In this issue of Blood, Song et al1 show that the transcription factors OBF1, OCT1, and OCT2 are essential for the germinal center (GC) B-cell program in a murine model. They directly regulate numerous key factors involved in GC B-cell development. They are also involved in maintaining proliferation of several types of B-cell lymphoma cell lines.
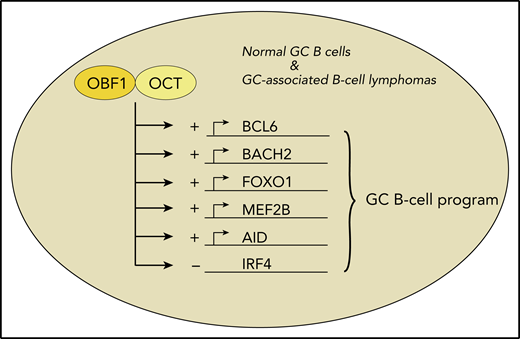
Among the ∼8000 genomic DNA-binding sites that are occupied by OBF1 in conjunction with OCT1 or OCT2, there are key factors for the GC B-cell gene expression program. Several of these are indicated here. These factors are induced by binding of OBF1 and OCT (+). Furthermore, OBF1/OCT1/OCT2 negatively regulate expression of IRF4 (−), thereby further suppressing an exit from the GC B-cell program both in normal B cells and in some types of B-cell lymphomas.
Among the ∼8000 genomic DNA-binding sites that are occupied by OBF1 in conjunction with OCT1 or OCT2, there are key factors for the GC B-cell gene expression program. Several of these are indicated here. These factors are induced by binding of OBF1 and OCT (+). Furthermore, OBF1/OCT1/OCT2 negatively regulate expression of IRF4 (−), thereby further suppressing an exit from the GC B-cell program both in normal B cells and in some types of B-cell lymphomas.
The GC reaction is the central process of T-cell–dependent humoral immune responses and is essential for affinity maturation of these responses and the generation of memory B cells and long-lived plasma cells.2 Twenty-five years ago, gene-knockout studies in the mouse by Matthias and colleagues3 and Kim et al4 revealed that the B-cell–specific transcription factor OBF1 is indispensable for the GC reaction. However, the way in which OBF1 functions in GC B cells remained largely unclear. OBF1 is a coactivating factor for the octamer motif-binding transcription factors OCT1 and OCT2 but lacks a DNA-binding domain.5 OCT1 is ubiquitously expressed, whereas OCT2 is largely B-cell specific.5,6 The 3 factors are expressed throughout B-cell development and differentiation, but in particular, OBF1 is most highly expressed in GC B cells.6
To elucidate the functions of OCT1, OCT2, and OBF1 in GC B cells, Song et al used an elegant approach involving transgenic mice expressing tagged versions of the 3 factors. The endogenously biotinylated factors could then be immunoprecipitated using streptavidin-coupled beads. This experimental strategy allowed for very efficient chromatin immunoprecipitation and, thereby, the first genome-wide comprehensive determination of the genomic DNA-binding sites of the three transcription factors in primary murine B cells. By combining these data with specific histone marks and comparative RNA-sequencing studies of wild-type and OBF1-deficient B cells, DNA binding of the factors was linked to chromatin activity status, and a map of direct OBF1 target genes was established. Several of the studies were extended to human GC B cells and lymphoma cell lines, so that findings directly relevant for human B cells are also presented. The majority of OBF1- and OCT1-binding sites were in promoter regions, whereas many OCT2-binding sites located to enhancers. The main finding was that OBF1, together with OCT1 and/or OCT2, promotes the expression of numerous key GC B-cell genes. Importantly, this includes BCL6, the master transcription factor of the GC B-cell gene expression program.7 Further key factors for GC B cells (eg, FOXO1, AID, MEF2B, and BACH2) were also induced (see figure). Moreover, IRF4, a main inducer of the differentiation of GC B cells into plasma cells, is suppressed by OBF1, which further stabilizes the GC B-cell program. Thus, a picture emerges in which BCL6, mainly by repressing non-GC B-cell genes, and OBF1 in conjunction with OCT1/OCT2, mainly by inducing expression of genes important for GC B cells, together orchestrate the GC B-cell gene expression program.
The detailed mapping of the genomic binding sites of OCT1, OCT2, and OBF1 in B cells revealed that nearly all OBF1-binding sites overlap with OCT1 and/or OCT2 binding sites, in line with the concept that OBF1 is dependent on the OCT factors for association with DNA. However, OCT1 and OCT2 showed many more binding sites than OBF1 (∼13 000 and 32 000 vs 8000). Although it cannot be excluded that this is partly due to a less efficient chromatin immunoprecipitation of OBF1, this indicates that OCT1 and OCT2 can bind to many genomic sites independent of OBF1. A further remarkable observation of the chromatin immunoprecipitation studies was that many DNA-binding sites of the 3 factors colocalize with binding sites for ETS family transcription factors. This at first glance indicates a frequent coregulation of target genes by members of the OCT and ETS families. However, knockdown experiments pointed to a largely independent gene regulation by these factors. This warrants further studies.
The inhibitory effect of OBF1 on IRF4 expression in GC B cells contrasts with the known coexpression of both factors in plasma cells. Moreover, whereas OBF1 in combination with OCT1 and OCT2 directly stimulates BCL6 expression in GC B cells, as shown by Song et al, one has to keep in mind that OBF1 and the OCT factors are expressed throughout B-cell development and differentiation (ie, also in many B-cell subsets that do not express BCL6). These observations indicate that the effects of OBF1/OCT1/OCT2 on target gene expression are strongly context dependent. Although these factors are essential for the GC B-cell program, they are not as GC specific as BCL6 is. The particularly high expression of OBF1 in GC B cells may influence the set of target genes induced by OBF1 in GC B cells. However, other, so-far-unrecognized transcription factors and/or epigenetic alterations may also influence which genes are activated by OBF1/OCT1/OCT2 in GC B cells. Clearly, more studies are needed to fully understand the target gene selection of the 3 factors in GC B cells.
A better understanding of the role of OBF1 and the OCT proteins in B cells is also of relevance for lymphoma biology, because it has already been shown earlier that OCT2 and OBF1 are essential for the proliferation and survival of diffuse large B-cell lymphoma cell lines.8,9 One factor for this is likely the inhibitory effect of OBF1 on IRF4 expression, as IRF4 inhibits proliferation of B-cell lymphoma cells.1 In a more general view, “freezing” of B cells at the proliferative GC B-cell differentiation stage has emerged as a major concept for the pathogenesis of GC-associated B-cell lymphomas, such as diffuse large B-cell and Burkitt lymphomas.10 Although strong expression and activity of OBF1 and OCT factors in lymphomas is only rarely due to genetic alterations,9 these factors may nevertheless represent vulnerabilities of the lymphoma cells that could be exploited for targeted therapy. In spite of the known difficulties to target transcription factors in cancer therapy, it has been proposed that interfering with the binding of OBF1 to the OCT factors could be an attractive approach or strategies to target OBF1 for degradation.9
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal