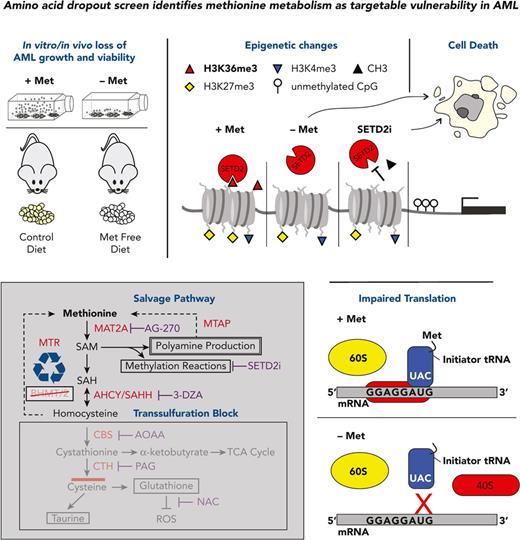
Key Points
Amino acid dropout screen reveals that AML cells highly depend on methionine, cysteine, arginine, glutamine, and lysine.
Dietary methionine removal impacts on the proteome, metabolome and epigenome, perturbing AML progression in vivo.
Abstract
Targeting altered tumor cell metabolism might provide an attractive opportunity for patients with acute myeloid leukemia (AML). An amino acid dropout screen on primary leukemic stem cells and progenitor populations revealed a number of amino acid dependencies, of which methionine was one of the strongest. By using various metabolite rescue experiments, nuclear magnetic resonance−based metabolite quantifications and 13C-tracing, polysomal profiling, and chromatin immunoprecipitation sequencing, we identified that methionine is used predominantly for protein translation and to provide methyl groups to histones via S-adenosylmethionine for epigenetic marking. H3K36me3 was consistently the most heavily impacted mark following loss of methionine. Methionine depletion also reduced total RNA levels, enhanced apoptosis, and induced a cell cycle block. Reactive oxygen species levels were not increased following methionine depletion, and replacement of methionine with glutathione or N-acetylcysteine could not rescue phenotypes, excluding a role for methionine in controlling redox balance control in AML. Although considered to be an essential amino acid, methionine can be recycled from homocysteine. We uncovered that this is primarily performed by the enzyme methionine synthase and only when methionine availability becomes limiting. In vivo, dietary methionine starvation was not only tolerated by mice, but also significantly delayed both cell line and patient-derived AML progression. Finally, we show that inhibition of the H3K36-specific methyltransferase SETD2 phenocopies much of the cytotoxic effects of methionine depletion, providing a more targeted therapeutic approach. In conclusion, we show that methionine depletion is a vulnerability in AML that can be exploited therapeutically, and we provide mechanistic insight into how cells metabolize and recycle methionine.
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous disease arising from the acquisition of mutations by hematopoietic stem or progenitor cells (HSPCs), resulting in the formation of leukemia-initiating and sustaining cells, termed leukemic stem cells (LSCs).1,2 Multiple genetically distinct subclones frequently co-occur within individual patients, further complicating treatment efficacy.3-7 Current standard of care consists of an induction regimen with chemotherapy followed by a consolidation therapy and/or a stem cell transplantation. Although they successfully de-bulk the blast population, these treatments poorly target LSC, and therefore most AML patients experience relapse.
The genetic heterogeneity is also observed at the metabolic level, where different mutations inflict specific metabolic alterations and reliances. For example, AMLs with an internal tandem duplication in the FLT3 gene (FLT3-ITD) are noncompliant with the so-called Warburg effect, relying also heavily on mitochondrial electron transport chain activity.8-10FLT3-ITD AMLs also display an enhanced dependence on the amino acid glutamine, particularly following use of FLT3-ITD inhibitors.11 Contrarily, FLT3 wild-type (wt) AMLs are often glycolytic and express elevated levels of pyruvate dehydrogenase kinase 1 (PDK1), which can be therapeutically targeted.9
LSCs have been proposed to have an overall higher level of amino acid metabolism compared to leukemic blasts, predominantly used to generate tricarboxylic acid (TCA) intermediates.12 Other studies in solid tumors reported serine synthesis to be frequently up-regulated, with increased phosphoglycerate dehydrogenase expression and serine levels promoting tumor growth.13 Colorectal cancers and lymphoma rely particularly on exogenous serine uptake, and dietary restrictions of serine reduced growth of these tumors.14,15 The Locasale group and others demonstrated that dietary methionine restriction improves the treatment outcome of colorectal soft tissue sarcoma and H3K27M mutant glioma rodent tumor models.16,17 Also, the translatability of such dietary restrictions to humans, achieving similar metabolic outcomes as in mice, has been demosntrated.16,18 Here, we performed an amino acid dropout screen in AML and identify dietary methionine removal as a potential therapeutic option.
Methods
Amino acid depletions
RPMI-1640 powder free of amino acids and glucose was purchased from USBio (#R9010-01). The powder was reconstituted in MilliQ water, before adding sodium bicarbonate (2 g/L) (Merck; #S5761) and the appropriate L-form amino acids at concentrations according to standard RPMI-1640 formulations. D-(+)-Glucose (Merck; #G8644) was added at 11.1 mM. pH was adjusted to 7.2 before adding HEPES (25 mM) (ThermoFisher; #15630106) and filter sterilizing. Subsequent methionine specific studies used methionine free RPMI-1640 liquid medium (ThermoFisher; #A1451701). Single amino acid depletion screens were performed in 96-well plates in triplicate. Cell lines were grown for 3/7 days at a density of 20 000/2000 cells, respectively. Primary AML cells and healthy CD34+ cells were grown for 7 days at 5000 wells per well in 200 μL medium. Amino acid dropouts on primary AMLs were performed on an MS5 supportive stromal layer. Cell counts were performed on a MacsQuantX (Miltenyi) or CytoFLEX (BeckmanCoulter) cytometer. Data were analyzed with FlowJo v10.7 software or CytExpert software, respectively.
Further information is provided in the supplemental Materials and Methods (available on the Blood website).
Results
Methionine, arginine, cysteine, glutamine, and lysine are strong AML dependencies
To study amino acid dependency in AML, we generated 20 media, 19 of which lacked 1 proteinogenic amino acid, and growth in these media was compared to a control amino acid complete medium (supplemental Table 1). Cell proliferation of HL60 and MOLM13 cells was strongly reduced upon depletion of methionine, arginine, cysteine, glutamine, and lysine (Figure 1A). Albeit to a lesser extent, depletion of branched chain amino acids leucine, isoleucine, valine, and threonine also reduced the proliferation rate. When repeated under hypoxia to mimic the bone marrow (BM) niche, few differences were observed, with the exception of reduced dependencies for branched chain amino acids as well as glutamine and tyrosine (supplemental Figure 1A). Alanine is typically absent from RPMI-1640, the medium of choice for many AML cell lines, but it is present in media used for primary AML or stem cell culture such as α−minimum essential medium/Iscove modified Dulbecco medium (α-MEM/IMDM). We compared growth of cell lines in liquid culture, as well as 3 primary AMLs and 1 healthy peripheral blood mononuclear cell (PBMC)−derived CD34+ sample grown both in liquid culture and on MS5 in media lacking and containing both physiological and α-MEM based concentrations of alanine. No differences in growth were observed, suggesting that the cells have sufficient de novo synthesis capacity to deal with low exogenous alanine supply (supplemental Figure 1B-D).
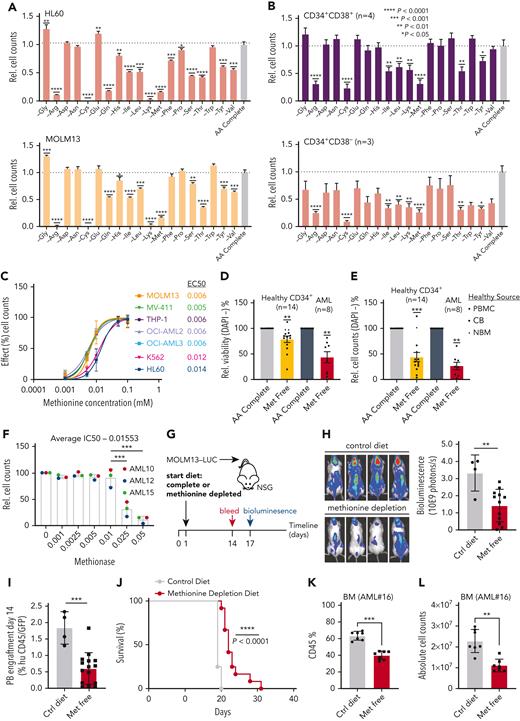
Methionine is a metabolic dependency of AML cells in vitro and in vivo. (A) Relative viable cell number of the AML lines MOLM13 and HL60 grown in amino acid dropout media compared to amino acid complete condition (n = 3). (B) Relative viable cell number of 4 primary AML samples (AML#1-4) divided into the presumed stem (CD34+CD38−) and progenitor (CD34+CD38+) populations and grown on MS5 cells for 7 days in amino acid dropout conditions (n = 4). One AML was entirely CD34+CD38+. (C) EC-50 of AML cell lines grown in a concentration range of methionine (n = 3-5). Effect is the percent stimulation in cell growth with methionine. (D) Relative viability (DAPI−) of healthy CD34+ cells derived from PBMCs (n = 8), normal bone marrow (NBM) (n = 2), cord blood (CB) (n = 4), and from 8 primary AML patient samples (CD34-enriched in the case of NPM1wt AML, mononuclear cells in the case of NPM1cyt AML) (AML#3, 5-11) grown in the absence and presence of methionine for 7 days. (E) Relative viable cell numbers of CD34+ cells derived from NBM and from 8 primary AML patient samples (AML#3, 5-11) grown in the absence and presence of methionine for 7 days. (F) Relative viable cell counts of 3 primary AML samples (AML#10-11, 15) grown in a concentration range of methionase (MGL). (G) Schematic depiction of a MOLM13luc xenograft mouse study in which methionine was depleted from the diet. (H) Bioluminescence examination of AML engraftment performed on day 17 following MOLM13Luc injection. (I) Human CD45 evaluation in peripheral blood samples to assess engraftment on day 14 following MOLM13Luc injection. (J) Kaplan-Meier plot of mouse survival (log-rank test was applied to compare curves). (K) PDX (AML#16) mouse bone marrow (BM) hCD45+ levels (%) at sacrifice. (L) Absolute human cell counts in BM of PDX (AML#16) mouse model. All PDX mice were sacrificed at 1 timepoint. All experiments: error bars represent mean ± standard error of the mean (SEM). Statistical analysis by Student t test or ordinary 1-way analysis of variance, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Methionine is a metabolic dependency of AML cells in vitro and in vivo. (A) Relative viable cell number of the AML lines MOLM13 and HL60 grown in amino acid dropout media compared to amino acid complete condition (n = 3). (B) Relative viable cell number of 4 primary AML samples (AML#1-4) divided into the presumed stem (CD34+CD38−) and progenitor (CD34+CD38+) populations and grown on MS5 cells for 7 days in amino acid dropout conditions (n = 4). One AML was entirely CD34+CD38+. (C) EC-50 of AML cell lines grown in a concentration range of methionine (n = 3-5). Effect is the percent stimulation in cell growth with methionine. (D) Relative viability (DAPI−) of healthy CD34+ cells derived from PBMCs (n = 8), normal bone marrow (NBM) (n = 2), cord blood (CB) (n = 4), and from 8 primary AML patient samples (CD34-enriched in the case of NPM1wt AML, mononuclear cells in the case of NPM1cyt AML) (AML#3, 5-11) grown in the absence and presence of methionine for 7 days. (E) Relative viable cell numbers of CD34+ cells derived from NBM and from 8 primary AML patient samples (AML#3, 5-11) grown in the absence and presence of methionine for 7 days. (F) Relative viable cell counts of 3 primary AML samples (AML#10-11, 15) grown in a concentration range of methionase (MGL). (G) Schematic depiction of a MOLM13luc xenograft mouse study in which methionine was depleted from the diet. (H) Bioluminescence examination of AML engraftment performed on day 17 following MOLM13Luc injection. (I) Human CD45 evaluation in peripheral blood samples to assess engraftment on day 14 following MOLM13Luc injection. (J) Kaplan-Meier plot of mouse survival (log-rank test was applied to compare curves). (K) PDX (AML#16) mouse bone marrow (BM) hCD45+ levels (%) at sacrifice. (L) Absolute human cell counts in BM of PDX (AML#16) mouse model. All PDX mice were sacrificed at 1 timepoint. All experiments: error bars represent mean ± standard error of the mean (SEM). Statistical analysis by Student t test or ordinary 1-way analysis of variance, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Next, 4 AML patient samples were sorted into immature CD34+CD38− and more committed CD34+CD38+ populations and cultured in the dropout media on MS5 BM stromal cells. Both populations shared the dependencies for methionine, cysteine, arginine, glutamine, consistent with AML cell lines, with the presumed LSC population having a higher dependency for tryptophan (Figure 1B). Confluent MS5 stromal cells were mostly not affected by the amino acid depletions, with the exception of cysteine depletion, which resulted in their detachment (data not shown).
As methionine depletion was one of the strongest dependencies, we focused our attention on this amino acid. AML cell lines showed a dose-dependent reduction in proliferation in response to methionine depletion, albeit with different sensitivities (Figure 1C). Furthermore, HL60, MOLM13, and OCI-AML2/3 showed drastically reduced viability, whereas viability of K562 and MV4-11 cells was not affected (supplemental Figure 3C-D).
Methionine dependence was then evaluated in an additional set of 7 primary AML samples and 1 AML from Figure 1B repeated (supplemental Table 2), which was compared to healthy CD34+ cells isolated from peripheral blood mononuclear cells (PBMCs), cord blood (CB), and normal bone marrow (NBM). Both cell proliferation and cell viability were reduced in the AML samples upon depletion of methionine (Figure 1D-E). The proliferation of healthy CD34+ controls was reduced upon methionine depletion (Figure 1E); however, their viability was much less affected (Figure 1D). Similar effects were observed upon use of the bacterial methionine-degrading enzyme, L-methionase (methionine-γ-lysase [MGL]) (Figure 1F; supplemental Figure 1E-H) or inhibition of MAT2A (supplemental Figure 1I-T). In vivo, we observed that a methionine depletion diet significantly reduced leukemic burden (Figure 1G-I) and enhanced overall survival in a MOLM13-luc xenograft model (Figure 1J). Leukemic burden was also decreased in a patient derived xenograph (PDX) model of AML, with both reduced BM CD45% and cell counts (supplemental Figure 2A-D; Figure 1K-L). Plasma methionine levels were significantly reduced in dietary mice (supplemental Figure 2E).
Because we noted slight differences in weight (supplementary Figure 2B), we analyzed the effects of methionine depletion on normal nonleukemic mice. Over a period of 8 weeks, healthy C57BL/6 mice showed similar reductions in total body weight (supplemental Figure 2F). Leucocyte counts were increased at week 4 but decreased at week 8; hemoglobin levels were also decreased at week 8, whereas platelet counts were unchanged (supplemental Figure 2G-I). Total T- and B-lymphocyte and natural killer cell frequencies were unchanged (supplemental Figure 2J). Looking to the HSPC compartment, megakaryocyte-erythroid progenitors (MEP) and multipotent progenitor (MPP) frequencies were increased at day 60, short-term hematopoietic stem cell (ST-HSC) frequencies were decreased, and the most immature long-term HSC (LT-HSC) or SLAM+ frequencies remained unchanged (supplemental Figure 2K-N).
Methionine deprivation induces a lower metabolic and cell cycle state in AML cells
The extracellular acidification rate (ECAR), an indicator of glycolytic lactate efflux, as well as oxygen consumption rate (OCR), were decreased in both AML cells and healthy CD34+ cells upon methionine depletion (Figure 2A-C). In contrast, arginine deprivation reduced glycolysis, but the effects on OCR were less pronounced, and depletion of glutamine in HL60 cells did not impact either of these (Figure 2A-B). Glucose consumption and lactate secretion rates were reduced upon methionine depletion (Figure 2D-E), whereas the mitochondrial membrane potential remained unaltered (Figure 2F). One–dimensional proton NMR (1D-1H-NMR) intracellular metabolite measurements revealed that, despite the presence of cysteine in methionine depletion conditions, the downstream metabolites taurine and glutathione were decreased as well as glutamine, glutamate, the TCA intermediates succinate and fumarate, and phosphocholine (Figure 2G). A reduction in myo-inositol levels was observed only in HL60 cells.
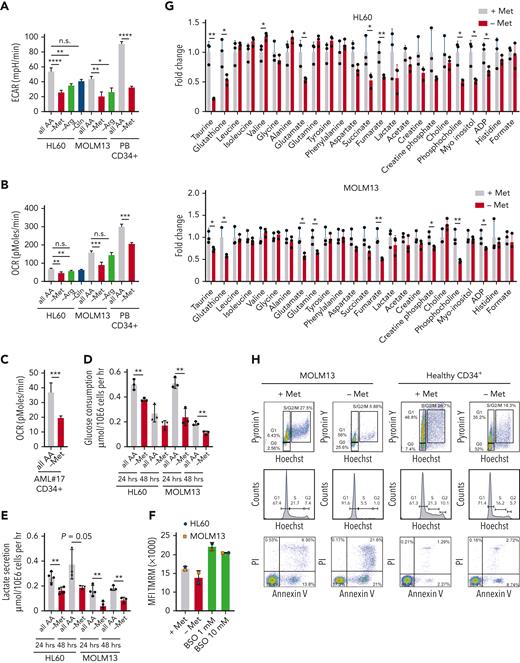
Methionine depletion induces a lower metabolic state and cell cycle block in ML cells. (A-C) The extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of AML cell lines (n = 3 biological replicates), PBMC-derived CD34+ cells (n = 1), as well as a patient AML sample (AML#17), as measured by Seahorse bioassay. (D-E) Forty-eight-hour glucose consumption and lactate production per cell per hour as determined enzymatic spectrophotometric assays (n = 3). (F) Tetramethylrhodamine, methyl ester (TMRM) mean fluorescent intensity (MFI) determined mitochondrial membrane potential (n = 3). (G) 1D 1H−nuclear magnetic resonance (NMR) spectroscopic metabolite intensity fold changes, control normalized to 1 (3 biological replicates measured once). (H) Hoechst-Pyronin Y cell cycle staining and Annexin-V PI staining of apoptosis of MOLM13 cells (representative plots of n = 3 experiments) and PBMC-derived CD34+ (n = 1) cells depleted of methionine for 24 hours. All experiments: error bars represent mean ± SEM. Statistical analysis by Student t test, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Methionine depletion induces a lower metabolic state and cell cycle block in ML cells. (A-C) The extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of AML cell lines (n = 3 biological replicates), PBMC-derived CD34+ cells (n = 1), as well as a patient AML sample (AML#17), as measured by Seahorse bioassay. (D-E) Forty-eight-hour glucose consumption and lactate production per cell per hour as determined enzymatic spectrophotometric assays (n = 3). (F) Tetramethylrhodamine, methyl ester (TMRM) mean fluorescent intensity (MFI) determined mitochondrial membrane potential (n = 3). (G) 1D 1H−nuclear magnetic resonance (NMR) spectroscopic metabolite intensity fold changes, control normalized to 1 (3 biological replicates measured once). (H) Hoechst-Pyronin Y cell cycle staining and Annexin-V PI staining of apoptosis of MOLM13 cells (representative plots of n = 3 experiments) and PBMC-derived CD34+ (n = 1) cells depleted of methionine for 24 hours. All experiments: error bars represent mean ± SEM. Statistical analysis by Student t test, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Hoechst-Pyronin Y staining revealed a G0-G1 cell cycle block upon methionine depletion, which was more pronounced in MOLM13 cells compared to healthy CB CD34+ cells (supplemental Figure 3A; Figure 2H). We also observed an overall reduction in Pyronin Y levels suggestive of a reduced total RNA pool (Figure 2H). Similar effects were observed in primary AML samples (supplemental Figure 3B). Apoptosis was induced in AML cell lines and primary AML samples upon methionine depletion, while healthy HSPCs were much less affected (Figure 2H; supplemental Figure 3C-D). Apoptosis occurred via the poly ADP ribose polymerase (PARP) cleavage pathway with increased caspase-3 cleavage (supplemental Figure 3E). Although cell proliferation was reduced in K562 cells upon methionine depletion (Figure 1C), apoptosis induction was not noted (supplemental Figure 3C-D).
Methionine deprivation is partially rescued by supplementation of S-adenosylmethionine
To determine how methionine is metabolized by AML cells, we performed supplementation experiments using metabolites derived from the methionine cycle and transsulfuration pathway in the absence of methionine. These included S-adenosylmethionine (SAM), its more cell permeable variant S-(5′-Adenosyl)-L-methionine p-toluenesulfonate salt (SAM-pTSA) that is hydrolysed to SAM intracellularly, as well as dimethyl-α-ketoglutarate (DiM-αKG), vitamin B12, folate, glutathione (GSH), taurine, and the polyamines spermine and spermidine (Figure 3A). Of these, SAM provided the only rescue (Figure 3B-G; supplemental Figure 4A-F). Despite intracellular GSH and taurine levels being reduced in the absence of methionine (Figure 2G), the lack of a rescue following GSH or taurine supplementation argued against a redox misbalance causing this phenotype. Furthermore, reactive oxygen species (ROS) levels were in fact decreased without methionine, suggesting a lower proliferative and metabolic state (Figure 3H). In addition, cysteine was present in our methionine-depleted medium, and further supplementation with the cysteine pro-drug and antioxidant N-acetylcysteine (NAC) did not rescue the phenotype (Figure 3I). In contrast, cysteine depletion resulted in increased ROS levels, which could be rescued by NAC, coinciding with restored cell proliferation (Figure 3J-L). Supplementation of cystathionine did not provide any rescue to methionine depletion but did give partial and full rescues to cysteine depletion, potentially dependent on expression of cystathionine-β-synthase (CBS) and cystathionine-γ-lyase (CTH), as their expression levels paralleled the levels of rescue (supplemental Figure 4G-M).
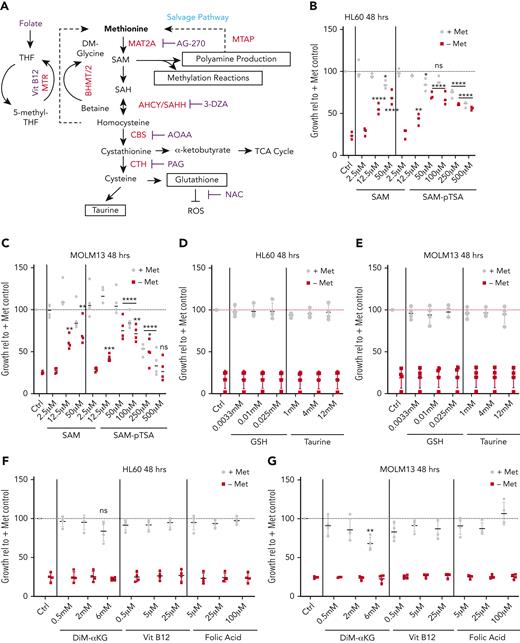
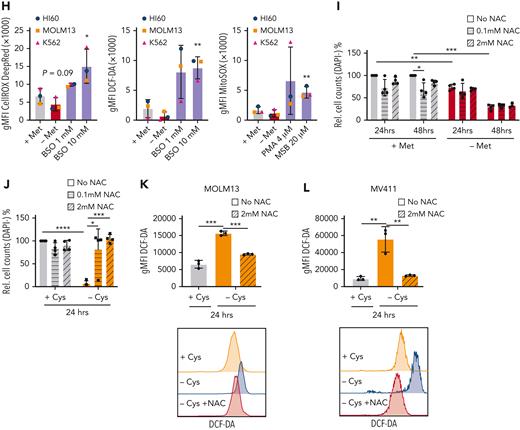
SAM rescues the depletion of methionine in an ROS-independent manner. (A) Schematic illustration of the methionine cycle and associated pathways. Homocysteine can be recycled back to methionine via the methyltransferase activity of MTR and/or BMHT/BHMT2. (B) Relative viable cell growth of HL60 cells grown for 48 hours in the absence of methionine and increasing concentrations of SAM and S-(5′-adenosyl)-L-methionine p-toluenesulfonate salt (SAM-pTSA) (n = 3). (C) Relative viable cell growth of HL60 cells grown for 6 days in the absence of methionine and increasing concentrations of SAM and SAM-pTSA. (D-E) Relative viable cell growth of HL60 and MOLM13 cells grown for 48 hours in the absence of methionine and supplemented with glutathione (GSH) and taurine (n = 3). (F-G) Relative viable cell growth of HL60 and MOLM13 cells grown for 48 hours in the absence of methionine and supplemented with dimethyl-α ketoglutarate (DiM-αKG) vitamin B12 and folic acid (n = 3). (H) ROS levels in the AML cell lines HL60, MOLM13, and K562 as determined by the total cellular ROS probes CellROX and DCF-DA and the superoxide probe MitoSOX.Buthionine sulfoximine (BSO) was used as positive control for CellROX and dichlorodihydrofluorescein diacetate (DCF-DA) and phorbol 12-myristate-13-acetate (PMA) and menadione sodium bisulfite (MSB) for MitoSOX. (I) Relative cell counts of MOLM13 cells grown in the presence or absence of methionine and N-acetylcysteine (NAC) for 24 and 48 hours (n = 4). (J) Absolute cell counts of MOLM13 cells grown with and without cysteine and in the absence or presence of 0.1 mM or 2 mM NAC for 24 hours (n = 4). (K) Graph and dotplot of ROS (DCF-DA) MFI in MOLM13 cells depleted of cysteine alone or also supplemented with 2 mM NAC (representative example from n = 3 experiments). (L) Bar graph and histogram of ROS (DCF-DA) MFI in MV4-11 cells depleted of cysteine alone or also supplemented with 2 mM NAC (representative example of n=3 experiments). Histogram representative of the 3 replicates. All experiments: error bars represent mean ± SEM. Statistical analysis by Student t test or ordinary 1-way analysis of variance (ANOVA), ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
SAM rescues the depletion of methionine in an ROS-independent manner. (A) Schematic illustration of the methionine cycle and associated pathways. Homocysteine can be recycled back to methionine via the methyltransferase activity of MTR and/or BMHT/BHMT2. (B) Relative viable cell growth of HL60 cells grown for 48 hours in the absence of methionine and increasing concentrations of SAM and S-(5′-adenosyl)-L-methionine p-toluenesulfonate salt (SAM-pTSA) (n = 3). (C) Relative viable cell growth of HL60 cells grown for 6 days in the absence of methionine and increasing concentrations of SAM and SAM-pTSA. (D-E) Relative viable cell growth of HL60 and MOLM13 cells grown for 48 hours in the absence of methionine and supplemented with glutathione (GSH) and taurine (n = 3). (F-G) Relative viable cell growth of HL60 and MOLM13 cells grown for 48 hours in the absence of methionine and supplemented with dimethyl-α ketoglutarate (DiM-αKG) vitamin B12 and folic acid (n = 3). (H) ROS levels in the AML cell lines HL60, MOLM13, and K562 as determined by the total cellular ROS probes CellROX and DCF-DA and the superoxide probe MitoSOX.Buthionine sulfoximine (BSO) was used as positive control for CellROX and dichlorodihydrofluorescein diacetate (DCF-DA) and phorbol 12-myristate-13-acetate (PMA) and menadione sodium bisulfite (MSB) for MitoSOX. (I) Relative cell counts of MOLM13 cells grown in the presence or absence of methionine and N-acetylcysteine (NAC) for 24 and 48 hours (n = 4). (J) Absolute cell counts of MOLM13 cells grown with and without cysteine and in the absence or presence of 0.1 mM or 2 mM NAC for 24 hours (n = 4). (K) Graph and dotplot of ROS (DCF-DA) MFI in MOLM13 cells depleted of cysteine alone or also supplemented with 2 mM NAC (representative example from n = 3 experiments). (L) Bar graph and histogram of ROS (DCF-DA) MFI in MV4-11 cells depleted of cysteine alone or also supplemented with 2 mM NAC (representative example of n=3 experiments). Histogram representative of the 3 replicates. All experiments: error bars represent mean ± SEM. Statistical analysis by Student t test or ordinary 1-way analysis of variance (ANOVA), ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
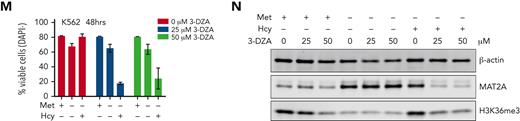
2D-NMR−based 13C methionine isotope tracing showed strong labeling of cystathionine but not metabolites further downstream such as cysteine, GSH, or taurine (supplemental Figure 5A-B). Next, we inhibited the enzymes S-adenosylhomocysteine hydrolase (AHCY/SAHH), CBS, and CTH (Figure 3A). Inhibition of SAHH with 3-deazaadenosine (3-DZA) exerted strong anti-leukemic effects in a number of AML cell lines and further reduced the effects of methionine depletion (supplemental Figure 5C-E). These effects, discussed in more detail further in the manuscript, highlight the relevance of S-adenosylhomocysteine to homocysteine progression, likely necessary to maintain the SAM:SAH ratio and methylation index. Inhibition of CBS or CTH showed anti-proliferative effects only at very high concentrations and did not accentuate the effects of methionine depletion (supplemental Figure 5F-K). Altogether, these data identify SAM as being the most essential methionine derivative in AML cells, indicative of an altered epigenome as a major contributor to the phenotype. Furthermore, we highlight a potential disconnect between the methionine cycle and transsulfuration pathway in AML, whereby de novo cysteine synthesis is absent.
Methionine depletion reduces histone methylation and is partially rescued by supplementation of SAM
We next examined the epigenetic consequences of methionine depletion. H3K36me3, H3K9me3, H3K79me2, H3K4me3, and H3K27me3 marks were reduced upon methionine depletion (Figure 4A,E). Of these, H3K36me3 was consistently the most strongly affected histone modification, for which inhibition of MAT2A phenocopied this effect (Figure 4B). Although treatment with low levels of SAM resulted in a slight further decrease in H3K36me3, at higher levels histone methylation could be restored upon treatment with SAM in a dose-dependent manner, coinciding with restored cell proliferation and viability (Figure 3B-C; Figure 4C-E). DNA methylation was also affected, albeit to a lesser extent, as we observed small but significant reductions in 5-methylcytosine (5mC) solely in HL60 cells (Figure 4F). Interestingly, 5-hydroxymethylcytostine (5hmC) levels were consistently increased in the absence of methionine (Figure 4G). Chromatin immunoprecipitation sequencing experiments in HL60 cells and CB−derived CD34+ cells revealed a dramatic decrease in H3K36me3 marks upon methionine depletion on actively transcribed gene bodies (Figure 4H-I). Around the transcription start sites, H3K4me3 levels were unchanged, whereas H3K4me1 marks actually accumulated (Figure 4H). Next, we ranked the genes with greater than 4-fold reduction in H3K36me3 levels and performed gene ontology analysis. These loci were enriched for processes such as protein translation, ribosome biogenesis, cell cycle, mitochondrial gene expression, histone modification, and anti-apoptosis (Figure 4I). Although H3K36me3 was also reduced at some of these loci in normal cells, it is clear that some of these genes such as BCL2L2 and MYC are much more highly expressed in AML vs normal CD34+ cells, and we anticipate that loss of these genes will have an even more detrimental effect on leukemic cells compared to healthy cells.
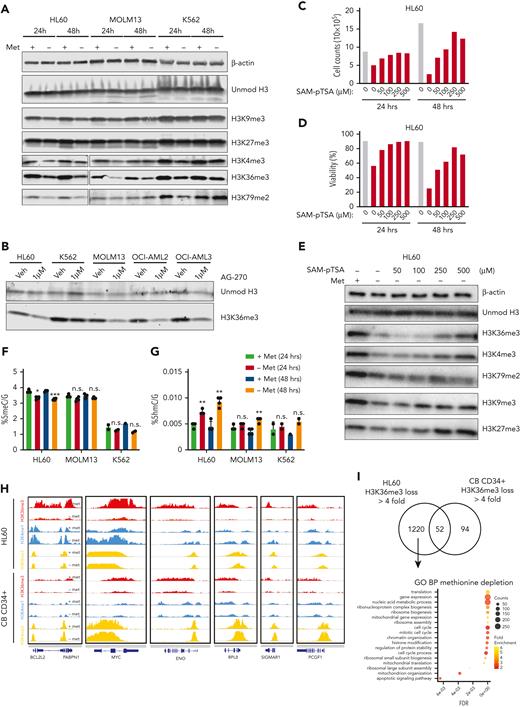
Methionine deprivation reduces histone methylation and is rescuable with SAM. (A) Western blot for histone methylation levels in AML lines HL60, MOLM13, and K562 grown for 24 and 48 hours in the presence or absence of methionine. β-Actin and unmodified histone H3 used as loading controls (B) Western blot for H3K36me3 levels in a panel of 4 AML cell lines grown with and without the MAT2A inhibitor AG-270 (1 μM) for 48 hours. (C-D) Cell counts and viability of HL60 cells grown for 24 hours in the absence or presence of methionine and without methionine but supplemented with increasing concentrations of S-(5′-Adenosyl)-L-methionine p-toluenesulfonate salt (SAM-pTSA). Graphs directly correspond to the western blot experiment shown in (E), and are replicated in Figure 3B-C with appropriate statistics. (E) Western blot for histone methyl marks levels in the AML line HL60 grown for 24 hours in the absence or presence of methionine and without methionine but supplemented with increasing concentrations of SAM-pTSA. β-Actin and unmodified histone H3 were used as loading controls (F-G) Mass spectrometeric quantification of methylated (5meC/G) and hydroxymethylated (5hmC∖G) cystosines in the cell lines HL60, MOLM13, and K562 grown in the presence or absence of methionine for 24 and 48 hours. (H) Chromatin immunoprecipitation sequencing tracks of HL60 and CB-derived CD34+ cells grown in the presence or absence of methionine for 24 hours. Shown are tracks for H3K36me3, H3K4me3, and H3K4me1 marks on a number of gene loci such as BCL2L2, MYC, ENO, etc. (I) Gene ontology analysis of processes enriched for in HL60 and CB CD34+ cells, determined from gene loci with a greater than 4-fold decrease in H3K36me3 levels. All experiments: error bars represent mean ± SEM. Statistical analysis by Student t test, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Methionine deprivation reduces histone methylation and is rescuable with SAM. (A) Western blot for histone methylation levels in AML lines HL60, MOLM13, and K562 grown for 24 and 48 hours in the presence or absence of methionine. β-Actin and unmodified histone H3 used as loading controls (B) Western blot for H3K36me3 levels in a panel of 4 AML cell lines grown with and without the MAT2A inhibitor AG-270 (1 μM) for 48 hours. (C-D) Cell counts and viability of HL60 cells grown for 24 hours in the absence or presence of methionine and without methionine but supplemented with increasing concentrations of S-(5′-Adenosyl)-L-methionine p-toluenesulfonate salt (SAM-pTSA). Graphs directly correspond to the western blot experiment shown in (E), and are replicated in Figure 3B-C with appropriate statistics. (E) Western blot for histone methyl marks levels in the AML line HL60 grown for 24 hours in the absence or presence of methionine and without methionine but supplemented with increasing concentrations of SAM-pTSA. β-Actin and unmodified histone H3 were used as loading controls (F-G) Mass spectrometeric quantification of methylated (5meC/G) and hydroxymethylated (5hmC∖G) cystosines in the cell lines HL60, MOLM13, and K562 grown in the presence or absence of methionine for 24 and 48 hours. (H) Chromatin immunoprecipitation sequencing tracks of HL60 and CB-derived CD34+ cells grown in the presence or absence of methionine for 24 hours. Shown are tracks for H3K36me3, H3K4me3, and H3K4me1 marks on a number of gene loci such as BCL2L2, MYC, ENO, etc. (I) Gene ontology analysis of processes enriched for in HL60 and CB CD34+ cells, determined from gene loci with a greater than 4-fold decrease in H3K36me3 levels. All experiments: error bars represent mean ± SEM. Statistical analysis by Student t test, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Protein translation is impaired upon methionine deprivation
Polysomal profiling (supplemental Figure 6A) in AML cell lines and PBMC-derived CD34+ cells, revealed reduced protein translation upon methionine depletion (Figure 5A-C; supplemental Figure 6B-D). These effects on translation were confirmed with puromycin pulsing of MOLM13 and HL60 cells depleted of methionine for 24 hours, followed by cell lysis and an antipuromycin immunoblot (Figure 5D; supplemental Figure 6E-F). Methionine depletion also resulted in reduced total RNA levels (Figure 5E), which was also observed in the input samples of our polysomal profiling assays (supplemental Figure 6G-I). These reductions in total RNA were also evident by reductions in Pyronin Y staining levels at 24 and 48 hours, as shown in Figure 2H and supplemental Figure 3A. Although the total protein levels were unchanged at time points up to 48 hours (Figure 5F), we observed a clear reduction of de novo protein synthesis such as for cyclin B1, MCL1, methionine synthase (MTR) and S-methyl-5'-thioadenosine phosphorylase (MTAP). Contrarily, abundant proteins such as histones with high stability remained unaffected (Figure 4A; supplemental Figure 3E and 8A).
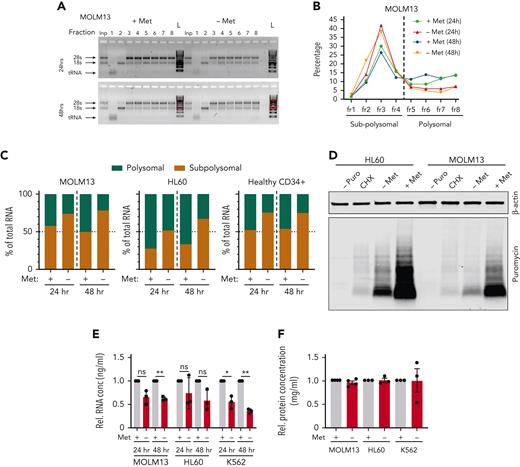
Methionine deprivation significantly impairs protein translation in AML and healthy HSPC. Polysomal profiling in the AML cell lines MOLM13 and HL60 along with PBMC-derived CD34+ cells. (A) Images of agarose gels loaded with RNA isolated from sucrose gradient input (Inp) sample (not run through gradient) and sucrose gradient− derived RNA fractions of MOLM13 cells grown for 24 and 48 hours with and without methionine. (B) Percentage of RNA in the sucrose gradients derived fractions as percentage of total RNA of all fractions in MOLM13 cells at 24 and 48 hours. (C) Percentage of polysomal and sub-polysomal RNA as a fraction of total cellular RNA in MOLM13, HL60, and PBMC CD34+ cells. Polysomal profiling was performed once for each cell type but in 3 experiments independent of each other. (D) Antipuromycin western blot performed on HL60 and MOLM13 cells following puromycin pulsing after 24 hours’ growth in the presence or absence of methionine. First lanes are a negative control with lysates from cells not exposed to puromycin and a cycloheximide-treated positive control (n = 3). Replicates in supplementary Figure 6E-F. β-Actin was used as loading control. (E) Relative total RNA of AML cell lines grown in the absence and presence of methionine for 24 and 48 hours. (F) Relative total protein concentration of AML cell lines grown with and without methionine for 48 hours. All experiments: error bars represent mean ± SEM. Statistical analysis by Student t test, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Methionine deprivation significantly impairs protein translation in AML and healthy HSPC. Polysomal profiling in the AML cell lines MOLM13 and HL60 along with PBMC-derived CD34+ cells. (A) Images of agarose gels loaded with RNA isolated from sucrose gradient input (Inp) sample (not run through gradient) and sucrose gradient− derived RNA fractions of MOLM13 cells grown for 24 and 48 hours with and without methionine. (B) Percentage of RNA in the sucrose gradients derived fractions as percentage of total RNA of all fractions in MOLM13 cells at 24 and 48 hours. (C) Percentage of polysomal and sub-polysomal RNA as a fraction of total cellular RNA in MOLM13, HL60, and PBMC CD34+ cells. Polysomal profiling was performed once for each cell type but in 3 experiments independent of each other. (D) Antipuromycin western blot performed on HL60 and MOLM13 cells following puromycin pulsing after 24 hours’ growth in the presence or absence of methionine. First lanes are a negative control with lysates from cells not exposed to puromycin and a cycloheximide-treated positive control (n = 3). Replicates in supplementary Figure 6E-F. β-Actin was used as loading control. (E) Relative total RNA of AML cell lines grown in the absence and presence of methionine for 24 and 48 hours. (F) Relative total protein concentration of AML cell lines grown with and without methionine for 48 hours. All experiments: error bars represent mean ± SEM. Statistical analysis by Student t test, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
MAT2A protein levels are increased following deprivation of SAM
We analyzed the expression of all enzymes that act in the methionine cycle and transsulfuration pathways upon methionine depletion; overall we noted no significant differences, with the exception of MAT2A, the expression of which increased significantly across multiple cell lines, both at RNA and protein level (supplemental Figure 7 and 8A-C). Previously, it was shown that reduced levels of SAM prevents the RNA methyltransferase METTL16 from methylating MAT2A transcripts, resulting in their stabilization.19 Indeed, SAM supplementation to methionine-depleted cells prevented the increase in MAT2A protein levels (supplemental Figure 8D). Also, as previously seen (Figure 4E), the reduction in H3K36me3 levels upon methionine depletion could be rescued by SAM supplementation (supplemental Figure 8D). These data show that reduced levels of SAM following methionine deprivation are likely responsible for the increase in MAT2A levels.
MTR maintains the methionine and SAM pool under methionine restriction via recycling of homocysteine
Although methionine falls into the class of essential amino acids in mammalian cells, recycling mechanisms do theoretically exist. Homocysteine (Hcy) can be re-methylated back to methionine via the enzymes methionine synthase (MTR) and/or the betaine-homocysteine S-methyltransferases (BHMT/BHMT2); alternatively, methionine can be regenerated from methylthioadenosine (MTA) via the enzyme S-methyl-5'-thioadenosine phosphorylase (MTAP) (Figure 3A). When we supplemented Hcy to AML cells in the absence of methionine, we observed a rescue in cell proliferation especially for K562 and MOLM13 cells (Figure 6A-C). Similar results were observed for ex vivo primary AML mononuclear cells (Figure 6D; supplemental Figure 9A-H). Concentrations beyond 1-2.5 mM were detrimental to cells. Furthermore, 1 mM Hcy also rescued H3K36me3 levels, evidence that it can be recycled back to methionine and further to SAM to provide methyl groups (Figure 6E). To estimate the level of baseline recycling of homocysteine to methionine, we returned to our 13C methionine tracing study data and determined the ratio of C4:C5 13C-labeled methionine molecules in the total methionine pool. As the intensity of 13C within a molecule is not always anticipated to be the same for all of the carbons within the molecule, we measured an NMR sample with uniformly 13C-labeled methionine (pure compound without cells) and took the 13C ratio between C4 and C5 as a reference (∼0.45) of non-recycled methionine. An equal ratio of C4:C5-labeled methionine was indicative of low to no recycling, at least when methionine is not restricted (Figure 6F). This is reasoned by the fact that if methionine with all 5 carbons being 13C labeled (C5) is supplemented to cells, this would be converted to homocysteine, a molecule then composed of 4 13C carbons (C4) for which an unlabeled carbon is donated by 5-methyltetrahydrofolate (via MTR) or betaine (via BHMT/2) to convert homocysteine to a 5-carbon methionine molecule of which only 4 carbons (C4) are 13C labeled. As homocysteine could rescue a methionine phenotype, likely by recycling via MTR or the BHMTs, these data indicate that homocysteine recycling occurs mostly on demand.
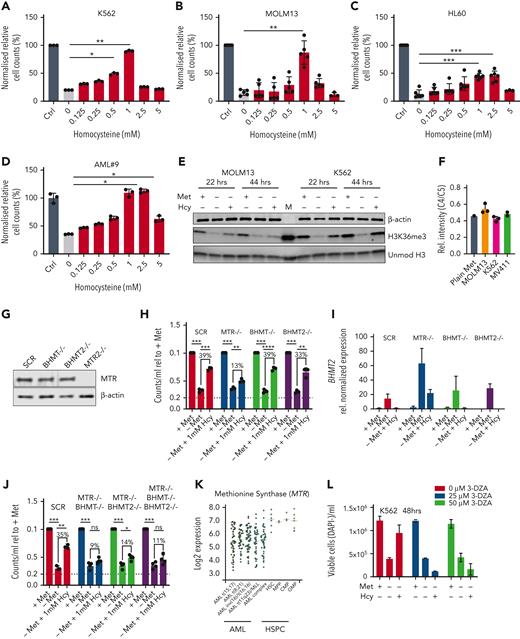
Homocysteine is recycled via MTR and not BHMTs in the absence of methionine. (A-D) Normalized relative cell counts of AML cell lines K562, MOLM13, and HL60, as well as 1 primary AML sample grown with and without methionine and in the absence of methionine but increasing concentrations of homocysteine (Hcy) (n = 3) (AML#9). (E) Western blot for H3K36me3, MAT2A, and MTR protein levels in lysates from MOLM13 and K562 cells grown for 22 and 44 hours in the presence and absence of methionine and without methionine but supplemented with 1 mM Hcy. β-Actin and unmodified histone H3 were used as loading controls. M = marker (F) Ratio of C4:C5 13C-labeled methionine in cell lines grown with universally labeled 13C methionine tracer. Data were generated by NMR spectroscopy (n = 2-3) biological replicates measured at 1 time point). (G) Western blot for MTR in K562 scrambled guide, BHMT KO, BHMT2 KO, and MTR KO cells. (H) Relative cell counts of K562 scrambled guide, BHMT KO, BHMT2 KO, and MTR KO cells grown with and without methionine and without methionine but with 1 mM Hcy. (I) Relative reverse transcription−polymerase chain reaction determined expression of BHMT2 in K562 scrambled guide, BHMT KO, BHMT2 KO, and MTR KO cells. (J) Relative cell counts of K562 scrambled guide, BHMT KO, BHMT2 KO, and MTR KO cells grown with and without methionine and without methionine but with 1 mM Hcy. (K) Transcriptome data of MTR expression in primary AML and healthy hematopoietic populations (BloodSpot.eu dataset). Probeset – 226969at. (L) Viable cell number/mL of DAPI− K562 cells grown in the presence or absence of 25 or 50 μM SAHH inhibitor (3-DZA) for 48 hours, and within the last 24 hours with and without methionine and with and without 1 mM Hcy. N = 2 experiments in K562 cells, representative of 2 independent experiments in HL60 cells. (M) Percent viable DAPI− K562 cells grown in the presence or absence of 25 or 50 μM SAHH inhibitor (3-DZA) for 48 hours, and within the last 24 hours with and without methionine and with and without 1 mM Hcy. N = 2 experiments in K562 cells, representative of 2 independent experiments in HL60 cells. (N) Western blot for H3K36me3 and MAT2A levels in K562 cells grown in the presence or absence of 25 or 50 μM SAHH inhibitor (3-DZA) for 72 hours, and within the last 24 hours with and without methionine and with and without 1 mM Hcy. β-Actin and unmodified histone H3 were used as loading controls. All experiments: error bars represent mean ± SEM. Statistical analysis: ordinary 1-way ANOVA or Student t test, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Homocysteine is recycled via MTR and not BHMTs in the absence of methionine. (A-D) Normalized relative cell counts of AML cell lines K562, MOLM13, and HL60, as well as 1 primary AML sample grown with and without methionine and in the absence of methionine but increasing concentrations of homocysteine (Hcy) (n = 3) (AML#9). (E) Western blot for H3K36me3, MAT2A, and MTR protein levels in lysates from MOLM13 and K562 cells grown for 22 and 44 hours in the presence and absence of methionine and without methionine but supplemented with 1 mM Hcy. β-Actin and unmodified histone H3 were used as loading controls. M = marker (F) Ratio of C4:C5 13C-labeled methionine in cell lines grown with universally labeled 13C methionine tracer. Data were generated by NMR spectroscopy (n = 2-3) biological replicates measured at 1 time point). (G) Western blot for MTR in K562 scrambled guide, BHMT KO, BHMT2 KO, and MTR KO cells. (H) Relative cell counts of K562 scrambled guide, BHMT KO, BHMT2 KO, and MTR KO cells grown with and without methionine and without methionine but with 1 mM Hcy. (I) Relative reverse transcription−polymerase chain reaction determined expression of BHMT2 in K562 scrambled guide, BHMT KO, BHMT2 KO, and MTR KO cells. (J) Relative cell counts of K562 scrambled guide, BHMT KO, BHMT2 KO, and MTR KO cells grown with and without methionine and without methionine but with 1 mM Hcy. (K) Transcriptome data of MTR expression in primary AML and healthy hematopoietic populations (BloodSpot.eu dataset). Probeset – 226969at. (L) Viable cell number/mL of DAPI− K562 cells grown in the presence or absence of 25 or 50 μM SAHH inhibitor (3-DZA) for 48 hours, and within the last 24 hours with and without methionine and with and without 1 mM Hcy. N = 2 experiments in K562 cells, representative of 2 independent experiments in HL60 cells. (M) Percent viable DAPI− K562 cells grown in the presence or absence of 25 or 50 μM SAHH inhibitor (3-DZA) for 48 hours, and within the last 24 hours with and without methionine and with and without 1 mM Hcy. N = 2 experiments in K562 cells, representative of 2 independent experiments in HL60 cells. (N) Western blot for H3K36me3 and MAT2A levels in K562 cells grown in the presence or absence of 25 or 50 μM SAHH inhibitor (3-DZA) for 72 hours, and within the last 24 hours with and without methionine and with and without 1 mM Hcy. β-Actin and unmodified histone H3 were used as loading controls. All experiments: error bars represent mean ± SEM. Statistical analysis: ordinary 1-way ANOVA or Student t test, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
To functionally test a potential role for MTR and BHMTs, we generated CRISPR-Cas9 knockouts (KOs) in K562 cells. KOs were confirmed by sequencing (data not shown) and by western blot for MTR (Figure 6G). BMHT and BHMT2 were undetected in our proteome and transcriptome data sets3,9,20 and were either very low or not detected in the CCLE AML cell lines data.21 Our western blot and quantitative polymerase chain reaction analyses confirmed very low expression of BHMT (data not shown) and BHMT2 (Figure 6I) in hematopoietic tissues. Therefore, we considered it highly unlikely that these proteins would be responsible for recycling homocysteine in AML cells. In homocysteine rescue experiments, only an MTR KO abrogated the rescue capacity of homocysteine (Figure 6H). BHMT2 expression was increased in the absence of methionine, particularly in the MTR KO setting, suggestive of BHMT2 compensating for a loss of MTR (Figure 6I). We therefore generated double and triple KOs of MTR with each of the BHMT isoforms, but KO of BHMTs additionally to MTR added no reduction to homocysteine rescue capacity (Figure 6J). Publicly available transcriptome datasets22 further highlighted the potential relevance of MTR in AML, as many AMLs have a much lower expression compared to healthy HSPC (Figure 6K). It is therefore possible that some AMLs are less well equipped to recycle homocysteine, raising it as an interesting target, particularly in the context of methionine deprivation or along with MAT2A inhibition.
As the conversion of S-adenosylhomocysteine (SAH) to homocysteine is reversible (Figure 3A), we further explored the inhibition of SAHH with 3-DZA in the context of homocysteine supplementation. For this we chose concentrations that verged on inducing cell death in K562 cells (supplemental Figure 5E) but that initiated a decrease in histone methylation. Homocysteine supplementation could rescue growth and viability of AML cells grown in the absence of methionine as shown before, but not when SAHH was inhibited (Figure 6L-M). Also, inhibition of SAHH was sufficient to prevent the homocysteine mediated rescue of H3K36me3 levels in the absence of methionine (Figure 6N). Given that SAH competitively inhibits methyltransferases, these data likely indicate that although MTR recycles homocysteine to methionine when methionine is restricted, the build-up of SAH caused by 3-DZA treatment prevents SAM regenerated from homocysteine from being used by methyltransferases. Interestingly, the combination of inhibiting SAHH in the context of high exogenous homocysteine levels proved rather toxic to cells (Figure 6L-M).
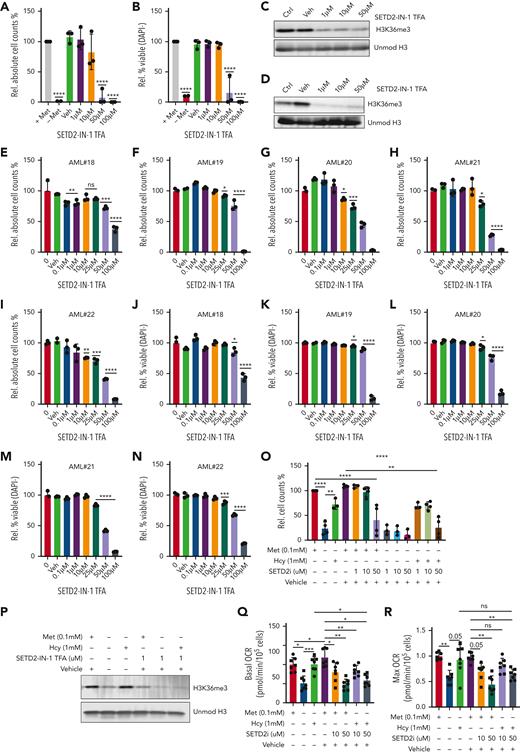
SETD2 inhibition mimics methionine depletion induced H3K36me3 reductions and reduces AML growth and viability
Given the strong impact on the epigenome following methionine depletion, with SAM and homocysteine being capable of rescuing cell growth, viability, and methylation levels, our data suggested the epigenetic effects, most notably reductions in H3K36me3, as a major contributor to the phenotype. We therefore hypothesized that targeting the H3K36-specific methyltransferase SETD2 would provide a more targeted therapeutic approach, phenocopying the epigenome effects of methionine depletion, and would aid in further pinpointing the specific source of reduced leukemic growth and viability. We tested two SETD2 inhibitors, STL135578 (Compound C13) and SETD2-IN-1 TFA. The efficacy of STL135578 could not be confirmed in our experiments, but SETD2-IN-1 TFA effectively reduced H3K36me3 levels in MOLM13 cells and led to a decrease in cell growth and survival in MOLM13 and primary ex vivo−treated AML mononuclear cells (Figure 7A-N, supplementary Figure 10). To determine whether other methionine depletion effects such as perturbed translation and lower metabolism also contribute to reduced AML growth, we took advantage of the fact that homocysteine can be recycled to methionine to presumably allow translation initiation, and farther downstream to SAM to rescue H3K36me3 levels. We supplemented homocysteine under methionine depletion in the presence and absence of the SETD2 inhibitor, with the idea that SETD2 inhibition would prevent the SAM generated via recycling of homocysteine to methionine from being loaded onto H3K36. Indeed, although homocysteine rescued the growth, viability, and H3K36me3 levels of methionine-depleted cells, as previously demonstrated, this rescue was lost by SETD2 inhibition (Figure 7O-P). Extracellular flux analysis revealed homocysteine as being able to completely rescue oxygen consumption in the absence of methionine. SETD2 inhibition perfectly phenocopied the reductions of OCR (Figure 7Q-R), suggesting that the reductions in cellular metabolism are linked to lower H3K36me3 levels. In line, the ability of homocysteine to rescue OCR in the absence of methionine was prevented by SETD2 inhibition.
SETD2 inhibition phenocopies methionine depletion inducing cell death in AML samples. (A-B) Relative cell counts and relative viability of MOLM13 cells grown for 72 hours with and without methionine and with increasing concentrations of the SETD2 inhibitor, SETD2-IN-1 TFA (n = 3). (C) Western blot for H3K36me3 levels in MOLM13 cells following 24 hours of exposure to SETD2-IN-1. (D) Western blot for H3K36me3 levels in MOLM13 cells following 72 hours of exposure to SETD2-IN-1. Unmodified histone H3 was used as loading control. (E-N) Relative viable cell counts and viability (DAPI−), both shown as percentages, in 5 primary AML samples (AML#18-22) grown for 7 days in liquid culture with a concentration range of SETD2-IN-1 TFA. The proliferation index of 4 of these 5 AMLs can be seen in supplemental Figure 9M-P, where the control conditions were the same and performed at the same time. (O) Relative cell counts for MOLM13 cells grown for 48 hours with or without methionine, with and without 1 mM homocysteine (Hcy), with SETD2-IN-1 both alone and in combination with Hcy supplementation. n = 3-4 experiments combined from the Seahorse experiments and independent homocysteine rescue experiments). (P) Western blot for H3K36me3 levels in a MOLM13 Hcy rescue experiment, with and without exposure to SETD-IN-1 TFA. β-Actin and unmodified histone H3 were used as loading controls. (Q) Basal OCR in a MOLM13 Hcy rescue of methionine depletion with and without SETD-IN-1 TFA exposure. Forty-eight-hour experiment: (R) Max OCR in a MOLM13 Hcy rescue of methionine depletion with and without SETD-IN-1 TFA exposure. All experiments: error bars represent mean ± SEM. Statistical analysis by ordinary 1-way ANOVA, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
SETD2 inhibition phenocopies methionine depletion inducing cell death in AML samples. (A-B) Relative cell counts and relative viability of MOLM13 cells grown for 72 hours with and without methionine and with increasing concentrations of the SETD2 inhibitor, SETD2-IN-1 TFA (n = 3). (C) Western blot for H3K36me3 levels in MOLM13 cells following 24 hours of exposure to SETD2-IN-1. (D) Western blot for H3K36me3 levels in MOLM13 cells following 72 hours of exposure to SETD2-IN-1. Unmodified histone H3 was used as loading control. (E-N) Relative viable cell counts and viability (DAPI−), both shown as percentages, in 5 primary AML samples (AML#18-22) grown for 7 days in liquid culture with a concentration range of SETD2-IN-1 TFA. The proliferation index of 4 of these 5 AMLs can be seen in supplemental Figure 9M-P, where the control conditions were the same and performed at the same time. (O) Relative cell counts for MOLM13 cells grown for 48 hours with or without methionine, with and without 1 mM homocysteine (Hcy), with SETD2-IN-1 both alone and in combination with Hcy supplementation. n = 3-4 experiments combined from the Seahorse experiments and independent homocysteine rescue experiments). (P) Western blot for H3K36me3 levels in a MOLM13 Hcy rescue experiment, with and without exposure to SETD-IN-1 TFA. β-Actin and unmodified histone H3 were used as loading controls. (Q) Basal OCR in a MOLM13 Hcy rescue of methionine depletion with and without SETD-IN-1 TFA exposure. Forty-eight-hour experiment: (R) Max OCR in a MOLM13 Hcy rescue of methionine depletion with and without SETD-IN-1 TFA exposure. All experiments: error bars represent mean ± SEM. Statistical analysis by ordinary 1-way ANOVA, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
Discussion
The notion that metabolism is frequently altered in cancer cells provides interesting alternative means for therapeutic targeting of leukemia.23-27 We set out to study the amino acid dependencies in human AML and uncover strong dependencies on cysteine, arginine, and methionine. Although the roles of cysteine28 and arginine29,30 in AML have previously been documented, we focused here on the role of methionine and show that methionine deprivation impacts on the proteome, metabolome, and epigenome, and that dietary methionine starvation perturbs AML progression in vivo.
We find that although the proliferation of healthy CD34+ cells is somewhat affected by depletion of methionine, the loss of viability upon its restriction is more predominant in leukemic cells. H3K36me3 across gene bodies is the most impacted epigenetic mark following methionine depletion. Although this mark was also reduced in healthy CD34+ cells upon methionine depletion, we observed much higher H3K36me3 levels on various genes such as BCL2L2 and MYC in AML cells, in line with their higher expression; and, given the high dependency of AML cells on these genes, a reduction in their expression might have more detrimental effects in AML compared to normal CD34+ cells. MAT2A converts methionine to SAM, the methyl group donor for all methylation reactions and also used in polyamine synthesis. Indeed, we find strong effects on the epigenome, in particular on histone methylation, which can be rescued by supplying exogenous SAM to cells. We hypothesize that the combined effects of methionine depletion on protein synthesis and epigenetic gene expression control are dominant factors that underlie the observed phenotypes. Supplementations of polyamines did not rescue the phenotype but were instead toxic, even at low concentrations, excluding this as a causative factor in the phenotype. The removal of a methyl group from SAM by methyltransferases generates SAH, a competitive inhibitor of methyltransferase reactions. SAH is further converted to homocysteine by the enzyme SAHH, where it feeds into the transsulfuration pathway from which cysteine and glutathione are derived (Figure 3A). Therefore, methionine is also viewed as important in ROS management. Despite this, impaired ROS management does not seem to play an important role in the phenotype. Although depletion of cysteine increased ROS levels and the observed phenotypes could be rescued by the ROS scavenger N-acetylcysteine (NAC), such rescues were not observed in methionine-depleted cells. ROS levels were also not increased following methionine depletion. Moreover, cysteine was also still present in the medium of methionine-depleted cells. In addition, our 13C-methionine tracing and cystathionine supplementation studies point to low conversion of methionine to cysteine, indicative that cysteine becomes an essential amino acid in these cells. The cleavage of cystathionine to cysteine generates α-ketobutyrate, which can be converted to succinyl-CoA to feed into the TCA cycle. Supplementation of DiM-αKG, another precursor to succinyl-CoA, did not provide a rescue, altogether arguing that the phenotype of methionine depletion does not result from lower TCA intermediate availability. 13C-methionine tracing and modulation of the expression of the enzymes CBS/CTH in the context of cysteine depletion will be important future steps for confirming the essentiality of cysteine in AML.
In line with previous studies, we find that MTR and not BHMT or BHMT2 recycles homocysteine in AML cells and that this occurs only when methionine becomes restricted.31,32 In contrast, however, we do not find supplementation of 5-methyltethrahydrofolate in cell culture as being necessary for the action of MTR. MTR expression levels appear to be lower in AML blasts compered to normal CD34+ cells, raising the possibility that AML cells have a lower capacity to recycle methionine compared to healthy CD34+ cells. Along similar lines, it was shown that reduced arginosuccinate synthetase-1 expression renders AML cells vulnerable to reduced exogenous arginine supply.30 Targeting MTR low-expressing AMLs, particularly in the context of methionine deprivation, may therefore be an interesting strategy, although no selective inhibitor yet exists. Despite a lack of availability of uniformly labeled 13C-homocysteine, we have been able to show the necessity of MTR in recycling homocysteine to methionine in AML.
Dietary strategies to target cancer are gaining increased interest.25,33-35 We show that dietary perturbation of methionine levels holds promise as a therapeutic option in delaying the progression of AML, as in solid cancers.16,17,36 Dietary restrictions are often already implemented for individuals with metabolic disorders, including those affecting amino acid metabolism such as phenylketonuria, homocystinuria, and even hypermethioninemia, for which Food and Drug Administration−approved food alternatives are already available.37-39 Although in our in vivo studies we opted for a complete removal of methionine from the diet, current clinical precedent for treating metabolic disorders as well as previous studies highlighting the clinical feasibility of methionine restriction and its ability to lower plasma methionine levels highlight that methionine restriction studies could be implemented as a therapeutic strategy for AML.16,18 Aside from dietary restrictions, the methionine-degrading enzyme MGL has proven to be safe in human trials, providing an easily implementable therapeutic strategy, and we and others have shown its efficacy in killing leukemic cells.40-42 Furthermore, knockout of MAT2A is an AML vulnerability, and we show that novel compounds such as AG-270 have potential for small molecule targeting of the methionine cycle in AML regardless of their MTAP status.43-46 We also identify that targeting the H3K36me3-specific methyltransferase SETD2 phenocopies the bulk of the anti-leukemic effect of methionine depletion, while providing a much more specific target.47 Our data also reveal the combination of SAHH inhibition along with supplementation of homocysteine as being highly cytotoxic to AML cells, warranting further in vivo testing. More studies will be required to further test and optimize such interventions, but our data highlight how amino acid metabolism can be manipulated to target genetically diverse AML cells.
Acknowledgments
The authors gratefully acknowledge Marcel de Vries and Hjalmar Permentier from the proteomics department of the ERIBA Institute Groningen for their help with methionine quantifications. The authors also thank Christina Muhs and Christian Richter for their help with the NMR spectroscopy measurements. The authors gratefully acknowledge Olav Rooyackers for his critical review of the manuscript. The authors are grateful to Marcel van Vugt for providing many of the cell cycle, apoptosis, and anti-puromycin antibodies.
These studies were financially supported by the EU (H2020-MSCA-ITN-2015-675790-HaemMetabolome) awarded to J.J.S. and U.G. In addition, A.C., A.E., and I.A. gratefully acknowledge receipt of a Marie Curie Fellowship and are participants in the same Initial Training Network. Work at BMRZ is supported by the state of Hesse.
Authorship
Contribution: A.C. and A.E. led the study from the beginning, contributed to the study design, and performed many of the experiments and data analysis, and prepared the figures for the manuscript; A.C. prepared the initial draft of the manuscript; A.E. critically reviewed the manuscript and final figures; at a later stage in the project, A.C. overtook implementation of experiments; I.A. ran all NMR spectroscopy samples and performed all of the associated analysis under the supervision of H.S.; M.G. and F.A.J.v.d.H. assisted throughout with experimental work, performing many of the metabolite supplementation experiments as well as western blots; M.P. contributed to the design of the AML mouse studies, their implementation and oversaw the planning, feeding, and sacrifice of mice; A.T.J.W. assisted with the design and cloning of short-hairpin RNAs, and participated in project discussions throughout; H.t.B. performed many of the cell line methionine titrations, worked on the methionine synthase (MTR) knockdown studies, and testing the combination of methionine depletion with decitabine; R.D. assisted with the design and performance of the polysomal profiling experiments under the supervision of C.W.; D.A.P.-M. provided samples and performed reverse transcription−polymerase chain reaction analysis as well as drug/methionine depletion combinations, the healthy mouse methionine dietary starvation study, as well as Seahorse and other experiments; I.W. helped conduct the healthy mouse dietary methionine starvation study; V.v.d.B. and S.M.H. performed chromatin immunoprecipitation (ChIP) reactions and sequencing analysis; R.K. ran all DNA methylation mass spectrometry samples as well their data analysis and interpretation under the supervision and financial support of J.H.J.; P.d.B. and M.R.H.-F. performed medium metabolite quantification in MTR genetic models, as well as the associated data analysis; U.L.G. was the awardee of the grant and partially supervised A.C.; E.M.R. provided the facilities for the healthy mouse methionine starvation study and cosupervised D.A.P.-M. and I.W.; J.H.A.M. performed the sequencing on chromatin immunoprecipitation sequencing samples; G.H. cosupervised A.C. and A.E., and was involved in discussions throughout; J.J.S. was responsible for the conceptualization of the study, supervision throughout, data analysis and interpretation of data, as well as preparation of the figures and manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Jacob Schuringa, Department of Experimental Hematology, University Medical Center Groningen, University of Groningen, Hanzeplein 1, 9700 RB, Groningen, The Netherlands; e-mail: j.j.schuringa@umcg.nl.
References
Author notes
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal