In this issue of Blood, Nadeu et al1 contribute to advancing our understanding of the so-called "intermediate" cases of chronic lymphocytic leukemia (i-CLL), one of the 3 epigenetic subgroups identified by DNA methylation analysis.
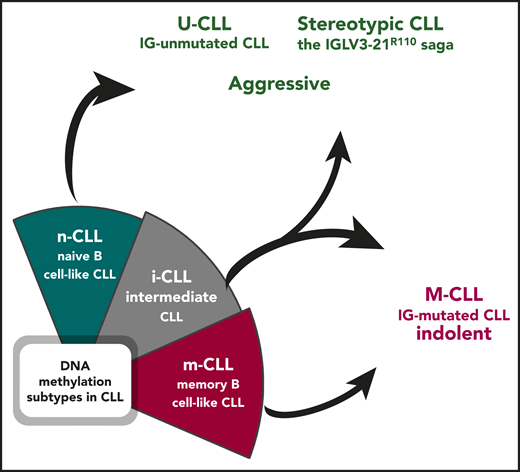
Epigenetically defined i-CLL can be reclassified based on immunogenetic features. Illustration by Andreas Agathangelidis, CERTH, Thessaloniki.
Epigenetically defined i-CLL can be reclassified based on immunogenetic features. Illustration by Andreas Agathangelidis, CERTH, Thessaloniki.
In the recent past, CLL has been a very fertile field for -omics studies, including epigenomics. Mature B lymphocytes are characterized by a complex although well-known pathway of differentiation and maturation. Therefore, it is not surprising that CLL, a prototypic malignancy of mature B cells, became a classic example of the fact that epigenetic marks not only are a consequence of the leukemogenic process, but also are associated with the cell of origin, for instance, representing a distinct differentiation stage.2,3 Based on DNA methylation profiles, CLL has been categorized in 3 distinct epigenetic subgroups associated with normal B-cell differentiation, namely naive B-cell–like CLL (n-CLL), memory B-cell–like CLL (m-CLL), and i-CLL.2,3 Notably, the 3 epigenetic groups of CLL show significantly different clinically relevant biological features, suggesting that the DNA methylation imprint might play a role in shaping the clinical outcome of a given patient.2,3
For at least a couple of decades, we have been used to interpreting the heterogeneity in CLL thanks to the dichotomous classification based on the somatic hypermutation (SHM) status of the immunoglobulin heavy variable (IGHV) genes. Immunoglobulin-mutated cases (M-CLL) tend to be associated with an indolent outcome, whereas immunoglobulin-unumutated cases (U-CLL) generally display a more aggressive behavior.4 More recently, the 2 immunogenetic subgroups appeared to be superseded by DNA methylation analysis, which allowed categorizing patients with CLL into 3 distinct epigenetic subgroups (m-CLL, n-CLL, i-CLL).2,3 Although it was obvious that m-CLL comprised immunoglobulin M-CLL, being epigenetically similar to memory, germinal-center experienced B cells, and n-CLL subgroup included U-CLL, with the signature of naive, pregerminal center B lymphocytes, the so-called i-CLL appeared for a long time to be an exception, with an overall clinical behavior seemingly at odds with their immunogenetic features. i-CLL showed an intermediate DNA methylation profile as well as intermediate clinicobiological features compared with the other 2 subsets. In addition, advanced prediction models with a more limited number of epigenetic markers validated the existence of the 3 subgroups, segregating patients with CLL based on distinct IGHV gene usage and SHM load.5 That said, it was somewhat overlooked that the i-CLL subgroup was enriched for immunoglobulin M-CLL, although with a relatively modest SHM imprint. A prior study had also shown that i-CLL cases were biased toward λ light chain usage, with ∼50% of them expressing the IGLV3-21 gene.6
By integrating whole-genome/exome sequencing and RNA sequencing data, Nadeu et al report that i-CLL display a highly restricted immunogenetic profile based on which they can be reassigned to existing immunogenetic categories (see figure). In particular, more than a third (38%) of the cases carry an SHM in the clonotypic rearranged IGLV3-21 gene, leading to a change at the VL-CL linker region position 110 from the germline glycine-to-arginine (R) (IGLV3-21R110). IGLV3-21R110 patients were found to have a poor clinical outcome, similar to patients with n-CLL. Conversely, the remaining cases of i-CLL that lack IGLV3-21R110 transcriptionally and clinically mirrored m-CLL, thus M-IGHV (see figure).4 These findings corroborate several published observations pointing to the seminal role of immunoglobulin light chains in antigen selection in CLL. This is especially true for the IGLV321 gene, whose expression in CLL has been reported to be associated with a distinct gene expression profile and a poor clinical outcome, regardless of IGHV gene usage, SHM status, and classic cytogenetic abnormalities.7 Moreover, the results by Nadeu et al are in line with the recent demonstration that IGLV3-21R110–expressing CLL likely represents a distinct subset with unfavorable prognosis independent of IGHV gene SHM load.8
Of note, IGLV3-21R110 light chains are ubiquitous in stereotyped subsets 2, the largest in CLL, and 169, the recently identified satellite to subset 2. Both subsets have an aggressive clinical courses regardless of IGHV gene SHM status.9 Importantly, in both stereotyped subsets, a relevant B-cell receptor (BCR)-BCR homotypic interaction leading to cell autonomous signaling is dependent on the IGLV3-21 light chains, as the critical residues are either encoded in the IGLV3-21 germline sequence or introduced by SHM (G-to-R at position 110) in the clonotypic IGLV3-21 gene rearrangements.9,10 Finally, both subsets display a remarkable enrichment for SF3B1 mutations,9 also reported by Nadeu et al in conjunction with enrichment for ATM mutations and a higher total number of driver alterations compared with non-IGLV3-21R110 i-CLL.4
In summary, the study by Nadeu et al reconciles most of our current notions about the role of the BCR in shaping the biology and the clinical behavior of CLL and brings back the spotlight on the immunogenetic characterization of CLL. DNA methylation profiling reveals that the epigenetic signatures strongly associate with the SHM status of the IGHV genes and/or with stereotypic BCR immunoglobulin features. In the end, m-CLL overlaps with M-CLL, n-CLL with U-CLL, and, finally, i-CLL can either be assigned to aggressive cases displaying stereotypic immunoglobulin features or regrouped with M-CLL based on the actual SHM load present. This supports the need for thorough immunogenetic analysis in patients with CLL, particularly assessing conserved BCR immunoglobulin features, such as the presence of IGLV3-21R110 mutation.
The epigenetically “intermediate” group has finally lost its aura of mystery, highlighting the centrality of the immunoglobulin for accurate risk stratification of patients with CLL. To quote T. S. Elliott: “the end of all our exploring will be to arrive where we started.”
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal