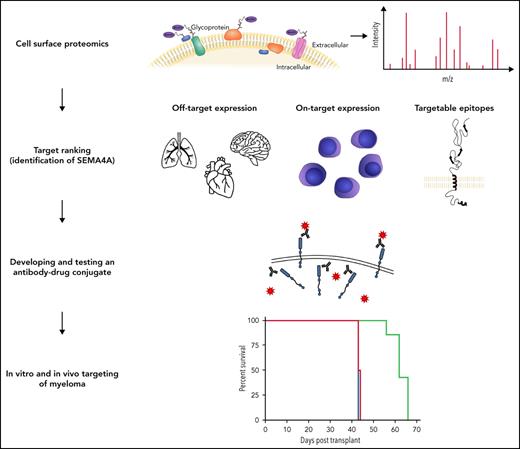
Abstract
The accessibility of cell surface proteins makes them tractable for targeting by cancer immunotherapy, but identifying suitable targets remains challenging. Here we describe plasma membrane profiling of primary human myeloma cells to identify an unprecedented number of cell surface proteins of a primary cancer. We used a novel approach to prioritize immunotherapy targets and identified a cell surface protein not previously implicated in myeloma, semaphorin-4A (SEMA4A). Using knock-down by short-hairpin RNA and CRISPR/nuclease-dead Cas9 (dCas9), we show that expression of SEMA4A is essential for normal myeloma cell growth in vitro, indicating that myeloma cells cannot downregulate the protein to avoid detection. We further show that SEMA4A would not be identified as a myeloma therapeutic target by standard CRISPR/Cas9 knockout screens because of exon skipping. Finally, we potently and selectively targeted SEMA4A with a novel antibody–drug conjugate in vitro and in vivo.
Key Points
Quantification of the primary myeloma cell surface proteome enables unbiased discovery of novel therapeutic targets.
Targeting SEMA4A using an antibody–drug conjugate potently and selectively eradicates myeloma cells both in vitro and in vivo.
Introduction
Multiple myeloma is a cancer of plasma cells. It causes fatigue, bone pain, pathologic fractures, immunosuppression, and renal failure. Despite the introduction of novel therapies in recent years, including monoclonal antibody therapy and chimeric antigen receptor (CAR) T-cell therapy, treatment resistance is inevitable. Myeloma therefore remains invariably fatal, and novel therapies are needed. Herein, we describe plasma membrane profiling1,2 of primary multiple myeloma samples and myeloma cell lines, leading to quantitation of a primary cancer cell surface proteome at unprecedented coverage. An algorithm to prioritize targets led to the identification of semaphorin-4A (SEMA4A), not previously implicated in myeloma biology. SEMA4A had desirable properties as a therapeutic target, and a tool antibody–drug conjugate (ADC) showed marked activity against myeloma in vitro and in vivo. We present our data both to highlight a valuable new target for myeloma therapy and as an exemplar of a generic approach that can be used to identify novel immunotherapy targets in other cancers.
Methods
Samples
Patients provided written informed consent according to the Declaration of Helsinki, under an established ethics protocol reviewed by a National Health Service Research Ethics Committee (reference 07/MRE05/44). Human myeloma cell lines were established to be mycoplasma-free and were genotyped by polymerase chain reaction of genomic DNA and Sanger sequencing.
Plasma membrane profiling
Plasma membrane profiling was performed as previously described,1,2 using a 1 × 108 cell line or 1 × 107 primary myeloma cells and a “one pot” oxidation and aminooxy-biotinylation reaction. Peptides were labeled with tandem mass tag reagents before pooling and fractionation using an UltiMate 3000 UHPLC system (Thermo Fisher Scientific). Samples were analyzed on a nano–liquid chromatography–mass spectrometry platform consisting of an UltiMate 3000 RSLC nano-UHPLC system (Thermo Fisher Scientific) coupled to an Orbitrap Fusion instrument (Thermo Fisher Scientific). The supplemental Methods (available on the Blood Web site) provide details on the extended protocol.
Flow cytometry and internalization assays
Flow cytometry was performed by using a Gallios (Beckman Coulter), a BD FACSCanto II (Becton Dickinson), or a BD LSRFortessa (Becton Dickinson) flow cytometer using antibodies shown in supplemental Tables 6 and 7. For the internalization assays, cells were removed at the indicated times, incubated with goat anti-mouse immunoglobulin G for 13 minutes, and fixed with 1% formaldehyde in phosphate-buffered saline (PBS) at 4°C for 30 minutes.
For immunofluorescence microscopy, NCI-H929 were incubated with unconjugated SEMA4A antibody for 30 minutes at 4°C. Unbound antibody was removed by PBS washes, and cells were incubated at 37°C. Samples were removed at time zero and at 3 hours and immediately fixed on ice in 4% formaldehyde solution for 30 minutes. Cells were then incubated with an Alexa Fluor 555 conjugated secondary antibody in 2% fetal bovine serum and 0.1% saponin (MilliporeSigma) in PBS for 1 hour at room temperature. Samples were washed and re-suspended in PBS/fetal bovine serum/saponin with lysosomal-associated membrane protein 1/Alexa Fluor 488 for 1 hour at room temperature. Cells were washed, nuclei stained using 4′,6-diamidino-2-phenylindole, and attached glass slides coated with poly-l-lysine (MilliporeSigma) for 30 minutes at 37°C. Slides were then mounted by using Prolong Diamond Antifade Mountant (Life Technologies) and images captured by using a Leica DFC7000T microscope and the Leica application suite software (Leica Microsystems). Captured images were analyzed by using ImageJ (National Institutes of Health).
Competition assays
For expression of short-hairpin RNA (shRNA), hairpins were cloned into pLKO.1 expressing green fluorescent protein (GFP). For constitutive expression of CAS9 or CAS9-KRAB, the vectors pHRSIN-3xFLAG-NLS-CAS9-NLS-WPRE-pSV40-Blast or pLKO5d.SFFV.dCas9-KRAB.P2A.BSD (Addgene, 90332) were used. For expression of single guide RNA (sgRNA), pKLV-U6-esgRNA-pGK-Puro2A-BFP was used. For CAS9/dCAS9, stable expression was achieved by selection with 10 µg/mL blasticidin (5 µg/mL for INA6). For the coculture competition assays, HS5 cells were treated with mitomycin (1 µg/mL) for 24 hours before the start of the assay, and NCI-H929 were supplemented with interleukin-6 (5 ng/mL; MilliporeSigma) where indicated. sgRNA/shRNA were transduced into cell lines using lentivirus at an infection efficiency of ∼50%. Cells were grown without selection, and the relative proportion of fluorescent (GFP or blue fluorescent protein) cells in the population was then measured over time by using flow cytometry.
ADC cytotoxicity assays
The SEMA4A antibody (5E3) was directly conjugated to either monomethyl auristatin E using a mc-vcPAB linker (maleimide-based linker, cysteine linked) or with mertansine (DM1) using an SMCC linker (N-hydroxysuccinimide–ester based, lysine linked) by Abzena. An isotype-matched control, trastuzumab, directly conjugated to monomethyl auristatin E (mc-vcPAB linker) was also provided by Abzena.
Cells were seeded at 5 × 103 per well in a 96-well plate in complete RPMI 1640. Antibody-ZAP complexes (Advanced Targeting Systems; produced according to manufacturer's instructions) or ADCs were added to the cells and plates incubated for 72 hours at 37°C and 5% carbon dioxide. Cell viability was determined by XTT assay (Biotium) as per the manufacturer's guidelines and calculated as a percentage of the control (media only) wells. For primary cells, CD138+ and CD14+ cells were isolated by using microbeads (Miltenyi Biotec), seeded at 5 to 10 × 103 per well and incubated for 72 hours with ADC or vehicle. Cell viability was determined by using CellTiter-Glo (Promega) per the manufacturer's guidelines or by flow cytometry using Annexin V and LIVE/DEAD (Thermo Fisher Scientific) staining.
In vivo testing of ADCs
For the in vivo dosing experiments, 1 million MM1.S (Luc+) or 3 million JK-6L (Luc+) cells were transplanted into sublethally (2.5 Gy) irradiated 8- to 12-week-old male NSG mice via tail vein injection. Disease dissemination was confirmed posttransplantation by IVIS bioluminescence imaging (PerkinElmer), and tumor burden was quantified by using Living Image software (version 4.7.2; PerkinElmer). In brief, d-luciferin (Perkin Elmer) was administered by intraperitoneal injection (10 µL/g, 15 mg/mL) followed by inhalation anesthesia (isoflurane) and IVIS bioluminescence imaging (7.5 minutes' post–luciferin injection for all animals per imaging sessions). Each animal then received a total of 4 doses (at 4 mg/kg for each dose) of 5E3, 5E3-vedotin, or trastuzumab-vedotin by means of tail vein injection. Mice were placed on tumor watch, and imaging sessions were repeated weekly (and at euthanasia). For cytotoxicity experiments, an identical dosing schedule with either 5E3-vedotin or vehicle (saline) was used in healthy 8- to 12-week-old male C57/BL6J mice. Mice were killed 14 days after the first injection.
Full blood counts were assessed from terminal bleeds by using a Mythic 18 analyzer (Orphée), and bone marrow single-cell suspensions were assessed using antibodies shown in supplemental Table 8 on a BD LSRFortessa cell analyzer. Data were analyzed by using FlowJo software (BD Biosciences). Tissue samples were automatically fixed in formalin by using the Leica ASP6025 Processor and embedded in paraffin using a Tissue-Tek embedding center (Sakura). Samples were then sectioned by using a Leica RM2255 Microtome (5 µm sections) and stained with hematoxylin and eosin (Harris Hematoxylin and Aqueous Eosin 1%; Leica) using a Leica ST5020 Autostainer. Scanned sections of key organs were examined by using the Aperio Image Scope system. Organs examined were the heart, lungs, liver, kidney, spleen, bone marrow in sternum and femur, eyes, and several transections of brain that included cerebrum, cerebellum, midbrain, and medulla.
Statistical analysis
GraphPad Prism 9 (GraphPad Software) and R version 3.5.2 (R Foundation for Statistical Computing) software were used for performing statistical analysis and generating graphs/plots. IC50 values were obtained from sigmoidal dose–response curves [log(inhibitor) vs response – variable slope (four parameters)]. Survival percentages were estimated by using Kaplan-Meier methods, and survival curves were compared by using the Cox proportional hazards model. Linear association between 2 variables was measured by using the Pearson correlation, and a two-tailed t test was used to determine statistical significance between 2 groups. Statistical significance was determined as P < .05.
Results
Characterization of the myeloma cell surface proteome by plasma membrane profiling
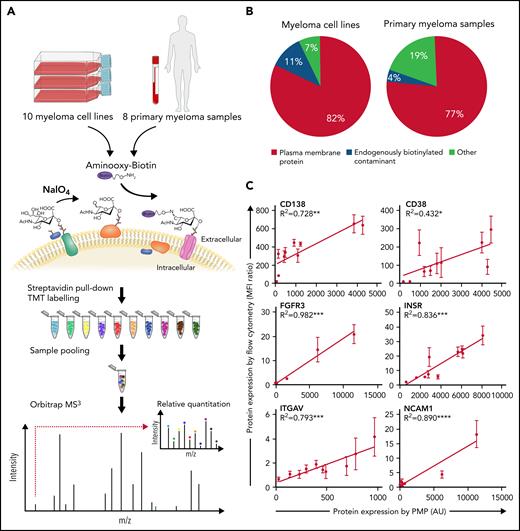
To characterize the surface proteome of myeloma, we used plasma membrane profiling1,2 (Figure 1A). We first profiled 10 genotyped human myeloma cell lines (supplemental Tables 1 and 2; supplemental Figure 1) to establish the feasibility of the technique in cancer cells, then profiled 8 primary myeloma samples (supplemental Table 3). This process identified a mean of 1791 cell surface proteins per cell line (supplemental Data 1) and 3206 cell surface proteins per primary sample (supplemental Data 2), according to published annotation strategies.3,4 Inclusion of 2 cell lines in this latter screen revealed that the surface proteomes of myeloma cell lines and primary myeloma are qualitatively very similar, with 3207 of 3213 proteins identified in both. We did not see any proteins differentially expressed between newly diagnosed or relapsed patients, although there were only 2 treated patients in our data set.
Plasma membrane profiling (PMP) of primary samples and cell lines leads to quantification of the cell surface proteome in myeloma at high coverage. (A) PMP overview. Ten human myeloma cell lines (HMCL) were profiled in a 10-plex and 8 primary myeloma samples plus 2 repeat HMCL were profiled in a second 10-plex. Sugar residues were oxidized with sodium periodate (NaIO4) to form aldehydes, which were then treated with aminooxy-biotin resulting in biotinylation via stable oxime bonds. This was followed by streptavidin pull-down, tandem mass tag (TMT) labeling, and mass spectrometry (MS3). (B) Identified proteins were annotated as plasma membrane proteins, as endogenously biotinylated contaminants, or unrecognized as plasma membrane proteins. Proportions within the pie charts refer to protein abundance. (C) Comparison of protein quantification by flow cytometry and PMP. Relative protein abundance by TMT labeling and mass spectrometry in arbitrary units (AU; x-axis) is compared with protein abundance by median fluorescence intensity (MFI) as determined by flow cytometry with validated antibodies relative to isotype control (y-axis). Error bars are ±SD of biological replicates; n = 3. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Plasma membrane profiling (PMP) of primary samples and cell lines leads to quantification of the cell surface proteome in myeloma at high coverage. (A) PMP overview. Ten human myeloma cell lines (HMCL) were profiled in a 10-plex and 8 primary myeloma samples plus 2 repeat HMCL were profiled in a second 10-plex. Sugar residues were oxidized with sodium periodate (NaIO4) to form aldehydes, which were then treated with aminooxy-biotin resulting in biotinylation via stable oxime bonds. This was followed by streptavidin pull-down, tandem mass tag (TMT) labeling, and mass spectrometry (MS3). (B) Identified proteins were annotated as plasma membrane proteins, as endogenously biotinylated contaminants, or unrecognized as plasma membrane proteins. Proportions within the pie charts refer to protein abundance. (C) Comparison of protein quantification by flow cytometry and PMP. Relative protein abundance by TMT labeling and mass spectrometry in arbitrary units (AU; x-axis) is compared with protein abundance by median fluorescence intensity (MFI) as determined by flow cytometry with validated antibodies relative to isotype control (y-axis). Error bars are ±SD of biological replicates; n = 3. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Enrichment for cell surface proteins was high, at 82% for cell lines and 77% for primary samples (Figure 1B). To confirm the reliability of our proteomic quantification, we selected 6 cell surface proteins known to be expressed in myeloma and found that expression by mass spectrometry and by flow cytometry were well correlated (Figure 1C). Correlation of RNA expression with protein expression was particularly poor at the cell surface, with R2 values ranging from 0.004 to 0.028 (supplemental Figure 2). Our proteomic data are readily available in processed (supplemental Data 1 and Data 2) and raw (Proteome Xchange; DOI 10.6019/PXD014165) formats.
A vector-based prioritization identifies SEMA4A as a novel therapeutic target in myeloma
To rank the cell surface proteins for therapeutic potential, we devised a single score that was defined as the magnitude of a vector whose components were protein expression in myeloma, lack of healthy tissue expression, and extracellular domain size (supplemental Figure 3). Extracellular domain size was used as a surrogate for number of discontinuous B-cell epitopes, as we showed that these features are well correlated (Poisson regression, P = 1.045 × 10−233) (supplemental Figure 4). Myeloma cell surface proteins were ranked according to the vector score, and the top-ranking proteins were filtered by using tissue microarray data (proteinatlas.org). The top 10 proteins are detailed in Table 1, and an extensive list of potential myeloma targets is provided in supplemental Table 4.
Prioritization of immunotherapy targets in primary myeloma
| Protein . | Gene . | Primary myeloma expression . | Extracellular size . | Off-tumor expression (1 = low, 0 = high) . | Score . |
|---|---|---|---|---|---|
| Q9H3S1 | SEMA4A | 0.827 | 0.443 | 0.846 | 1.263 |
| Q96LA5 | FCRL2 | 0.668 | 0.260 | 1.000 | 1.230 |
| Q96RD9 | FCRL5 | 0.707 | 0.568 | 0.811 | 1.217 |
| Q02223 | TNFRSF17 (BCMA) | 0.644 | 0.037 | 1.000 | 1.190 |
| P28908 | TNFRSF8 | 0.588 | 0.245 | 1.000 | 1.186 |
| Q9NQ25 | SLAMF7 | 0.761 | 0.139 | 0.866 | 1.161 |
| O15389 | SIGLEC5 | 0.511 | 0.289 | 1.000 | 1.159 |
| Q7Z6M3 | MILR1 | 0.546 | 0.141 | 1.000 | 1.148 |
| P28907 | CD38 | 0.970 | 0.175 | 0.558 | 1.132 |
| Q7Z6A9 | BTLA | 0.696 | 0.086 | 0.867 | 1.115 |
| Protein . | Gene . | Primary myeloma expression . | Extracellular size . | Off-tumor expression (1 = low, 0 = high) . | Score . |
|---|---|---|---|---|---|
| Q9H3S1 | SEMA4A | 0.827 | 0.443 | 0.846 | 1.263 |
| Q96LA5 | FCRL2 | 0.668 | 0.260 | 1.000 | 1.230 |
| Q96RD9 | FCRL5 | 0.707 | 0.568 | 0.811 | 1.217 |
| Q02223 | TNFRSF17 (BCMA) | 0.644 | 0.037 | 1.000 | 1.190 |
| P28908 | TNFRSF8 | 0.588 | 0.245 | 1.000 | 1.186 |
| Q9NQ25 | SLAMF7 | 0.761 | 0.139 | 0.866 | 1.161 |
| O15389 | SIGLEC5 | 0.511 | 0.289 | 1.000 | 1.159 |
| Q7Z6M3 | MILR1 | 0.546 | 0.141 | 1.000 | 1.148 |
| P28907 | CD38 | 0.970 | 0.175 | 0.558 | 1.132 |
| Q7Z6A9 | BTLA | 0.696 | 0.086 | 0.867 | 1.115 |
The targets of immunotherapies approved by the US Food and Drug Administration and European Medicines Agency in myeloma are in bold. Primary myeloma expression refers to median expression in primary myeloma by plasma membrane profiling, standardized to a maximum expression of 1. Extracellular size is number of amino acid residues in the extracellular domain, standardized to a maximum size of 1. Off-tumor expression relates to expression across multiple tissues in the Human Proteome Map (www.humanproteomemap.org). It is taken as 1 if there is no expression in any of the tissues or (negative log of the maximum tissue expression in arbitrary units)/10 otherwise. Thus, each of myeloma expression, extracellular size, and off-tumor expression have a maximum possible value of 1. The score is the magnitude of the vector of standardized on-tumor expression, extracellular size, and off-tumor expression and has a maximum possible value of (∼1.732).
B-cell maturation antigen (BCMA; TNFRSF17), CD38, and SLAMF7, all approved targets in myeloma,5-8 were found within the top 10 hits (Table 1), confirming the validity of our approach. Our top-ranked target, SEMA4A, has not been previously identified as a target in myeloma. We therefore prioritized it for further investigation. SEMA4A is a class 4 semaphorin that acts as a soluble ligand involved in embryonic and pathologic vascularization and fine-tuning of the immune response, and it may have a role in cancer.9-11 It can also act as a receptor, signaling via its cytoplasmic domain to mediate cell migration.12 Our data showed that SEMA4A was expressed on all profiled cell lines and primary samples, at a higher level than SLAMF7 or BCMA, although at a lower level than CD38 (Figure 2A). We also examined SEMA4A transcript expression in monoclonal gammopathy of uncertain significance and asymptomatic myeloma. No difference was noted in SEMA4A expression between these premalignant conditions and myeloma (supplemental Figure 5A), nor was SEMA4A expression associated with underlying cytogenetic abnormalities (supplemental Figure 5B).
Prioritization of immune therapy targets leads to the identification of SEMA4A as a potential ADC target in myeloma. (A) Expression of SEMA4A by plasma membrane profiling (PMP) in myeloma cell lines and primary myeloma cells. Log relative expression is standardized to the range 0 to 1 for each sample. The median expressions of SLAMF7 (the target of elotuzumab), CD38 (the target of daratumumab), and BCMA (the target of CAR T cells) are also indicated. (B) Comparison of SEMA4A expression by flow cytometry and by PMP. Relative protein abundance by tandem mass tag labeling and mass spectrometry in arbitrary units (AU; x-axis) is compared with protein abundance by median fluorescence intensity (MFI) as determined by flow cytometry with the SEMA4A 5E3 clone antibody relative to isotype control (y-axis). Error bars are ±SD of biological replicates; n = 3. (C) Representative images of healthy (non–multiple myeloma [MM]) and MM bone marrow after duplex immunohistochemistry staining with CD138 (yellow) and SEMA4A (purple). CD138+ cells in the non-MM bone marrow are indicated by arrows. (D) Comparison of SEMA4A expression by flow cytometry in CD138+ cells in non-MM (n = 3) and in MM (n = 5) as described in panel B. (E) Representative histograms of flow cytometric profiling of SEMA4A expression in hematopoietic cells from a single patient. Markers used are CD66b (granulocytes), CD14 (monocytes), CD138 (myeloma cells), CD3 (T cells), CD19 (B cells), and CD34 (primitive hematopoietic cells). Black lines show intensity of isotype control, and blue lines show the intensity of SEMA4A. (F) Summary of the flow cytometric profiling of hematopoietic cells as shown in panel E. SEMA4A expression for each patient was calculated as the MFI of the 5E3 clone relative to the isotype control and then normalized to SEMA4A expression on CD19+ cells; n = 12. (G) Expression of SEMA4A in normal healthy tissues by whole-cell proteomic profiling (Human Proteome Map, www.humanproteomemap.org; left) and by tissue microarray (Human Protein Atlas, proteinatlas.org; right). NK, natural killer. ***P < .001.
Prioritization of immune therapy targets leads to the identification of SEMA4A as a potential ADC target in myeloma. (A) Expression of SEMA4A by plasma membrane profiling (PMP) in myeloma cell lines and primary myeloma cells. Log relative expression is standardized to the range 0 to 1 for each sample. The median expressions of SLAMF7 (the target of elotuzumab), CD38 (the target of daratumumab), and BCMA (the target of CAR T cells) are also indicated. (B) Comparison of SEMA4A expression by flow cytometry and by PMP. Relative protein abundance by tandem mass tag labeling and mass spectrometry in arbitrary units (AU; x-axis) is compared with protein abundance by median fluorescence intensity (MFI) as determined by flow cytometry with the SEMA4A 5E3 clone antibody relative to isotype control (y-axis). Error bars are ±SD of biological replicates; n = 3. (C) Representative images of healthy (non–multiple myeloma [MM]) and MM bone marrow after duplex immunohistochemistry staining with CD138 (yellow) and SEMA4A (purple). CD138+ cells in the non-MM bone marrow are indicated by arrows. (D) Comparison of SEMA4A expression by flow cytometry in CD138+ cells in non-MM (n = 3) and in MM (n = 5) as described in panel B. (E) Representative histograms of flow cytometric profiling of SEMA4A expression in hematopoietic cells from a single patient. Markers used are CD66b (granulocytes), CD14 (monocytes), CD138 (myeloma cells), CD3 (T cells), CD19 (B cells), and CD34 (primitive hematopoietic cells). Black lines show intensity of isotype control, and blue lines show the intensity of SEMA4A. (F) Summary of the flow cytometric profiling of hematopoietic cells as shown in panel E. SEMA4A expression for each patient was calculated as the MFI of the 5E3 clone relative to the isotype control and then normalized to SEMA4A expression on CD19+ cells; n = 12. (G) Expression of SEMA4A in normal healthy tissues by whole-cell proteomic profiling (Human Proteome Map, www.humanproteomemap.org; left) and by tissue microarray (Human Protein Atlas, proteinatlas.org; right). NK, natural killer. ***P < .001.
To further characterize SEMA4A, we tested a commercially available monoclonal antibody, clone 5E3, a murine antibody reactive against mouse and human SEMA4A. Expression according to flow cytometry correlated with expression by plasma membrane profiling in the cell lines (R2= 0.870; P = 8.15 × 10−5) (Figure 2B). We therefore used this antibody to perform flow cytometry on a further 53 newly diagnosed and relapsed bone marrow myeloma samples. High SEMA4A expression was seen in 49 (92.5%) of 53 samples, with a distribution of expression similar to that of the original proteomic data set (supplemental Figure 6). Costaining of SEMA4A with CD138 was observed in bone marrow trephine sections from patients with myeloma (Figure 2C). SEMA4A was also expressed on CD138+ plasma cells from healthy control subjects (Figure 2C-D). In a subset of the patients with myeloma, SEMA4A expression was assessed in other hematopoietic cell lineages. Lower expression was seen on monocytes and neutrophils, whereas expression was minimal on B-, T-, and CD34+ cells (Figure 2E-F). A small population of T cells expressed SEMA4A (Figure 2E), and further phenotyping suggested that these were principally CD8+ T cells, with increased representation in the CD8+/CD45RA–/CCR7– effector memory population (supplemental Figure 7). Within the B-cell lineage, SEMA4A is expressed only in healthy and malignant plasma cells, which are CD20–, but not in earlier CD20+ B cells, suggesting that it is a marker of late B-cell differentiation.
Both whole-cell proteomic data and tissue microarray data indicated low expression outside the hematopoietic system (Figure 2G). To check the sensitivity of the tissue microarray, we made paraffin-embedded cell pellets of several of the cell lines already profiled and stained these using the same antibody (supplemental Figure 8). This suggested that the limit of detection of SEMA4A by tissue microarray was very similar to the limit of detection by flow cytometry. We therefore concluded that SEMA4A is expressed at high levels on myeloma cells but exhibits low off-tumor expression, making it a promising therapeutic target.
SEMA4A is required for normal myeloma cell growth
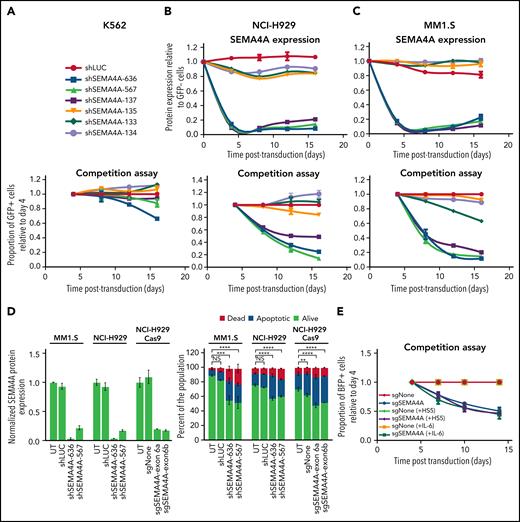
Downregulation of cell surface targets is a potential mechanism of immunotherapy resistance.13 We therefore tested the effect of downregulating SEMA4A using lentivirally delivered shRNA, with six shRNAs directed against SEMA4A plus a control shRNA targeting luciferase. These shRNAs had no effect on the growth of K562 cells, which do not express SEMA4A (Figure 3A), implying that none of the shRNAs had an off-target effect on cellular proliferation. In the myeloma cell lines NCI-H929 and MM1.S, three of the SEMA4A-specific shRNAs efficiently reduced cell surface expression of SEMA4A, whereas the control shRNA and the three remaining SEMA4A shRNAs had little effect on expression (Figure 3B-C; supplemental Figure 9). SEMA4A-specific shRNAs that reduced target expression also markedly delayed cell growth in a GFP competition assay, whereas shRNAs that did not affect SEMA4A expression had minimal effect on cell growth (Figure 3B-C). Loss of SEMA4A expression preceded cell death (supplemental Figure 10). CRISPR/Cas9 targeting of SEMA4A also resulted in a competitive growth disadvantage in H929 and INA6 cells but not in MM1.S cells (supplemental Figure 11A-B). However, RNA-sequencing revealed that MM1.S cells compensated for SEMA4A loss by exon skipping (supplemental Figure 11C-E). This compensation is probably an artifact of CRISPR/Cas9 targeting, as has been described previously.14,15 Indeed, only 8 (0.86%) of 934 samples in the CoMMpass dataset (https://themmrf.org/finding-a-cure/our-work/the-mmrf-commpass-study/) showed any evidence of this novel splice variant. Furthermore, support for those reads was minimal, with 1 to 3 reads per sample (supplemental Table 5). To re-confirm that SEMA4A was essential in MM1.S cells, we performed CRISPR interference experiments, which showed that abrogation of expression was associated with a competitive growth disadvantage (supplemental Figure 11F). Loss of SEMA4A expression was associated with apoptosis; both early apoptotic and dead cell fractions were increased in myeloma cells treated with shRNA targeting SEMA4A (Figure 3D). Interestingly, neither coculture of myeloma cell lines with bone marrow stromal cells nor supplementation with interleukin-6 had any effect on SEMA4A abrogation (Figure 3E), suggesting that SEMA4A signals independently from these pathways.
Expression of shRNA against SEMA4A is associated with reduced growth in myeloma cells but not K562 erythroleukemia cells. The erythroleukemia cell line, K562 (A), and the human myeloma cell lines, NCI-H929 (B) and MM1.S (C), were lentivirally transduced with six shRNA predicted to target SEMA4A plus a control shRNA directed against luciferase (shLUC). All shRNA vectors expressed GFP. Transduction efficiency was deliberately maintained at ∼50%. Top panels: SEMA4A expression was measured at the cell surface by flow cytometry and is expressed relative to expression in GFP-negative (GFP-) cells. K562 cells do not express SEMA4A, and thus expression is not shown. Bottom panels: GFP-positive (GFP+)/total cell ratio was measured from day 4 after transduction. GFP+/total cell ratio is then plotted relative to that ratio at day 4 and normalized to the ratio for the control hairpin, shLUC. Error bars indicate ±SD from a minimum of 2 replicates for each shRNA. (D) MM1.S, NCI-H929, and NCI-H929 Cas9 were lentivirally transduced as described in panel A with a control (shLUC/sgNone) or shRNA/sgRNA targeting SEMA4A. Left panel: at 144 hours' post–viral transduction, SEMA4A expression was measured at the cell surface by flow cytometry and is expressed relative to expression in untransduced cells (UT). Right: Cell viability was also assessed by flow cytometry to determine the percentages of dead (Annexin V and LIVE/DEAD Fixable Violet positive), early apoptotic (Annexin V positive and LIVE/DEAD Fixable Violet negative), and alive (Annexin V and LIVE/DEAD Fixable Violet negative) cells. A one-way analysis of variance comparing MM1.S alive cells F(3,8) = 42.45, P < .0001, NCI-H929 alive cells F(3,8) = 101.2, P < .0001, and NCI-H929 Cas9 alive cells F(3,8) = 56.84, P < .0001 was performed. Dunnett's multiple comparisons correction is shown. (E) NCI-H929 constitutively expressing Cas9 were lentivirally transduced with sgRNA targeting exon six of SEMA4A (sgSEMA4A) or a nontargeting control sgRNA (sgNone). All sgRNA vectors expressed blue fluorescent protein (BFP), and transduction efficiency was deliberately maintained at ∼50%. After lentiviral transduction, cells were cultured ± the stromal cell line, HS5, or were supplemented ± interleukin-6 (IL-6). The BFP-positive (BFP+)/total cell ratio was measured from day 4 after transduction and is plotted relative to proportion at day 4 and normalized to the ratio of each control sgRNA, sgNone. Error bars are ±SD of replicates; n = 3. **P < .01; ***P < .001; ****P < .0001. NS, not significant.
Expression of shRNA against SEMA4A is associated with reduced growth in myeloma cells but not K562 erythroleukemia cells. The erythroleukemia cell line, K562 (A), and the human myeloma cell lines, NCI-H929 (B) and MM1.S (C), were lentivirally transduced with six shRNA predicted to target SEMA4A plus a control shRNA directed against luciferase (shLUC). All shRNA vectors expressed GFP. Transduction efficiency was deliberately maintained at ∼50%. Top panels: SEMA4A expression was measured at the cell surface by flow cytometry and is expressed relative to expression in GFP-negative (GFP-) cells. K562 cells do not express SEMA4A, and thus expression is not shown. Bottom panels: GFP-positive (GFP+)/total cell ratio was measured from day 4 after transduction. GFP+/total cell ratio is then plotted relative to that ratio at day 4 and normalized to the ratio for the control hairpin, shLUC. Error bars indicate ±SD from a minimum of 2 replicates for each shRNA. (D) MM1.S, NCI-H929, and NCI-H929 Cas9 were lentivirally transduced as described in panel A with a control (shLUC/sgNone) or shRNA/sgRNA targeting SEMA4A. Left panel: at 144 hours' post–viral transduction, SEMA4A expression was measured at the cell surface by flow cytometry and is expressed relative to expression in untransduced cells (UT). Right: Cell viability was also assessed by flow cytometry to determine the percentages of dead (Annexin V and LIVE/DEAD Fixable Violet positive), early apoptotic (Annexin V positive and LIVE/DEAD Fixable Violet negative), and alive (Annexin V and LIVE/DEAD Fixable Violet negative) cells. A one-way analysis of variance comparing MM1.S alive cells F(3,8) = 42.45, P < .0001, NCI-H929 alive cells F(3,8) = 101.2, P < .0001, and NCI-H929 Cas9 alive cells F(3,8) = 56.84, P < .0001 was performed. Dunnett's multiple comparisons correction is shown. (E) NCI-H929 constitutively expressing Cas9 were lentivirally transduced with sgRNA targeting exon six of SEMA4A (sgSEMA4A) or a nontargeting control sgRNA (sgNone). All sgRNA vectors expressed blue fluorescent protein (BFP), and transduction efficiency was deliberately maintained at ∼50%. After lentiviral transduction, cells were cultured ± the stromal cell line, HS5, or were supplemented ± interleukin-6 (IL-6). The BFP-positive (BFP+)/total cell ratio was measured from day 4 after transduction and is plotted relative to proportion at day 4 and normalized to the ratio of each control sgRNA, sgNone. Error bars are ±SD of replicates; n = 3. **P < .01; ***P < .001; ****P < .0001. NS, not significant.
SEMA4A is internalized and is thus a potential ADC
Both cellular and humoral immunity are frequently impaired in myeloma; thus, ADCs, which do not rely on a functioning immune system, provide an attractive therapeutic modality. We therefore investigated whether the SEMA4A antibody was internalized on binding to myeloma cells, a prerequisite for an ADC. Upon initial labeling, SEMA4A was detected at the cell surface (Figure 4A). After 3 hours' incubation at 37°C, the majority of antibody was internalized. Internalized protein was associated with the protein lysosomal-associated membrane protein 1, which is expressed primarily in lysosomal membranes and is one of the major constituents of those membranes.16 It is therefore a useful marker to show that an internalized protein traffics to the lysosomal compartment, a requirement for the successful activity of ADCs.17 Flow cytometry experiments suggested that ∼50% of SEMA4A was internalized at 2 hours and that this was a temperature-dependent process (Figure 4B). Importantly, internalization was seen on all cells, indicated by a left shift of the entire cell population at 37°C compared with 4°C (Figure 4C). These data suggest that SEMA4A antibodies can be internalized and trafficked to the lysosomal compartment and that SEMA4A is a potential ADC target.
SEMA4A antibody is internalized in a temperature-dependent manner and localizes to the lysosomal compartment. (A) Immunofluorescence microscopy in NCI-H929 cells demonstrating localization of SEMA4A (green) and lysosomal-associated membrane protein 1 (LAMP-1) (red) before and after 3 hours of incubation at 37°C. Below the photomicrographs are plots showing fluorescence intensity as a function of distance from a reference point and confirming colocalization of SEMA4A with LAMP-1 after incubation. (B) Flow cytometry of NCI-H929 cells exhibiting the dynamics of SEMA4A internalization, as indicated by loss of antibody from the cell surface over time when cultured at 37°C but not at 4°C. (C) Histogram of cells from panel B, at the beginning and end of the time course. Internalization of SEMA4A from the cell surface affects the entire cell population. IgG1, immunoglobulin G1.
SEMA4A antibody is internalized in a temperature-dependent manner and localizes to the lysosomal compartment. (A) Immunofluorescence microscopy in NCI-H929 cells demonstrating localization of SEMA4A (green) and lysosomal-associated membrane protein 1 (LAMP-1) (red) before and after 3 hours of incubation at 37°C. Below the photomicrographs are plots showing fluorescence intensity as a function of distance from a reference point and confirming colocalization of SEMA4A with LAMP-1 after incubation. (B) Flow cytometry of NCI-H929 cells exhibiting the dynamics of SEMA4A internalization, as indicated by loss of antibody from the cell surface over time when cultured at 37°C but not at 4°C. (C) Histogram of cells from panel B, at the beginning and end of the time course. Internalization of SEMA4A from the cell surface affects the entire cell population. IgG1, immunoglobulin G1.
To check the degree of shedding of SEMA4A from the cell surface into the bloodstream, we developed an enzyme-linked immunosorbent assay and measured serum levels of soluble SEMA4A in patients with myeloma and in healthy control subjects. Dilution studies showed that the assay had a limit of detection of 2 ng/mL (supplemental Figure 12A). The median serum concentrations of soluble SEMA4A were 3.3 ng/mL and 8.1 ng/mL in healthy control subjects and patients with myeloma, respectively (supplemental Figure 12B). The difference between the 2 groups was significant (t test, P = .0022). However, we did not see any association between myeloma tumor load or International Staging System stage and serum levels (supplemental Figure 12C-D). The absolute levels of soluble SEMA4A in myeloma are highly unlikely to disrupt the activity of a SEMA4A ADC in myeloma. For example, targeting of TNFRSF17 (BCMA) in myeloma by belantamab mafodotin or by CAR T cells has not been impaired by median serum expression levels of 176 ng/mL of soluble BCMA.18 Taken together, these data suggest that SEMA4A antibodies can be internalized and trafficked to the lysosomal compartment and that the level of shedding of SEMA4A from the cell surface would be unlikely to compromise the activity of an ADC.
A novel SEMA4A ADC kills myeloma cells in vitro
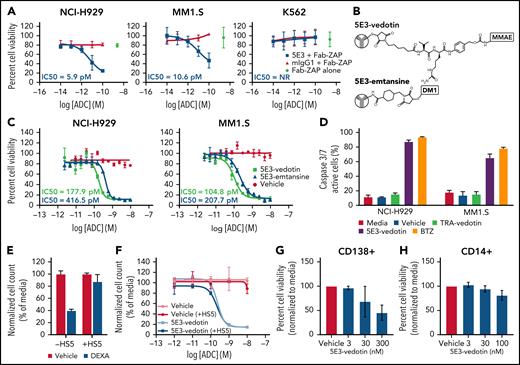
To explore SEMA4A further as an ADC target, we first used a Fab-ZAP assay19 with clone 5E3 SEMA4A antibody. Myeloma cell lines incubated with 5E3, but not un-incubated cells or cells incubated with an isotype control antibody, were efficiently killed by anti-mouse secondary antibodies conjugated to saporin, with an 50% inhibitory concentration (IC50) in the low picomolar range (Figure 5A). K562 erythroleukemia cells, which do not express SEMA4A, were not affected when incubated with 5E3 antibody and exposed to Fab-ZAP. We then proceeded to directly conjugate the 5E3 clone antibody to 2 intracellular toxins, monomethyl auristatin E and mertansine (DM1), using established linker chemistry to produce 5E3-vedotin and 5E3-emtansine, respectively (Figure 5B). 5E3-emtansine was toxic to all myeloma cell lines, with an IC50 of 207.7 pM to 39.6 nM. 5E3-vedotin was toxic to 8 of 10 myeloma cell lines, with an IC50 of 98.0 pM to 1.4 µM (Figure 5C; supplemental Figures 13 and 14). For both conjugates, the IC50 correlated with SEMA4A surface expression (supplemental Figure 15). As expected, treatment with the ADC led to an increase in caspase 3/7 cleavage, confirming that it induced apoptosis (Figure 5D). Bone marrow stroma can provide protection to myeloma cells, and this action can be modeled in vitro by using coculture of myeloma cells with bone marrow stromal cells. Coculture protects myeloma cells against cytotoxicity induced by certain drugs, such as corticosteroids.20 We used this stromal cell coculture system and confirmed that it indeed rescued dexamethasone-induced cytotoxicity (Figure 5E). Interestingly, bone marrow stromal coculture had no effect on 5E3-vedotin–induced killing (Figure 5F). We also tested 5E3-vedotin against primary cells ex vivo and found that 5E3-vedotin was active against those cells (Figure 5G). Finally, because of its high expression in monocytes, 5E3-vedotin was tested against primary monocytes. As anticipated, the ADC did show some effects against these cells at higher concentrations (Figure 5H).
ADCs against SEMA4A are potent and selective in vitro. (A) Fab-ZAP assay in NCI-H929 cells, MM1.S cells, and K562 cells, which do not express SEMA4A, as a control. Cells were incubated with Fab-ZAP alone or with clone 5E3 anti-SEMA4A or with an isotype control. Cell viability was measured by XTT assay at 72 hours. (B) Linker chemistry for 5E3-vedotin and 5E3-emtansine. (C) Dose–response curves for 5E3-vedotin and 5E3-emtansine in NCI-H929 and MM1.S cells. Citrate buffer was used as a control. Cell viability was measured by XTT assay at 72 hours. IC50 values are specified, where the calculation was possible. NR = IC50 was not reached. (D) NCI-H929 and MM1.S cells were treated with media only, citrate buffer (vehicle), 10 nM trastuzumab-vedotin (TRA-vedotin), 10 nM 5E3-vedotin, or 10 nM bortezomib (BTZ) and incubated with IncuCyte Caspase-3/7 apoptosis assay reagent for 72 hours and imaged by using an IncuCyte Live Cell Analysis system. (E) NCI-H929 cells were cocultured ± HS5 and treated with vehicle or dexamethasone (DEXA, 10 µM) for 72 hours. Total cell counts were determined by using Flow-Count Fluorospheres (Beckman Coulter) by flow cytometry and normalized to media-treated controls. (F) NCI-H929 were cocultured with HS5 and treated with either vehicle or 5E3-vedotin. Cell counts were determined as in panel E at 72 hours. CD138+ cells (G) or CD14+ cells (H) from myeloma patients were isolated by using microbeads (Miltenyi Biotec) and incubated with vehicle or 5E3-vedotin. Cell viability was measured by CellTiter-Glo or Annexin V and LIVE/DEAD staining by flow cytometry. Error bars indicate ±SD of a minimum of 3 replicates.
ADCs against SEMA4A are potent and selective in vitro. (A) Fab-ZAP assay in NCI-H929 cells, MM1.S cells, and K562 cells, which do not express SEMA4A, as a control. Cells were incubated with Fab-ZAP alone or with clone 5E3 anti-SEMA4A or with an isotype control. Cell viability was measured by XTT assay at 72 hours. (B) Linker chemistry for 5E3-vedotin and 5E3-emtansine. (C) Dose–response curves for 5E3-vedotin and 5E3-emtansine in NCI-H929 and MM1.S cells. Citrate buffer was used as a control. Cell viability was measured by XTT assay at 72 hours. IC50 values are specified, where the calculation was possible. NR = IC50 was not reached. (D) NCI-H929 and MM1.S cells were treated with media only, citrate buffer (vehicle), 10 nM trastuzumab-vedotin (TRA-vedotin), 10 nM 5E3-vedotin, or 10 nM bortezomib (BTZ) and incubated with IncuCyte Caspase-3/7 apoptosis assay reagent for 72 hours and imaged by using an IncuCyte Live Cell Analysis system. (E) NCI-H929 cells were cocultured ± HS5 and treated with vehicle or dexamethasone (DEXA, 10 µM) for 72 hours. Total cell counts were determined by using Flow-Count Fluorospheres (Beckman Coulter) by flow cytometry and normalized to media-treated controls. (F) NCI-H929 were cocultured with HS5 and treated with either vehicle or 5E3-vedotin. Cell counts were determined as in panel E at 72 hours. CD138+ cells (G) or CD14+ cells (H) from myeloma patients were isolated by using microbeads (Miltenyi Biotec) and incubated with vehicle or 5E3-vedotin. Cell viability was measured by CellTiter-Glo or Annexin V and LIVE/DEAD staining by flow cytometry. Error bars indicate ±SD of a minimum of 3 replicates.
A novel SEMA4A ADC kills myeloma cells in vivo
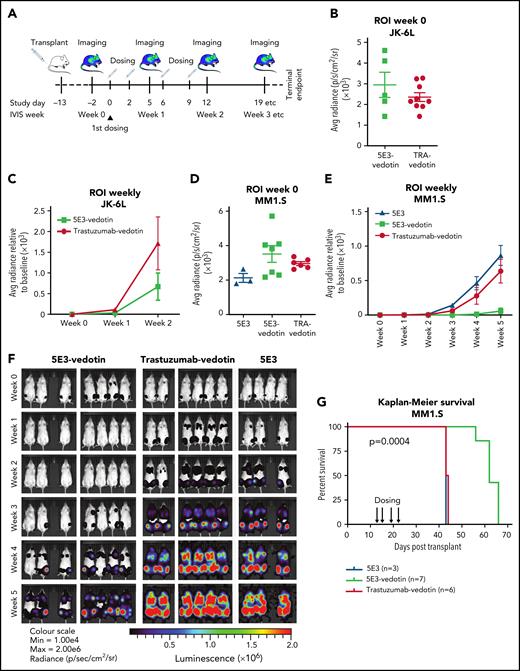
To model in vivo treatment, we used 2 independent murine xenograft models using tail vein injection of myeloma cells.21 At day −13, luciferase-expressing human myeloma cell lines were injected via the tail vein into nonobese, diabetic gamma mice with severe combined immunodeficiency, and the establishment of medullary myeloma was confirmed by in vivo imaging (Figure 6A). We tested the efficacy of 5E3-vedotin because this conjugation was more stable than 5E3-emtansine. Trastuzumab-vedotin served as an isotype control for nonspecific binding and internalization as trastuzumab is a humanized immunoglobulin G1 antibody against ERBB2, which is not expressed on myeloma cells. The unconjugated 5E3 clone antibody provided an additional control to ensure that activity was a consequence of antibody internalization and release of conjugated toxin. The initial luciferase signal intensity was equivalent between 5E3-vedotin–injected and control mice (Figure 6B,D). For practical reasons, including limited ADC, we confined dosing to 4 tail vein injections, delivered twice weekly for 2 weeks. Mice injected with 5E3-vedotin, but not control mice, experienced a remission with suppression of luciferase signal to background after the first 2 injections (Figure 6C,E-F). This resulted in a significant increase in median overall survival compared with control mice (61 days vs 42 days for the MM1.S model; Cox proportional hazards model, P = .0004) (Figure 6G). Even though clone 5E3 is cross-reactive with murine tissue (supplemental Figure 16), we observed no overt toxicity or treatment-related weight loss in the mice. Nor did we observe hematologic or solid organ cellular toxicity (supplemental Figure 17; supplemental Data 3). These data show that 5E3-vedotin is delivered efficiently to the bone marrow and targets myeloma specifically in vivo.
An ADC against SEMA4A is potent in vivo. (A) Xenograft model to test in vivo efficacy of 5E3-vedotin. Mice were injected with MM1.S-luciferase or JK-6L luciferase cells by tail vein injection at day −13 and established disease confirmed by luciferase intensity, measured by using an in vitro imaging system. 5E3-vedotin was injected via the tail vein at 4 mg/kg twice weekly for a total of 4 doses on days 0, 2, 6, and 9. Controls were trastuzumab-vedotin (isotype; TRA-vedotin) and nonconjugated 5E3 antibody. Mice were imaged weekly and at euthanasia. (B) Luciferase intensity in the regions of interest (ROI) of the JK-6L model before dosing, exhibiting similar disease dissemination levels between treatment and control arms. (C) Time course of luciferase intensity for the same study as in panel B. ROI intensity was normalized to the baseline ROI (week 0) for each mouse. (D) Luciferase intensity in the ROI of the MM1.S model before dosing. (E) Time course of luciferase intensity for the same study as in panel D. ROI intensity was normalized to the baseline ROI (week 0) for each mouse. (F) Images of luciferase intensity of the MM1.S model. (G) Kaplan-Meier curve for the MM1.S model comparing survival of mice in the 5E3-vedotin treatment group vs that of mice in the control groups. Survival was significantly increased in the treatment group compared with isotype control (Cox proportional hazards model, P = .0004). Error bars are ±SD.
An ADC against SEMA4A is potent in vivo. (A) Xenograft model to test in vivo efficacy of 5E3-vedotin. Mice were injected with MM1.S-luciferase or JK-6L luciferase cells by tail vein injection at day −13 and established disease confirmed by luciferase intensity, measured by using an in vitro imaging system. 5E3-vedotin was injected via the tail vein at 4 mg/kg twice weekly for a total of 4 doses on days 0, 2, 6, and 9. Controls were trastuzumab-vedotin (isotype; TRA-vedotin) and nonconjugated 5E3 antibody. Mice were imaged weekly and at euthanasia. (B) Luciferase intensity in the regions of interest (ROI) of the JK-6L model before dosing, exhibiting similar disease dissemination levels between treatment and control arms. (C) Time course of luciferase intensity for the same study as in panel B. ROI intensity was normalized to the baseline ROI (week 0) for each mouse. (D) Luciferase intensity in the ROI of the MM1.S model before dosing. (E) Time course of luciferase intensity for the same study as in panel D. ROI intensity was normalized to the baseline ROI (week 0) for each mouse. (F) Images of luciferase intensity of the MM1.S model. (G) Kaplan-Meier curve for the MM1.S model comparing survival of mice in the 5E3-vedotin treatment group vs that of mice in the control groups. Survival was significantly increased in the treatment group compared with isotype control (Cox proportional hazards model, P = .0004). Error bars are ±SD.
Discussion
Herein, we present an unbiased approach that enabled the discovery of a novel immunotherapy target in myeloma. One of the key features of the approach was quantitation of the cell surface proteome of primary myeloma at unprecedented coverage. This may prove especially important in the discovery of novel CAR T-cell targets, which can be expressed at relatively low levels on the cancer cell. Another important feature was a ranking algorithm, validated by its capture of the 3 approved immunotherapy targets in myeloma within the top 10 hits. This algorithm could be modified to incorporate further features, if desired. For example, it could incorporate the results from a whole-genome CRISPR/Cas9 screen so that it promoted cell surface proteins essential to the survival of the myeloma cell. However, we note that SEMA4A would not have been ranked so highly had we done this with publicly available data sets. This is because we showed that targeting exons with CRISPR/Cas9 caused artifactual exon skipping and a false-negative result. Therefore, if protein essentiality was to be incorporated, we would recommend a combination of knock-down and knock-out screens. Our algorithm can also be modified to favor individual features over others. For example, to pursue a CAR T-cell target, it would be important to place more weight on low off-tumor expression and less weight on on-tumor expression of the target.
Our approach identified SEMA4A, which has not previously been implicated as a potential target in myeloma. Our use of a tool ADC that was cross-reactive with human and murine tissue allowed us to simultaneously assess potential on-target/off-tumor toxicity as well as efficacy in vivo. We saw no evidence of in vivo toxicity, which suggests that our selected dose of 4 mg/kg for these studies was below the maximally tolerated dose in mice. A higher dose and/or longer exposure may have shown much greater antimyeloma effect. Although we were unable to investigate whether targeting SEMA4A affected normal primary plasma cells, we anticipate that plasma cells would be affected by SEMA4A targeting. However, loss of healthy plasma cells is well tolerated in patients treated with BCMA CAR T cells, and we do not anticipate that this would prevent therapeutic targeting of SEMA4A. We do also see toxicity against primary human monocytes in vitro. However, monocytopenia is a common sequela of chemotherapy, including high-dose corticosteroids, used extensively in myeloma treatment, and is generally well tolerated.
As well as being a novel target, SEMA4A was shown here to play a role in myeloma biology, as loss of expression leads to apoptosis of myeloma cells. In terms of SEMA4A as a target, this is important, as it limits the potential for downregulation or selection of low-expressing clones as a means of antigen escape. Understanding the molecular mechanism by which SEMA4A promotes myeloma cell growth will be important. This may suggest novel pathways that could be exploited therapeutically, or it may provide a rational basis for choosing existing drugs to use alongside a SEMA4A ADC to provide maximum synergy. This mechanism is under study. However, the results of the competition assays suggest that depletion of SEMA4A leads to a cell-intrinsic defect rather than a loss of a trans-acting ligand. This may be due to loss of reverse signaling through SEMA4A, as has been previously described,12 or it may be due to loss of a cis-acting ligand function.22-24
In summary, we illustrate the value of quantifying the cell surface proteome of a primary cancer cell at high coverage and present an approach to yield novel therapeutic targets. We have made our data set publicly accessible for the benefit of the pharmaceutical industry and the myeloma research community.
Acknowledgments
The authors are very grateful to the following individuals: Bridget Manasse from the Haematology-Oncology Diagnostic Service at Addenbrooke's Hospital; Mark Frigerio, Claudio Dagostin, and Joao Nunes from Abzena; and Bridget Lu from Thermo Fisher Scientific. The authors also thank the Cambridge Blood and Stem Cell BioBank for providing the human samples. Finally, they express their gratitude to the patients with myeloma at Addenbrooke's Hospital who kindly donated samples to support this research.
This research was supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre (BRC-1215-20014). G.S.F.A. was supported by a Cancer Research UK (CRUK) studentship (C48525/A18347) and by the MRC Toxicology Unit (MC_UU_00025/10). Work in the laboratory of B.J.P.H. was supported by a CRUK program grant (C18680/A25508), a European Research Council Consolidator award (COMAL 647685), and the NIHR Cambridge Biomedical Research Centre. Work in the laboratory of P.J.L. was supported by a Wellcome Trust Principal Research Fellowship (210688/Z/18/Z). Work in the laboratory of M.M. was funded by the MRC Toxicology Unit (MC_UU_00025/4). Work in the laboratory of J.E.T. was funded by the MRC Toxicology Unit (MC_UU_0025/12). Work in the laboratory of M.A.C. was funded by a Wellcome-Beit Intermediate Fellowship (098638/Z/12/Z), a Multiple Myeloma Research Foundation Senior Research Grant, a CRUK Biotherapeutic Drug Discovery Project Award (A24724), and by the MRC Toxicology Unit (MC_UU_00025/10).
The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Authorship
Contribution: G.S.F.A., S. Surget, J.C.W., and P.J.L. performed plasma membrane profiling; G.S.F.A., J.B.-B., J.A.G., S. Surget, and A.A. performed other experimental work in support of the project; G.S.F.A., J.B.-B., S. Surget, J.A.G., A.A., I.W., J.P.R., J.C.W., S. Skerget, S.N., J.J.K., and M.A.C. performed data analysis; E.J.S. performed fluorescence in situ hybridization on immunohistochemistry samples; G.S.F.A., J.B.-B., S. Surget, J.A.G., G.G., and D.B. performed flow cytometry; G.S.F.A., J.B.B., and S. Surget performed internalization assays; J.B.-B. and L.J. performed enzyme-linked immunosorbent assays; G.S.F.A. and J.B.B. performed in vitro toxicity assays; J.B.-B., G.G., R.A., T.S., J.E.T., and B.J.P.H. performed xenograft assays; Y.-T.T. and K.C.A. supported the ex vivo toxicity assay design; A.T., C.F., L.O.-J., M.M., and P.G. performed the preparation and analysis of the mouse histology for the toxicology study and bone marrow section staining; B.J.P.H., P.J.L., and M.A.C. provided project leadership; and all authors wrote the manuscript.
Conflict-of-interest disclosure: A patent application has been filed in relation to this work. J.E.T. received an honorarium from Gilead for speaking at an educational meeting. M.A.C. was on an advisory board for Bristol Myers Squibb. The remaining authors declare no competing financial interests.
Michael A. Chapman, MRC Toxicology Unit, Tennis Court Rd, Cambridge CB2 1QR, United Kingdom; e-mail: MAC54@cam.ac.uk.
REFERENCES
Author notes
The plasma membrane profiling data generated during this study are included in this published article (in supplemental Data 1 and supplemental Data 2). Raw data are accessible via the Proteome Xchange database, under accession PXD014165 and DOI 10.6019/PXD014165. SEMA4A expression in normal healthy tissues by whole cell proteomic profiling was taken from the Human Proteome Map (www.humanproteomemap.org) and immunohistochemistry images obtained from The Human Protein Atlas database (proteinatlas.org, version 18).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Immunotherapy for multiple myeloma shows great promise, but the optimal target for antibody-based therapies is uncertain. Anderson and colleagues describe a novel strategy for prioritizing potential candidate targets using mass-spectrometry-based plasma membrane profiling. Their work identifies a new membrane-associated antigen, SEMA4A. Anti-SEMA4A antibodies conjugated to toxins, eradicated myeloma cells in vitro and in vivo, and may clinically have therapeutic potential.

![Prioritization of immune therapy targets leads to the identification of SEMA4A as a potential ADC target in myeloma. (A) Expression of SEMA4A by plasma membrane profiling (PMP) in myeloma cell lines and primary myeloma cells. Log relative expression is standardized to the range 0 to 1 for each sample. The median expressions of SLAMF7 (the target of elotuzumab), CD38 (the target of daratumumab), and BCMA (the target of CAR T cells) are also indicated. (B) Comparison of SEMA4A expression by flow cytometry and by PMP. Relative protein abundance by tandem mass tag labeling and mass spectrometry in arbitrary units (AU; x-axis) is compared with protein abundance by median fluorescence intensity (MFI) as determined by flow cytometry with the SEMA4A 5E3 clone antibody relative to isotype control (y-axis). Error bars are ±SD of biological replicates; n = 3. (C) Representative images of healthy (non–multiple myeloma [MM]) and MM bone marrow after duplex immunohistochemistry staining with CD138 (yellow) and SEMA4A (purple). CD138+ cells in the non-MM bone marrow are indicated by arrows. (D) Comparison of SEMA4A expression by flow cytometry in CD138+ cells in non-MM (n = 3) and in MM (n = 5) as described in panel B. (E) Representative histograms of flow cytometric profiling of SEMA4A expression in hematopoietic cells from a single patient. Markers used are CD66b (granulocytes), CD14 (monocytes), CD138 (myeloma cells), CD3 (T cells), CD19 (B cells), and CD34 (primitive hematopoietic cells). Black lines show intensity of isotype control, and blue lines show the intensity of SEMA4A. (F) Summary of the flow cytometric profiling of hematopoietic cells as shown in panel E. SEMA4A expression for each patient was calculated as the MFI of the 5E3 clone relative to the isotype control and then normalized to SEMA4A expression on CD19+ cells; n = 12. (G) Expression of SEMA4A in normal healthy tissues by whole-cell proteomic profiling (Human Proteome Map, www.humanproteomemap.org; left) and by tissue microarray (Human Protein Atlas, proteinatlas.org; right). NK, natural killer. ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/139/16/10.1182_blood.2021015161/5/m_bloodbld2021015161f2.jpeg?Expires=1770327184&Signature=L9Us8uhdWCdRhK1vToNI42K0~oXhEbC2i7vO6n2knhsmwnMtKeqfnKz3PViKpqeoMN90UryVvxON418tePmrwEyr65SpraQUSk7O11gg~ZXryu7lLvb7tbtAhsqucLO7tEBCTU~Clw5JLl~SUnkdl3nLdF3wbvjGCrmCxt8B3u-YFOb7gTwnj-fq6IamPQnzW-EN~pJfa3b4JjOtp5S1EXQU4u1S3R-7WICU1SRrAHp9DhatcCqvlqb4UJrie~DggVjEsjQg5dutq1deOJRajYomOA4sUsZJL3CWunUwZexRZPyjtl42bbYJ-p9Fy610ebd8GHv~z05LSBzf061P4g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal