Key Points
Single cell multiomics in PLZF-RARA leukemic cells resolves the retinoic acid resistance network.
Targeting pan-EZH2 activities (canonical/noncanonical) eradicates leukemia relapse-initiating cells.
Abstract
Cancer cell heterogeneity is a major driver of therapy resistance. To characterize resistant cells and their vulnerabilities, we studied the PLZF-RARA variant of acute promyelocytic leukemia, resistant to retinoic acid (RA), using single-cell multiomics. We uncovered transcriptional and chromatin heterogeneity in leukemia cells. We identified a subset of cells resistant to RA with proliferation, DNA replication, and repair signatures that depend on a fine-tuned E2F transcriptional network targeting the epigenetic regulator enhancer of zeste homolog 2 (EZH2). Epigenomic and functional analyses validated the driver role of EZH2 in RA resistance. Targeting pan-EZH2 activities (canonical/noncanonical) was necessary to eliminate leukemia relapse-initiating cells, which underlies a dependency of resistant cells on an EZH2 noncanonical activity and the necessity to degrade EZH2 to overcome resistance. Our study provides critical insights into the mechanisms of RA resistance that allow us to eliminate treatment-resistant leukemia cells by targeting EZH2, thus highlighting a potential targeted therapy approach. Beyond RA resistance and acute promyelocytic leukemia context, our study also demonstrates the power of single-cell multiomics to identify, characterize, and clear therapy-resistant cells.
Introduction
Acute promyelocytic leukemia (APL) is a class of acute myeloid leukemia (AML) that accounts for 10% to 15% of all cases and is characterized by recurrent chromosomal translocations involving invariably the gene encoding the retinoic acid receptor alpha (RARA) (17q21) with several fusion partners, such as PML (15q22) or PLZF (11q23) (for a review, see Dos Santos et al1). The resulting X-RARA fusion proteins were among the first transcription factors (TFs) to be identified as drivers of cancer.2 They behave as RARA signaling repressors due to their ability to oligomerize and to recruit epigenetic repressors at cis-regulatory DNA regions of RARA target genes and initiate oncogenic gene expression signatures.3 APL patients with PML-RARA fusion are exquisitely sensitive to retinoic acid (RA) treatment, a sensitivity that has made APL one of the most successful examples of targeted therapy to mutational events.4,5 However, APL variant cases, such as the t(11;17), which represent 1% of APL cases, still pose clinical challenges,6 as they respond poorly to RA and remain clinically resistant.7
Contrary to PML-RARA APL, pharmacological doses of RA, although inducing partial differentiation of the PLZF-RARA blasts, do not clear the leukemia-initiating cell of this APL variant.8,9 This has highlighted the uncoupling between blast differentiation and tumor eradication. The PLZF moiety of the fusion is thought to play a determinant role in RA resistance. PLZF is a potent transcriptional repressor that can interact on its own with epigenetic complexes.3,10 Given that PLZF binding sites with co-repressors are conserved in the PLZF-RARA fusion, one hypothesis is that inappropriate recruitment of functional epigenetic repressors, such as the polycomb repressor complex 1, would trigger epigenetic imbalance at very specific genomic loci in an RA insensitive manner.11 This is consistent with the observed degradation of PLZF-RARA under RA treatment,12 suggesting a persistent mechanism even in the absence of detectable fusion involving chromatin modifications. However, the molecular basis for the resistance of PLZF-RARA-expressing cells and why some cells retain their self-renewal capacity while others do not after RA treatment remain unknown.
To identify actionable vulnerabilities that will prevent resistance and facilitate treatment response, we studied PLZF-RARA APL cellular heterogeneity by integrating single-cell RNA sequencing (scRNA-seq) and scATAC-seq data obtained from an RA-treated PLZF-RARA mouse model. Establishing cellular clusters and arranging them in hierarchies helped us to identify a subpopulation of transformed promyelocytes that were insensitive to RA-induced differentiation and characterized by a high expression of enhancer of zeste homolog 2 (EZH2), the catalytic subunit of polycomb repressive complex 2 (PRC2). Because EZH2 is an interesting cancer target,13 we further explored EZH2 function in APL development and RA treatment response. We discovered a dual role of EZH2 in APL onset and RA response, suggesting the need to target the nonhistone methyltransferase activity of EZH2 for leukemia clearance.
Methods
A complete description of all methods is presented in the supplemental Methods, available on the Blood website.
Mouse models and purification of leukemic cells
The APL mouse model (named here PLZF-RARA model) was previously described by Pandolfi (He et al14). For transplantation, Cd45.2 APL cells were transplanted in sublethally irradiated (1.5 Gy) NOD scid gamma (NSG) mice. All-trans RA and GSK126 (GSK) (MedChemExpress) were intraperitoneally administered; RA: 0.8 to 1 mg per mice for 3 or 7 consecutive days; GSK126 (GSK): 1.25 mg per mice for 10 consecutive days. Leukemic myeloid progenitors (promyelocytes: Cd45.2+, C-Kit+, Gr1+; ProReP: Cd45.2+, C-Kit+, Gr1+, Cd48+, Cd11b−; NeuRA: Cd45.2+, C-Kit+, Gr1+, Cd48−, Cd11b+) were purified using the FACSAria III cell sorter (Beckman Dickinson). APL models were bred and maintained in the CRCM mouse facility (Marseille, France) in accordance with institutional guidelines and approved by the French authority (authorization number: 23893).
Single-cell analyses
Single-cell analyses were performed on the 10X Genomics platform.
For scRNA-seq, promyelocytes were processed with the Chromium Next GEM Single Cell 3′ GEM, Library & Gel bead Kit v2 according to the manufacturer’s instructions, at a target capture rate of 6000 individual cells per sample.
For scATAC-seq, promyelocyte nuclei were purified according 10X Genomics instructions and processed using the Chromium Next GEM Single Cell ATAC Reagent Kits v1.1 following the manufacturer’s protocols at a target capture rate of 10 000 nuclei per sample.
scRNA-seq and scATAC-seq were sequenced using a 75-nt single-end and a 150-nt paired-end protocol, respectively. Data were processed using the 10X software Cell Ranger (v4.0) and the 10X software Cell Ranger ATAC (v1.2.0). The mm10 genome was used as reference.
Results
Single-cell transcriptomic analysis identifies a subset of RA-resistant leukemic cells
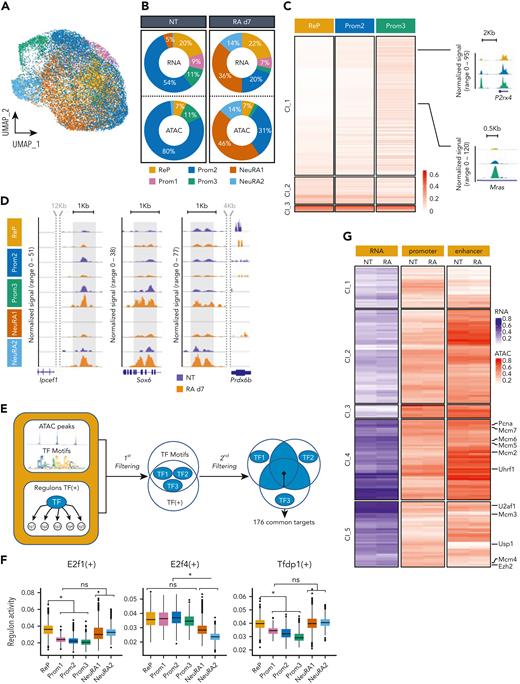
To decipher mechanisms linked to APL t(11;17) RA resistance and identify features of RA-resistant leukemic cells, we performed scRNA-seq on promyelocytes (cKit+; Gr1+) obtained from bone marrow (BM) of PLZF-RARA transgenic (TG) transplanted mice, treated or not treated with RA for 7 days. The impact of RA treatment was measured by the expression of cell surface markers and the morphology of fluorescence-activated cell sorter (FACS)–sorted promyelocytes (supplemental Figure 1A-C). We identified 6 clusters that were annotated by enrichment analysis (Figure 1A-B and supplemental Figure 1D). Five of the clusters had neutrophil associated signatures (Prom1, Prom2, Prom3, NeuRA1, NeuRA2) consistent with the promyelocytic stage of induced leukemia, and 1 cluster was characterized by genes involved in DNA replication associated with DNA repair and proliferation processes (ReP) (Figure 1B, supplemental Figure 1E, and supplemental Table 1A-C). Prom1, Prom2, and Prom3 were enriched with untreated (NT) cells suggesting their fading upon RA treatment (Figure 1C), and NeuRA1 and NeuRA2 appeared almost exclusively in RA-treated cells. The terminal myeloid differentiation status of these 2 groups was supported by their gene signature (supplemental Figure 1F). By contrast the ReP cluster was characterized by the expression of genes involved in homologous recombination and DNA replication such as Rad51, Pcna, and Mcm3 (supplemental Figure 1G) and was equally composed of NT and RA-treated cells (Figure 1C), suggesting that RA treatment did not impact on the cell identity of these promyelocytes.
Single-cell transcriptome analysis identifies a subset of RA-resistant PLZF-RARA leukemic cells with a specific signature. (A-B) Uniform Manifold Approximation and Projection (UMAP) visualization of the scRNA-seq data set colored by (A) condition (NT and RA day 7 [d7]) and by (B) cluster (total integrated cells: 11 900). Six clusters were identified: ReP, DNA repair/Replication/Proliferation; Prom1-3, Promyelocyte (Neutrophil) 1-3; NeuRA1-2, Neutrophil RA1-2. (C) (Bottom) NT and RA cell distribution in each cluster. The black line indicates expected NT and RA cell proportions according to the data set size. (Top) Absolute cell number per cluster and per condition. Names of the clusters for which the proportion of NT or RA cells is significantly higher than expected (P < .01) are purple for NT cells and orange for RA cells. (D) Differentiation trajectory. (Top) Schematic representation of the different states (A-E) in each trajectory (trajectory 1: A-C-D, trajectory 2: A-C-E, trajectory 3: A-B). (Bottom) UMAP is colored according to the pseudotime value of each cell. (E) Cluster distribution in each state of the pseudotime. (F) NT and RA cell distribution (bottom: percentage; top: absolute) in each state of the pseudotime. (C and F) Names of the clusters for which the proportion of NT or RA cells was significantly higher than expected (P < .01) are purple for NT cells and orange for RA cells. (G) Violin plots showing PML-RARA response and (H) E2f signature scores in each cluster. (G and H) The signature score represents the global expression of annotated genes for the selected signature. Gray/black arrows pointing down/up: significant lower/higher expression in the cluster against all the others (average score difference > 0.02 and adjusted P < .05). (I) Violin plot showing Usp37 expression in each cluster. Black arrows pointing up: significant higher expression in the cluster against all the others (average log2|FC|> .1 and adjusted P < .05). (J) Gating strategy for isolating ProReP cells (Prom1-3 and ReP) and NeuRA (NeuRA1-2). The donut plot represents the proportion of Prom1-3 and ReP cells into the ProReP population according to the scRNA-seq analysis. (K) PLZF-RARA (black arrow: full length; black star: degraded form) levels in untreated (NT) and treated promyelocytes (RA d7). Actin is used as loading control. (L) Survival rate of mice transplanted with ProReP or NeuRA cells. Each cohort contains at least 5 mice. ∗∗∗P < .001.
Single-cell transcriptome analysis identifies a subset of RA-resistant PLZF-RARA leukemic cells with a specific signature. (A-B) Uniform Manifold Approximation and Projection (UMAP) visualization of the scRNA-seq data set colored by (A) condition (NT and RA day 7 [d7]) and by (B) cluster (total integrated cells: 11 900). Six clusters were identified: ReP, DNA repair/Replication/Proliferation; Prom1-3, Promyelocyte (Neutrophil) 1-3; NeuRA1-2, Neutrophil RA1-2. (C) (Bottom) NT and RA cell distribution in each cluster. The black line indicates expected NT and RA cell proportions according to the data set size. (Top) Absolute cell number per cluster and per condition. Names of the clusters for which the proportion of NT or RA cells is significantly higher than expected (P < .01) are purple for NT cells and orange for RA cells. (D) Differentiation trajectory. (Top) Schematic representation of the different states (A-E) in each trajectory (trajectory 1: A-C-D, trajectory 2: A-C-E, trajectory 3: A-B). (Bottom) UMAP is colored according to the pseudotime value of each cell. (E) Cluster distribution in each state of the pseudotime. (F) NT and RA cell distribution (bottom: percentage; top: absolute) in each state of the pseudotime. (C and F) Names of the clusters for which the proportion of NT or RA cells was significantly higher than expected (P < .01) are purple for NT cells and orange for RA cells. (G) Violin plots showing PML-RARA response and (H) E2f signature scores in each cluster. (G and H) The signature score represents the global expression of annotated genes for the selected signature. Gray/black arrows pointing down/up: significant lower/higher expression in the cluster against all the others (average score difference > 0.02 and adjusted P < .05). (I) Violin plot showing Usp37 expression in each cluster. Black arrows pointing up: significant higher expression in the cluster against all the others (average log2|FC|> .1 and adjusted P < .05). (J) Gating strategy for isolating ProReP cells (Prom1-3 and ReP) and NeuRA (NeuRA1-2). The donut plot represents the proportion of Prom1-3 and ReP cells into the ProReP population according to the scRNA-seq analysis. (K) PLZF-RARA (black arrow: full length; black star: degraded form) levels in untreated (NT) and treated promyelocytes (RA d7). Actin is used as loading control. (L) Survival rate of mice transplanted with ProReP or NeuRA cells. Each cohort contains at least 5 mice. ∗∗∗P < .001.
To determine the potential differentiation journey of the RA-treated transformed promyelocytes, we computationally reconstructed the differentiation trajectory of NT and RA-treated cells (Figure 1D-F). We revealed 3 trajectories split into 5 different states of promyelocytes (segments A, B, C, D, E) and defined the departure of the trajectories at the extremity of state A, which was mostly composed of ReP cells (Figure 1D). Trajectories 1 (states A-C-D) and 2 (states A-C-E) ended toward RA-differentiated cells, because their final states C, D, and E were largely composed of RA-treated cells grouped in NeuRA1 and NeuRA2, respectively (Figure 1E-F). These data confirmed a pronounced differentiating effect of RA on a portion of leukemic cells consistently with a stronger expression of differentiation marker in NeuRA2 (supplemental Figure 1F). Interestingly, the third trajectory (state B), which went far into the pseudotime, was composed mostly of NT cells grouped in Prom2 and Prom3 (Figure 1E-F), suggesting a spontaneous differentiation program in leukemic cells. This analysis showed a pronounced but partial differentiating effect of RA on PLZF-RARA–expressing cells and designated cells in the ReP cluster as the treatment-persistent cells.
To further investigate the RA (un)responsiveness of the identified clusters, we took advantage of available transcriptional signatures reflecting RA sensitivity of PML-RARA vs RA-resistant PLZF-RARA murine APL.12 We found that the NeuRA1 and NeuRA2 clusters highly expressed a computed PML-RARA RA-responsive signature confirming that the 2 clusters were composed of RA-responsive blasts (Figure 1G). By contrast, the ReP cluster highly or specifically expressed the proliferative E2F signature (Figure 1H) and the deubiquitinase Usp37 involved in PLZF-RARA fusion stability15 (Figure 1I). E2F and Usp37 expressions persisted upon RA treatment, likely supporting the malignancy and RA insensitivity of these cells (supplemental Figure 1H).
We attempted to purify the ReP cluster after RA treatment based on the expression of specific surface markers identified in our scRNA-seq data set. Taking into account Cd48 and Itgam (Cd11b), we were limited to isolate Prom1-3 and ReP (ProReP) from NeuRA1 and NeuRA2 (NeuRA) cells (Figure 1J and supplemental Figure 1I). After RA treatment, whereas the PLZF-RARA fusion was detected neither in the bulk nor in NeuRA populations, ProReP cells did maintain PLZF-RARA expression (Figure 1K and supplemental Figure 1J) and kept the potential to develop leukemia in transplanted mice (Figure 1L).
Altogether, these results reveal transcriptional heterogeneity and differentiation states within the PLZF-RARA transformed cells and identify the ReP cluster, which exhibits no differentiation features, high E2F signature, and PLZF-RARA residual expression as the potential driver of RA resistance of PLZF-RARA leukemia.
Single-cell integrative multiomics analysis highlights chromatin genes as responsible for RA resistance in PLZF-RARA–expressing cells
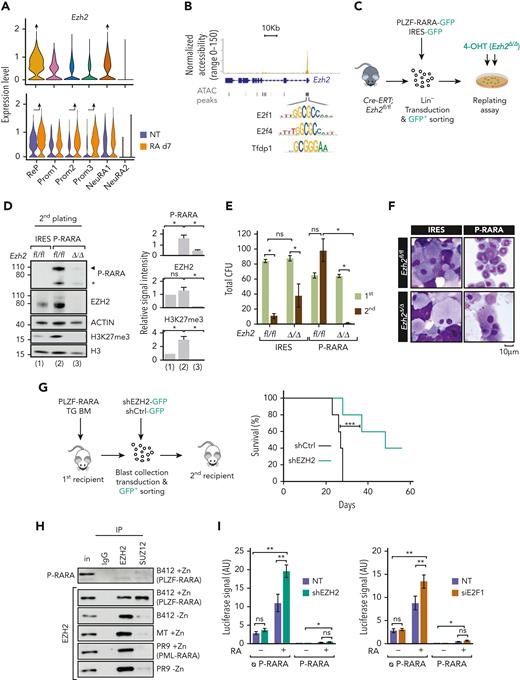
To characterize at the regulatory/epigenomic level the heterogeneity underlying PLZF-RARA transformation and RA response, we generated a chromatin accessibility profile (sc-ATACseq) of untreated and RA-treated cells in our PLZF-RARA TG mouse model. To link transcriptome variations with changes in epigenome, we defined a new low-dimensional shared space by mapping cells from our scATAC-seq data to the 6 scRNA-seq defined clusters16 (Figure 2A-B and supplemental Figure 2A). We showed an overall good concordance between the 2 levels of information (scATAC-seq and scRNA-seq), especially high on RA-treated promyelocytes (Figure 2A-B), confirming the known effect of RA on differentiation through remodeling of the chromatin landscape.19 Interestingly, untreated ReP cells differed from other cells in their chromatin state, including more closed sites. This suggests that in addition to transcriptional reprogramming, epigenetic alterations may predispose the cells to resist to RA (Figure 2C). Besides, consistent with our scRNA-seq data, RA treatment had a low impact on the ratio of scATAC-seq cells in the ReP cluster (7% in both conditions) (Figure 2B) and on chromatin opening (Figure 2D), confirming the little impact of RA on the ReP cluster at both RNA and chromatin levels.
Single-cell integrative multiomics analysis highlights chromatin genes as responsible for RA resistance in PLZF-RARA–expressing cells. (A) UMAP visualization of integrated scRNA-seq (11 900 cells) and scATAC-seq (7367 cells) data set colored by cluster. (B) Donut plots showing the distribution of scRNA- and scATAC-seq cells in each cluster for NT and RA d7 conditions. (C) (Left) Heat map showing the ATAC signal in the NT condition on the differentially accessible peaks between ReP and Prom2-3 clusters. Hierarchical clustering is done according to the ReP data set. (Right) Coverage plot of ATAC signal at selected genes. (D) Coverage plot of ATAC signal per condition in each cluster associated with Ipcef1, Sox6, and Prdx6b genes. (E) Computational scheme to identify key regulon targets in the ReP cluster. The first filtering consists to select TFs with high activity in the ReP cluster by considering the accessibility of their DNA motifs in this cluster. TF motif accessibility scores are calculated with Signac,17 and TF motif markers are identified for each cluster (supplemental Table 2A). Selected ReP TFs are cross-referenced with master transcriptional regulators identified from scRNA-seq data using SCENIC18 (supplemental Table 2B). After this filtering, 3 TFs remain. Target genes shared at least by 2 TFs are taken into account for further filtering (second filtering). Target genes considered are: (i) found in all pySCENIC run, (ii) linked with positive regulons, and (iii) filtered based on the sum of normalized importance (>0.35). One hundred seventy-six genes are conserved (supplemental Table 3). (F) Box plots showing E2f1, E2f4, and Tfdp1 (TFs obtained after the first filtering) regulon activity in each cluster. ∗P < .05. (G) Heat map showing the mRNA expression (left), the promoter accessibility (middle, ±3 kb from the TSS), and the enhancer accessibility (right, ±50 kb from the TSS minus the ±3-kb promoter region) of the 176 target genes in the ReP cluster (obtained after the second filtering). Results are expressed as normalized log (mean gene activity). Hierarchical clustering is done according to the NT data set.
Single-cell integrative multiomics analysis highlights chromatin genes as responsible for RA resistance in PLZF-RARA–expressing cells. (A) UMAP visualization of integrated scRNA-seq (11 900 cells) and scATAC-seq (7367 cells) data set colored by cluster. (B) Donut plots showing the distribution of scRNA- and scATAC-seq cells in each cluster for NT and RA d7 conditions. (C) (Left) Heat map showing the ATAC signal in the NT condition on the differentially accessible peaks between ReP and Prom2-3 clusters. Hierarchical clustering is done according to the ReP data set. (Right) Coverage plot of ATAC signal at selected genes. (D) Coverage plot of ATAC signal per condition in each cluster associated with Ipcef1, Sox6, and Prdx6b genes. (E) Computational scheme to identify key regulon targets in the ReP cluster. The first filtering consists to select TFs with high activity in the ReP cluster by considering the accessibility of their DNA motifs in this cluster. TF motif accessibility scores are calculated with Signac,17 and TF motif markers are identified for each cluster (supplemental Table 2A). Selected ReP TFs are cross-referenced with master transcriptional regulators identified from scRNA-seq data using SCENIC18 (supplemental Table 2B). After this filtering, 3 TFs remain. Target genes shared at least by 2 TFs are taken into account for further filtering (second filtering). Target genes considered are: (i) found in all pySCENIC run, (ii) linked with positive regulons, and (iii) filtered based on the sum of normalized importance (>0.35). One hundred seventy-six genes are conserved (supplemental Table 3). (F) Box plots showing E2f1, E2f4, and Tfdp1 (TFs obtained after the first filtering) regulon activity in each cluster. ∗P < .05. (G) Heat map showing the mRNA expression (left), the promoter accessibility (middle, ±3 kb from the TSS), and the enhancer accessibility (right, ±50 kb from the TSS minus the ±3-kb promoter region) of the 176 target genes in the ReP cluster (obtained after the second filtering). Results are expressed as normalized log (mean gene activity). Hierarchical clustering is done according to the NT data set.
To decipher specific TF activity that might be associated with RA resistance, we inferred information from ATAC-seq and RNA-seq data in the ReP cells (Figure 2E). Doing so and in line with our previous annotation, we identified 3 TFs, linked to E2F (E2F1, E2F4, TFDP1) with high transcriptional activity (Figure 2F and supplemental Figure 2B-C). We performed clustering on their 176 shared targets and highlighted 5 clusters of genes (Figure 2G and supplemental Table 3) with a stronger average ATAC signal at enhancer regions than at promoters. Two clusters of genes (Cl_4 and Cl_5) were highly expressed in comparison with the others and not impacted by RA treatment. These genes were not highly expressed but modified by RA treatment in the RA-responsive NeuRA2 cells (supplemental Figure 2D and supplemental Table 3), suggesting a role of these genes in RA resistance. In line with our previous analysis, these highly expressed genes (Pcna, Mcm2-7, and Ezh2) were related to DNA repair, replication, and chromatin.
The results show that RA resistance depends on E2F transcriptional activity and relies on differences in chromatin accessibility, mainly at enhancer regions.
EZH2 is necessary for PLZF-RARA transformation activity
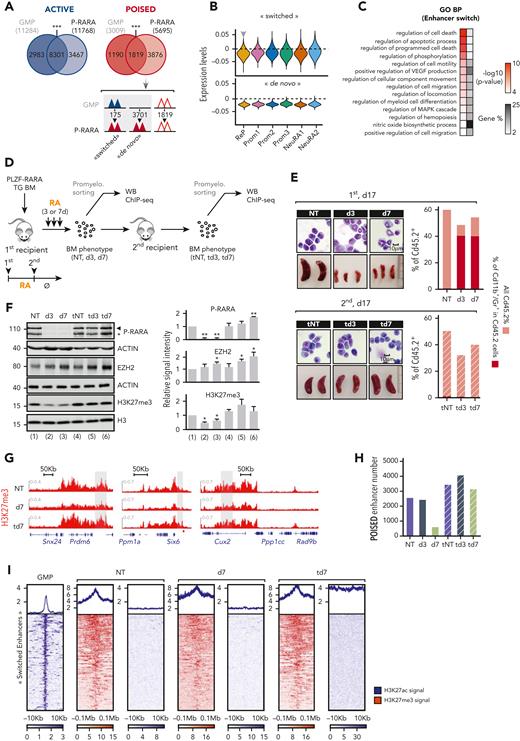
EZH2 is a component of PRC2 whose canonical enzymatic activity induces the repressive histone mark H3K27me3, frequently involved in AML.20 To ascertain the functional relevance of EZH2 in PLZF-RARA APL cells, we first confirmed its high expression in the ReP cluster and after RA treatment (Figure 3A), and the presence of E2F1, E2F4, and TFDP1 binding motifs in its promoter (Figure 3B). Next, we analyzed the clonogenic activity of PLZF-RARA in the absence of EZH2 ex vivo by transducing BM lineage-negative cells of a conditional KO Ezh2 mouse model21 with a PLZF-RARA-IRES-GFP retroviral construct and performing replating assay (Figure 3C). Consistent with our single-cell data, PLZF-RARA transduction in lineage-negative cells induced, as soon as the second plating, an overall increase in both EZH2 and H3K27me3 levels (Figure 3D) while sustaining replatings (supplemental Figure 3A). Ezh2 deletion (Δ/Δ), even in the absence of RA, reduced the cell number and dramatically altered the replating capacity of the PLZF-RARA-expressing cells (Figure 3E) and promoted their terminal differentiation (Figure 3F). Ezh2 deletion and the consecutive H3K27me3 loss were associated with a PLZF-RARA decrease (Figure 3D and supplemental Figure 3B), which was consistent with the altered replating capacity. This loss in replating capacity was also observed when Ezh2 deletion was achieved in vivo before PLZF-RARA transduction (supplemental Figure 3C) or ex vivo after the transformation process (supplemental Figure 3D). Finally, we showed that knocking down EZH2 in PLZF-RARA TG BM decreased PLZF-RARA expression, significantly delayed the development of leukemia, and prolonged the survival of the transplanted mice (Figure 3G and supplemental Figure 3E). These assays suggest that EZH2 is required for the initiation and maintenance of PLZF-RARA oncogenic activity.
EZH2 relevance in PLZF-RARA APL. (A) Violin plot showing Ezh2 expression per cluster (top) and in NT or RA-treated cell conditions per cluster (bottom). The black arrow pointing up indicate significant higher expression in the cluster against all the other clusters (average log2|FC| > .25 and adjusted P < .05). (B) Coverage plot of ATAC signal on Ezh2 gene in the ReP cluster. E2f1, E2f4, and Tfdp1 motifs are detected under the highlighted peak. (C) Experimental scheme to ascertain whether Ezh2 activity is required for PLZF-RARA transformation. Lineage negative (Lin−) cells purified from Cre-ERT;Ezh2fl/fl are purified and transduced with an empty-IRES-GFP (IRES) or a PLZF-RARA-IRES-GFP (P-RARA) retroviral construct. Lin− green fluorescent protein (GFP)–positive cells are purified by FACS and Ezh2 deletion in Lin-GFP positive cells is obtained by adding 150 nM 4-hydroxytamoxifen (4-OHT) in the methylcellulose. (D) (Left) Global levels of PLZF-RARA (black arrow: full length, black star: degraded form), Ezh2 and H3K27me3 detected by western blotting after 4-OHT-induced Ezh2 deletion in the second round of plating. Actin and H3 are used as loading controls. (Right) Bar plots representing the signal intensity of PLZF-RARA (P-RARA), EZH2, and H3K27me3 normalized to the loading control (for P-RARA) and to the IRES condition (for EZH2 and H3K27me3). ns, not significant. ∗P < .05 (2 replicates). (E) Replating efficiency is monitored by counting the total colony-forming units (CFU) of nontransformed (IRES) and PLZF-RARA–transformed (P-RARA) cells in presence (fl/fl) or absence (Δ/Δ) of Ezh2. Results are expressed as a mean standard deviation of 3 experiments (n = 3). ∗P < .05. (F) Cell morphology of P-RARA or IRES-transduced cells in presence (fl/fl) or absence (Δ/Δ) of Ezh2. Representative colonies of indicated conditions after 2 rounds of plating. Cells are cytospun and observed after May-Grünwald Giemsa (MGG) staining. Magnification 64×; bar 10 μm. (G) (Left) Experimental scheme to ascertain whether Ezh2 activity is required for PLZF-RARA leukemia development in vivo. PLZF-RARA TG BM is transduced with an shCtrl-GFP or an shEZH2-GFP retroviral construct. GFP-positive cells are purified by FACS and reinjected into recipient mice. (Right) Survival rate of mice transplanted with shCtrl (gray curve) or shEZH2 cells (green curve). Each cohort contains 5 mice. ∗∗∗P < .001. (H) Nuclear extracts of U937 cells treated or not with ZnSO4 (Zn) immunoprecipitated with anti-immunoglobulin G, anti-EZH2, or anti-SUZ12 antibodies. Immunoprecipitations (IPs) are immunoblotted with anti-PLZF (top) or anti-EZH2 antibodies (bottom). Inputs (in) represent 2% of samples processed in each IP. U937 B412: PLZF-RARA Zn-inducible; U937 MT: parental cells; U937 PR9: PML-RARA Zn-inducible. (I) Relative luciferase intensity monitored in HEK293T transduced or not (NT) with a shEZH2 (left) or transfected with a siE2F1 or siCtrl (NT) (right). Cells were transfected with the RARE-Luc and with a GFP (ᴓ P-RARA) or PLZF-RARA (P-RARA) construct. Cells are treated or not for 48 hours with RA (1 μM). AU, arbitrary units. ∗P < .05, ∗∗P < .01 (n = 6).
EZH2 relevance in PLZF-RARA APL. (A) Violin plot showing Ezh2 expression per cluster (top) and in NT or RA-treated cell conditions per cluster (bottom). The black arrow pointing up indicate significant higher expression in the cluster against all the other clusters (average log2|FC| > .25 and adjusted P < .05). (B) Coverage plot of ATAC signal on Ezh2 gene in the ReP cluster. E2f1, E2f4, and Tfdp1 motifs are detected under the highlighted peak. (C) Experimental scheme to ascertain whether Ezh2 activity is required for PLZF-RARA transformation. Lineage negative (Lin−) cells purified from Cre-ERT;Ezh2fl/fl are purified and transduced with an empty-IRES-GFP (IRES) or a PLZF-RARA-IRES-GFP (P-RARA) retroviral construct. Lin− green fluorescent protein (GFP)–positive cells are purified by FACS and Ezh2 deletion in Lin-GFP positive cells is obtained by adding 150 nM 4-hydroxytamoxifen (4-OHT) in the methylcellulose. (D) (Left) Global levels of PLZF-RARA (black arrow: full length, black star: degraded form), Ezh2 and H3K27me3 detected by western blotting after 4-OHT-induced Ezh2 deletion in the second round of plating. Actin and H3 are used as loading controls. (Right) Bar plots representing the signal intensity of PLZF-RARA (P-RARA), EZH2, and H3K27me3 normalized to the loading control (for P-RARA) and to the IRES condition (for EZH2 and H3K27me3). ns, not significant. ∗P < .05 (2 replicates). (E) Replating efficiency is monitored by counting the total colony-forming units (CFU) of nontransformed (IRES) and PLZF-RARA–transformed (P-RARA) cells in presence (fl/fl) or absence (Δ/Δ) of Ezh2. Results are expressed as a mean standard deviation of 3 experiments (n = 3). ∗P < .05. (F) Cell morphology of P-RARA or IRES-transduced cells in presence (fl/fl) or absence (Δ/Δ) of Ezh2. Representative colonies of indicated conditions after 2 rounds of plating. Cells are cytospun and observed after May-Grünwald Giemsa (MGG) staining. Magnification 64×; bar 10 μm. (G) (Left) Experimental scheme to ascertain whether Ezh2 activity is required for PLZF-RARA leukemia development in vivo. PLZF-RARA TG BM is transduced with an shCtrl-GFP or an shEZH2-GFP retroviral construct. GFP-positive cells are purified by FACS and reinjected into recipient mice. (Right) Survival rate of mice transplanted with shCtrl (gray curve) or shEZH2 cells (green curve). Each cohort contains 5 mice. ∗∗∗P < .001. (H) Nuclear extracts of U937 cells treated or not with ZnSO4 (Zn) immunoprecipitated with anti-immunoglobulin G, anti-EZH2, or anti-SUZ12 antibodies. Immunoprecipitations (IPs) are immunoblotted with anti-PLZF (top) or anti-EZH2 antibodies (bottom). Inputs (in) represent 2% of samples processed in each IP. U937 B412: PLZF-RARA Zn-inducible; U937 MT: parental cells; U937 PR9: PML-RARA Zn-inducible. (I) Relative luciferase intensity monitored in HEK293T transduced or not (NT) with a shEZH2 (left) or transfected with a siE2F1 or siCtrl (NT) (right). Cells were transfected with the RARE-Luc and with a GFP (ᴓ P-RARA) or PLZF-RARA (P-RARA) construct. Cells are treated or not for 48 hours with RA (1 μM). AU, arbitrary units. ∗P < .05, ∗∗P < .01 (n = 6).
We next investigated a potential interaction between the PLZF-RARA fusion protein and EZH2 using a myeloid cell line inducible for PLZF-RARA (U937-B412) (Figure 3H) and HEK293T cells overexpressing a Flag-tagged EZH2 and PLZF-RARA (supplemental Figure 3F). In both systems, we could not demonstrate any interaction between PLZF-RARA and EZH2 or SUZ12, another PRC2 component. However, we evidenced a stronger interaction between EZH2 and SUZ12 in the presence of PLZF-RARA than without (Figure 3H). The increase in interaction was not found in the presence of PML-RARA, demonstrating a specificity of the stabilizing effect of PLZF-RARA on the EZH2/SUZ12 complex. To test whether the E2F-EZH2 axis was involved in RA signaling, we knocked down EZH2 or E2F1 and quantified RA response by using a reporter luciferase assay under a retinoic acid response element (RARE). We showed that reduction of either EZH2 or E2F1 increased the RA response in the absence of PLZF-RARA but had no impact on PLZF-RARA repression activity (Figure 3I), suggesting that EZH2 per se limits signaling to RA.
Collectively, these results reveal a dependence of leukemic cells expressing PLZF-RARA on EZH2, whose activity may itself be modified by the presence of the fusion protein.
PLZF-RARA induced H3K27me3 level at specific enhancer genes that marks RA relapse-initiating cells
Next, we investigated the effect of PLZF-RARA expression on EZH2 chromatin activity. We compared the epigenetic landscape of PLZF-RARA promyelocytes with the granulocyte-monocyte-progenitor (GMP) compartment (public data set GSE124190),22 which is the closest cell compartment to promyelocytes according to its transcriptional signature (supplemental Figure 4A). Because our scATAC-seq data revealed stronger chromatin opening at enhancer than promoter regions (Figure 2G) and since cis-regulatory enhancer elements are known to influence the development of leukemia,23 we mapped the 4 histone marks (H3K27ac, H3K27me3, H3K4me1, and H3K4me3) that allow to discriminate active (H3K27ac-enriched) and poised (H3K27me3-enriched) enhancers22 (supplemental Figure 4B). By comparing PLZF-RARA promyelocytes with GMP genomic enhancer distribution, we found that PLZF-RARA expression modified poised (H3K27me3-enriched) enhancer distributions (68% of them were specific to PLZF-RARA condition) or active (H3K27ac-enriched) enhancer (30% of them were specific to PLZF-RARA condition) distributions (Figure 4A). We decided to focus on the new poised enhancers as they reflected a functional difference in H3K27me3 enrichment, shifting from enhancers regulating developmental processes to those regulating kinase signaling and bloodstream systems (supplemental Figure 4C).
PLZF-RARA-induced H3K27me3 level at specific enhancers genes that marks RA relapse-initiating cells. (A) Enhancer distribution in PLZF-RARA promyelocytes and normal GMPs. Overlap of Active (left) and Poised enhancers (right) in GMP and PLZF-RARA (P-RARA) conditions. ∗∗∗P < .001. The Poised enhancer dynamics upon PLZF-RARA expression is schematized below. Triangles represent enhancers; colors indicate their activity (Active: blue, Poised: red). Empty triangles: no change in enhancer activity between the 2 conditions. (B) Violin plots showing “switched” and “de novo” Poised signature scores per cluster. Signature score represents the global expression of annotated genes for the signature identified by scRNA-seq. Gray arrow pointing down: significantly lower expression in the cluster against all the others (average score difference > 0.005 and adjusted P < .05). (C) GO analysis of enhancer nearby genes. Gene %, number of genes observed/total number of genes within each GO term, BP, biological processes. (D) Experimental scheme to assess chromatin events associated with RA resistance. PLZF-RARA TG BM is transplanted into recipient mice. Ten days after, mice are injected with corn oil (NT) or with RA for 3 or 7 days (d3 and d7) and sacrificed. Treated BMs are immunophenotyped and reinjected into new recipient mice (tNT, td3, td7). Secondary transplanted mice are not treated (Ø) and sacrificed at day 17 posttransplantation. (E) Leukemia evolution analyzed by MGG staining (magnification 64×), spleen size (bar in centimeter), and FACS (Cd45.2, Cd11b, and Gr1 are monitored). (Top) Impact of RA on PLZF-RARA leukemia (NT, d3, d7). (Bottom) Leukemia relapse evaluation of transplanted untreated (tNT) and RA-treated BM (td3, td7). (F) (Left) Global levels of PLZF-RARA (black arrow: full length, black star: degraded form), EZH2, and H3K27me3. Actin and H3 are used as loading controls. Signal intensity is normalized according to the loading control and to the NT. (Right) Bar plots representing the signal intensity of PLZF-RARA (P-RARA), EZH2, and H3K27me3 normalized to the loading control and to the NT condition. ∗P < .05, ∗∗P < .01 (2 biological replicates). (G) Representative integrative genomics viewer (IGV) tracks of H3K27me3 in NT, d7, and td7 conditions. The gray box underlines enhancer coordinates. (H) Total number of poised enhancers in each condition. (I) Plot heat map of H3K27ac (blue) and H3K27me3 (red) signals in GMP, NT, d7, and td7 conditions at switched enhancers. Signals are plotted 10 Kb (for H3K27ac) and 0.1 Mb (for H3K27me3) upstream and downstream the enhancer center.
PLZF-RARA-induced H3K27me3 level at specific enhancers genes that marks RA relapse-initiating cells. (A) Enhancer distribution in PLZF-RARA promyelocytes and normal GMPs. Overlap of Active (left) and Poised enhancers (right) in GMP and PLZF-RARA (P-RARA) conditions. ∗∗∗P < .001. The Poised enhancer dynamics upon PLZF-RARA expression is schematized below. Triangles represent enhancers; colors indicate their activity (Active: blue, Poised: red). Empty triangles: no change in enhancer activity between the 2 conditions. (B) Violin plots showing “switched” and “de novo” Poised signature scores per cluster. Signature score represents the global expression of annotated genes for the signature identified by scRNA-seq. Gray arrow pointing down: significantly lower expression in the cluster against all the others (average score difference > 0.005 and adjusted P < .05). (C) GO analysis of enhancer nearby genes. Gene %, number of genes observed/total number of genes within each GO term, BP, biological processes. (D) Experimental scheme to assess chromatin events associated with RA resistance. PLZF-RARA TG BM is transplanted into recipient mice. Ten days after, mice are injected with corn oil (NT) or with RA for 3 or 7 days (d3 and d7) and sacrificed. Treated BMs are immunophenotyped and reinjected into new recipient mice (tNT, td3, td7). Secondary transplanted mice are not treated (Ø) and sacrificed at day 17 posttransplantation. (E) Leukemia evolution analyzed by MGG staining (magnification 64×), spleen size (bar in centimeter), and FACS (Cd45.2, Cd11b, and Gr1 are monitored). (Top) Impact of RA on PLZF-RARA leukemia (NT, d3, d7). (Bottom) Leukemia relapse evaluation of transplanted untreated (tNT) and RA-treated BM (td3, td7). (F) (Left) Global levels of PLZF-RARA (black arrow: full length, black star: degraded form), EZH2, and H3K27me3. Actin and H3 are used as loading controls. Signal intensity is normalized according to the loading control and to the NT. (Right) Bar plots representing the signal intensity of PLZF-RARA (P-RARA), EZH2, and H3K27me3 normalized to the loading control and to the NT condition. ∗P < .05, ∗∗P < .01 (2 biological replicates). (G) Representative integrative genomics viewer (IGV) tracks of H3K27me3 in NT, d7, and td7 conditions. The gray box underlines enhancer coordinates. (H) Total number of poised enhancers in each condition. (I) Plot heat map of H3K27ac (blue) and H3K27me3 (red) signals in GMP, NT, d7, and td7 conditions at switched enhancers. Signals are plotted 10 Kb (for H3K27ac) and 0.1 Mb (for H3K27me3) upstream and downstream the enhancer center.
Next, we focused on switched enhancers, which were marked by H3K27ac in GMP condition but gained H3K27me3 with PLZF-RARA expression (Figure 4A and supplemental Figure 4D). Although these switched enhancers were in the minority (175 of a total of 3876 PLZF-RARA–induced poised enhancers), they exerted a specific effect on the expression of their nearby genes in the ReP cluster, in which the expression was lower compared with the other clusters (Figure 4B, top). In contrast, de novo enhancer-associated genes were equally weakly expressed in all scRNA-seq clusters (Figure 4B, bottom). Gene Ontology (GO) analysis revealed that switched enhancer-related genes were associated with myeloid cell differentiation and proapoptotic terms (Figure 4C). In addition, their expression was decreased in the presence of PLZF-RARA (supplemental Figure 4E). This indicates that PLZF-RARA expression resulted in a redistribution of repressed enhancers that may be related to RA unresponsiveness.
To determine the role of EZH2 in RA resistance, we investigated the effect of RA on EZH2 chromatin activity after sequential transplantations (Figure 4D). In line with the literature,11 RA treatment, although inducing a clear differentiation of blasts (Figure 4E, top), did not eradicate relapse-initiating cells, since relapse was observed at day 17 posttransplant in mice transplanted with RA-treated BM (Figure 4E, bottom). RA treatment decreased the overall level of H3K27me3 as early as day 3 of treatment (Figure 4F), while increasing EZH2 protein level, which was consistent with our scRNA-seq data (Figure 3A). Interestingly, leukemia relapse of RA-treated BM was characterized with a restoration of high H3K27me3 levels in the leukemic cells (Figure 4F). This shows that RA had a contrasting effect on EZH2 and suggests that RA-induced differentiated cells maintained EZH2 independently of its methyltransferase activity. More precisely, RA decreased H3K27me3 signal at poised enhancers (Figure 4G) and the numbers of these enhancers after 7 days of treatment (Figure 4H). Clearly, leukemia relapse was characterized by both the restoration of high levels of H3K27me3 at the enhancer regions and the high number of H3K27me3-enriched enhancers (Figure 4G-H). Because PLZF-RARA cells respond differently to RA and the ReP cluster may be responsible for RA resistance, we measured the levels of H3K27me3/H3K27ac after RA treatment and at relapse at the 175 switched enhancers (described in Figure 4A). We found that neither RA treatment (3 or 7 days) nor the following transplantations erased H3K27me3 levels or restored H3K27ac signal at these enhancers (Figure 4I and supplemental Figure 5A). Consistently, the signature of the switched enhancers was not impacted by RA in our scRNA-seq data (supplemental Figure 5B).
Collectively, these results show that PLZF-RARA modifies EZH2 activity by redirecting the H3K27me3 on a subset of enhancers that are not affected by RA treatment. We also highlighted a discrepancy between EZH2 protein level and its methyltransferase activity during RA-induced differentiation.
Elimination of EZH2 eradicates relapse leukemia-initiating cells
Because EZH2 is necessary for PLZF-RARA–induced transformation and high H3K27me3 level at specific loci is a marker of resistance, we questioned the pertinence of targeting EZH2 with GSK, an S-adenosyl-methionine competitor that inhibits EZH2 methyltransferase activity,24 in combination with RA to overcome resistance. To test this hypothesis, we treated PLZF-RARA TG transplanted mice with GSK and/or with RA and transplanted the treated BMs into new recipients to follow disease progression according to the pretreatment (Figure 5A). At first, the inhibitory effect of GSK on H3K27me3 level in the treated BM, which was more pronounced when the drug was administered in combination with RA, was confirmed (Figure 5B). However, GSK treatment did not affect PLZF-RARA protein level (Figure 5B) nor the percent of PLZF-RARA TG cells (percent of Cd45.2-positive cells) in total BM and, in contrast to RA, had no impact on disease progression and blast differentiation, and no synergistic effect of GSK with RA was observed (Figure 5C, left, and supplemental Figure 6A). At day 13 after secondary transplantation (Figure 5C, middle), GSK-treated BM expanded as untreated BM, suggesting that GSK alone did not impact the relapse-initiating cells. A delay in leukemia progression was observed in the RA-treated BM, but addition of the GSK did not change in any instance the leukemia progression (Figure 5C, right). To rule out accessibility and dosage problems that could be faced using animal models, we compared the effect of GSK alone or in combination with RA on PLZF-RARA replating capacity (supplemental Figure 6B, left). As for the in vivo experiment, GSK alone or in combination with RA had no effect on the replating capacity of PLZF-RARA ex vivo (supplemental Figure 6B, right). These data showed that despite a strong dependency of PLZF-RARA cell transformation on EZH2, inhibition of its catalytic activity is not sufficient to promote APL clearance and further suggested that PLZF-RARA APL depends on a noncanonical activity of EZH2. To explore this possibility, we took advantage of a new commercially available EZH2 degrader, MS1943 (MS).25 PLZF-RARA TG BM was treated with MS and/or RA and retransplanted in new recipient mice (Figure 5D). Concomitant with its ability to degrade EZH2, MS effectively erased the global level of H3K27me3 (supplemental Figure 6C), decreased PLZF-RARA expression at the protein (Figure 5E and supplemental Figure 6C) and mRNA levels (supplemental Figure 6D), induced cell differentiation (Figure 5F), and reduced cell viability (Figure 5G). Despite low synergistic effect of MS with RA in vitro (supplemental Figure 6E), reinjecting alive MS and MS-RA–treated PLZF-RARA BM cells in recipient mice significantly prolonged the survival of the mice, confirming the importance of EZH2 noncanonical activity in PLZF-RARA leukemia development and RA resistance (Figure 5H and supplemental Figure 6F). To link this loss of leukemic potential upon EZH2 degradation to a transcriptional reprogramming, we performed bulk RNA-seq on PLZF-RARA TG BM treated with MS or GSK (Figure 5I, supplemental Figure 6G, and supplemental Table 4). MS treatment was associated with a more pronounced and specific decrease in the expression of ReP cluster marker genes in comparison with GSK treatment (Figure 5I). GO analysis on genes modified by MS further confirmed the importance to degrade EZH2 to target the biological processes (cell cycle and DNA-dependent DNA replication) associated with RA resistance (supplemental Figure 6G). Interestingly, the gene signature resulting from the targeting of methyltransferase activity of EZH2 (named methyl targeting) as well as the ReP marker genes were positively correlated to PLZF-RARA patients, and genes associated with the targeting of EZH2 nonmethyltransferase activity (named nonmethyl targeting) and the NeuRA marker genes were correlated to PML-RARA patients (Figure 5J). These data not only show that our RA (un)response signatures identified in our mouse model could be used for APL patients but also confirm that targeting EZH2 nonmethyltransferase activities is necessary to promote RA sensitivity in APL patients.
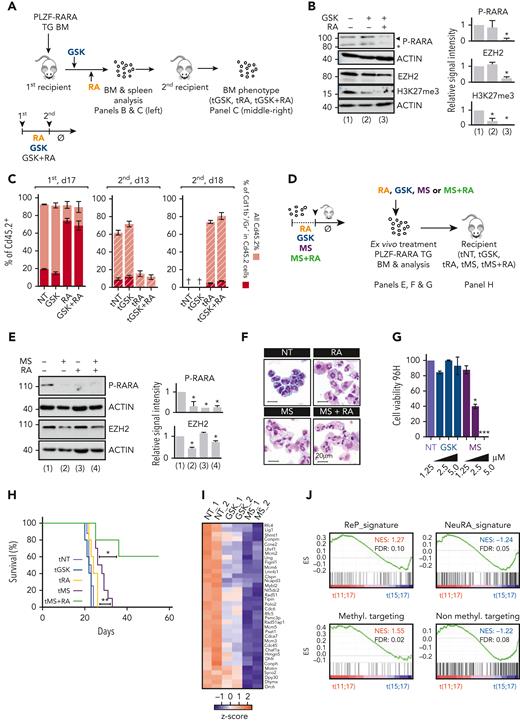
Impact of targeting EZH2 on APL progression. (A) Experimental scheme of the analysis of RA, GSK, or combination-treated BM. PLZF-RARA TG BM is transplanted into recipient mice. Seven days after, mice are injected for 3 days with GSK or with corn oil (NT) followed by 7 consecutive days of RA (RA). After treatments (NT: corn oil, GSK, RA, GSK and RA) mice are sacrificed, and treated BM are immunophenotyped and reinjected into new recipient mice (tNT, tGSK, tRA, tGSK+RA). At this time treatments are stopped (Ø). Mice are sacrificed 15 to 20 days after the secondary transplantation. (B) (Left) Global levels of PLZF-RARA (black arrow: full length, black star: degraded form), Ezh2, and H3K27me3. Actin is used as loading control. (Right) Bar plots representing the signal intensity of PLZF-RARA (P-RARA), EZH2, and H3K27me3 normalized to the loading control and to the NT condition. ∗P < .05 (3 biological replicates). (C) (Left) Impact of GSK, RA, and combo treatments on PLZF-RARA leukemia at day 17 after the first transplantation (NT, GSK, RA, GSK+RA). (Middle and right) Evaluation of leukemia relapse monitored by FACS analysis (Cd45.2, Cd11b, and Gr1 markers are monitored) at day 13 (d13) and day 18 (d18) after the secondary transplantation (tNT, tGSK, tRA, tGSK+RA). (D) Scheme resuming the protocol to evaluate the effect of MS treatment on PLZF-RARA BM. (E) (Left) Global levels of PLZF-RARA and Ezh2 after RA (0.1 μM) and/or MS (1.25 μM) treatments. Actin is used as loading controls. (Right) Bar plots representing the signal intensity of PLZF-RARA (P-RARA) and EZH2 normalized to the loading control and to the NT condition. ∗P < .05 (2 biological replicates). (F) Cell morphology analyzed by MGG staining (magnification 64×; bar 20 μm). (G) Cell viability of PLZF-RARA TG cells upon GSK or MS treatments monitored by bioluminescence. Results are expressed by percent of living cells and normalized to the untreated condition (NT). Results are expressed as the mean standard deviation of 3 independent experiments (n = 3). ∗P < .05, ∗∗∗P < .001. (H) Survival rate of mice transplanted with untreated (tNT, gray curve, 8 mice) GSK (tGSK, 2.5 μM, blue curve, 5 mice), RA (tRA, 0.1 μM, 5 mice), MS (tMS, 2.5 μM, purple curve, 10 mice), MS and RA (tMS+RA, 1.25 μM MS and 0.1 μM RA, green curve, 5 mice) pretreated PLZF-RARA TG BM. ∗P < .05, ∗∗P < .01. (I) RNA-seq (2 biological replicates) performed on promyelocytes purified from PLZF-RARA TG mice and treated or not (NT) with MS (MS, 2.5 μM), or GSK (2.5 μM) (in vitro treatments). Impact of MS (MS) and GSK on the expression of the most differentially overexpressed genes in the ReP cluster compared with the other clusters. Results are expressed as gene counts z score. (J) Gene set enrichment analysis of t(11;17) APL patients (PLZF-RARA) compared with t(15;17) APL patients (PML-RARA). ReP, NeuRA, the targeting of nonmethyltransferase activities of EZH2 (Non methyl.targeting) and the targeting of methyltransferase activities of EZH2 (Methyl. targeting) enrichment signatures (ESs) are computed. FDR, false discovery rate; NES, normalized enrichment score.
Impact of targeting EZH2 on APL progression. (A) Experimental scheme of the analysis of RA, GSK, or combination-treated BM. PLZF-RARA TG BM is transplanted into recipient mice. Seven days after, mice are injected for 3 days with GSK or with corn oil (NT) followed by 7 consecutive days of RA (RA). After treatments (NT: corn oil, GSK, RA, GSK and RA) mice are sacrificed, and treated BM are immunophenotyped and reinjected into new recipient mice (tNT, tGSK, tRA, tGSK+RA). At this time treatments are stopped (Ø). Mice are sacrificed 15 to 20 days after the secondary transplantation. (B) (Left) Global levels of PLZF-RARA (black arrow: full length, black star: degraded form), Ezh2, and H3K27me3. Actin is used as loading control. (Right) Bar plots representing the signal intensity of PLZF-RARA (P-RARA), EZH2, and H3K27me3 normalized to the loading control and to the NT condition. ∗P < .05 (3 biological replicates). (C) (Left) Impact of GSK, RA, and combo treatments on PLZF-RARA leukemia at day 17 after the first transplantation (NT, GSK, RA, GSK+RA). (Middle and right) Evaluation of leukemia relapse monitored by FACS analysis (Cd45.2, Cd11b, and Gr1 markers are monitored) at day 13 (d13) and day 18 (d18) after the secondary transplantation (tNT, tGSK, tRA, tGSK+RA). (D) Scheme resuming the protocol to evaluate the effect of MS treatment on PLZF-RARA BM. (E) (Left) Global levels of PLZF-RARA and Ezh2 after RA (0.1 μM) and/or MS (1.25 μM) treatments. Actin is used as loading controls. (Right) Bar plots representing the signal intensity of PLZF-RARA (P-RARA) and EZH2 normalized to the loading control and to the NT condition. ∗P < .05 (2 biological replicates). (F) Cell morphology analyzed by MGG staining (magnification 64×; bar 20 μm). (G) Cell viability of PLZF-RARA TG cells upon GSK or MS treatments monitored by bioluminescence. Results are expressed by percent of living cells and normalized to the untreated condition (NT). Results are expressed as the mean standard deviation of 3 independent experiments (n = 3). ∗P < .05, ∗∗∗P < .001. (H) Survival rate of mice transplanted with untreated (tNT, gray curve, 8 mice) GSK (tGSK, 2.5 μM, blue curve, 5 mice), RA (tRA, 0.1 μM, 5 mice), MS (tMS, 2.5 μM, purple curve, 10 mice), MS and RA (tMS+RA, 1.25 μM MS and 0.1 μM RA, green curve, 5 mice) pretreated PLZF-RARA TG BM. ∗P < .05, ∗∗P < .01. (I) RNA-seq (2 biological replicates) performed on promyelocytes purified from PLZF-RARA TG mice and treated or not (NT) with MS (MS, 2.5 μM), or GSK (2.5 μM) (in vitro treatments). Impact of MS (MS) and GSK on the expression of the most differentially overexpressed genes in the ReP cluster compared with the other clusters. Results are expressed as gene counts z score. (J) Gene set enrichment analysis of t(11;17) APL patients (PLZF-RARA) compared with t(15;17) APL patients (PML-RARA). ReP, NeuRA, the targeting of nonmethyltransferase activities of EZH2 (Non methyl.targeting) and the targeting of methyltransferase activities of EZH2 (Methyl. targeting) enrichment signatures (ESs) are computed. FDR, false discovery rate; NES, normalized enrichment score.
Altogether, these results demonstrate that targeting EZH2 methyltransferase activity is not sufficient to eradicate relapse leukemia-initiating cells and suggest elimination of EZH2 to overcome RA resistance.
Discussion
Here, we focused on the effect of RA treatment in APL cells expressing PLZF-RARA to address leukemic cell heterogeneity and its consequence on therapy resistance. The unique differentiating properties of RA make it a promising drug for the treatment of non-APL AML26 and solid tumors,27 and a better understanding of resistance mechanisms could widen its use.
Clonal diversity and evolution patterns of AML by high-throughput single-cell genomics (scRNA-seq and scDNA-seq) have revealed hierarchies in the AML with heterogeneity correlating with their underlying genetic alterations.28,29 Here, we analyzed the effect of a single oncogenic event and identified different subgroups of PLZF-RARA–transformed promyelocytes, which reflect the APL hierarchy as well as spontaneous differentiation. Among them, we identified a cell subgroup, named ReP, which remained transcriptionally and at the chromatin level unperturbed by RA and retained leukemic activity. What could make the Rep cells the RA-resistant cells? The first striking observation was that this group of cells maintains PLZF-RARA expression after RA treatment, which is known to act as a competitive transcriptional repressor of RARA30 and block RA signalization.31 This suggests that in our experimental setting, as it was shown for PML-RARA model,8 maintaining PLZF-RARA expression is key for RA resistance. This also suggests that RA resistance is not acquired by RA treatment but inherent to this particular group of transformed promyelocytes. Thus, mechanism of resistance maybe different than the acquired RA resistance observed in PML-RARA patients, which has been associated to additional oncogenic events.32-34 These transformed promyelocytes were different from the others with a marked replication/repair program and high E2F activity; this oncogenic activity35 may support the RA resistance of the cells considering its role in promoting leukemic cell survival,36 escape from apoptosis,37 and resistance to cisplatin treatment.38 In addition, by studying our scATAC-seq data, we specifically probe the activity of E2F in RA-resistant cells and identify an E2F oncogenic regulatory network involving the PRC2 methyltransferase EZH2. Deregulations of EZH2 are linked to cancer initiation, metastasis, immunity, metabolism, and drug resistance in a wide variety of cancer39 and may contribute to RA resistance. Thus, although we cannot determine whether the maintenance of PLZF-RARA is the cause or consequence of RA unresponsiveness, we revealed a E2F/EZH2 axis, which can be the key to overcome RA resistance.
EZH2 represents one of the most promising epigenetic anticancer therapeutic targets, although its mechanism of action is incompletely understood.40 Indeed, due to its tissue and mode of action complexity,20 targeting EZH2 in leukemia is challenging. Within the AML entity, EZH2 exerts an oncogenic function by enhancing differentiation blockade during AML maintenance,41 and it acts as a tumor suppressor during leukemia initiation.42 Here, we demonstrated the dependence of PLZF-RARA leukemia on EZH2 activity, suggesting an oncogenic role of EZH2 in this type of leukemia. However, we clearly showed little effect of inhibiting its methyltransferase activity on PLZF-RARA leukemia progression and relapse, and we demonstrated the need to deplete EZH2 to clear leukemia. This means that PLZF-RARA leukemic cells depend on an oncogenic activity that goes beyond EZH2 catalytic activity.
Molecularly, we showed that PLZF-RARA redirects EZH2 methyltransferase activity on a subset of apoptotic genes and that this PLZF-RARA H3K27me3-induced signature marks the RA-persistent cells. Interestingly, we could not find a direct interaction between EZH2 and PLZF-RARA as we previously did for PLZF.43 However, we clearly evidenced a stronger association of EZH2 and SUZ12 upon PLZF-RARA induction. This PRC2 stabilization could either account for a modification of the methyltransferase activity of EZH244,45 or favor its noncanonical activity.46 Decreasing EZH2 did not alter the repressive activity of PLZF-RARA, although it was able to increase the RARA response, in agreement with the literature showing that EZH2, as part of the PRC2 complex, inhibits RARA signaling during development.47,48 Many cancers have been shown to rely on an EZH2 oncogenic effect that is not solely based on its histone transferase activity. This is the case of estrogen receptor-negative basal-like breast cancers,49 AR-dependent prostate cancers,45,50 or SWI/SNF mutant tumors that rely on both the catalytic and noncatalytic activities of EZH2.46 Interestingly, for the latter tumors, as in PLZF-RARA leukemic cells, GSK has shown only limited efficacy.46
By leveraging our own single-cell data, we reveal a novel mechanism of resistance, involving the multifaceted enzyme EZH2, opening up a therapeutic opportunity. We provide evidence, by using an EZH2-selective degrader,25 which significantly reduced the growth of PLZF-RARA-expressing leukemia cells and increased the survival of transplanted mice, of the importance of eliminating the target on which the tumor depends. Our study supports the development of EZH2-based PROTACs (PROteolysis TArgeting Chimera) to degrade the PRC2 complex, which targets the enzymatic and nonenzymatic activities of EZH251 and holds great promise for the future treatment of cancers dependent on the noncatalytic activity of EZH2. A very recent study clearly demonstrated that EZH2 depletion inhibits neuroblastoma and small cell carcinoma tumor formation by potentiating MYC degradation.52 In addition, while revising this work a study came out showing that a PROTAC EZH2, which degrades almost more efficiently its oncogenic partner MYC, is efficient in reducing MLL-positive leukemia.53
In conclusion, we demonstrated the power of single-cell multiomics to understand cancer cell heterogeneity and to identify treatment-resistant cells and characterize their activity. We also revealed a novel mechanism of RA resistance dependent on EZH2 noncatalytic activity, which should be considered when developing targeted therapeutic approaches.
Acknowledgments
The authors thank Pier Pandolfi for sharing the PLZF-RARA TG mouse model; Valerie Lallemand-Breitenbach for facilitating the access to the PLZF-RARA TG bone marrow; Christophe Lachaud for helpful discussion and for reading the manuscript; and members of the Duprez laboratory for helpful discussions. The authors thank the animal support facility of the Centre de Recherche en Cancérologie de Marseille, the flow cytometry and cell sorting platform, the microscopy platform, and the CIBI and DISC platforms for computational analyses and support.
This work was supported by the Ligue Nationale contre le Cancer (E.D.), the Association Laurette Fugain (E.D.), the Fondation Recherche Médicale (E.D.), the Fondation A∗MIDEX (E.D.), the Fondation de France (M.P.), l’Institut National du Cancer grant # 20141PLBIO-06-1 (M.P. and E.D.), the Japan Society for the Promotion of Sciences (JSPS) (M.P.), and the Cancéropôle Provence Alpes Côte d’Azur (M.P.). L.H. was supported by a PhD grant from Aix Marseille University. The study was supported by the collaborative IMSUT joint grant between A.I. and E.D. This study was also supported partly by a grant from International Joint Usage/Research Center, The Institute of Medical Science, The University of Tokyo.
Authorship
Contribution: M.P. and E.D. developed the original hypothesis and designed experiments; M.P., with help from N.P., S.K., W.K., L.N., L. Haboub, Y.N.-T., M.O., and N.C., performed experiments and/or analyzed data; A.M. and L. Hérault carried out computational analyses of the scRNA-seq and scATAC-seq; L.Hérault developed the pipeline for scRNA-seq and scATAC-seq data integration; M.P. and J.V. conducted ChIP-seq analyses; J.V. analyzed RNA-seq; A.I. and D.B. provided reagents and discussed results; M.P., E.D., and A.M. designed the figures; M.P. and E.D. wrote the manuscript; and all coauthors proofread the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mathilde Poplineau, Aix Marseille University CRCM, CNRS UMR7258, INSERM U1068, Institut Paoli-Calmettes, Marseille, France; e-mail: mathilde.poplineau@inserm.fr; and Estelle Duprez, Aix Marseille University CRCM, CNRS UMR7258, INSERM U1068, Institut Paoli-Calmettes, Marseille, France; e-mail: estelle.duprez@inserm.fr.
References
Author notes
∗A.M. and L.H. contributed equally to this study.
The raw ChIP-seq, scATAC-seq, and scRNA-seq data (fastq files) are deposited under the accession number GSE181190. The repository also contains the bigwig, the peak matrix, and the count matrix for the ChIP-seq, scATAC-seq, and scRNA-seq, respectively. All R and python codes used for data analysis are integrated in a global Snakemake workflow available at https://gitcrcm.marseille.inserm.fr/herault/GMP_PRARA.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Single-cell transcriptome analysis identifies a subset of RA-resistant PLZF-RARA leukemic cells with a specific signature. (A-B) Uniform Manifold Approximation and Projection (UMAP) visualization of the scRNA-seq data set colored by (A) condition (NT and RA day 7 [d7]) and by (B) cluster (total integrated cells: 11 900). Six clusters were identified: ReP, DNA repair/Replication/Proliferation; Prom1-3, Promyelocyte (Neutrophil) 1-3; NeuRA1-2, Neutrophil RA1-2. (C) (Bottom) NT and RA cell distribution in each cluster. The black line indicates expected NT and RA cell proportions according to the data set size. (Top) Absolute cell number per cluster and per condition. Names of the clusters for which the proportion of NT or RA cells is significantly higher than expected (P < .01) are purple for NT cells and orange for RA cells. (D) Differentiation trajectory. (Top) Schematic representation of the different states (A-E) in each trajectory (trajectory 1: A-C-D, trajectory 2: A-C-E, trajectory 3: A-B). (Bottom) UMAP is colored according to the pseudotime value of each cell. (E) Cluster distribution in each state of the pseudotime. (F) NT and RA cell distribution (bottom: percentage; top: absolute) in each state of the pseudotime. (C and F) Names of the clusters for which the proportion of NT or RA cells was significantly higher than expected (P < .01) are purple for NT cells and orange for RA cells. (G) Violin plots showing PML-RARA response and (H) E2f signature scores in each cluster. (G and H) The signature score represents the global expression of annotated genes for the selected signature. Gray/black arrows pointing down/up: significant lower/higher expression in the cluster against all the others (average score difference > 0.02 and adjusted P < .05). (I) Violin plot showing Usp37 expression in each cluster. Black arrows pointing up: significant higher expression in the cluster against all the others (average log2|FC|> .1 and adjusted P < .05). (J) Gating strategy for isolating ProReP cells (Prom1-3 and ReP) and NeuRA (NeuRA1-2). The donut plot represents the proportion of Prom1-3 and ReP cells into the ProReP population according to the scRNA-seq analysis. (K) PLZF-RARA (black arrow: full length; black star: degraded form) levels in untreated (NT) and treated promyelocytes (RA d7). Actin is used as loading control. (L) Survival rate of mice transplanted with ProReP or NeuRA cells. Each cohort contains at least 5 mice. ∗∗∗P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/22/10.1182_blood.2022015668/5/m_blood_bld-2022-015668-gr1.jpeg?Expires=1763463158&Signature=cplKAchaOdKRpCPlLzJqN5FLsV8XHWe0~qG-phJXozona5NiwpZLX6FkOU0uFWz3Va7zq4LuPg4bv8V2MtcS9tt8UANcsDBwsySPx75Tjeora9mAFsG6JRyY65cDRR4Ggrx75Mqbox7ycR8ceNMNe-v2DNBSEiYTL7esZGNnKavqMMf72NiqXu-C6oqOjckSnuEByVFHptyrxs~6zX7LioEi4mKcIx0YmKNJVrXH~x5-qn6~EorjRbGCOQeRChcohZXFdofSYBbZIVys5MoTrwCooVjM3U118VU3LFqRFM0CKymo3FhMzprA9~5pFVHXORA1W82YxyW~XISW6Nh1Rw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal