and 1 MM. The 19 cases (Tables 1 and 2, patients 177-195) with only somatic variants included 10 AML, 8 MDS, and 1 cytopenia. Overall, 82 distinct presumed germline variants were identified, among which 39 were classified as CV (red in Figure 1B) and 43 as VUS (blue in Figure 1B; supplemental Table 3) according to the proposed classification criteria. Loss of function variants, recurrent missense variants in association with a low MAF with specific exceptions (eg, p.M155I and p.P510S), and novel missense variants accompanied by pathogenic somatic DDX41 variants (Figure 1A) were generally considered CV. Here we reported 53 novel germline (15 CV and 38 VUS) and 13 novel somatic variants (4 CV and 9 VUS, underlined in Figure 1B) among which the 5 new missense germline CVs (Table 1, 58, 65, 89, 90, and 93) were all accompanied by previously characterized somatic pathogenic variants (p.R525H, p.G530D, or p.E345D).
The previously reported germline and somatic DDX41 variants in HM,7,10,11,28,30-34 together with those in the current study, are summarized in Figure 2A. Most germline CV (63% in this study and 68% by literature review) were loss of function mutations, including start codon loss (p.M1I), nonsense, frameshift, or mutations disrupting splicing sites (Figures 1B and 2A), concentrated upstream to the DEAD box domain. p.M1I and p.D140fs, the most common germline CVs (19% and 15% of all, respectively), were primarily identified in White patients (Figure 2B). Missense germline CV, although less common and most often classified as VUS according to unmodified ACMG guidelines, were reported in this study and literature accompanied by pathogenic somatic variants.7,10,11,28-34 Interestingly, approximately half of Asian patients with HM carried missense germline CV (Figure 2C), with Y259C being the most common hotspot (Figure 2D), whereas only 1 p.M1I28 and 0 p.D140fs variants were documented in Asian patients (Figure 2B). Data obtained from the gnomAD database showed similar ethnic differences (not shown). This unique ethnic difference29 in DDX41 CV was also highlighted by p.A500fs, seen exclusively in Asian patients (Figure 2D) as the second most common germline CV and more frequent somatic variants alone (Figure 2E).
Concomitant somatic variants were detected by NGS in patients with germline DDX41 CV (red) and VUS (blue in Figure 3). Beyond somatic DDX41 variants, ASXL1 was the second most commonly mutated gene concomitant with germline DDX41 CV (28%), followed by DNMT3A (13%) and TET2 (11%), similar to those in HM without germline DDX41 variants.35 In stark contrast, the most frequent concomitant variant in patients with HM with DDX41 VUS was the JAK2 p.V617F mutation (18%; Figure 3), and most (82%, 9 of 11) exhibited a leading VAF (Table 1), suggestive of a disease driver mutation. The genetic profiles in 19 patients with somatic DDX41 CV alone appeared similar to those with somatic DDX41 VUS (Figure 3; supplemental Figure 1). Interestingly, the median age of patients with HM with CV (68 years; Table 3) was greater than patients with VUS (63 years; P = .01). Similar to previous reports,7,8,10,32 there was a striking male predominance of patients with CV (74%, Table 3) compared with WT control cohorts (50%, P < .0001), which was markedly diminished in patients with VUS (62%).
Integrated genetic profiles of the 195 HM patients with epidemiologic characteristics grouped by different HM diagnoses. A total of 176 patients with presumed germline (gl DDX41) 116 CV and 60 VUS are grouped (CV in red and VUS in blue, respectively), along with the associated somatic DDX41 (s DDX41), concomitant somatic variants, and cytogenetics. In addition, 19 patients with HM with somatic DDX41 variants in the absence of germline variants are appended to the right of the variant table, 13 CV in red and 6 VUS in blue. Each column represents 1 patient. The concomitant variants are grouped into 6 categories based on gene function: epigenetic, epigenetic regulators, genes involving DNA methylation or histone acetylation, and deacetylation (light green); SFs, RNA splicing factors (purple); TFs, transcription factors (orange); signaling, molecules in tyrosine kinase pathway or RAS/MAPK pathways (pink); C, cohesins (light blue); and others (dark blue), genes with function beyond the above categories. Each bar represents 1 variant, and split bars indicate 2 or more variants in the same gene. A&B, AML and breast cancer; CLL & B, chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) and breast cancer; complex, complex karyotype; DLBCL, diffuse large B-cell lymphoma; gl DDX41, germline DDX41 variants; LPL, lymphoplasmacytic lymphoma (MYD88 negative); LR, low risk; MBL, monoclonal B-cell lymphocytosis; MM, multiple myeloma; NI, no information; NL, normal; NM, no mutation; RCA, recurrent cytogenetic abnormalities in AML; s DDX41, somatic DDX41 variants.
Integrated genetic profiles of the 195 HM patients with epidemiologic characteristics grouped by different HM diagnoses. A total of 176 patients with presumed germline (gl DDX41) 116 CV and 60 VUS are grouped (CV in red and VUS in blue, respectively), along with the associated somatic DDX41 (s DDX41), concomitant somatic variants, and cytogenetics. In addition, 19 patients with HM with somatic DDX41 variants in the absence of germline variants are appended to the right of the variant table, 13 CV in red and 6 VUS in blue. Each column represents 1 patient. The concomitant variants are grouped into 6 categories based on gene function: epigenetic, epigenetic regulators, genes involving DNA methylation or histone acetylation, and deacetylation (light green); SFs, RNA splicing factors (purple); TFs, transcription factors (orange); signaling, molecules in tyrosine kinase pathway or RAS/MAPK pathways (pink); C, cohesins (light blue); and others (dark blue), genes with function beyond the above categories. Each bar represents 1 variant, and split bars indicate 2 or more variants in the same gene. A&B, AML and breast cancer; CLL & B, chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) and breast cancer; complex, complex karyotype; DLBCL, diffuse large B-cell lymphoma; gl DDX41, germline DDX41 variants; LPL, lymphoplasmacytic lymphoma (MYD88 negative); LR, low risk; MBL, monoclonal B-cell lymphocytosis; MM, multiple myeloma; NI, no information; NL, normal; NM, no mutation; RCA, recurrent cytogenetic abnormalities in AML; s DDX41, somatic DDX41 variants.
Summary of ages and sexes of patients with HM with DDX41 CV, and VUS and controls
| Disease . | Germline DDX41 . | DDX41+ CV (reference) . | DDX41+ VUS . | P (a) . | DDX41− WT . | P (b) . |
|---|---|---|---|---|---|---|
| HM | n | 111 | 60 | 4307 | ||
| Median age (y, range) | 68 (37-90) | 63 (7-92) | .01* | 67 (18-100) | .35 | |
| Sex male/female (M%) | 82/29 (74%) | 37/23(62%) | .10 | 2154/2153 (50%) | <.0001**** | |
| AML | n | 66 | 18 | 1365 | ||
| Median age (y, range) | 69 (47-90) | 62 (7-84) | .02* | 64 (18-90) | .002** | |
| Sex male/female (M%) | 49/17 (71%) | 10/8 (56%) | .12 | 737/628 (54%) | .001** | |
| MDS | n | 28 | 12 | 1109 | ||
| Median age (y, range) | 72 (60-88) | 69 (50-92) | .49 | 74 (36-98) | 0.64 | |
| Sex male/female (M%) | 21/7 (75%) | 9/3 (75%) | >.99 | 555/554 (50%) | .009** | |
| MPN | n | 4 | 11 | 470 | ||
| Median age (y, range) | 59 (40-85) | 54 (7-85) | .64 | 66 (18-95) | .74 | |
| Sex male/female (M%) | 1/3 (25%) | 5/6 (45%) | .33 | 192/287 (40%) | .36 |
| Disease . | Germline DDX41 . | DDX41+ CV (reference) . | DDX41+ VUS . | P (a) . | DDX41− WT . | P (b) . |
|---|---|---|---|---|---|---|
| HM | n | 111 | 60 | 4307 | ||
| Median age (y, range) | 68 (37-90) | 63 (7-92) | .01* | 67 (18-100) | .35 | |
| Sex male/female (M%) | 82/29 (74%) | 37/23(62%) | .10 | 2154/2153 (50%) | <.0001**** | |
| AML | n | 66 | 18 | 1365 | ||
| Median age (y, range) | 69 (47-90) | 62 (7-84) | .02* | 64 (18-90) | .002** | |
| Sex male/female (M%) | 49/17 (71%) | 10/8 (56%) | .12 | 737/628 (54%) | .001** | |
| MDS | n | 28 | 12 | 1109 | ||
| Median age (y, range) | 72 (60-88) | 69 (50-92) | .49 | 74 (36-98) | 0.64 | |
| Sex male/female (M%) | 21/7 (75%) | 9/3 (75%) | >.99 | 555/554 (50%) | .009** | |
| MPN | n | 4 | 11 | 470 | ||
| Median age (y, range) | 59 (40-85) | 54 (7-85) | .64 | 66 (18-95) | .74 | |
| Sex male/female (M%) | 1/3 (25%) | 5/6 (45%) | .33 | 192/287 (40%) | .36 |
P value (a) applies to DDX41+ CV vs DDX41+ VUS, and P value (b) applies to DDX41+ CV vs DDX41− WT.
P < .05; **P < .01; ****P < .0001.
AML/MDS with germline DDX41 CV is a distinct and the most common HM
By the proposed variant classification framework, patients with AML/MDS with DDX41-presumed germline CV and VUS were readily distinguished by differing genetic characteristics, epidemiologic features, and OS. Seventy-nine percent of patients with CV developed later-onset AML/MDS, whereas approximately 52% of those carrying VUS manifested with AML/MDS including a subset of early-onset AML (Tables 2 and 3). Specifically, the median age at the time of AML diagnosis was 69 years in patients carrying CV in contrast to those with VUS, in which some were children or young adults (median, 62 years; P = .02), and sporadic AML in adults (median, 64 years; P = .002; Table 3). Interestingly, the median age at MDS diagnosis (72 years) was similar to that in patients with AML with germline CV (Table 3; P > .05), whereas the median ages of patients with MDS carrying either VUS or WT DDX41 was older than those in patients with AML with the same genotype (Table 3; 69 in MDS vs 62 in AML with VUS, P = .08; 74 in MDS vs 64 in AML with WT, P < .0001), as MDS is primarily a disease of the elderly.
More frequent somatic DDX41 variants (Figure 4A) and a lower somatic mutation burden (Figure 4B) were observed in patients with AML and MDS with germline DDX41 CV compared with those with VUS or without germline DDX41 variants (Figure 4B). Mutated NPM1, rarely seen in patients with AML with CV (1.5%), was the most common concomitant variant, some associated with FLT3-ITD, in AML with VUS (37%; P < .0001; Figures 3 and 4C-D). In addition, t(8;21), inv(16), and biallelic CEBPA mutations were identified in 4 patients with AML with VUS, whereas none were seen in cases with CV (Figures 3 and 4E; P < .0001). Furthermore, mutations involved in tyrosine kinase and RAS/MAPK pathways were significantly more frequent in patients with AML/MDS with VUS than those with CV (Figures 3 and 4C-E; P < .0001). Germline CV-related AML/MDS cases shared similar mutational profiles; however, a higher somatic mutation burden was observed in patients with AML (Figure 4B, red bars; 1.6 in AML vs 0.7 in MDS; P = .0016). Mutations in other splicing factors, although previously reported to be mutually exclusive to DDX41 variants,36 were seen in patients with CV and enriched in those with VUS (Figures 3 and 4E). TP53 mutations were infrequent, seen in 7% of AML/MDS with CV, 12.5% with VUS, and 9% with WT DDX41 (Figure 4D-E; P > .05). In patients with AML/MDS with CV with cytogenetic results, 80% (53 of 66) were associated with a normal karyotype, and 20% (13 of 66) had an abnormal karyotype (9 low-risk and 4 complex karyotypes; Figures 3 and 5A; Table 1).
Genetic characteristics of patients with AML/MDS with germline variants in DDX41. (A) The occurrence of somatic DDX41 variants in patients with AML is closely linked to the presence of germline DDX41 CV (79%; 52 of 66), in comparison with patients with VUS (5.6%, 1 of 18, P < .0001) or patients not carrying germline DDX41 variants (DDX41−, 0.2%; 3 of 1365, P < .0001). A similar trend is seen in patients with MDS (79% in CV, 0% in VUS, and 0.1% in DDX41−, P < .0001). (B) A lower somatic mutation burden, calculated by the number of total concomitant somatic variants (excluding somatic DDX41 variants) per case, is seen in patients with AML with CV (mean ± standard error of the mean: 1.6 ± 0.2) compared with patients with AML with wild-type DDX41 (DDX41−, 3.7 ± 0.07, P < .0001) and those with VUS (2.6 ± 0.4, P = .03). Similarly, in MDS, a lower somatic mutation burden is seen in patients with CV (0.7 ± 0.1) in contrast to those with wild-type DDX41 (DDX41−, 2.7 ± 0.06, P < .0001) or VUS (3.2 ± 0.5, P < .0001). (C-D) Circos plot diagrams illustrate the pairwise co-occurrence of somatic variants and cytogenetic abnormalities in 94 patients with AML/MDS with germline CV (C) and 30 with VUS (D). Genetic variants and cytogenetic events listed in Figure 3 appear in descending order clockwise, starting at 12 o’clock. Each link (ribbon) indicates pairwise co-occurrence of mutational events, and the width of the ribbons indicates the frequency of the co-occurrent events. The occurrence of germline and somatic DDX41 variants is indicated in red and yellow ribbons, respectively. Variants in signaling and RAS/MAPK pathways are labeled in pink; NPM1 and TP53 variants are labeled in black and green, respectively; the remaining variants are labeled in gray. s (yellow) in D, somatic DDX41; SF (purple), RNA splicing factors; TF (orange), transcription factors; S (pink) in panel C, signaling; O (gray), others; # (blue), cohesin; ! (black), NPM1; @ (green), TP53; nl CG, normal cytogenetics; a CG, abnormal cytogenetics. (E) Frequencies of somatic DDX41 and other concurrent variants in AML/MDS patients. For each gene or genetic category, the percentage of mutations is displayed, associated with either germline CV (red bars, gl DDX41+ CV), VUS (blue bars, gl DDX41+ VUS), or wild-type DDX41 (green bars, gl DDX41−). s DDX41, somatic DDX41 variants; epigenetics, genes involving DNA methylation or histone acetylation and deacetylation; signaling transduction, molecules in tyrosine kinase pathway or RAS/MAPK pathways; RCA, recurrent cytogenetic abnormalities in AML. *P < .05; ****P < .0001.
Genetic characteristics of patients with AML/MDS with germline variants in DDX41. (A) The occurrence of somatic DDX41 variants in patients with AML is closely linked to the presence of germline DDX41 CV (79%; 52 of 66), in comparison with patients with VUS (5.6%, 1 of 18, P < .0001) or patients not carrying germline DDX41 variants (DDX41−, 0.2%; 3 of 1365, P < .0001). A similar trend is seen in patients with MDS (79% in CV, 0% in VUS, and 0.1% in DDX41−, P < .0001). (B) A lower somatic mutation burden, calculated by the number of total concomitant somatic variants (excluding somatic DDX41 variants) per case, is seen in patients with AML with CV (mean ± standard error of the mean: 1.6 ± 0.2) compared with patients with AML with wild-type DDX41 (DDX41−, 3.7 ± 0.07, P < .0001) and those with VUS (2.6 ± 0.4, P = .03). Similarly, in MDS, a lower somatic mutation burden is seen in patients with CV (0.7 ± 0.1) in contrast to those with wild-type DDX41 (DDX41−, 2.7 ± 0.06, P < .0001) or VUS (3.2 ± 0.5, P < .0001). (C-D) Circos plot diagrams illustrate the pairwise co-occurrence of somatic variants and cytogenetic abnormalities in 94 patients with AML/MDS with germline CV (C) and 30 with VUS (D). Genetic variants and cytogenetic events listed in Figure 3 appear in descending order clockwise, starting at 12 o’clock. Each link (ribbon) indicates pairwise co-occurrence of mutational events, and the width of the ribbons indicates the frequency of the co-occurrent events. The occurrence of germline and somatic DDX41 variants is indicated in red and yellow ribbons, respectively. Variants in signaling and RAS/MAPK pathways are labeled in pink; NPM1 and TP53 variants are labeled in black and green, respectively; the remaining variants are labeled in gray. s (yellow) in D, somatic DDX41; SF (purple), RNA splicing factors; TF (orange), transcription factors; S (pink) in panel C, signaling; O (gray), others; # (blue), cohesin; ! (black), NPM1; @ (green), TP53; nl CG, normal cytogenetics; a CG, abnormal cytogenetics. (E) Frequencies of somatic DDX41 and other concurrent variants in AML/MDS patients. For each gene or genetic category, the percentage of mutations is displayed, associated with either germline CV (red bars, gl DDX41+ CV), VUS (blue bars, gl DDX41+ VUS), or wild-type DDX41 (green bars, gl DDX41−). s DDX41, somatic DDX41 variants; epigenetics, genes involving DNA methylation or histone acetylation and deacetylation; signaling transduction, molecules in tyrosine kinase pathway or RAS/MAPK pathways; RCA, recurrent cytogenetic abnormalities in AML. *P < .05; ****P < .0001.
Common clinical, hematologic, pathologic, and genetic features and superior OS in AML/MDS patients with germline DDX41 CV. (A) Both patients with AML and MDS with germline DDX41 CV present a similarly indolent and chronic course of cytopenia years before the diagnosis of an overt myeloid neoplasm. Furthermore, the bone marrow examination shows predominantly normal to hypocellular marrow in AML (86%) and MDS (75%, P > .05), and a borderline increase in blasts is seen in patients with AML (31% in AML vs 8% in MDS, P < .0001). Most patients with AML (79%) and MDS (85%, P > .05) carry normal karyotypes with similar germline DDX41 variant subtypes and somatic mutation profiles. (B) The median OS of 57 patients with AML with CV (red line, CV AML, not reached) is significantly longer than that of 13 patients with AML with VUS (blue line, VUS AML, 613 days, P = .02) or 158 patients with DDX41 wild-type AML (dark green line, DDX41− or WT AML, 630 days, P < .0001) in the current study and 1040 patients documented in cBioPortal (lavender line, DDX41− or WT AML R 433 days, P < .0001). Similarly, the median OS of 24 patients with MDS with CV (orange line, CV MDS, not reached) is significantly longer than that of 7 patients with MDS with DDX41 VUS (green line, VUS MDS, 468 days, P = .003) or 87 patients with DDX41 WT MDS (purple line, DDX41− or WT MDS 1425 days, P = .03) in this study and 3128 patients reported recently (navy blue line, DDX41− or WT MDS R, 1116 days, P = .003).27 (C-D) Statistical characteristics of the median OS in each genotype and disease group (C) and P values in pairwise comparisons (D) are listed in the tables. (E) The results of univariate analysis for different factors predicting OS in patients with AML/MDS with DDX41 CV show that the superior OS is not impacted by patient’s age, duration or severity of cytopenia, blast count, presence of abnormal cytogenetics, somatic DDX41 or other concomitant variants, somatic mutation burden, or different types of germline DDX41 CV. Each circle represents the mean HR calculated by Cox proportional hazards regression, and the horizontal lines represent the 95% confidence interval (CI) for the subgroup’s HR. Right of the dashed vertical line (HR = 1), unfavorable OS; left of the dashed line, favorable OS. WBC, white blood cells; Hgb, hemoglobin; PLT, platelet count; BM, bone marrow; gl DDX41, germline DDX41 CV, s DDX41, somatic DDX41 variants. *P < .05; **P < .01; ***P < .001; ****P < .0001; NS, not significant, P > .05.
Common clinical, hematologic, pathologic, and genetic features and superior OS in AML/MDS patients with germline DDX41 CV. (A) Both patients with AML and MDS with germline DDX41 CV present a similarly indolent and chronic course of cytopenia years before the diagnosis of an overt myeloid neoplasm. Furthermore, the bone marrow examination shows predominantly normal to hypocellular marrow in AML (86%) and MDS (75%, P > .05), and a borderline increase in blasts is seen in patients with AML (31% in AML vs 8% in MDS, P < .0001). Most patients with AML (79%) and MDS (85%, P > .05) carry normal karyotypes with similar germline DDX41 variant subtypes and somatic mutation profiles. (B) The median OS of 57 patients with AML with CV (red line, CV AML, not reached) is significantly longer than that of 13 patients with AML with VUS (blue line, VUS AML, 613 days, P = .02) or 158 patients with DDX41 wild-type AML (dark green line, DDX41− or WT AML, 630 days, P < .0001) in the current study and 1040 patients documented in cBioPortal (lavender line, DDX41− or WT AML R 433 days, P < .0001). Similarly, the median OS of 24 patients with MDS with CV (orange line, CV MDS, not reached) is significantly longer than that of 7 patients with MDS with DDX41 VUS (green line, VUS MDS, 468 days, P = .003) or 87 patients with DDX41 WT MDS (purple line, DDX41− or WT MDS 1425 days, P = .03) in this study and 3128 patients reported recently (navy blue line, DDX41− or WT MDS R, 1116 days, P = .003).27 (C-D) Statistical characteristics of the median OS in each genotype and disease group (C) and P values in pairwise comparisons (D) are listed in the tables. (E) The results of univariate analysis for different factors predicting OS in patients with AML/MDS with DDX41 CV show that the superior OS is not impacted by patient’s age, duration or severity of cytopenia, blast count, presence of abnormal cytogenetics, somatic DDX41 or other concomitant variants, somatic mutation burden, or different types of germline DDX41 CV. Each circle represents the mean HR calculated by Cox proportional hazards regression, and the horizontal lines represent the 95% confidence interval (CI) for the subgroup’s HR. Right of the dashed vertical line (HR = 1), unfavorable OS; left of the dashed line, favorable OS. WBC, white blood cells; Hgb, hemoglobin; PLT, platelet count; BM, bone marrow; gl DDX41, germline DDX41 CV, s DDX41, somatic DDX41 variants. *P < .05; **P < .01; ***P < .001; ****P < .0001; NS, not significant, P > .05.
Similar to that reported previously,8 indolent courses of cytopenia (Figure 5A) were seen prior to an overt MN, and there was a borderline increase in blasts in patients with AML with DDX41 CV (31% in AML vs 8% in MDS; P < .0001). Similar genetic features and DDX41 CV types were seen in both normo/hypocellular AML/MDS with CV (Figure 5A). Fifty-seven patients with AML with CV had a favorable OS (Figure 5B-D; supplemental Figure 2A; median OS not reached) compared with 13 with VUS (613 days; P = .02) or 158 WT patients; a similar trend was seen in patients with MDS (Figure 5B-D). This superior OS, similar to a previous study7 (supplemental Figure 2A), appeared independent of blasts (Figure 5B-E; P = .30), patient age (Figure 5E; supplemental Figure 2B; P = .69), sex (P = .61), somatic variant burden, presence of somatic DDX41 variants (Figure 5E; supplemental Figure 2C; P = .95), and other concomitant variants (P = .50) including TP53 (supplemental Figure 2D; P = .39), regardless of cytogenetic abnormalities (P = .91) or type of germline DDX41 CV (Figure 5E).
Among the 50 patients with AML/MDS with CV and available FH, only 18% (9 of 50) had FH of MN, and 2% (1 of 50) had FH of lymphoma, whereas nonhematologic tumors were rather common (32%, 16 of 50; Table 2). In contrast, none of the patients with VUS had FH of myeloid or lymphoid neoplasms (Table 2). In this study, 2 patients with MN with CV had concomitant lymphoid or solid tumors, similar to previous reports (Table 1, patients 60 and 76).7,19
Germline DDX41 CV predisposing to MPN and lymphoma
We further focused on 15 patients (4 CV and 11 VUS) with MPN. Male predominance was not observed here in contrast to patients with AML/MDS (Table 3). A similar tendency for more frequent somatic DDX41 mutation (Figure 6A;P < .0001) and lower somatic mutation burden (Figure 6B;P = .05) was seen in patients with CV compared with those with WT DDX41. Interestingly, JAK2 V617F and CALR mutations, absent in all patients with CV, were identified in 72% of patients with VUS (Figures 3 and 6C), most (7 of 8) being a leading clone (Table 1; VAFs at 45%, 85%, 44%, 26%, 18%, 47%, and 26%, respectively), whose frequency was almost identical to that in patients with DDX41 WT (Figure 6C-E). Karyotypes in patients with MPN with CV were normal, and TP53 mutations were not identified (Figures 3 and 6D).
MPN and lymphoma predisposed by germline DDX41 CV. (A) The occurrence of somatic DDX41 variants in MPN patients is more frequent in patients with germline CV (25%) compared with patients with VUS (9%; 1/11, P = .4) or patients not carrying germline DDX41 variants (DDX41−, 0%, P < .0001). (B) There appears to be a lower concomitant somatic mutation burden in patients with CV (mean ± standard error of the mean: 0.5 ± 0.5), compared with those with WT DDX41 (DDX41−, 2.6 ± 0.1, P = .05) and VUS (1.5 ± 0.4, P > .05). (C) No mutations in JAK2 or CALR were seen in MPN with CV, whereas these canonical variants are seen in 73% (8 of 11) of MPN patients with VUS and 82% in the WT cohort (P < .0001). (D-E) Circos plot diagrams illustrate the pairwise co-occurrence of variants and cytogenetic events in MPN patients with germline CV (D) and VUS (E). Genetic variants and cytogenetic events listed in Figure 2 appear in descending order clockwise starting at 12 o’clock. Each link (ribbon) indicates the pairwise co-occurrence of mutational events, and the width of the ribbons indicates the frequency of the co-occurrent events. TF (orange), transcription factors; nl CG (green), normal cytogenetics; s (yellow), somatic DDX41; E (light green), epigenetic modulators; SF (purple), RNA splicing factors; C (pink), CALR. (F) HM predisposed by germline DDX41 CV. AML (55%) and MDS (24%) are the most common entities predisposed by DDX41 CV, followed by cytopenia (16%), MPN (3%), and lymphoma (3%). ****P < .0001.
MPN and lymphoma predisposed by germline DDX41 CV. (A) The occurrence of somatic DDX41 variants in MPN patients is more frequent in patients with germline CV (25%) compared with patients with VUS (9%; 1/11, P = .4) or patients not carrying germline DDX41 variants (DDX41−, 0%, P < .0001). (B) There appears to be a lower concomitant somatic mutation burden in patients with CV (mean ± standard error of the mean: 0.5 ± 0.5), compared with those with WT DDX41 (DDX41−, 2.6 ± 0.1, P = .05) and VUS (1.5 ± 0.4, P > .05). (C) No mutations in JAK2 or CALR were seen in MPN with CV, whereas these canonical variants are seen in 73% (8 of 11) of MPN patients with VUS and 82% in the WT cohort (P < .0001). (D-E) Circos plot diagrams illustrate the pairwise co-occurrence of variants and cytogenetic events in MPN patients with germline CV (D) and VUS (E). Genetic variants and cytogenetic events listed in Figure 2 appear in descending order clockwise starting at 12 o’clock. Each link (ribbon) indicates the pairwise co-occurrence of mutational events, and the width of the ribbons indicates the frequency of the co-occurrent events. TF (orange), transcription factors; nl CG (green), normal cytogenetics; s (yellow), somatic DDX41; E (light green), epigenetic modulators; SF (purple), RNA splicing factors; C (pink), CALR. (F) HM predisposed by germline DDX41 CV. AML (55%) and MDS (24%) are the most common entities predisposed by DDX41 CV, followed by cytopenia (16%), MPN (3%), and lymphoma (3%). ****P < .0001.
Beyond MN, 3 unrelated patients with B-cell lymphomas were linked by an identical presumably germline DDX41 variant, p.R164W (Tables 1 and 2, patients 172-174). Two patients (patients 173 and 174) carrying this variant developed earlier-onset lymphoma at age 51 and 52 years, respectively. Both patients had affected family members diagnosed with either myelofibrosis or follicular lymphoma (Table 2), adding further support to this likely CV predisposing to lymphoma. A third patient with this variant (pt 172) developed MYD88-negative lymphoplasmacytic lymphoma (LPL) and pancytopenia at age 77 years without significant FH. A somatic SF3B1 variant was also identified in this patient, which might potentially contribute to the patient’s reported pancytopenia. Importantly, p.R164W was previously reported in a family with LPD, in which all 5 affected individuals developed lymphoma (4) and multiple myeloma (1), whereas all 3 unaffected individuals of similar age did not.19
The prevalence of disease entities in patients with HM with germline DDX41 CV can be summarized as follows: AML/MDS (Figure 6F, 79%), as a distinct clinical entity, is the most common disease, followed by cytopenia (15%), MPN (3%), and lymphoma (3%). Per the data collected at ARUP Laboratories, approximately 3.0% (41 of 1406, 29 CV and 12 VUS) of patients with AML, 1.4% (16 of 1125, 9 CV and 7 VUS) of patients with MDS, and 2.0% (10 of 489, 3 CV and 7 VUS) of patients with MPN carried a presumed germline DDX41 variant. The prevalence of DDX41-related lymphoma remains uncertain, as this disease is not fully acknowledged, and NGS testing for patients with LPD is not yet a standard of care.
Asymptomatic carriers with germline CV
Six asymptomatic individuals with normal CBC and germline DDX41 CV who were related to patients with HM in this study underwent tumor surveillance (Tables 1 and 2, patients 196-201); their median age (51 years; range, 28-69 years) was significantly lower compared with patients with overt diseases (supplemental Figure 3A; P < .0001). No somatic DDX41 or other mutations were identified by NGS testing (supplemental Figure 3B), and all 5 patients who underwent cytogenetic testing showed a normal karyotype (Table 1). Furthermore, 4 patients with HSCT for previously diagnosed AML were found to have donor-derived DDX41 variants (2 CV and 2 VUS) during surveillance (Table 1, patients 202-205). All 4 had unremarkable CBC and complete engraftment confirmed by 100% donor chimerism with a median follow-up of 30 months in surveillance (Table 2) without biopsy proving recurrent/residual AML.
Discussion
In this study, we analyzed the genetic, epidemiologic and hematologic features, and clinical outcomes of 116 patients with HM with germline DDX41 CV and 60 with VUS identified by NGS. Using the proposed DDX41-specific variant classification framework, we identified a phenotype encompassing primarily AML/MDS and rarely MPN and lymphoma associated with germline CV. A complete germline CV landscape is critical to direct appropriate clinical management, preventive care, and family screening.
In this largest cohort to date of 176 patients with HM with DDX41 germline variants, we proposed that the acquisition of a pathogenic somatic DDX41 variant is a compelling criterion for causality in germline variant interpretation. The marked segregation in genetic profiles, epidemiologic features, and clinical behavior separating patients with AML/MDS with germline CV from those with VUS provided validation for this modified variant classification strategy.7,20,29 Patients with HM with germline DDX41 VUS behaved similarly to patients who were WT, in which canonical somatic mutations or recurrent genetic alterations in other genes were common as drivers of tumorigenesis.
Patients with AML and MDS, the most common entities associated with germline CV, present during their late 60s or early 70s with indolent cytopenia years before overt myeloid neoplasia, with a male predominance.7,8,10,29,32,37 Furthermore, both were characterized by frequent somatic DDX41 variants, infrequent other somatic mutations, largely normal karyotype, normo/hypocellular marrow, and a favorable OS.7,8,10,29,32,37 This superior OS was independent of blast counts or additional genetic abnormalities, regardless of the patients’ age, sex, or specific germline CV. This unique disease was also characterized by near mutual exclusion of recurrent cytogenetic abnormalities and canonical mutations in FLT3, NPM1, and CEPBA in sporadic AML. Thus, AML/MDS caused by DDX41 CV appear to be a spectrum of the same disease, unlike sporadic de novo AML and MDS, caused by completely different pathogenic mechanisms. Beyond AML/MDS, unrelated patients with MPN and B-cell lymphoma were linked to germline DDX41 CV. Further studies identifying more germline CV are warranted to provide more insights into disease prevalence, characteristic pathologic features, the underlying mechanisms, and genotype-phenotype correlation in MPN and lymphoma19 to further refine clinical management.
Unique ethnic differences were highlighted by different recurrent CVs seen nearly exclusively in White or Asian patients and more common missense CV in Asian patients.7,10,11,28-34 There is also an urgent need for gene-specific classification guidelines by expert panels, without which a large number of missense CV are classified as VUS, and for international collaboration to fully characterize the CV landscape. Further studies are needed to address the currently uncertain significance of rare missense germline variants, especially those accompanied by rare noncanonical somatic DDX41 variants (patient 150, Table 1). Germline confirmation of DDX41 variants was limited in this study, partially because of the setting of a national reference laboratory. Creating gene-specific diagnostic and management guidelines could raise awareness of this disease and provide necessary guidance for germline confirmation.
This is the first study to expand the link of germline DDX41 CV to MPN and lymphoma beyond AML/MDS by outlining the CV landscape in unselected and unrelated patients. AML/MDS caused by germline CV is 1 distinct clinical entity with relative indolent course and favorable outcomes, as shown by this and other studies. Our study presents the first and most complete characterization of germline CV profiles to date and highlights the need for guidelines addressing variant classification, patient management, carrier surveillance, and stem cell donor selection.
Authorship
Contribution: P.L. designed the study and drafted the manuscript; T.W., S.B., M.W., W.X., W.C., D.P., H.-Y.W., L.L., and C.A.K. collected patients’ clinical and family history and cytogenetic and molecular data; J.V. and T.K. examined patients and performed DDX41 germline testing; P.L., T.W., W.X., W.C., D.P., H.-Y.W., S.S.M., L.L., C.A.K., and S.B. interpreted and classified all variants by NGS testing; P.L. and M.W. examined the bone marrow biopsies for healthy individuals in cancer surveillance; and all authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peng Li, University of Utah, 500 Chipeta Way, Salt Lake City, UT 84108; e-mail, pengl.li@aruplab.com.
For original and additional data, please contact Peng Li via peng.li@aruplab.com or peng.li@hsc.utah.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

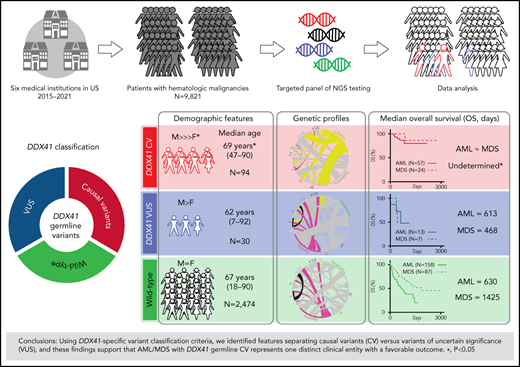
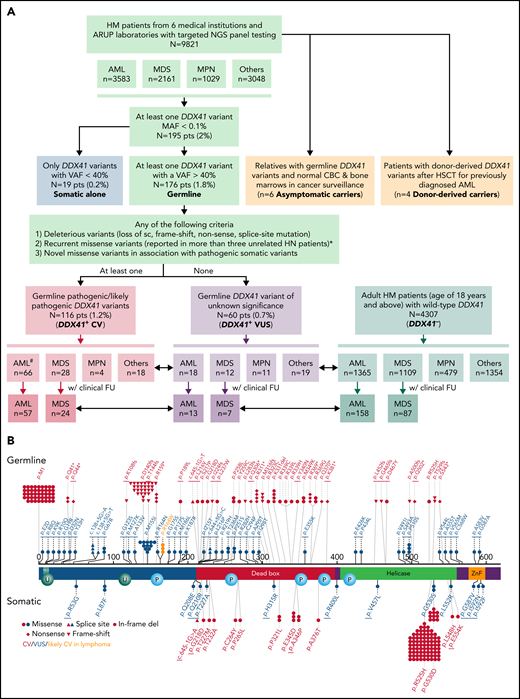
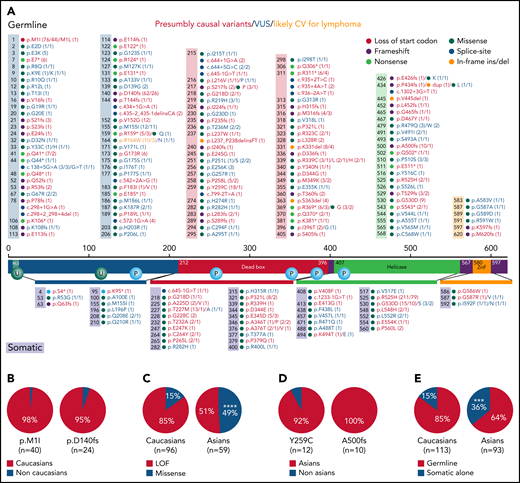
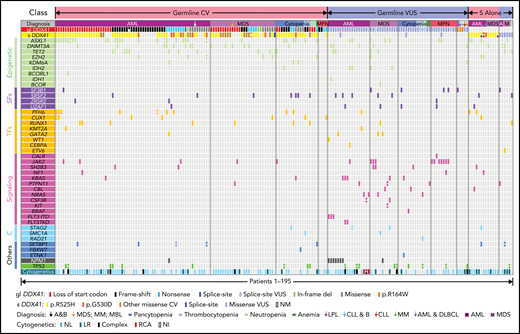
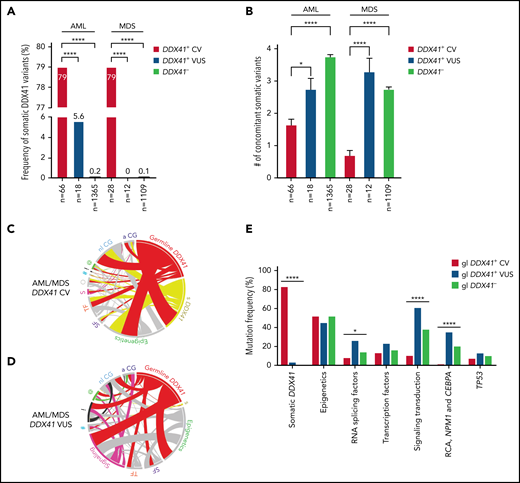
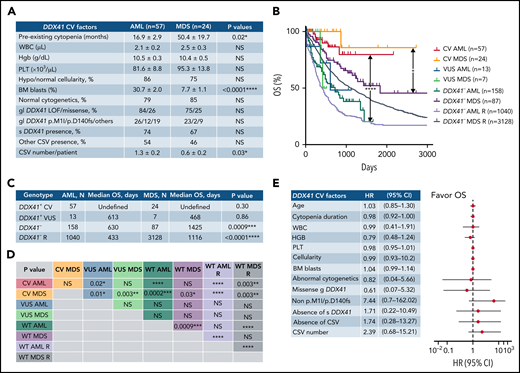

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal