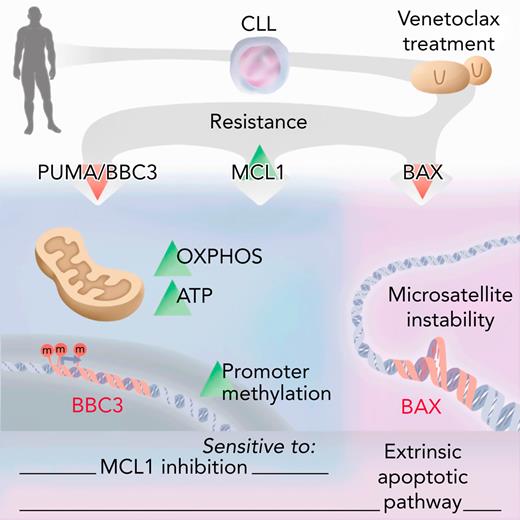
Key Points
Venetoclax resistance is mediated by methylation and silencing of PUMA.
Treatment algorithms should consider the PUMA, MCL1, and BAX status.
Abstract
The BCL2 inhibitor venetoclax has been approved to treat different hematological malignancies. Because there is no common genetic alteration causing resistance to venetoclax in chronic lymphocytic leukemia (CLL) and B-cell lymphoma, we asked if epigenetic events might be involved in venetoclax resistance. Therefore, we employed whole-exome sequencing, methylated DNA immunoprecipitation sequencing, and genome-wide clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 screening to investigate venetoclax resistance in aggressive lymphoma and high-risk CLL patients. We identified a regulatory CpG island within the PUMA promoter that is methylated upon venetoclax treatment, mediating PUMA downregulation on transcript and protein level. PUMA expression and sensitivity toward venetoclax can be restored by inhibition of methyltransferases. We can demonstrate that loss of PUMA results in metabolic reprogramming with higher oxidative phosphorylation and adenosine triphosphate production, resembling the metabolic phenotype that is seen upon venetoclax resistance. Although PUMA loss is specific for acquired venetoclax resistance but not for acquired MCL1 resistance and is not seen in CLL patients after chemotherapy-resistance, BAX is essential for sensitivity toward both venetoclax and MCL1 inhibition. As we found loss of BAX in Richter’s syndrome patients after venetoclax failure, we defined BAX-mediated apoptosis to be critical for drug resistance but not for disease progression of CLL into aggressive diffuse large B-cell lymphoma in vivo. A compound screen revealed TRAIL-mediated apoptosis as a target to overcome BAX deficiency. Furthermore, antibody or CAR T cells eliminated venetoclax resistant lymphoma cells, paving a clinically applicable way to overcome venetoclax resistance.
Introduction
Because the recent approval of venetoclax (VEN) for treatment of patients with chronic lymphocytic leukemia (CLL) and acute myeloid leukemia, the number of patients with VEN resistance is increasing, demanding in-depth analysis of resistance mechanisms.1-7 The B-cell lymphoma 2 (BCL2) protein family, consisting of pro- and antiapoptotic proteins regulating mitochondrial apoptosis, plays an important role in resistance toward VEN. Acquired mutations in BCL2 associated protein (BAX) and BCL2 were found in hematopoietic cell lines with acquired resistance toward BH3 mimetics.8 Whereas proapoptotic BAX is a mediator of apoptosis, leading to permeabilization of the mitochondrial outer membrane upon induction of apoptosis, antiapoptotic BCL2 sequesters proapoptotic BIM. Upon binding of VEN, BIM is released and can induce apoptosis. Interestingly, mutations in BAX can also occur in the myeloid compartment of VEN-treated CLL patients and are associated with clonal hematopoiesis, indicating lineage-specific adaptation to VEN.9 Moreover, recurrent mutations in BCL2 (G101V) occur in patients on a subclonal level, leading to resistance due to decreased affinity of BCL2 for VEN.10 Functionally relevant mutations also occur in the proximity of G101V (eg, D103 and F104).11,12 Recurrent mutations/deletions in the cell cycle regulators BTG1 and CDKN2A contribute to resistance particularly in CLL, where resistance occurs earlier and is associated with a more aggressive phenotype.13,14 Beside mutations, especially upregulated MCL1 due to amplification of chromosome 1 (amp[1q]) confers resistance toward VEN.15-19 MCL1, an antiapoptotic protein, can be targeted pharmacologically. Similar to BCL2, MCL1 interacts with proapoptotic proteins like BAX or BAK to block their apoptotic function, an interaction that can be disturbed by proapoptotic BH3-only proteins like PUMA or NOXA.20
Although genetic reasons for VEN resistance have been explored in the last years, epigenetic causes regulating gene expression are poorly understood. We aim to understand the genetic and epigenetic mechanisms of VEN resistance in high-risk CLL and various B-cell non-Hodgkin lymphoma (B-NHL) models.
Methods
Patient sampling
The material of 6 CLL patients from the M13 982 trial were investigated as reported earlier.13 T-cell prolymphocytic leukemia (T-PLL) patients were diagnosed according to World Health Organization criteria. Samples were obtained from patients under institutional review board–approved protocols following written informed consent. All patient samples were collected according to the Declaration of Helsinki, and collection and use of patient material was approved by the ethics committee of the University Hospital of Cologne (EudraCT-Nr.: #2008-001421-34 and AZ11-319).
Experimental mice
The generation of the single alleles for Cd19Cre, Eμ-TCL1tg, Baxfl/wt, and Baxfl/fl have been described before.21-23 The sex of the examined mice was balanced. Animals were housed in a specific pathogen-free facility, and animal breeding and experiments were approved by the local animal care committee and the relevant authorities (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, 81-02.04.2019.A009).
Cell lines
B-cell lymphoma cell lines derived from diffuse large B-cell lymphoma (DLBCL), acute lymphoblastic leukemia, follicular lymphoma, CLL, and unspecified B-cell lymphoma were used. Detailed information is given in the supplemental data (supplemental Table 1; supplemental Materials and methods, available on the Blood website).
Generation of VEN- and S63845-resistant cell lines
VEN and S63845 resistance was established by long-term exposure to VEN and S63845, respectively. The initial dose was 1 nM for VEN and 0.15 μM for S63845. As soon as the treated cells displayed viability and growth rate similar to the parental lines, the drug dose was doubled until the final dose of 8 μM VEN or S63845 was reached. Forty-eight hours before experiments were performed, cell lines were transferred to VEN-/S63845-free medium.
CRISPR/Cas9 screening and data analysis
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) screen was performed in the murine activated B-cell diffuse large B-cell lymphoma cell line BSQ12.4 as published.26 Experimental details are given in supplemental Methods.
Compounds
S63845 was purchased from APExBIO. Tumor necrosis factor–related apoptosis-inducing ligand was purchased from Enzo Life Sciences. All other compounds (supplemental Table 2) were purchased from Selleck Chemicals.
WES and analysis
Whole-exome sequencing (WES) of genomic DNA from VEN-/S63845-sensitive and -resistant B-cell lymphoma cell lines was performed as reported earlier.13 Details on DNA extraction are given in supplemental Methods.
The WES data were deposited to https://dataview.ncbi.nlm.nih.gov/object/PRJNA716141?reviewer=amsdmumopsfn00r1smcc0hnbqg.
MeDIP-seq and analysis
Genome-wide methylation analysis was performed using methylated DNA immunoprecipitation (MeDIP-seq)27 of 9 cell line pairs: DOHH-2, DB, KARPAS-422, P30-OH-KUBO, WSU-NHL, HBL-1, 697, and OCI-LY-19.
Raw Illumina 450k files were downloaded from the Gene Expression Omnibus (supplemental Table 3), and β values were computed with the R/bioconductor package Minfi (minfi, RRID:SCR_012830).28 Please refer to supplemental Methods for details.
Pyrosequencing assay
Targeted methylation analysis was performed using bisulfite conversion and pyrosequencing. Converted DNA was amplified with the Pyromark PCR Kit (Qiagen).
Pyrosequencing was performed on a PSQHS96A (Qiagen) with Pyromark Gold Q96 reagents (Qiagen), and methylation percentages were calculated using the PSQHS96A 1.2 software. For details on preparation of DNA and sequences of used oligos, please refer to supplemental Methods.
Metabolic flux analysis
Seahorse XFe96 Analyzer (Seahorse Bioscience, Agilent) was used to assess oxygen consumption rate (OCR) in the described cell lines following manufacturer instructions. For details, please refer to supplemental Methods. The Seahorse XF Cell Mito Stress Test Report Generator (Seahorse Bioscience, Agilent) was used to analyze the above-mentioned parameters.
Immunohistochemistry
Three micromolars of formalin-fixed, paraffin-embedded sections were immunostained for BAX (anti-human BAX clone D2E11, Cell Signaling), pretreated as indicated by the manufacturer using a Laboratory Vision Autostainer 480S (Thermo Fisher Scientific), and counterstained with hematoxylin.
Bispecific antibodies against CD3:CD19
Peripheral blood mononuclear cells (PBMCs) from healthy donors were incubated with cell lines at a tumor:effector ratio of 10:1 after stimulation with anti-CD3 antibody (200 ng/mL) and anti-CD28 antibody (50 ng/mL) at a density of 1 × 106 cells per mL for 72 hours.
Cells were stained with antibodies against CD4 and CD8 (both Miltenyi Biotech) and annexin-V (Immunotools) and analyzed by flow cytometry.
CAR T-cell experiments
VEN-sensitive and -resistant Nalm6 cells as well as OSU-CLL wild-type and BAX−/− clones were incubated with different proportions of anti-CD19 chimeric antigen receptor (CAR) T cells. CAR T-cell preparation was performed as described earlier.29 The retroviral expression cassettes for the chimeric antigen receptors used in this study were generated by replacing the single-chain variable fragment (scFv) binding domain of the anti-CEA CAR BW431/26scFv-Fc-CD3ζ (439),30 with the FMC63 scFv31 to obtain the CD19-specific CAR. CD3+ T cells were isolated by magnetic-activated cell sorting to purities >98% using human anti-CD3 MicroBeads (Miltenyi Biotec). CD3+ T cells were retrovirally transduced to express the CAR. Afterward cell viability was determined by an XTT assay.
Statistical analyses
GraphPad Prism 7 (GraphPad Prism, RRID:SCR_002798) was used for data analyses. Data were presented as mean ± standard deviation (SD). Comparison between groups was performed using 2-tailed Student t test or 1-way analysis of variance; P < .05 was considered as a significant difference.
Results
VEN-resistant cell lines and patient samples feature distinct regulation of PUMA, BAX, and MCL1
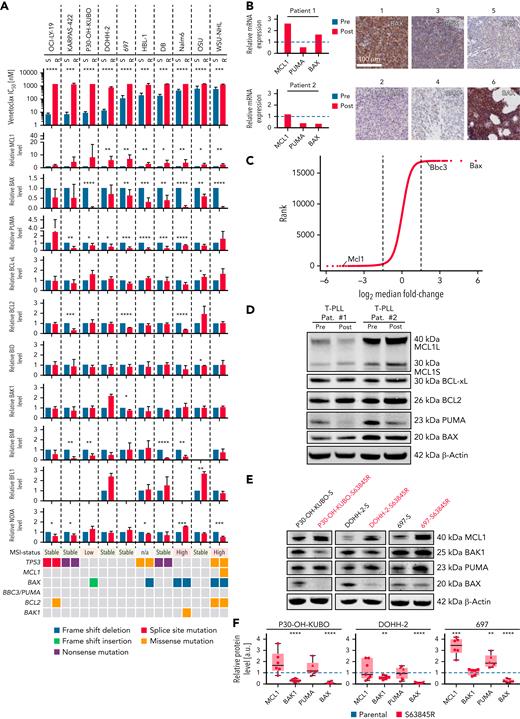
Ten cell lines from different leukemia/lymphoma entities with acquired resistance toward VEN were generated. Resistance was validated by toxicity assays and analysis of PARP cleavage and was stable even after 3 months without VEN treatment (supplemental Figure 1C). We identified MCL1, BAX, and PUMA (BBC3) protein level recurrently affected in cell lines with acquired resistance toward VEN. MCL1 was significantly upregulated in 7 of 10 resistant cell lines. Our data reveal a significant reduction or loss of BAX in 6 of 10 resistant cell lines and, unexpectedly, a significantly decreased expression of PUMA in 8 of 10 cell lines (Figure 1A; supplemental Figure 1A-C). MCL1 upregulation as well as downregulation of BAX and PUMA could also be detected in primary CLL patient samples at RNA level (Figure 1B). We performed a genome-wide CRISPR/Cas9 knockout screen26 with and without low-dose VEN as selective pressure in another activated B-cell diffuse large B-cell lymphoma mouse model.32,33 guide RNAs against Bax and Bbc3 were strongly selected in the presence of VEN (mean fold change, 56 and 3.84, respectively). In contrast, loss of the guide RNA against Mcl1 was detrimental for the cells after VEN treatment (mean fold change, 0.04, Figure 1C).
Downregulation of BAX and PUMA and upregulation of MCL-1 in vitro and in vivo. (A) Top: IC50 values for VEN in 10 sensitive (blue) vs VEN-resistant (red) B-cell lymphoma cell lines: WSU-NHL, OCI-LY-19, DOHH-2, DB, KARPAS-422, HBL-1, 697, P30-OH-KUBO, Nalm6, and OSU. Middle: densitometric analyses of immunoblots of BCL2 proteins (supplemental Figure 1A-B). Mean ± SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, compared with parental (VEN naïve) cells, Students t test. Lower: results from WES. Genomic alterations are annotated according to the color panel below the image. (B) Immunohistochemistry of BAX in 6 primary CLL samples. Scale bar, 100 μm. Pictures 1 through 5: lymph node sections posttherapy; picture 6: bone marrow post–VEN therapy. Relative MCL1 (left panel) and BBC3 (PUMA) (right panel) and BAX mRNA expression level for CLL patients 1 and 2 pre- (blue) and post (red)-VEN therapy determined by bulk 3′RNA-seq. (C) Results for Bax, Bbc3, and Mcl1 guide RNAs from CRISPR/Cas9-Screen in murine lymphoma cell line after 28 days with/without VEN (10 nM). (D) Immunoblot for MCL1, BCL-xL, BCL2, PUMA, and BAX in 2 T-PLL patients before and after VEN resistance. (E) Immunoblot for MCL1, BAK1, PUMA, and BAX in 3 sensitive and S63845-resistant B-cell lymphoma cell lines, respectively. (F) Densitometric analyses of immunoblots against MCL1, BAK1, PUMA, and BAX normalized to β-actin. Data illustrated as mean ± SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .001, compared with parental (blue) cells, Student t test. 3′RNA-seq, 3’RNA-sequencing; IC50, median inhibition concentration.
Downregulation of BAX and PUMA and upregulation of MCL-1 in vitro and in vivo. (A) Top: IC50 values for VEN in 10 sensitive (blue) vs VEN-resistant (red) B-cell lymphoma cell lines: WSU-NHL, OCI-LY-19, DOHH-2, DB, KARPAS-422, HBL-1, 697, P30-OH-KUBO, Nalm6, and OSU. Middle: densitometric analyses of immunoblots of BCL2 proteins (supplemental Figure 1A-B). Mean ± SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, compared with parental (VEN naïve) cells, Students t test. Lower: results from WES. Genomic alterations are annotated according to the color panel below the image. (B) Immunohistochemistry of BAX in 6 primary CLL samples. Scale bar, 100 μm. Pictures 1 through 5: lymph node sections posttherapy; picture 6: bone marrow post–VEN therapy. Relative MCL1 (left panel) and BBC3 (PUMA) (right panel) and BAX mRNA expression level for CLL patients 1 and 2 pre- (blue) and post (red)-VEN therapy determined by bulk 3′RNA-seq. (C) Results for Bax, Bbc3, and Mcl1 guide RNAs from CRISPR/Cas9-Screen in murine lymphoma cell line after 28 days with/without VEN (10 nM). (D) Immunoblot for MCL1, BCL-xL, BCL2, PUMA, and BAX in 2 T-PLL patients before and after VEN resistance. (E) Immunoblot for MCL1, BAK1, PUMA, and BAX in 3 sensitive and S63845-resistant B-cell lymphoma cell lines, respectively. (F) Densitometric analyses of immunoblots against MCL1, BAK1, PUMA, and BAX normalized to β-actin. Data illustrated as mean ± SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .001, compared with parental (blue) cells, Student t test. 3′RNA-seq, 3’RNA-sequencing; IC50, median inhibition concentration.
Because BAX plays an important role as a direct executioner of apoptosis, we analyzed samples from 6 VEN (single agent)-treated high-risk CLL patients.13,34 At the time of VEN resistance, 3 patients (numbers 1, 3, and 6) had a regular CLL morphology, whereas 2 (numbers 2 and 4) showed morphological signs of Richter’s transformation. One patient (number 5) showed a high Ki67 index (∼50%) without signs of transformation. Three patients (numbers 2, 4, and 5) did not show measurable BAX expression (Figure 1B), supporting our finding that VEN resistance is frequently associated with loss of BAX in cell lines and in primary patient samples. We could not detect mutations in the BAX gene in these 6 CLL samples by WES.13 We assessed expression of MCL1, BCL2, BCL-xL, BAX, and PUMA in primary material of 2 T-PLL patients before and after VEN failure. T-PLL, a high-risk hematologic malignancy, is susceptible to VEN, with encouraging clinical data.35,36 Decreased PUMA levels were the most remarkable changes detected at the time of clinically acquired VEN resistance (Figure 1D).
Resistance toward S63845 is associated with BAX mutations but independent of PUMA
Because acquired resistance toward MCL1 inhibitors has not been studied so far, we compared acquired resistance toward VEN with acquired resistance against the MCL1 inhibitor S63845 in 5 B-NHL cell lines (Figure 1A,E-F; supplemental Figures 1A-B and 2A-C). S63845-resistant cells showed an MCL1 upregulation and a reduction of BAX (Figure 1E-F; supplemental Figure 2C). WES of S63845- and VEN-resistant lines revealed genetic alterations in the coding part of BAX. In 2 cell lines (Nalm6, WSU-NHL), we observed an enrichment of a preexisting frameshift deletion in BAX (p.M38fs) upon VEN resistance. In 2 further cell lines (HBL-1, P30-OH-KUBO), we observed de novo mutations/deletions of BAX (Figure 1A; supplemental Table 4). For S63845-resistant cells, the same frameshift deletion in BAX p.M38fs was either enriched (WSU-NHL) or developed de novo (P30-OH-KUBO) (supplemental Figure 2D; supplemental Table 5). Similar to solid cancers,37,38 B-NHL cell lines with micro satellite instability (MSI) showed BAX M38fs frameshift deletions upon resistance (cancer.sanger.ac.uk/cell lines; cosmic.org) (Figure 1A; supplemental Figure 1E). Expression of BAK1, which mediates apoptosis induced by MCL1 inhibitors, was significantly reduced in P30-OH-KUBO, DOHH2, and WSU-NHL cell lines. PUMA expression remained unaltered in 4 of 5 MCL1i-resistant cell lines (Figure 1E-F; supplemental Figure 2C), indicating that PUMA downregulation is specific for VEN resistance but less for S63845 resistance.
BBC3/PUMA expression in VEN-resistant lymphoma cell lines and primary CLL cells is mediated by DNA methylation
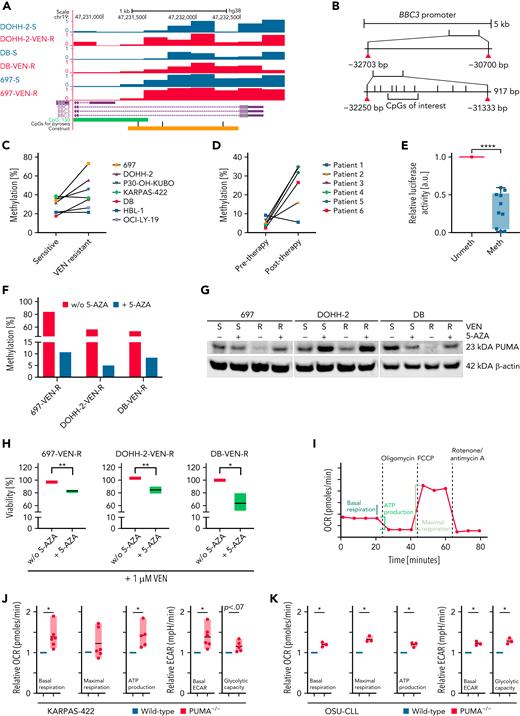
PUMA expression was the most striking difference between VEN- and S63845-resistant cell lines (Figure 1A,E; supplemental Figures 1A-B and 2D). A knockout for PUMA in KARPAS-422 cells and in mouse embryonic fibroblasts with Puma−/− showed significantly reduced sensitivity toward VEN (supplemental Figure 3D-G). Because PUMA mRNA expression was reduced (supplemental Figure 3A), MeDIP-seq was performed. A principal component analysis showed no global alterations of methylation upon VEN exposure (supplemental Figure 3B). However, the promoter region of BBC3 of the cell lines with initially high sensitivity toward VEN contained a region with an increase in DNA methylation after acquired resistance (Figure 2A; log2 fold change, 0.99; P < .001). For specific analyses of CpG methylation in the PUMA promoter region, we examined 3 CpGs by pyrosequencing (Figure 2B). One CpG (chr19: 47 231 698) revealed an increased methylation level after VEN treatment in 4 of 7 cell lines by 10.8% to 39.8% (mean, 21.8%; SD, 9%; P30-OH-KUBO, DOHH-2, 697, DB) (Figure 2C). All 4 cell lines with increased methylation level showed a decrease of PUMA on transcript and protein level upon VEN treatment (Figure 1A; supplemental Figures 1A-B and 3A). In contrast to VEN-resistant cell lines, changes in BBC3 promoter methylation could not be detected in cell lines with acquired resistance toward S63845 (supplemental Figure 2E). In primary patient samples before and after VEN treatment, we observed an increase in DNA methylation by 10% to 30% (Figure 2D) in 5 of 6 cases, suggesting that also in the clinical setting PUMA promoter methylation occurs at resistance.
Effect of VEN on the expression of BBC3 in B-cell lymphoma cell lines and primary CLL cells. (A) Schematic representation of MeDip-seq results. (B) Schematic drawing of BBC3 promoter region. For the Dual-Glo Luciferase Assay (Figure 2E), a 917 bp big region containing the CpGs of interest was cloned in a CpG-free vector, followed by the luciferase reporter. (C-D) Methylation changes detected by pyrosequencing for the CpG of interest in cell lines (C) and primary CLL cells before and after VEN resistance (D). (E) Dual-Glo Luciferase Assay analysis of methylated (meth) and unmethylated (unmeth) versions of the promoter region of BBC3. Mean ± SD, N = 10. ∗∗∗∗P < .0001, compared with unmethylated reporter construct, Student t test. (F) Methylation of BBC3 promoter region in 5′AZA-treated (5 passages) VEN-resistant B-cell lymphoma cell lines, determined by pyrosequencing. (G) Immunoblot for PUMA in 3 VEN-sensitive and -resistant B-cell lymphoma lines treated with or without 5′AZA (0.1 μM) for 5 passages. (H) Viability assay of 3 VEN-treated cell lines (24 hours, 1 μM) after incubation with 5′AZA (5 passages). Mean ± SD of 3 independent experiments, viability determined by flow cytometry. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, compared with untreated (-5-AZA) cells, Student t test. (I) Schematic analysis of OCR analysis. (J-K) Mitochondrial respiration and glycolysis in PUMA-KO KARPAS-422 (J) and OSU (K) cells upon injection of the Seahorse Mito Stress test drugs. Data are shown as floating bars (min. to max.) and are representative of 3 to 6 independent experiments. Paired 2-tailed Student t test: ∗P < .05; ∗∗P < .01. ECAR, extracellular acidification rate; KO, knockout; w/o, without.
Effect of VEN on the expression of BBC3 in B-cell lymphoma cell lines and primary CLL cells. (A) Schematic representation of MeDip-seq results. (B) Schematic drawing of BBC3 promoter region. For the Dual-Glo Luciferase Assay (Figure 2E), a 917 bp big region containing the CpGs of interest was cloned in a CpG-free vector, followed by the luciferase reporter. (C-D) Methylation changes detected by pyrosequencing for the CpG of interest in cell lines (C) and primary CLL cells before and after VEN resistance (D). (E) Dual-Glo Luciferase Assay analysis of methylated (meth) and unmethylated (unmeth) versions of the promoter region of BBC3. Mean ± SD, N = 10. ∗∗∗∗P < .0001, compared with unmethylated reporter construct, Student t test. (F) Methylation of BBC3 promoter region in 5′AZA-treated (5 passages) VEN-resistant B-cell lymphoma cell lines, determined by pyrosequencing. (G) Immunoblot for PUMA in 3 VEN-sensitive and -resistant B-cell lymphoma lines treated with or without 5′AZA (0.1 μM) for 5 passages. (H) Viability assay of 3 VEN-treated cell lines (24 hours, 1 μM) after incubation with 5′AZA (5 passages). Mean ± SD of 3 independent experiments, viability determined by flow cytometry. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001, compared with untreated (-5-AZA) cells, Student t test. (I) Schematic analysis of OCR analysis. (J-K) Mitochondrial respiration and glycolysis in PUMA-KO KARPAS-422 (J) and OSU (K) cells upon injection of the Seahorse Mito Stress test drugs. Data are shown as floating bars (min. to max.) and are representative of 3 to 6 independent experiments. Paired 2-tailed Student t test: ∗P < .05; ∗∗P < .01. ECAR, extracellular acidification rate; KO, knockout; w/o, without.
In a CpG-free luciferase reporter assay, luciferase activity revealed that methylation of this region results in lower promoter activity, explaining reduction of transcript level (Figure 2E). For validation, treatment with demethylating 5-azacytidine (5′AZA) resulted in demethylation of the region in 3 independent cell lines (Figure 2F). PUMA protein expression of resistant cell lines after 5′AZA treatment increased, indicating that local methylation level is causal for the aberrant expression (Figure 2G; supplemental Figure 3C). Accordingly, VEN-resistant cell lines were resensitized toward VEN upon 5′AZA treatment (Figure 2H).
Loss of PUMA results in metabolic reprogramming of lymphoma cells
Previous findings showed that mitochondrial metabolism is increased in VEN-resistant cells.19 We asked if disbalancing pro- and antiapoptotic molecules by PUMA knockout is sufficient to rewire metabolic properties of lymphoma cells. In accordance with our assumption, respiration and glucose metabolism was increased in both PUMA-depleted cell lines (Karpas-422PUMA−/− and OSUPUMA−/− cells) (Figure 2J-K; supplemental Figure 3F,H), resembling the changes upon VEN resistance (supplemental Figure 4A-B). This was also confirmed in murine settings upon knockout of Puma in mouse embryonic fibroblasts (supplemental Figure 4C). The increase in basal and maximal respiration level and oxidative phosphorylation–dependent ATP production was similar in cell lines with PUMA knockout and acquired VEN resistance (Figure 2I-K; supplemental Figure 4A). Moreover, these cell lines exhibited a higher level of basal glycolysis, as assessed by ECARs, and increased glycolytic capacity upon injection of the ATPase inhibitor oligomycin (supplemental Figure 4B). Absence of glucose from the medium did not display significant differences (supplemental Figure 4A). Upon VEN acute injection in KARPAS-422 and DOHH-2 cell lines during metabolic assays (supplemental Figure 4D-K), our data revealed an immediate yet transitory increase in ECAR upon VEN injection (supplemental Figure 4G,K). Although confirming that the drug treatment strongly decreased OCR in sensitive cells (supplemental Figure 4D-E,H-I), the absence of glucose from the medium decreased maximal respiration even in the resistant settings (supplemental Figure 4D,F,H,J).
Furthermore, cell cycle analysis of PUMA-depleted cell lines was performed. No changes could be detected between PUMA-depleted cells and control cells (supplemental Figure 3J-K).
Overall, our data demonstrate that VEN resistance leads to an increased cellular metabolism and show that these metabolic changes can be mediated by loss of PUMA.
VEN resistance can be overcome by MCL1 inhibition in BAX-proficient cell lines
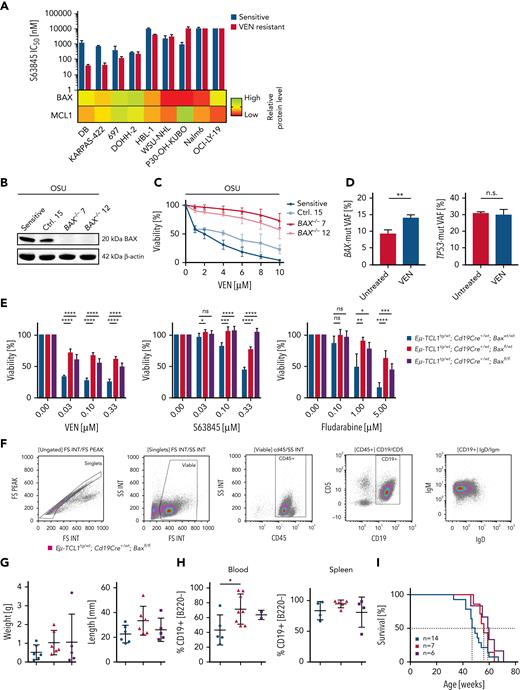
Because MCL1 was upregulated in all resistant cell lines, we analyzed whether MCL1 inhibitors like S63845 can overcome VEN resistance. Biomarkers predicting treatment efficacy toward MCL1 inhibitors are urgently needed.39,40 Five out of 9 VEN-resistant cell lines were equally or even significantly more susceptible toward MCL1 inhibition than their parental lines (Figure 3A), with 4 of them at low nanomolar IC50s (supplemental Figure 5A). However, the remaining 4 VEN-resistant cell lines were as resistant or even more resistant toward MCL1 inhibition (supplemental Figure 5A). These results indicate that VEN resistance can be overcome by MCL1 inhibition in some instances. As most cell lines showed significantly increased MCL1 protein, this did not differentiate the MCL1 inhibitor responders. In contrast to this, low or absent BAX expression predicts insensitivity toward S63845 (Figure 3A).
MCL1 inhibition (S63845) cannot eliminate BAX-deficient VEN-resistant cells. (A) Top: mean IC50 for S63845 of 9 VEN-sensitive (blue) and VEN-resistant cell lines (red) determined by flow cytometry after 48 hours. N ≥3. Lower part: heat map with relative BAX and MCL1 protein level in VEN-resistant cell lines. (B) Validation of BAX KO in OSU cells by immunoblot. N = 3. (C) Sensitivity of OSU KO cells toward VEN determined by flow cytometry after VEN treatment for 48 hours. Mean ± SD of 4 experiments. (D) Allelic fraction of TP53 (c.515T>A) and BAX (c.361del) before and after VEN treatment (24 hours, 5 nM) in a high-risk CLL patient. Mean plus SD, 3 technical replicates; P = .0061 (E) Viability of purified, malignant splenic B cells of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxwt/wt (blue), Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red) and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple) mice treated with VEN, S63845 (24 hours), or fludarabine (48 hours), determined by MTT assays. Mean ± SD of 2 experiments; 3 technical replicates each. (F) Immunophenotyping of splenocytes of a 50-week-old Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl mouse. Gating strategy of viable, single cells on FSC/SSC dot plot and FSC-area/FSC-height dot plot. Analysis for Cd45, Cd19, IgM, and IgD expression. (G) Spleen weight and length of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxwt/wt (blue; n = 6), Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red; n = 6), and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple; n = 5) mice. Mean ± SD. ∗P < .05, compared with Bax wild-type mice, Student t test. (H) Determination of the amount of Cd19+/B220dim/neg-positive cells in the blood (left panel; 32-week-old animals; n = 5, n = 8, and n = 2, respectively) and splenocytes (right panel; 44 weeks old animals; n = 4, n = 5, and n = 4, respectively) of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxwt/wt (blue), Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red), and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple) mice. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001, compared with Bax wild-type mice, Student t test. (I) Kaplan-Meier curves of overall survival of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxwt/wt (blue; 48 weeks; n = 14), Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red; 56 weeks; n = 7), and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple; 59.5 weeks; n = 6) mice. Survival of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red), and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple) compared with the respective controls (log-rank test; P = .0914). FS, forward scatter integral; FSC, forward scatter; IgM, immunoglobulin M; INT, integral; KO, knockout; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromid; ns, not significant; SSC, side scatter integral.
MCL1 inhibition (S63845) cannot eliminate BAX-deficient VEN-resistant cells. (A) Top: mean IC50 for S63845 of 9 VEN-sensitive (blue) and VEN-resistant cell lines (red) determined by flow cytometry after 48 hours. N ≥3. Lower part: heat map with relative BAX and MCL1 protein level in VEN-resistant cell lines. (B) Validation of BAX KO in OSU cells by immunoblot. N = 3. (C) Sensitivity of OSU KO cells toward VEN determined by flow cytometry after VEN treatment for 48 hours. Mean ± SD of 4 experiments. (D) Allelic fraction of TP53 (c.515T>A) and BAX (c.361del) before and after VEN treatment (24 hours, 5 nM) in a high-risk CLL patient. Mean plus SD, 3 technical replicates; P = .0061 (E) Viability of purified, malignant splenic B cells of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxwt/wt (blue), Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red) and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple) mice treated with VEN, S63845 (24 hours), or fludarabine (48 hours), determined by MTT assays. Mean ± SD of 2 experiments; 3 technical replicates each. (F) Immunophenotyping of splenocytes of a 50-week-old Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl mouse. Gating strategy of viable, single cells on FSC/SSC dot plot and FSC-area/FSC-height dot plot. Analysis for Cd45, Cd19, IgM, and IgD expression. (G) Spleen weight and length of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxwt/wt (blue; n = 6), Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red; n = 6), and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple; n = 5) mice. Mean ± SD. ∗P < .05, compared with Bax wild-type mice, Student t test. (H) Determination of the amount of Cd19+/B220dim/neg-positive cells in the blood (left panel; 32-week-old animals; n = 5, n = 8, and n = 2, respectively) and splenocytes (right panel; 44 weeks old animals; n = 4, n = 5, and n = 4, respectively) of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxwt/wt (blue), Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red), and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple) mice. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001, compared with Bax wild-type mice, Student t test. (I) Kaplan-Meier curves of overall survival of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxwt/wt (blue; 48 weeks; n = 14), Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red; 56 weeks; n = 7), and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple; 59.5 weeks; n = 6) mice. Survival of Eμ-TCL1tg/wt; Cd19Cre+/wt; Baxfl/wt (red), and Eμ-TCL1tg; Cd19Cre+/wt; Baxfl/fl (purple) compared with the respective controls (log-rank test; P = .0914). FS, forward scatter integral; FSC, forward scatter; IgM, immunoglobulin M; INT, integral; KO, knockout; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromid; ns, not significant; SSC, side scatter integral.
Loss of BAX mediates drug resistance but does not alter aggressiveness and kinetics of CLL development in a mouse model
To define the role of BAX deficiency in CLL, we employed 3 different systems. First, we generated OSU cell lines with BAX knockout and tested for susceptibility toward VEN. Loss of BAX induced resistance toward VEN in OSU cells (Figure 3B-C). Next, we investigated cells of the CLL patient with mutated BAX and TP53 alleles. These primary CLL cells were treated with VEN for 24 hours and surviving cells were isolated and subjected to next generation sequencing. Although the mean variant allelic frequency for BAX (c.361del) remained at 9.3% in untreated cells, it increased to 14% in the surviving fraction after VEN exposure (P = .0061), indicating that cells carrying the BAX mutation were enriched during VEN treatment. At the same time, the variant allele frequency of TP53 mutations remained stable (Figure 3D; supplemental Figure 5B).
As we could show that BAX loss was found in CLL patients with more aggressive course of disease after VEN resistance (Figure 1B), we generated a compound-mutant mouse to define the role of Bax for CLL development and drug resistance. Baxfl (www.jax.org) mice were crossbred with Cd19Cre/wt and EμTCL1tg/wt mice to achieve a conditional Bax knockout in malignant B cells (supplemental Figure 5C). We purified CLL-like cells from spleens of diseased animals and treated these cells ex vivo with VEN, S63845, and fludarabine. Bax loss resulted in severe drug crossresistance not only against VEN but also against MCL1 inhibition and fludarabine (Figure 3E). Surprisingly, the reduction of Bax in EμTCL1tg/wt;Cd19Cre+/wt;Baxfl/wt or EμTCL1tg/wt;Cd19Cre+/wt;Baxfl/fl mice did not result in altered kinetics of the disease (Figure 3I). In contrast to our findings in TP53-deleted mice, we did not find signs of transformation, because all cells exhibited the canonical IgM+/Cd19+/Cd5+ immunophenotype (Figure 3F).41 Spleen sizes, blood cell parameters, and amount of malignant B cells within the spleen were similar, only the CLL cell count in the peripheral blood was higher (Figure 3G-H; supplemental Figure 5D-F). In contrast to loss of Tp53 or constitutively active Akt signaling, Bax loss is no driver of transformation in EμTCL1 mice.41,42 However, in contrast to Tp53 mutations, Bax is crucial for susceptibility toward VEN (Figure 3C-D). In addition, in TCL1 wild-type mice, we did not find any signs of B-cell expansion or unphysiological B-cell subsets in Bax knockouts until the age of 83 weeks (supplemental Figure 5G-J).
Cells with acquired VEN resistance are susceptible toward extrinsic apoptosis
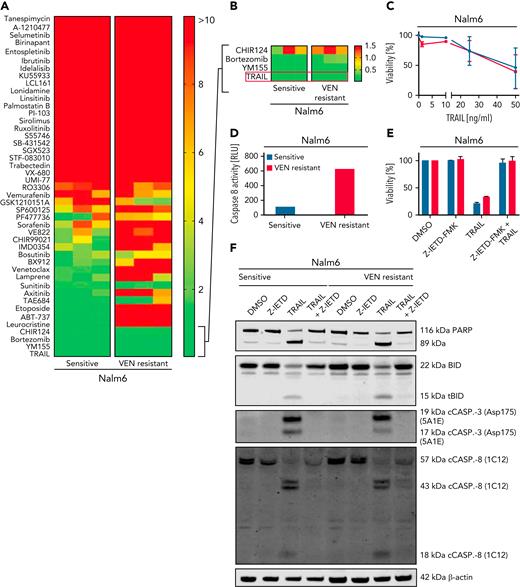
To analyze crossresistance of VEN, we performed a compound screen with 45 compounds in Nalm6 VEN-sensitive or -resistant cells (Figure 4A; supplemental Table 2). Nalm6 cells with VEN resistance showed crossresistance toward DNA-damaging drugs and different tyrosine kinase inhibitors (Figure 4A-B). Results were similar to compound screens performed in VEN-resistant OSU and DB cells, respectively (supplemental Figure 6A).
VEN resistance can by overcome by activation of the extrinsic apoptotic pathway. (A) Heat map showing IC50 values of sensitive and VEN-resistant Nalm6 cells for 45 compounds: red, IC50 ≥10 μM; yellow, IC50 ≥5 μM; and green, IC50 ≤1 μM. Cumulative results from 3 experiments. (B) Close-up view of the 3 most potent drugs identified for Nalm6 and corresponding Nalm6-VEN–resistant cells. (C) Cell death assay of sensitive and VEN-resistant Nalm6 for TRAIL (48 hours) determined by flow cytometry. Mean ± SD. N = 3. (D) Results from Caspase-Glo 8 Luminescent Assay in sensitive and VEN-resistant Nalm6 cells after treatment with TRAIL (50 ng/mL, 4 hours). N = 1. (E) Viability of sensitive and VEN-resistant Nalm6 cells after incubation with caspase-8 inhibitor Z-IETD-FMK (25 μM, 6 hours) and/or TRAIL treatment (200 ng/mL, 4 hours) determined by flow cytometry. N = 2. (F) Immunoblots for (t)BID, caspase-3, caspase-8, and PARP isoforms in Z-IETD-FMK and/or TRAIL treated cells. N = 2. DMSO, dimethyl sulfoxide.
VEN resistance can by overcome by activation of the extrinsic apoptotic pathway. (A) Heat map showing IC50 values of sensitive and VEN-resistant Nalm6 cells for 45 compounds: red, IC50 ≥10 μM; yellow, IC50 ≥5 μM; and green, IC50 ≤1 μM. Cumulative results from 3 experiments. (B) Close-up view of the 3 most potent drugs identified for Nalm6 and corresponding Nalm6-VEN–resistant cells. (C) Cell death assay of sensitive and VEN-resistant Nalm6 for TRAIL (48 hours) determined by flow cytometry. Mean ± SD. N = 3. (D) Results from Caspase-Glo 8 Luminescent Assay in sensitive and VEN-resistant Nalm6 cells after treatment with TRAIL (50 ng/mL, 4 hours). N = 1. (E) Viability of sensitive and VEN-resistant Nalm6 cells after incubation with caspase-8 inhibitor Z-IETD-FMK (25 μM, 6 hours) and/or TRAIL treatment (200 ng/mL, 4 hours) determined by flow cytometry. N = 2. (F) Immunoblots for (t)BID, caspase-3, caspase-8, and PARP isoforms in Z-IETD-FMK and/or TRAIL treated cells. N = 2. DMSO, dimethyl sulfoxide.
Our screening identified TRAIL, which induces extrinsic apoptosis, as a promising candidate to induce BAX-independent cell death (Figure 4A-F). We determined the activity of soluble TRAIL in all resistant cell lines (supplemental Figure 6B-F). TRAIL was efficient in cell lines with significantly decreased BAX levels, suggesting that TRAIL kills the respective cells independently of BAX (supplemental Figure 6B). To confirm this observation, TRAIL-mediated cell death was measured in the BAX-knockout OSU cells. Data demonstrate an equal sensitivity and caspase-8 activation independent of BAX expression (supplemental Figure 6E-F). Hence, TRAIL seems a promising salvage strategy to overcome VEN resistance. Similar to VEN-resistant cell lines, 3 of 5 S63845-resistant cell lines were sensitive toward TRAIL independent of their BAX level (supplemental Figure 6G).
The compound screen revealed another substance with high sensitivity in VEN-resistant cell lines, YM155, a BIRC3/survivin inhibitor. YM155 diminishes BAX-induced apoptosis and induces a downregulation of MCL1.43 In line with this, BAX-proficient cell lines exhibited susceptibility toward YM155, whereas IC50s in BAX-deficient cell lines were significantly higher (supplemental Figure 6H-I). To rule out that defective MCL1 downregulation was the reason for inefficiency in BAX-deficient cell lines, we investigated the expression of MCL1 after YM155 treatment. In fact, MCL1 was also efficiently downregulated independent of caspases in P30-OH-KUBO cells (supplemental Figure 6J). These data support our prior finding that loss of BAX can hardly be overcome by MCL1 inhibition.
Because death receptors and caspase-8 are also involved in necroptosis, we asked whether VEN resistance affects this type of programmed cell death. We used tumor necrosis factor α (TNFα) or TRAIL in combination with inhibitors of caspase-8 (emricasan) and cellular inhibitors of apoptosis (cIAPs; birinapant) to induce necroptosis.44 As proof of concept, we inhibited receptor-interacting protein kinase 1 (GSK’963), a mediator of necroptosis.45 VEN-resistant Nalm6 cells are sensitive toward TRAIL- and TNFα-mediated apoptosis but not necroptosis. The same observation was made in DOHH-2 cell line (supplemental Figure 7).
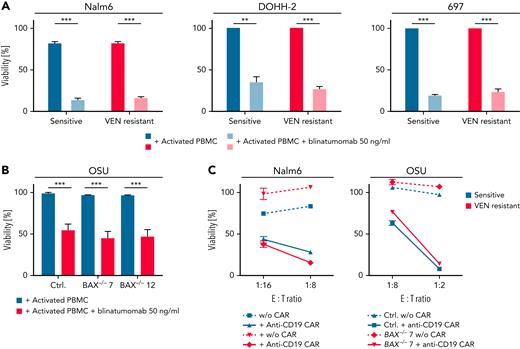
In a next step, we investigated if CAR T cells and CD3:CD19 bispecific antibodies, both promising agents to target (refractory) leukemia and lymphoma, can overcome VEN resistance. T cells from healthy donors were costimulated with CD3/CD28 and then incubated with and without the CD3:CD19 bispecific antibody blinatumomab and the target cells. In both VEN-resistant cell lines Nalm6, DOHH-2, 697, and the OSU-BAX knockout cells, highly effective and specific killing could be observed. (Figure 5A-B). There was no difference in the efficacy in killing of sensitive and resistant or BAX-proficient and -deficient cells (Figure 5A-B).
Immunotherapeutic approaches are able to overcome BAX-dependent and -independent VEN resistance. (A-B) Viability assays of Nalm6, DOHH-2, 697, and OSU cells (Ctrl, BAX−/− #7, BAX−/− #12) incubated with activated PBMCs (tumor:effector ratio of 10:1) and blinatumomab (50 ng/mL, 24/48 hours). Percentage of annexin-V+, CD4, and CD8− cells was determined by flow cytometry, normalized to PBMC cocultured, untreated cells. N = 4; P < .001; paired t test. (C) Viability of sensitive and VEN-resistant Nalm6 cells and OSU-CLL wild-type and BAX−/− clones after treatment with anti-CD19 CAR T cells determined by an XTT assay. Representative experiment of N = 3 experiments shown. Susceptibility toward CAR T-cell killing was not statistically different between sensitive and resistant cells. ∗∗P < .01; ∗∗∗P < .001. Ctrl, control; w/o, without.
Immunotherapeutic approaches are able to overcome BAX-dependent and -independent VEN resistance. (A-B) Viability assays of Nalm6, DOHH-2, 697, and OSU cells (Ctrl, BAX−/− #7, BAX−/− #12) incubated with activated PBMCs (tumor:effector ratio of 10:1) and blinatumomab (50 ng/mL, 24/48 hours). Percentage of annexin-V+, CD4, and CD8− cells was determined by flow cytometry, normalized to PBMC cocultured, untreated cells. N = 4; P < .001; paired t test. (C) Viability of sensitive and VEN-resistant Nalm6 cells and OSU-CLL wild-type and BAX−/− clones after treatment with anti-CD19 CAR T cells determined by an XTT assay. Representative experiment of N = 3 experiments shown. Susceptibility toward CAR T-cell killing was not statistically different between sensitive and resistant cells. ∗∗P < .01; ∗∗∗P < .001. Ctrl, control; w/o, without.
In a second approach, we generated CAR T cells against CD19 and incubated them with Nalm6 (sensitive/resistant) and OSU cells (BAX wild-type and knockout) (Figure 5C). Again, we observed equal frequencies in cell death in both sensitive and resistant cell lines.
Together, our data highlight that immunotherapeutic approaches inducing extrinsic apoptosis can efficiently target VEN-resistant cells with BAX deficiency.
Discussion
Although VEN is increasingly used for the therapy of hematologic malignancies, the frequency of resistance to VEN is also increasing. The underlying mechanisms for VEN resistance are not completely understood, and strategies to overcome this resistance are needed. Here, we investigated mechanisms of resistance against VEN in high-risk CLL, T-PLL, and human and murine B-cell lymphoma in more detail.
Resistance toward VEN was mediated by methylation of the PUMA promoter and subsequent downregulation of PUMA in cell lines from different lymphoma and leukemia entities. Aberrant methylation patterns were also observed in relapsed CLL samples with 2 to 6 prior treatments that were almost unmethylated at this particular CpG before VEN treatment. Interestingly, PUMA regulation is specific for VEN and not due to acquired resistance toward MCL1i or chemotherapy. Indeed, the relevance of PUMA loss is supported by findings in a mouse model for Burkitt’s lymphoma, where Puma deletion resulted in accelerated lymphomagenesis mediated by reduced apoptosis.46,47 Moreover, it was shown that substances (eg, statins) that are able to increase PUMA expression lead to higher susceptibility toward VEN.48,49 5′AZA treatment reverted methylation of the identified CpG with upregulation of the PUMA protein and resensitized resistant cells toward VEN. In line with our data, a recently published manuscript by Fresquet et al suggests a mechanistic rationale for synergistic effects of hypomethylating agents and VEN.50
We identified increased respiration and glucose metabolism in 3 different models with PUMA loss. This finding stands in contrast to a recent report where high-level of PUMA was shown to enhance glycolysis by suppressing pyruvate-driven oxidative phosphorylation in hepato-cellular carcinoma.51 The discrepancy may be explained by different roles of PUMA in hematological malignancies,46 where it is often deleted, and hepatocellular carcinoma, where PUMA shows robust expression.51,52 Our data implies that the role of PUMA for treatment resistance is far more complex as it results from specific treatment and is independent of the TP53 status of the cells. Indeed, PUMA is able to induce apoptosis in Bid−/−, Bim−/−, Bid−/−;Bim−/−, Bax−/−, and Bak−/− lines with equal efficacy.53
We and others reported earlier that MCL1 upregulation is a common observation in VEN-resistant settings.18,19 Although we identified altered phospho-p38 signaling as a reason for MCL1 upregulation, others reported cytogenetic aberrations like amplification 1q18,19,54. In acute myeloid leukemia, sustained MAPK/extracellular signal-regulated kinase signaling led to increased levels of MCL1 and confers resistance to BCL2 inhibitors.55,56 In follicular lymphoma, increased MAPK/extracellular signal-regulated kinase signaling led to resistance without alterations in MCL1 expression.57 BAX expression has not been investigated in CLL under VEN treatment so far. VEN-resistant cell lines show equal or significantly higher sensitivity toward S63845 compared with the corresponding original cell lines, supporting the strategy to overcome VEN resistance by MCL1 as a mono substance or in addition to VEN.58 A common feature of the cell lines that responded worse to S63845 was a very low or absent BAX level. We conclude that BAX is a central mediator of resistance toward BCL2 and MCL1 inhibition. We identified enrichment of preexisting or de novo aberrations in BAX in B-cell lymphoma cell lines and a BAX mutation in 1 CLL patient. These observations are consistent with the occurrence of deleterious mutations BAX in a cohort of patients with progressive CLL from early-phase VEN monotherapy trials as well as other single reports (in both CLL and mantle cell lymphoma) in the literature.50,59,60
In our cell lines, the MSI status correlated with M38fs frameshift deletion after VEN resistance. Therefore, the correlation of MSI and VEN response in DLBCL is worth to be determined in trials as up to 10% of DLBCLs might harbor MSIlow and up to 3% MSIhigh.61 Furthermore, the role of BAX for DLBCL in molecular subgroups and for novel treatment options62-64 needs to be investigated.
Because we found downregulation of BAX in VEN-resistant CLL patients with Richteŕs transformation or more aggressive course of diseases, we introduced a CLL mouse model with B-cell–specific Bax loss. Intriguingly, our data suggest that Bax-associated apoptosis resistance is a major cause for therapy resistance but not for disease progression or transformation, which stands in contrast to the roles of Tp53 or constitutive active Akt signaling in TCL1 mice.41,42 Because we did not find an expansion of B cells in Bax knockout (TCL1 wild-type) mice until the age of 83 weeks, oncogenic stimulation or genomic instability seem to be necessary for development of CLL and other B-cell lymphoma. Our findings on the role of BAX are supported by data from the myeloid compartment, where BAX mutated clonal hematopoiesis occurred after VEN treatment but was not related to therapy-related myeloid neoplasms.9
Furthermore, we showed that VEN-resistant cells are prone to TRAIL- and TNFα-induced apoptosis but not necroptosis. However, resistance mechanisms toward substances used for treatment upon VEN resistance need to be studied in the future. Ultimately, our data provide evidence that immunotherapies are suitable strategies to overcome VEN resistance.65,66 Indeed, recently presented clinical data showed that VEN- and ibrutinib-resistant CLL patients were successfully treated with CAR T cells.67,68
In summary, we identified a novel resistance mechanism for VEN that is induced by epigenetic silencing of PUMA in relapsed/refractory high-risk CLL and high-grade lymphoma. These findings suggest the use of MCL1 targeting agents in patients with functional BAX. Moreover, agents that induce extrinsic apoptosis such as TRAIL, BiTE, or CAR T–redirected T cells overcome BAX deficiency in VEN-resistant lymphoid malignancies.
Acknowledgments
The authors are indebted to their patients, who provided primary material.
This work was supported by the German-Israeli Foundation for Research and Development (I-65-412.20-2016) (H.C.R.), the Deutsche Forschungsgemeinschaft (KFO-286-RP2 [L.P.F. and H.C.R.], RE 2246/13-1 [H.C.R.], and HE3552/3-2 [M. Herling]), the Stiftung Kölner Krebsforschung (L.P.F.) and Köln Fortune (grant 479/2019, L.P.F.), the Deutsche Jose Carreras Leukämie Stiftung (R12/08) (L.P.F. and H.C.R.), the Else Kröner-Fresenius Stiftung (EKFS-2014-A06 [H.C.R.] and 2016_Kolleg.19 [H.C.R.]), the Deutsche Krebshilfe (70114055, 1117240, and 70113041 [H.C.R.] and 70172788 [M. Herling]), and the German Ministry of Education and Research (BMBF e:Med 01ZX1303A [H.C.R.] and CLL-CLUE [B.E.]). R.T.U. received funding from Deutsche Krebshilfe (70113009), the Thyssen Foundation (10.16.1.028MN), the Nachwuchsforschungsgruppen NRW grant (1411ng005), and the Deutsche Forschungsgemeinschaft (UL-379/5 and SFB-1530). Deutsche Forschungsgemeinschaft (SFB-1530) (R.B., C.P.P., H.C.R., H.K., M. Hallek, and L.P.F.).
Authorship
Contribution: L.P.F. and M.R.S. conducted the research plan; D.T., L.B., M.O., C.D.H., P.N., T.F., O.M., L.M., E.L., A.d.P.G., J.v.J., I.K., E.W., J.C., E.F.-M., and J.A. performed experiments; D.T., L.B., C.G., M.O., P.N., T.-P.Y., M.L., R.H., C.P.P., S.C.S., J.H., H.A., M.P., H.K., G.K., M. Herling, H.C.R., M. Hallek, and L.P.F. analyzed the data; P.N., K.-A.K., C.P.P., R.B., B.E., R.T.U., and M. Hallek provided material, reagents, and equipment; and L.B., C.G., M.O., H.C.R., M.R.S., and L.P.F. wrote the paper.
Conflict-of-interest disclosure: L.P.F. received research funding from Abbvie, Hofmann-La Roche, and Gilead; obtained consulting and/or speaker’s honoraria and travel support from AbbVie. H.C.R. received consulting and lecture fees from Abbvie, AstraZeneca, Vertex, and Merck; received research funding from Gilead Pharmaceuticals; is a cofounder of CDL Therapeutics GmbH. M. Herling received honoraria and research funding unrelated to the data presented here by Abbvie, EDO-Mundipharma, Janpix, Janssen-Cilag, Jazz, Novartis, Roche Stemline Therapeutics, and Takeda; holds nonexclusive licenses to clone 1-21 of diagnostic TCL1A antibodies. The remaining authors declare no competing financial interest.
Correspondence: L. Beckmann, Joseph-Stelzmann-Str 26, 50937 Köln, Germany; e-mail: laura.beckmann@uk-koeln.de; and L. P. Frenzel, Joseph-Stelzmann-Str 26, 50937 Köln, Germany; e-mail: lukas.frenzel@uk-koeln.de.
References
Author notes
∗D.T., L.B., C.G., and M.O. contributed equally to this study.
†M.R.S. and L.P.F. are joint senior authors.
The MeDIP-seq data were deposited to the Gene Expression Omnibus (GEO) database (accession number GSE166577; password: kjahakyeptejvaj).
Send data sharing requests via email to the corresponding author.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal